Submitted:
06 November 2023
Posted:
07 November 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
3. Results
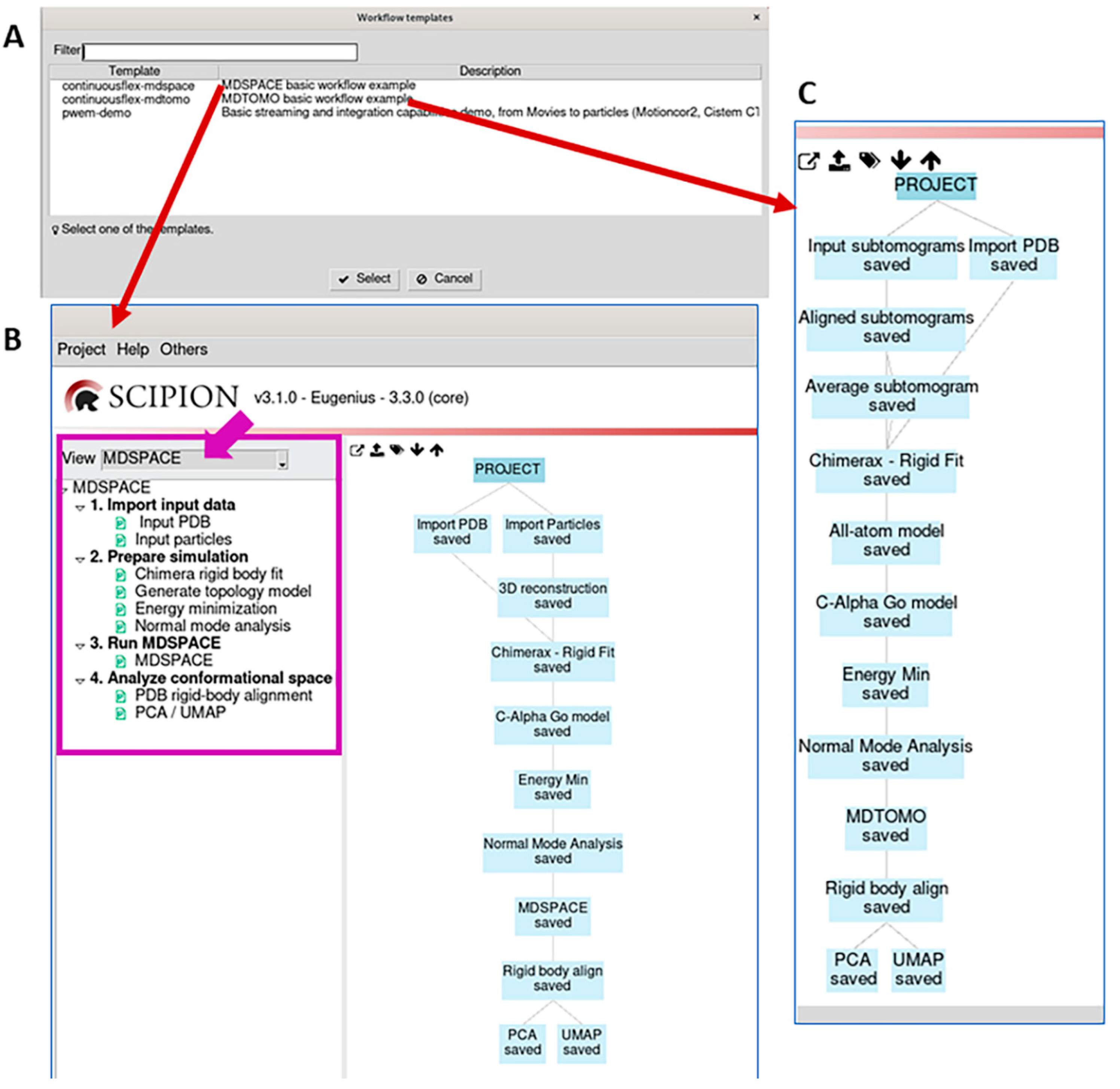
3.1. Import input data
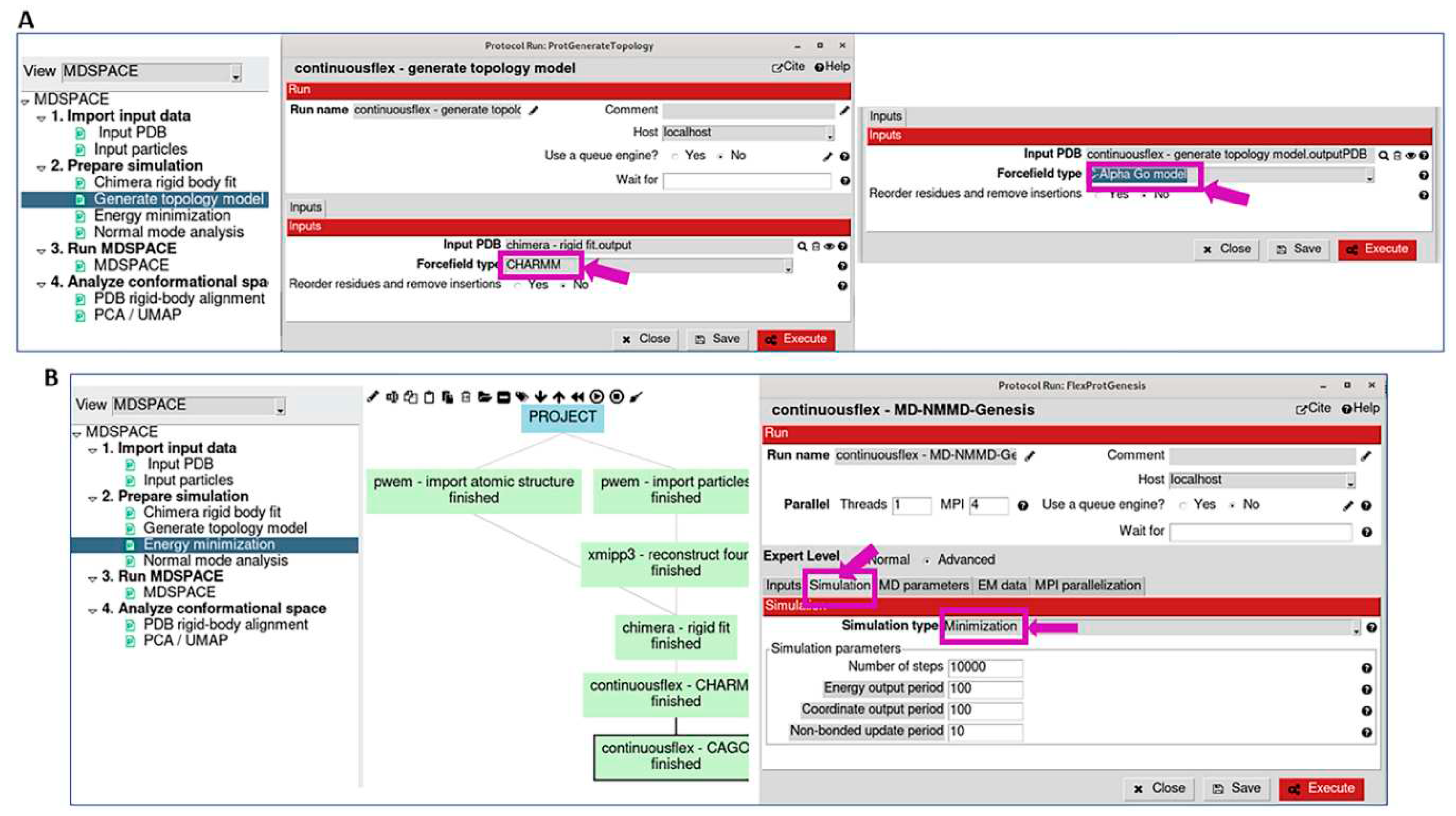
3.2. Prepare simulation
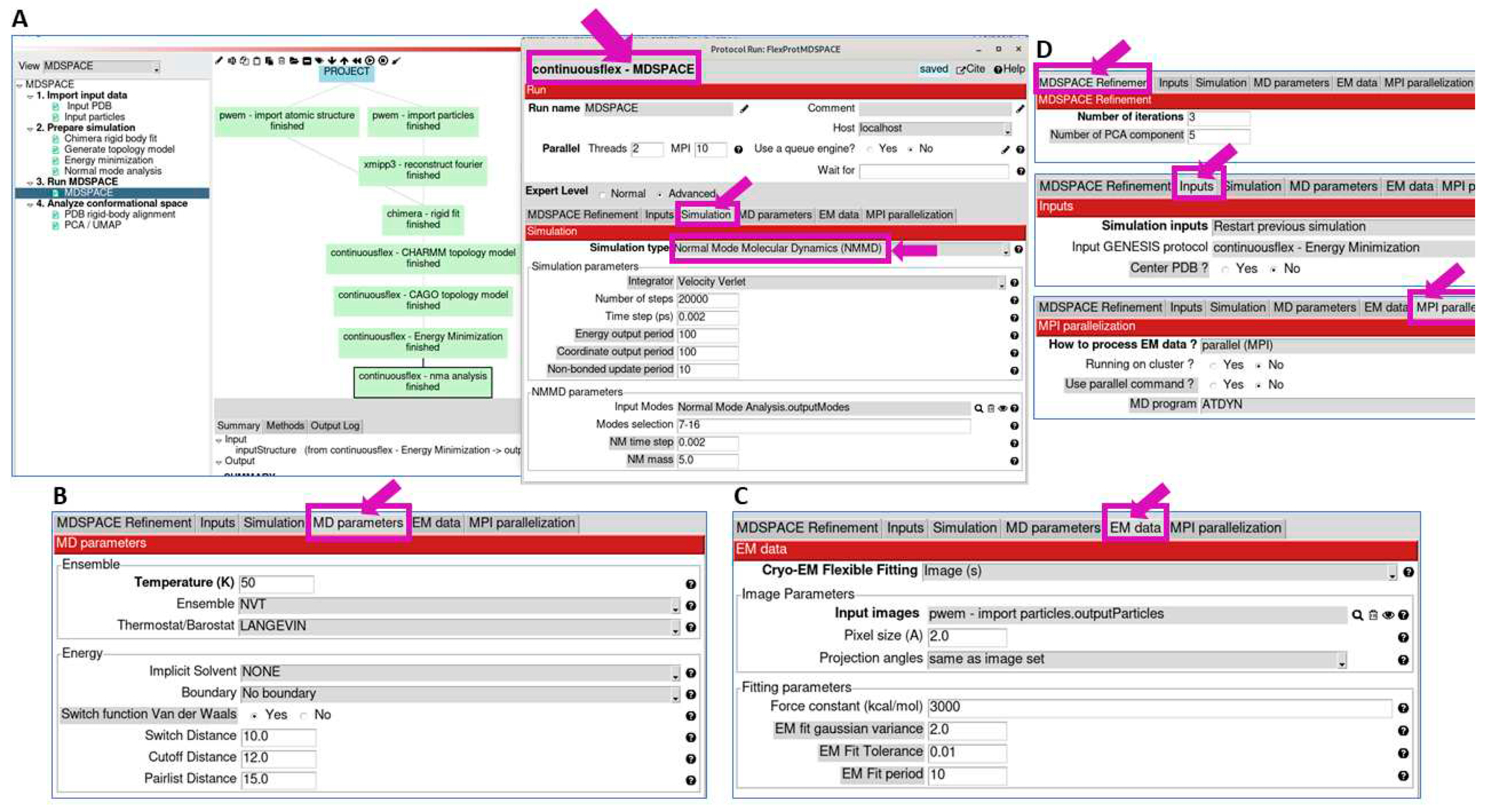
3.3. Run MDSPACE/MDTOMO
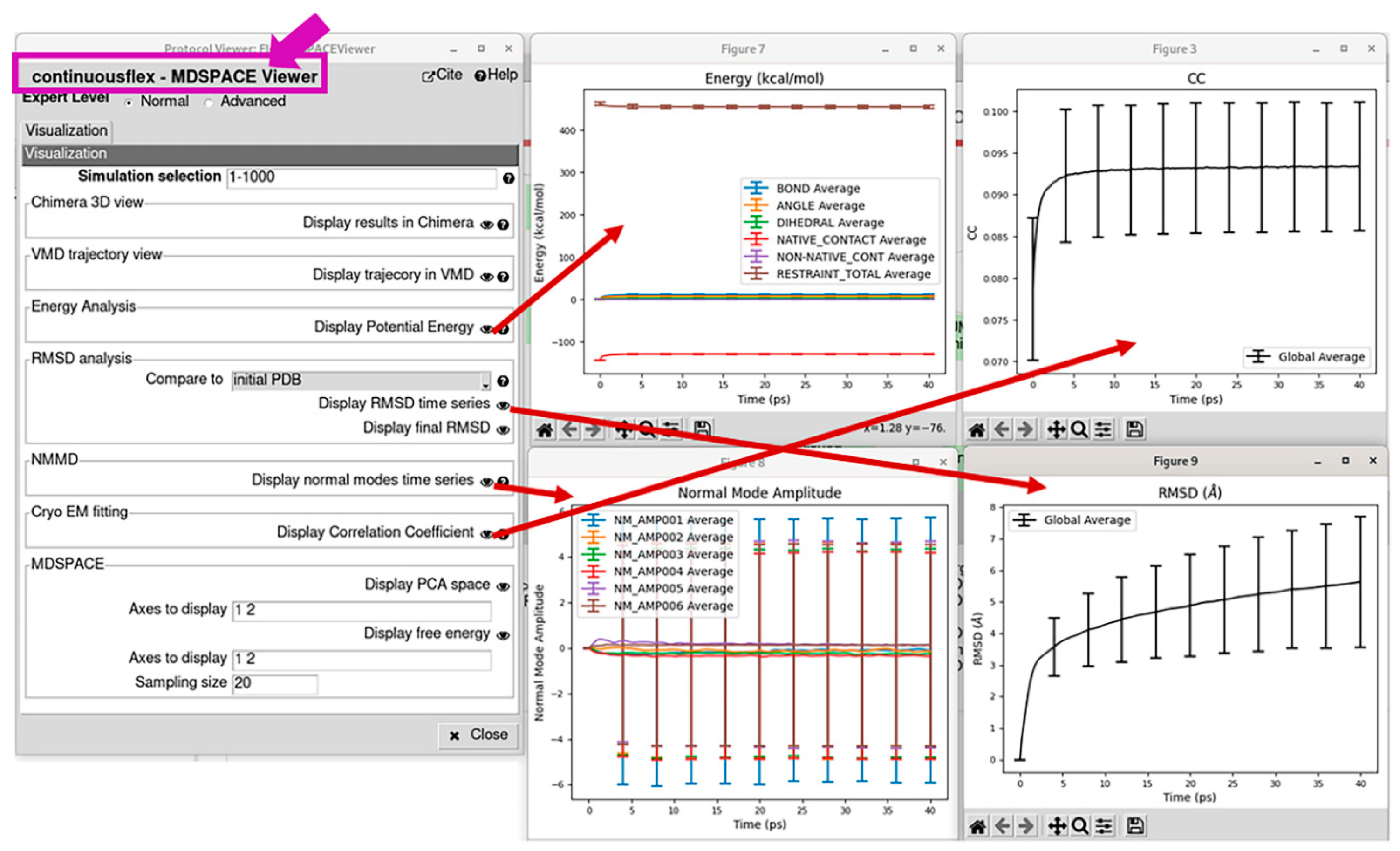
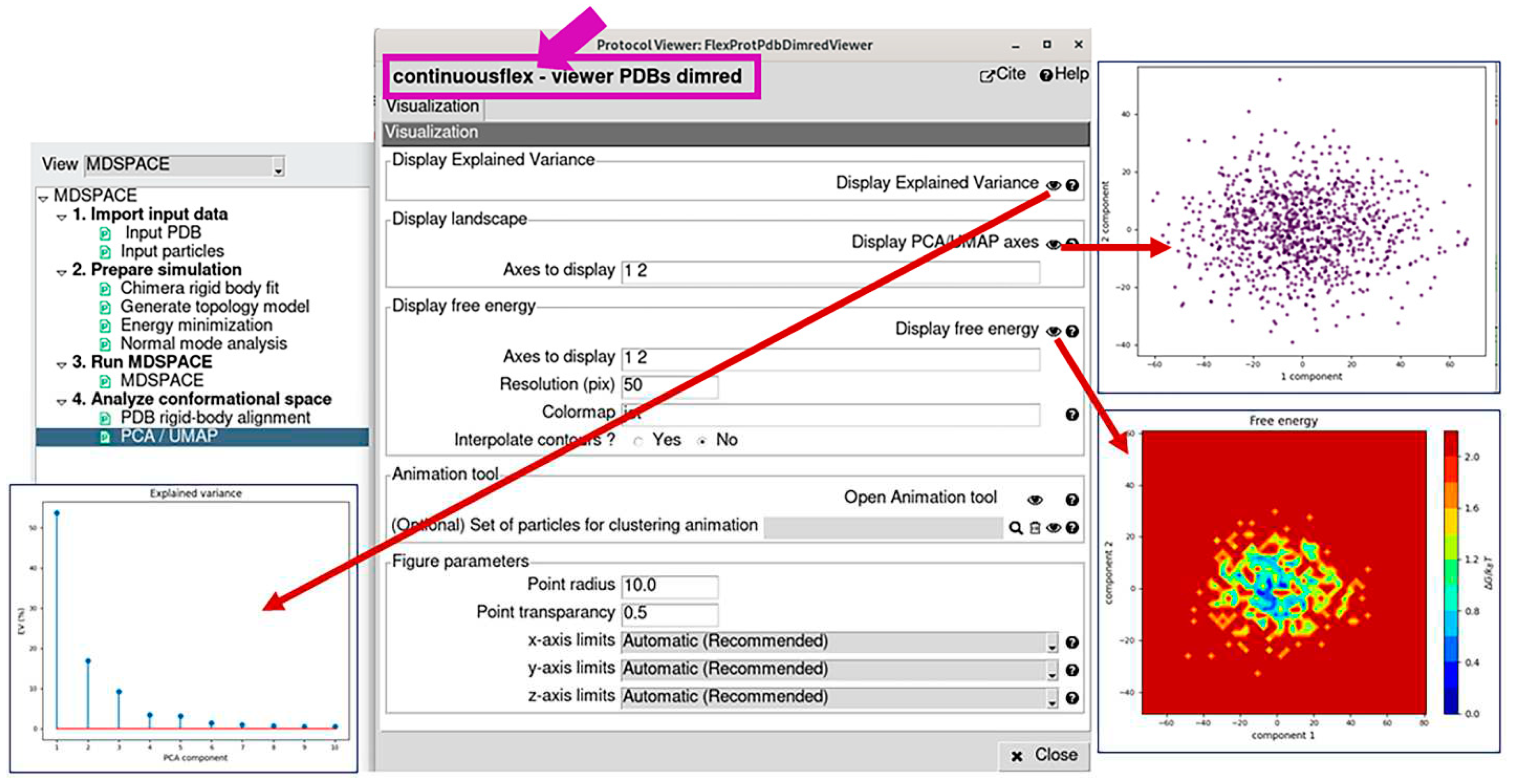
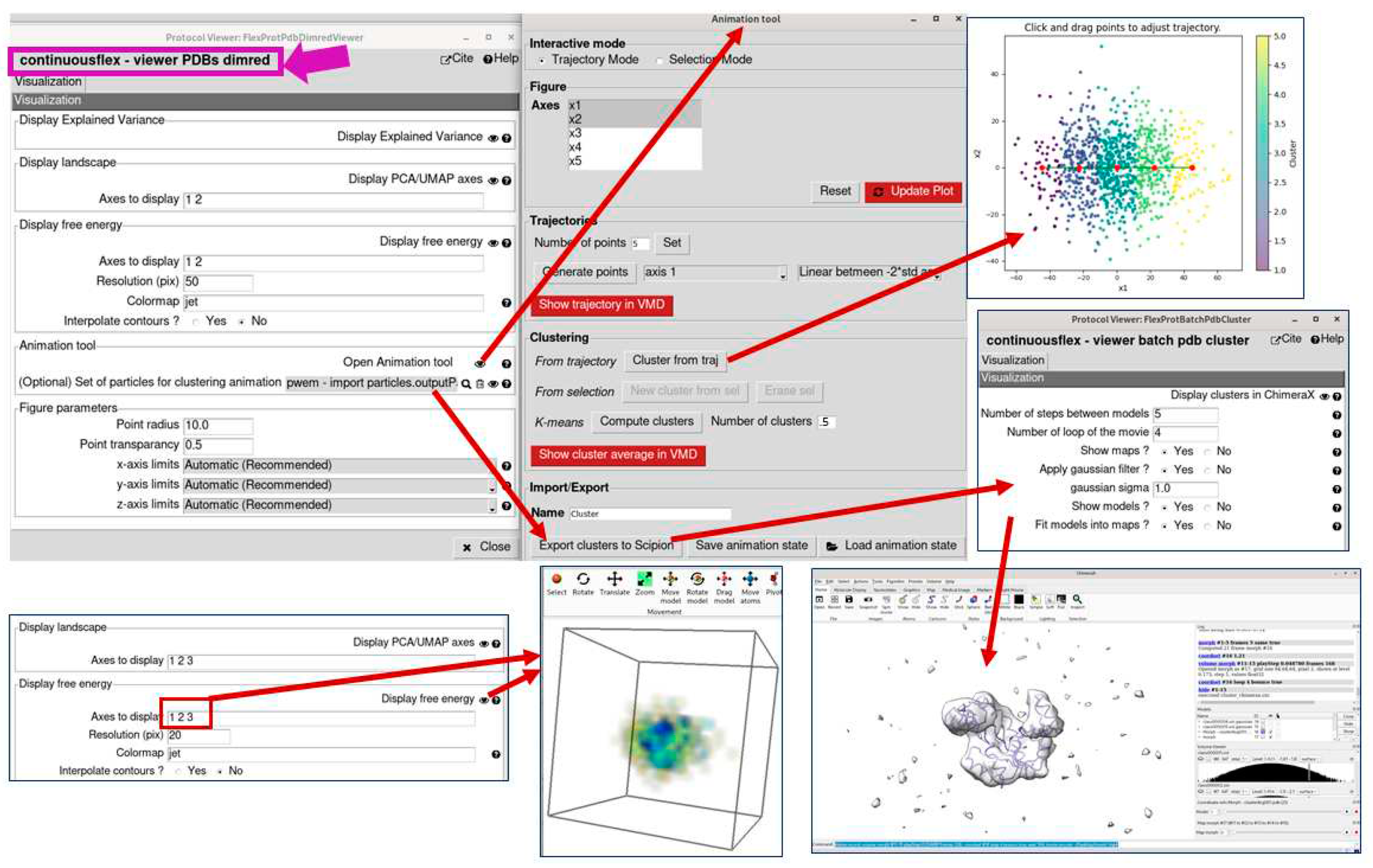
3.4. Analyze conformational space
