1. Introduction
The escalating worldwide public health issue of skin cancer is a pressing concern, with millions of individuals being impacted annually. In order to attain a successful outcome, prompt identification and precise diagnosis are imperative. Dermatologists can utilize computer-aided analytical techniques [
1] to examine images of skin cancer. Despite the considerable progress made in the area of skin cancer detection, the current feature extraction and classification approach to skin cancer images is limited [
2]. Previous research has mainly focused on CT’s individual applications, but incorporating other transforms like LBP for additional feature extraction may enhance classification performance. Selecting the best features from skin cancer images through optimization techniques [
3] is critical for improving the efficiency and effectiveness of the detection system. Once an image’s optimum input feature set is identified, classification methods should be employed further to enhance the detection process regarding time and space complexity.
Our Contributions:
To enhance the manual feature extraction process, We adopted an improved combination of CT and LBP image transform techniques to accurately recognise borders, contrast changes, and shapes of skin cancer images from the International Skin Imaging Collaboration (ISIC) data set.
We applied advanced PSO to optimise the feature extraction process to convert the initial images into a reduced set of input data vectors.
The reduced set of input feature vectors is applied to three advanced ML classification methods: SVM, RF, and NN for skin cancer detection. These models were evaluated based on various metrics, including accuracy, precision, recall, and F1-score, as well as the time and space complexity of each model’s training process.
This paper is structured as follows. Section II introduces Preliminaries. Section III focuses on the proposed methods. Section IV provides simulation results and discussion details. Finally, the conclusion is noted in section V.
2. Preliminaries
2.1. Skin cancer
There are three primary types of skin cancer lesions [
4]: Basal cell carcinoma (BCC), Squamous cell carcinoma (SCC), and Melanoma. BCC is prevalent in regions exposed to the sun, SCC is characterised by firmness, and Melanoma [
5] is associated with aggressiveness. Different types of skin cancer lesions exhibit distinct features and characteristics, which aid in their identification and diagnosis. Here are some key features of the three main types of skin cancer shown in
Table 1.
Dermatologists employ the ABCDE rule to assess which type of cancer moles and lesions. For instance, Melanoma was identified using the ABCDE rule [
6], as shown in
Table 2. While these features and identification methods provide valuable guidance, it is essential to consult a dermatologist for a definitive diagnosis.
2.2. Contourlet Transform (CT)
The Contourlet Transform [
7,
8] is a sophisticated, multi-faceted image transform that operates on multiple scales and directions. It is widely utilized in various image processing tasks, including but not limited to image compression, denoising, and feature extraction. This transform builds upon the fundamental principles of the Wavelet Transform, but goes a step further by incorporating directional information. The Contourlet Transform involves a series of steps, which are hereby elucidated below in a more academic fashion:
Preprocessing: To analyze an input image, ensure it is preprocessed, possibly resizing or denoising, to improve its quality.
Decomposition into Sub bands:The Contourlet Transform involves decomposing an image into sub bands using a filter bank, which is crucial for determining the properties of the extracted features.
Directional Filter Banks: The Contourlet Transform employs directional filter banks, unlike the standard Wavelet Transform, which uses only horizontal and vertical filters, enabling the representation of contours in the image.
Sub band Decomposition: The image is divided into multiple sub bands, each containing specific frequency and direction information, resulting in a more accurate representation of the data.
Quantization and Feature Extraction: Obtaining sub bands allows for quantization or feature extraction techniques, such as histogram-based statistics and texture analysis, depending on the specific application.
Feature Selection: Feature selection is a method used to select the most relevant features for a task, which can reduce dimensionality and enhance the efficiency of subsequent processing steps.
