Submitted:
03 November 2023
Posted:
06 November 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
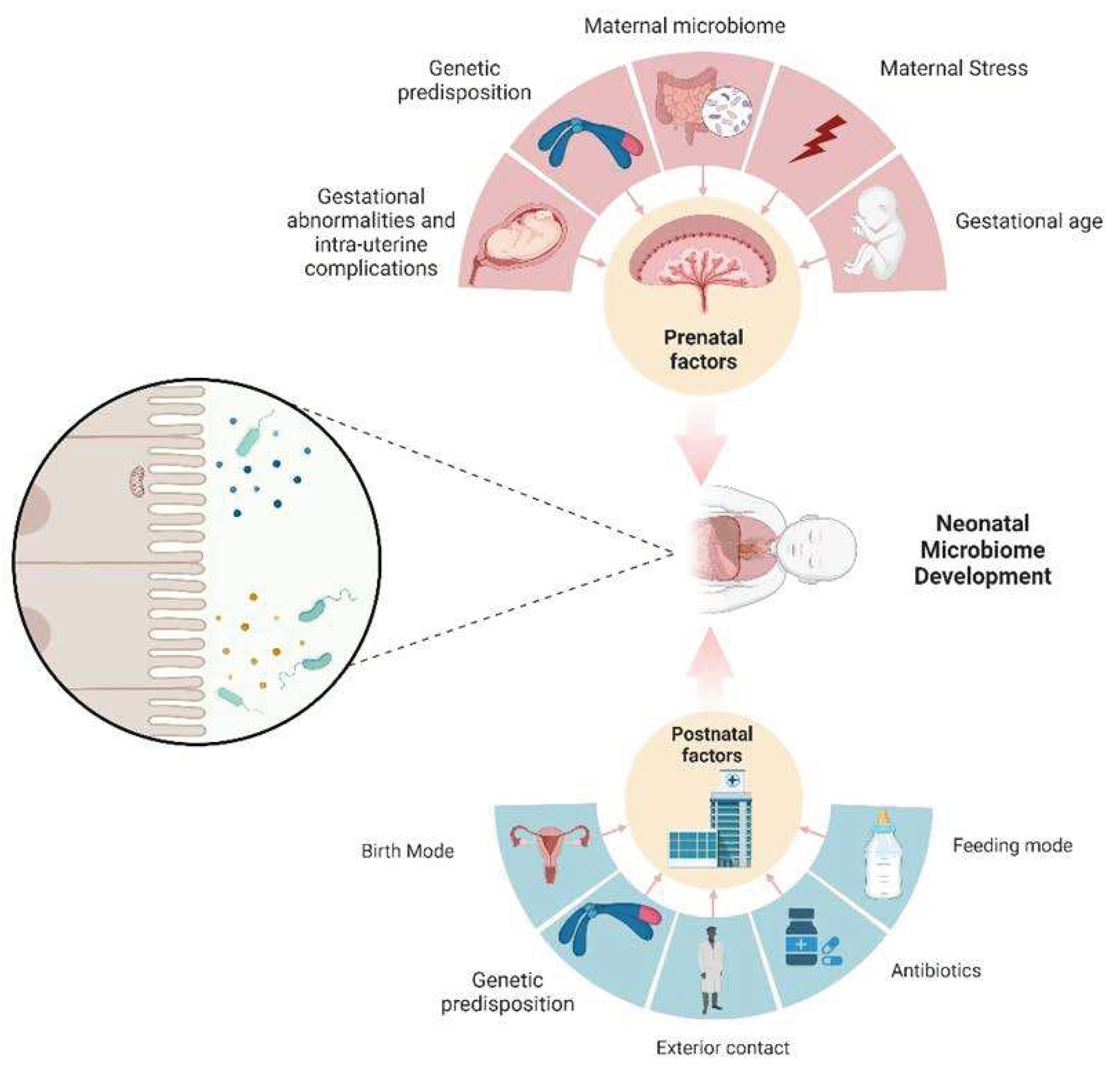
2. The Neonatal Microbiome
3. Microbial Transfer & Postnatal influences
4. Prenatal Influences on Neonatal Microbiome
5. Interpreting Varied Microbiome Profiles
6. Case Studies and Animal Models
7. Conclusions
8. Future Directions
References
- Costello, E.K. et al., Bacterial community variation in human body habitats across space and time. Science 2009, 326, 1694–1697. [Google Scholar] [CrossRef] [PubMed]
- Proctor, L.M. et al., The Integrative Human Microbiome Project. Nature 2019, 569, 641–648. [Google Scholar]
- Wang, L. et al., Gastrointestinal microbiota and metabolite biomarkers in children with autism spectrum disorders. Biomark Med 2014, 8, 331–344. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y. et al., Gut microbiota influence tumor development and Alter interactions with the human immune system. Journal of Experimental & Clinical Cancer Research 2021, 40, 42. [Google Scholar]
- Gopalakrishnan, V. et al., Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 2018, 359, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Byrd, A.L. and J.A. Segre, Elucidating microbial codes to distinguish individuals. Proc Natl Acad Sci U S A 2015, 112, 6778–6779. [Google Scholar] [CrossRef] [PubMed]
- Franzosa, E.A. et al., Identifying personal microbiomes using metagenomic codes. Proc Natl Acad Sci U S A 2015, 112, E2930–8. [Google Scholar] [CrossRef] [PubMed]
- Faith, J.J. et al., The long-term stability of the human gut microbiota. Science 2013, 341, 1237439. [Google Scholar] [CrossRef]
- Gajer, P. et al., Temporal Dynamics of the Human Vaginal Microbiota. Science Translational Medicine 2012, 4, 132ra52–132ra52. [Google Scholar] [CrossRef]
- Azevedo, M.J. et al., The contribution of maternal factors to the oral microbiota of the child: Influence from early life and clinical relevance. Japanese Dental Science Review 2023, 59, 191–202. [Google Scholar] [CrossRef]
- Yeramilli, V. et al., A Review of the Impact of Maternal Prenatal Stress on Offspring Microbiota and Metabolites. Metabolites 2023, 13, 535. [Google Scholar] [CrossRef] [PubMed]
- Enav, H. F.Bäckhed, and R.E. Ley, The developing infant gut microbiome: A strain-level view. Cell Host Microbe 2022, 30, 627–638. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, K.M. et al., Fetal meconium does not have a detectable microbiota before birth. Nat Microbiol 2021, 6, 865–873. [Google Scholar] [CrossRef] [PubMed]
- Warner, B.B. et al., Gut bacteria dysbiosis and necrotising enterocolitis in very low birthweight infants: a prospective case-control study. Lancet 2016, 387, 1928–1936. [Google Scholar] [CrossRef]
- Sender, R. S.Fuchs, and R. Milo, Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol 2016, 14, e1002533. [Google Scholar] [CrossRef] [PubMed]
- Bordenstein, S.R. and K.R. Theis, Host Biology in Light of the Microbiome: Ten Principles of Holobionts and Hologenomes. PLoS Biol 2015, 13, e1002226. [Google Scholar] [CrossRef] [PubMed]
- Zilber-Rosenberg, I. and E. Rosenberg, Microbial-driven genetic variation in holobionts. FEMS Microbiology Reviews 2021, 45. [Google Scholar] [CrossRef] [PubMed]
- Blanton, L.V. et al., Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children. Science 2016, 351, aad3311. [Google Scholar] [CrossRef] [PubMed]
- Singh, A. and M. Mittal, Neonatal microbiome - a brief review. J Matern Fetal Neonatal Med 2020, 33, 3841–3848. [Google Scholar] [CrossRef]
- Bäckhed, F. et al., Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe 2015, 17, 852. [Google Scholar] [CrossRef]
- Martin, R. et al., Early-Life Events, Including Mode of Delivery and Type of Feeding, Siblings and Gender, Shape the Developing Gut Microbiota. PLoS One 2016, 11, e0158498. [Google Scholar] [CrossRef] [PubMed]
- Yatsunenko, T. et al., Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef] [PubMed]
- Chong, C.Y.L. F.H. Bloomfield, and J.M. O’Sullivan, Factors Affecting Gastrointestinal Microbiome Development in Neonates. Nutrients 2018, 10, 274. [Google Scholar] [CrossRef] [PubMed]
- Turunen, J. et al., Investigating prenatal and perinatal factors on meconium microbiota: a systematic review and cohort study. Pediatric Research 2023. [Google Scholar]
- Gosalbes, M.J. et al., Meconium microbiota types dominated by lactic acid or enteric bacteria are differentially associated with maternal eczema and respiratory problems in infants. Clinical & Experimental Allergy 2013, 43, 198–211. [Google Scholar]
- Dudley, D. The placental microbiome: yea, nay or maybe? BJOG: An International Journal of Obstetrics & Gynaecology 2020, 127, 170–170. [Google Scholar]
- Aagaard, K. et al., The placenta harbors a unique microbiome. Sci Transl Med 2014, 6, 237ra65. [Google Scholar] [PubMed]
- D’Argenio, V. The Prenatal Microbiome: A New Player for Human Health. High Throughput 2018, 7, 38. [Google Scholar] [CrossRef]
- de Goffau, M.C. et al., Human placenta has no microbiome but can contain potential pathogens. Nature 2019, 572, 329–334. [Google Scholar] [CrossRef]
- Sterpu, I. et al., No evidence for a placental microbiome in human pregnancies at term. Am J Obstet Gynecol 2021, 224, 296–e1. [Google Scholar] [CrossRef]
- Younge, N. et al., Fetal exposure to the maternal microbiota in humans and mice. JCI Insight 2019, 4. [Google Scholar] [CrossRef]
- Jakobsson, H.E. et al., Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by caesarean section. Gut 2014, 63, 559–566. [Google Scholar] [CrossRef] [PubMed]
- Nagpal, R. et al., Sensitive Quantitative Analysis of the Meconium Bacterial Microbiota in Healthy Term Infants Born Vaginally or by Cesarean Section. Front Microbiol 2016, 7, 1997. [Google Scholar] [CrossRef]