2.3. Local Binary Pattern (LBP)
The Local Binary Pattern (LBP) [
9] is renowned for its exceptional discriminatory capabilities as a texture descriptor. It assigns a binary number to each pixel in an image by comparing its Gray level with that of its neighboring pixels. In a predefined patch, neighboring pixels with a higher Gray level than the center pixel are given a value of unity, whereas those with a lower Gray level are assigned a value of zero. As a result, the center pixel is assigned a binary number. The original LBP operator operates on a
patch, yielding an 8-digit binary number derived from the surrounding pixels. Once all pixels in an image have been labeled, the LBP feature map and a histogram with 256 bins are generated. The LBP histogram can be employed as a feature vector for classification, where each bin denotes a unique feature.
2.4. Particle Swarm Optimization (PSO)
CT images often contain numerous features, leading to high computational costs and over-fitting issues. PSO [
10], a meta-heuristic optimization algorithm inspired by bird flocking or fish schooling, aims to select informative and discriminative features for accurate classification. PSO [
11] searches for an optimal subset of features, with each particle representing a potential feature subset. The particles are binary vectors with values representing different feature subsets. The fitness function evaluates each particle’s performance, and based on these evaluations, particles adjust their positions within the search space.
Two factors guide the movement Personal Best (pBest): A particle has found the best solution (feature subset) so far. Global Best (gBest): The best solution among all the particles in the swarm. Particles are attracted to their pBest and gBest positions, and acceleration coefficients and random factors control this movement. The optimization process involves multiple iterations where particles continue to move and update their statuses. The strategy aims to converge to an optimal solution, i.e., the best feature subset that maximizes the classification performance. The PSO optimization terminates after a predefined number of iterations or when a convergence criterion is met. After the PSO optimization, the particle that achieved the best feature subset is selected as the final reduced feature subset.
2.5. Classification algorithms
With the reduced feature subset obtained from PSO, a classification algorithm [
12] such as SVM, Random forest, Neural network is trained using this subset as input features. The reduced feature space typically results in faster training and improved generalization performance since it reduces the risk of over-fitting. By leveraging the PSO optimizer to select the most informative features from CT images, the feature reduction process helps improve the efficiency and accuracy of classification algorithms. It enables more focused and efficient training, essential for handling large-scale medical image data sets and improving the overall performance of computer-aided diagnosis systems for CT image analysis.
2.5.1. Support Vector Machines (SVM)
Support-vector machines (SVMs) [
13] referred to as support-vector networks in their early stages. SVMs are a binary classification algorithm that implements the concept of non-linearly mapping vectors to a high-dimensional feature space to devise a linear decision surface (hyperplane) within this feature space. Consequently, SVMs are proficient in solving separable and non-separable problems, as the hyperplane is optimal in the sense of being a maximal margin classifier with respect to the training data. The Structural Risk Minimization (SRM) principle, which SVMs adhere to, equips them with enhanced generalization abilities. As a result of these notable advantages, SVMs were quickly applied to classification and regression problems. Furthermore, SVMs embody the attributes of small sample sizes, non-linear problems, and the curse of dimensionality, which have always been of concern .
2.5.2. Random forest (RF)
Random Forests [
14] is a machine learning ensemble learning algorithm that uses multiple decision trees to create robust, accurate models, reducing variance and over-fitting in classification and regression tasks. This generates bootstrap samples by randomly selecting data points, which are then used to train a distinct decision tree. They consider a random subset of features, introducing diversity and reducing the likelihood of a dominant feature. Decision Tree construction involves growing multiple trees using bootstrap samples and random feature subsets. Voting or averaging is used for classification and regression tasks. Ensemble aggregation reduces over-fitting and improves generalization performance, with randomness in feature selection and bootstrapping introducing diversity for a robust model.
2.5.3. Neural network (NN)
Neural networks [
15,
16], which are machine learning algorithms, have been inspired by the human brain and are currently utilized in diverse domains such as natural language processing, image and speech recognition. They are classified as transformers, which operate by modifying input data through activation functions and weighted connections. The architecture of the network comprises of input, hidden, and output layers, consisting of weight initialization and loss function. During the feed forward process, input data is multiplied by weights and subsequently passed through activation functions. Optimization methods like stochastic gradient descent, are applied to iteratively adjust weights to minimize loss function and enhance prediction precision. The network undergoes numerous iterations of training, loss calculation, and back propagation to refine weights and biases. Regularization and hyper-parameter tuning are also executed to prevent over-fitting and enhance generalization.