- Erick, M. Breast milk is conditionally perfect. Med Hypotheses 2018, 111, 82–89. [Google Scholar] [CrossRef]
- Groer, M.W. et al., Development of the preterm infant gut microbiome: a research priority. Microbiome 2014, 2, 38. [Google Scholar] [CrossRef]
- Kalbermatter, C. et al., Maternal Microbiota, Early Life Colonization and Breast Milk Drive Immune Development in the Newborn. Front Immunol 2021, 12, 683022. [Google Scholar] [CrossRef] [PubMed]
- Dritsakou, K. et al., The impact of maternal- and neonatal-associated factors on human milk’s macronutrients and energy. J Matern Fetal Neonatal Med 2017, 30, 1302–1308. [Google Scholar] [CrossRef] [PubMed]
- Vaidya, Y.H. et al., Human milk microbiome in urban and rural populations of India. Meta Gene 2017, 13, 13–22. [Google Scholar] [CrossRef]
- Li, S.-W. et al., Bacterial Composition and Diversity in Breast Milk Samples from Mothers Living in Taiwan and Mainland China. Frontiers in Microbiology 2017, 8. [Google Scholar]
- Gianni, M.L. P. Roggero, and F. Mosca, Human milk protein vs. formula protein and their use in preterm infants. Curr Opin Clin Nutr Metab Care 2019, 22, 76–81. [Google Scholar] [CrossRef]
- Ouyang, R. et al., Crosstalk between Breast Milk N-Acetylneuraminic Acid and Infant Growth in a Gut Microbiota-Dependent Manner. Metabolites 2023, 13, 846. [Google Scholar] [CrossRef] [PubMed]
- Khodayar-Pardo, P. et al., Impact of lactation stage, gestational age and mode of delivery on breast milk microbiota. Journal of Perinatology 2014, 34, 599–605. [Google Scholar] [CrossRef] [PubMed]
- Gasparrini, A.J. et al., Persistent metagenomic signatures of early-life hospitalization and antibiotic treatment in the infant gut microbiota and resistome. Nat Microbiol 2019, 4, 2285–2297. [Google Scholar] [CrossRef] [PubMed]
- Russell, J.T. et al., Antibiotics and the developing intestinal microbiome, metabolome and inflammatory environment in a randomized trial of preterm infants. Sci Rep 2021, 11, 1943. [Google Scholar] [CrossRef]
- Dardas, M. et al., The impact of postnatal antibiotics on the preterm intestinal microbiome. Pediatr Res 2014, 76, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Cetinbas, M. et al., Long-term dysbiosis and fluctuations of gut microbiome in antibiotic treated preterm infants. iScience 2023, 26, 107995. [Google Scholar] [CrossRef] [PubMed]
- Selma-Royo, M. et al., Perinatal environment shapes microbiota colonization and infant growth: impact on host response and intestinal function. Microbiome 2020, 8, 167. [Google Scholar] [CrossRef] [PubMed]
- Combellick, J.L. et al., Differences in the fecal microbiota of neonates born at home or in the hospital. Scientific Reports 2018, 8, 15660. [Google Scholar] [CrossRef]
- Stojanov, M. et al., Home or hospital birth: the neonatal microbiota perspective. The Lancet Microbe 2022, 3, e247. [Google Scholar] [CrossRef]
- Rooks, M.G. et al., Gut microbiome composition and function in experimental colitis during active disease and treatment-induced remission. Isme J 2014, 8, 1403–1417. [Google Scholar] [CrossRef]
- Tohi, M. et al., The Developmental Origins of Health and Disease: Adolescence as a Critical Lifecourse Period to Break the Transgenerational Cycle of NCDs-A Narrative Review. Int J Environ Res Public Health 2022, 19, 6024. [Google Scholar] [CrossRef] [PubMed]
- Khambadkone, S.G. Z.A. Cordner, and K.L.K. Tamashiro, Maternal stressors and the developmental origins of neuropsychiatric risk. Front Neuroendocrinol 2020, 57, 100834. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Twinn, D.S. et al., Intrauterine programming of obesity and type 2 diabetes. Diabetologia 2019, 62, 1789–1801. [Google Scholar] [CrossRef] [PubMed]
- Power, M.L. and J. Schulkin, Maternal obesity, metabolic disease, and allostatic load. Physiol Behav 2012, 106, 22–28. [Google Scholar] [CrossRef]
- Cheddadi, R. et al., The impact of maternal stress on the development of necrotizing enterocolitis: A comprehensive review. Seminars in Pediatric Surgery 2023, 32, 151324. [Google Scholar] [CrossRef]
- Kramer, A.C. et al., Maternal-fetal cross-talk via the placenta: influence on offspring development and metabolism. Development 2023, 150. [Google Scholar] [CrossRef] [PubMed]
- Jašarević, E. et al., Stress during pregnancy alters temporal and spatial dynamics of the maternal and offspring microbiome in a sex-specific manner. Sci Rep 2017, 7, 44182. [Google Scholar] [CrossRef]
- Lyte, M. L. Vulchanova, and D.R. Brown, Stress at the intestinal surface: catecholamines and mucosa-bacteria interactions. Cell Tissue Res 2011, 343, 23–32. [Google Scholar] [CrossRef]
- Winter, S.E. and A.J. Bäumler, Why related bacterial species bloom simultaneously in the gut: principles underlying the ‘Like will to like’ concept. Cell Microbiol 2014, 16, 179–184. [Google Scholar] [CrossRef]
- Robertson, B.R. et al., Mucispirillum schaedleri gen. nov., sp. nov., a spiral-shaped bacterium colonizing the mucus layer of the gastrointestinal tract of laboratory rodents. Int J Syst Evol Microbiol 2005, 55 Pt 3, 1199–1204. [Google Scholar] [CrossRef]
- Berry, D. et al., Phylotype-level 16S rRNA analysis reveals new bacterial indicators of health state in acute murine colitis. Isme j 2012, 6, 2091–2106. [Google Scholar] [CrossRef]
- Friswell, M.K. et al., Site and strain-specific variation in gut microbiota profiles and metabolism in experimental mice. PLoS One 2010, 5, e8584. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J. et al., Organismal, genetic, and transcriptional variation in the deeply sequenced gut microbiomes of identical twins. Proceedings of the National Academy of Sciences 2010, 107, 7503–7508. [Google Scholar] [CrossRef]
- Yang, J. et al., Comparison of Meconium Microbiome in Dizygotic and Monozygotic Twins Born by Caesarean Section (CS). Front Microbiol 2020, 11, 1139. [Google Scholar]