3. Proposed methods
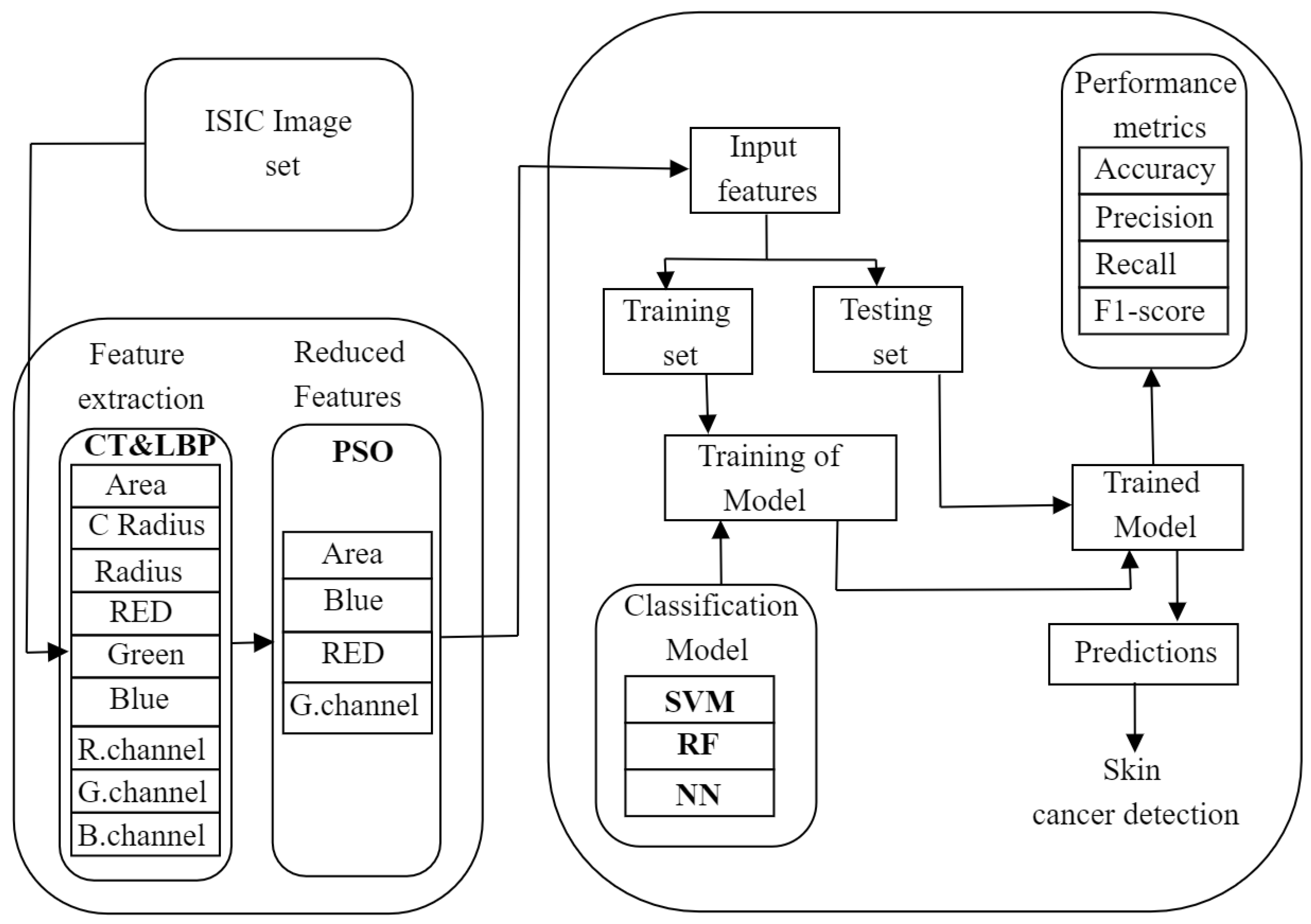
This section presents our approach to feature extraction and reduction and classification of skin cancer images, as shown in
Figure 1. We applied the ISIC image set for the image preprocessing tools like CT and LBP for feature extraction of an image. After obtaining an extensive feature set, we adopted PSO to obtain the reduced optimal feature set. We gave this reduced feature set as input to three classification algorithms: Support Vector Machine (SVM), Random Forest (RF), and Neural Networks (NN). Our goal was to evaluate the performance of each classification algorithm using the reduced feature sets and determine the most effective approach for image classification.
Table 3.
Image File Properties.
Table 3.
Image File Properties.
| Image File |
Radius |
Area |
Contour Radius |
Color Features |
Green Channel |
Red Channel |
Blue Channel |
| (1).jpg |
123 |
2533.5 |
28.39 |
[170 161 188] |
135.72 |
130.73 |
162.00 |
| (10).jpg |
10 |
94 |
5.47 |
[181 167 221] |
151.45 |
142.93 |
203.205 |
| (100).jpg |
361 |
128722 |
202.41 |
[159 157 223] |
161.72 |
160.82 |
228.54 |
| (101).jpg |
374 |
249447 |
281.78 |
[138 130 195] |
137.99 |
141.28 |
199.43 |
| (102).jpg |
15 |
157.5 |
7.08 |
[147 139 170] |
128.27 |
125.96 |
162.17 |
| (103).jpg |
175 |
29871 |
97.51 |
[129 128 200] |
119.43 |
115.25 |
203.72 |
| (104).jpg |
374 |
268128 |
292.14 |
[107 92 153] |
124.22 |
133.55 |
208.22 |
| (105).jpg |
130 |
12405.5 |
62.83 |
[161 144 181] |
156.10 |
151.9 |
190.90 |
| (106).jpg |
123 |
19065 |
77.90 |
[156 152 193] |
135.85 |
131.33 |
185.97 |
4. Simulation results and its discussion
For the simulations, we employed a Kaggle dataset [
17] focused on skin cancer. This collection comprises a total of 2357 images depicting both malignant and benign oncological ailments. These images were derived from The International Skin Imaging Collaboration (ISIC). The sorting of the images was based on the classification provided by ISIC, and each subset was divided into an equal number of images, except for melanomas and moles, which exhibited a slight dominance in terms of the number of images. To better understand the distribution of images within this dataset, please refer to
Table 4 for a detailed breakdown of benign and malignant classifications.
4.1. Extracted Features
The provided data contains information about different images and their corresponding color channel features - Green, Red, and Blue Channels. As shown in
Table 3, the provided data gives us insights into the color characteristics of different images [
18]. The intensity values of the three color channels indicate the presence and dominance of specific colors in each image. These features are obtained after application of CT and LBP methods, and these can be useful for further image analysis tasks, such as color-based image segmentation, object recognition, and classification. Additionally, this data can be utilized as feature vectors for machine learning models to perform image-related tasks based on color channel information.
4.2. Particle Swarm Optimization for Feature reduction
After applying PSO model [
19] to obtain strong features, the number of features has been reduced from 9 to 4. From the
Table 5, the optimization process has successfully identified the most relevant and informative features, leading to a significant reduction in both time and space complexity. The time complexity reduction achieved is 0.00204, indicating that the optimized feature set allows for faster computation and processing compared to using all 9 features. This reduction in time complexity is crucial in enhancing the efficiency of the model or algorithm. Furthermore, the space complexity has been reduced by a factor of 5. This means that the optimized feature set requires much less memory, making it more memory-efficient and suitable for applications with limited resources. After the PSO feature reduction, the selected features are [’Area’, ’Green Channel’, ’Blue’, ’RED’]. These features have been identified as the most influential in making predictions or decisions within the given context. By focusing on these key features, the model’s performance can be maintained or even improved, while simplifying the model and reducing potential over-fitting.