- Blumfield, M.L. et al., Dietary balance during pregnancy is associated with fetal adiposity and fat distribution. Am J Clin Nutr 2012, 96, 1032–1041. [Google Scholar] [CrossRef]
- Rautava, S. et al., Probiotics modulate host-microbe interaction in the placenta and fetal gut: a randomized, double-blind, placebo-controlled trial. Neonatology 2012, 102, 178–184. [Google Scholar] [CrossRef] [PubMed]
- Yan, F. et al., Soluble proteins produced by probiotic bacteria regulate intestinal epithelial cell survival and growth. Gastroenterology 2007, 132, 562–575. [Google Scholar] [CrossRef]
- Tsamantioti, E.S.; Hashmi, M.F. Teratogenic Medications, in StatPearls. 2023, StatPearls Publishing. Copyright © 2023, StatPearls Publishing LLC.: Treasure Island (FL) ineligible companies. Disclosure: Muhammad Hashmi declares no relevant financial relationships with ineligible companies.
- Harshaw, C. et al., Maternal antibiotics disrupt microbiome, behavior, and temperature regulation in unexposed infant mice. Dev Psychobiol 2022, 64, e22289. [Google Scholar] [CrossRef] [PubMed]
- Derrien, M. A.-S. Alvarez, and W.M. de Vos, The Gut Microbiota in the First Decade of Life. Trends in Microbiology 2019, 27, 997–1010. [Google Scholar] [CrossRef]
- Roswall, J. et al., Developmental trajectory of the healthy human gut microbiota during the first 5 years of life. Cell Host and Microbe 2021, 29, 765–776. [Google Scholar] [CrossRef]
- Yeramilli, V. et al., The Impact of Stress, Microbial Dysbiosis, and Inflammation on Necrotizing Enterocolitis. Microorganisms 2023, 11, 2206. [Google Scholar] [CrossRef] [PubMed]
- Shelton, C.D. et al., An early-life microbiota metabolite protects against obesity by regulating intestinal lipid metabolism. Cell Host & Microbe 2023, 31, 1604–1619. [Google Scholar]
- Pammi, M. E.Hollister, and J. Neu, Gut Injury and the Microbiome in Neonates. Clin Perinatol 2020, 47, 369–382. [Google Scholar] [CrossRef] [PubMed]
- Ng, P.C. An update on biomarkers of necrotizing enterocolitis. Seminars in Fetal and Neonatal Medicine 2018, 23, 380–386. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y. et al., Exploration of pathogenic microorganism within the small intestine of necrotizing enterocolitis. World Journal of Pediatrics 2023. [Google Scholar]
- Sun, Z. et al., A review of neuroendocrine immune system abnormalities in IBS based on the brain-gut axis and research progress of acupuncture intervention. Front Neurosci 2023, 17, 934341. [Google Scholar] [CrossRef]
- Peppas, S. et al., The Brain-Gut Axis: Psychological Functioning and Inflammatory Bowel Diseases. J Clin Med 2021, 10, 377. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q. et al., Metabolomics changes in brain-gut axis after unpredictable chronic mild stress. Psychopharmacology (Berl) 2022, 239, 729–743. [Google Scholar] [CrossRef]