4.3. Performance analysis of various Classification Models
Three classification models: Support Vector Machine (SVM), Random Forest (RF), and Neural Network(NN) were applied to a reduced feature set. As shown in
Table 6, the models were evaluated based on various metrics, including accuracy, precision, recall, and F1-score, as well as the time and space complexity of each model’s training process. All three models achieved the same accuracy, precision, recall, and F1- score. This suggests that the predictive capabilities of the models are comparable for the given skin cancer classification task.
The time complexity of the model training phase indicates the time taken to fit the model to the training data. The SVM has the lowest time complexity of 0.0458 seconds, followed by the Neural Network at 0.0873 seconds, and the Random Forest model with the highest time complexity of 0.1622 seconds. The SVM and Neural Network models are faster to train than the Random Forest, making them more suitable for real-time or latency-sensitive applications. The space complexity refers to the memory required to store the model and its associated parameters. The Neural Network model has the highest space complexity of 816 units, while the SVM and Random Forest models have the same space complexity of 448 units. This difference in space complexity could be attributed to the architecture and size of the Neural Network.
Considering time and space complexity is essential, especially when deploying models to resource-constrained environments such as mobile devices or embedded systems. The SVM might be preferred in such cases due to its faster training time and lower memory requirements.
4.3.1. Comparative analysis
Table 7 compares proposed model with existing methods for skin cancer classification. The model outperforms existing Machine learning and Deep learning models models as shown references, providing the highest test accuracy and significantly higher results in other evaluation metrics like precision, recall, and F1-score. This results highlight the superiority of our model in skin cancer detection.
5. Conclusion
The Contourlet Transform, optimized with Particle Swarm Optimization (PSO), emerges as the most promising feature extraction mechanism for skin cancer detection, offering high accuracy and robustness in classification. This research work contributes to the growing body of knowledge in medical image analysis and provides valuable insights for the development of more reliable and precise diagnostic tools in the medical field.
References
- Dorrell, Deborah N., and Lindsay C. Strowd, “Skin cancer detection technology ”, Dermatologic clinics,37, no. 4 (2019): 527-536. [CrossRef]
- Bhuiyan, Md Amran Hossen, Ibrahim Azad, and Md Kamal Uddin, “Image processing for skin cancer features extraction,”International Journal of Scientific and Engineering Research 4.2” (2013): 1-6.
- Elgamal, Mahmoud, “Automatic skin cancer images classification,”International Journal of Advanced Computer Science and Applications 4.3, 2013. [CrossRef]
- Magnus, Knut, “The Nordic profile of skin cancer incidence. A comparative epidemiological study of the three main types of skin cancer.,” International journal of cancer 47, no. 1 (1991): 12-19. [CrossRef]
- Alquran, Hiam, Isam Abu Qasmieh, Ali Mohammad Alqudah, Sajidah Alhammouri, Esraa Alawneh, Ammar Abughazaleh, and Firas Hasayen, “The melanoma skin cancer detection and classification using support vector machine,” 2017 IEEE Jordan Conference on Applied Electrical Engineering and Computing Technologies (AEECT), IEEE, 2017. [CrossRef]
- H. R. Firmansyah, E. M. Kusumaningtyas and F. F. Hardiansyah, “Detection melanoma cancer using ABCD rule based on mobile device”, 2017 International Electronics Symposium on Knowledge Creation and Intelligent Computing (IES-KCIC), Surabaya, Indonesia, 2017, pp. 127-131. [CrossRef]