- Cryan, J.F. et al., The Microbiota-Gut-Brain Axis. Physiol Rev 2019, 99, 1877–2013. [Google Scholar] [CrossRef]
- Chernikova, M.A. et al., The Brain-Gut-Microbiome System: Pathways and Implications for Autism Spectrum Disorder. Nutrients 2021, 13, 4497. [Google Scholar] [CrossRef]
- Taniya, M.A. et al., Role of Gut Microbiome in Autism Spectrum Disorder and Its Therapeutic Regulation. Front Cell Infect Microbiol 2022, 12, 915701. [Google Scholar] [CrossRef]
- Sivamaruthi, B.S. et al., The Role of Microbiome, Dietary Supplements, and Probiotics in Autism Spectrum Disorder. Int J Environ Res Public Health 2020, 17, 2647. [Google Scholar] [CrossRef]
- Yap, C.X. et al., Autism-related dietary preferences mediate autism-gut microbiome associations. Cell 2021, 184, 5916–5931. [Google Scholar] [CrossRef]
- Alamoudi, M.U. et al., Comparing the Gut Microbiome in Autism and Preclinical Models: A Systematic Review. Front Cell Infect Microbiol 2022, 12, 905841. [Google Scholar] [CrossRef] [PubMed]
- Golubeva, A.V. et al., Microbiota-related Changes in Bile Acid & Tryptophan Metabolism are Associated with Gastrointestinal Dysfunction in a Mouse Model of Autism. EBioMedicine 2017, 24, 166–178. [Google Scholar] [PubMed]
- Chaidez, V. R.L. Hansen, and I. Hertz-Picciotto, Gastrointestinal problems in children with autism, developmental delays or typical development. J Autism Dev Disord 2014, 44, 1117–1127. [Google Scholar] [CrossRef] [PubMed]
- Laue, H.E. M.O. Coker, and J.C. Madan, The Developing Microbiome From Birth to 3 Years: The Gut-Brain Axis and Neurodevelopmental Outcomes. Front Pediatr 2022, 10, 815885. [Google Scholar] [CrossRef]
- Thevaranjan, N. et al., Age-Associated Microbial Dysbiosis Promotes Intestinal Permeability, Systemic Inflammation, and Macrophage Dysfunction. Cell Host & Microbe 2017, 21, 455–466. [Google Scholar]
- Rera, M., R.I. Clark, and D.W. Walker, Intestinal barrier dysfunction links metabolic and inflammatory markers of aging to death in <i>Drosophila</i>. Proceedings of the National Academy of Sciences 2012, 109, 21528–21533.
- Rera, M. et al., Modulation of Longevity and Tissue Homeostasis by the Drosophila PGC-1 Homolog. Cell Metabolism 2011, 14, 623–634. [Google Scholar] [CrossRef]
- Stiernagle, T. Maintenance of C. elegans. WormBook, 2006; pp. 1–11. [Google Scholar]
- Garigan, D. et al., Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Genetics 2002, 161, 1101–1112. [Google Scholar] [CrossRef] [PubMed]
- Félix, M.A. and F. Duveau, Population dynamics and habitat sharing of natural populations of Caenorhabditis elegans and C. briggsae. BMC Biol 2012, 10, 59. [Google Scholar]
- Brenner, S. The genetics of Caenorhabditis elegans. Genetics 1974, 77, 71–94. [Google Scholar] [CrossRef]
- Komura, T. et al., Mechanism underlying prolongevity induced by bifidobacteria in Caenorhabditis elegans. Biogerontology 2013, 14, 73–87. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