- Do, Minh N., and Martin Vetterli. “The contourlet transform: an efficient directional multiresolution image representation.” IEEE Transactions on image processing 14, no. 12 (2005): 2091-2106. [CrossRef]
- Chitaliya, N. G., and A. I. Trivedi, “An efficient method for face feature extraction and recognition based on Contourlet transforms and principal component analysis,” Procedia Computer Science 2 (2010): 52-61. [CrossRef]
- Yang, Bo, and Songcan Chen. “A comparative study on local binary pattern (LBP) based face recognition: LBP histogram versus LBP image.” Neurocomputing 120 (2013): 365-379. [CrossRef]
- Tran, Binh, Bing Xue, and Mengjie Zhang, “Overview of particle swarm optimization for feature selection in classification.,” Simulated Evolution and Learning: 10th International Conference, SEAL 2014, Dunedin, New Zealand, December 15-18, 2014. Proceedings 10. Springer International Publishing, 2014. [CrossRef]
- Imran, Muhammad, Rathiah Hashim, and Noor Elaiza Abd Khalid, “An overview of particle swarm optimization variants.,” Procedia Engineering 53 (2013): 491-496. [CrossRef]
- Kotsiantis, Sotiris B., Ioannis Zaharakis, and P. Pintelas, “Supervised machine learning: A review of classification techniques.” Emerging artificial intelligence applications in computer engineering 160.1 (2007): 3-24.
- Pisner, Derek A., and David M. Schnyer. “Support vector machine.” Machine learning. Academic Press, 2020. 101-121. [CrossRef]
- Paul, Angshuman, Dipti Prasad Mukherjee, Prasun Das, Abhinandan Gangopadhyay, Appa Rao Chintha, and Saurabh Kundu. “Improved random forest for classification.” IEEE Transactions on Image Processing 27, no. 8 (2018): 4012-4024. [CrossRef]
- Abiodun, Oludare Isaac, Aman Jantan, Abiodun Esther Omolara, Kemi Victoria Dada, Abubakar Malah Umar, Okafor Uchenwa Linus, Humaira Arshad, Abdullahi Aminu Kazaure, Usman Gana, and Muhammad Ubale Kiru. “Comprehensive review of artificial neural network applications to pattern recognition.” IEEE access 7 (2019): 158820-158846. [CrossRef]
- Tumpa, Priyanti Paul, and Md Ahasan Kabir, “An artificial neural network based detection and classification of melanoma skin cancer using hybrid texture features,” Sensors International 2 (2021): 100128. [CrossRef]
-
https://www.kaggle.com/datasets/nodoubttome/skin-cancer9-classesisic.
- Ilea, Dana E., and Paul F. Whelan. “Image segmentation based on the integration of colour–texture descriptors—A review.” Pattern Recognition 44, no. 10-11 (2011): 2479-2501. [CrossRef]
- Gad, Ahmed G, “Particle swarm optimization algorithm and its applications: a systematic review.,” Archives of computational methods in engineering 29.5 (2022): 2531-2561. [CrossRef]
- Kawahara, Jeremy, and Ghassan Hamarneh,“Multi-resolution-tract CNN with hybrid pretrained and skin-lesion trained layers.,” In International workshop on machine learning in medical imaging, pp. 164-171. Cham: Springer International Publishing, 2016. [CrossRef]
- Lopez, Adria Romero, Xavier Giro-i-Nieto, Jack Burdick, and Oge Marques, “Skin lesion classification from dermoscopic images using deep learning techniques. ” In 2017 13th IASTED international conference on biomedical engineering (BioMed), pp. 49-54. IEEE, 2017. [CrossRef]
- Brinker, Titus J., Achim Hekler, Alexander H. Enk, and Christof von Kalle, “ Enhanced classifier training to improve precision of a convolutional neural network to identify images of skin lesions.”, PloS one 14, no. 6 (2019): e0218713. [CrossRef]
- Wu, Jing, Wei Hu, Yuan Wen, WenLi Tu, and XiaoMing Liu, “ Skin lesion classification using densely connected convolutional networks with attention residual learning”, Sensors 20, no. 24 (2020): 7080. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).