4. Discussion
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Svidritskiy E, Brilot AF, Koh CS, Grigorieff N, Korostelev AA. Structures of yeast 80S ribosome-tRNA complexes in the rotated and nonrotated conformations. Structure. 2014, 22, 1210–1218.
- Zhou A, Rohou A, Schep DG, Bason JV, Montgomery MG, Walker JE, et al. Structure and conformational states of the bovine mitochondrial ATP synthase by cryo-EM. Elife. 2015, 4, e10180.
- Bai XC, Rajendra E, Yang G, Shi Y, Scheres SH. Sampling the conformational space of the catalytic subunit of human gamma-secretase. Elife. 2015, 4.
- Abeyrathne PD, Koh CS, Grant T, Grigorieff N, Korostelev AA. Ensemble cryo-EM uncovers inchworm-like translocation of a viral IRES through the ribosome. Elife. 2016, 5.
- Banerjee S, Bartesaghi A, Merk A, Rao P, Bulfer SL, Yan Y, et al. 2.3 A resolution cryo-EM structure of human p97 and mechanism of allosteric inhibition. Science. 2016, 351, 871–875.
- Hofmann S, Januliene D, Mehdipour AR, Thomas C, Stefan E, Brüchert S, et al. Conformation space of a heterodimeric ABC exporter under turnover conditions. Nature. 2019, 571, 580–583.
- Nakane T, Kotecha A, Sente A, McMullan G, Masiulis S, Brown PMGE, et al. Single-particle cryo-EM at atomic resolution. Nature. 2020, 587, 152–156.
- Kato K, Miyazaki N, Hamaguchi T, Nakajima Y, Akita F, Yonekura K, et al. High-resolution cryo-EM structure of photosystem II reveals damage from high-dose electron beams. Communications Biology. 2021, 4, 382.
- Schur FK, Obr M, Hagen WJ, Wan W, Jakobi AJ, Kirkpatrick JM, et al. An atomic model of HIV-1 capsid-SP1 reveals structures regulating assembly and maturation. Science. 2016, 353, 506–508.
- Wan W, Kolesnikova L, Clarke M, Koehler A, Noda T, Becker S, et al. Structure and assembly of the Ebola virus nucleocapsid. Nature. 2017, 551, 394–397.
- Himes BA, Zhang P. emClarity: software for high-resolution cryo-electron tomography and subtomogram averaging. Nature Methods. 2018, 15, 955–961.
- von Kügelgen A, Tang H, Hardy GG, Kureisaite-Ciziene D, Brun YV, Stansfeld PJ, et al. In Situ Structure of an Intact Lipopolysaccharide-Bound Bacterial Surface Layer. Cell. 2020, 180, 348–358.
- Scheres SH, Gao H, Valle M, Herman GT, Eggermont PP, Frank J, et al. Disentangling conformational states of macromolecules in 3D-EM through likelihood optimization. Nat Methods. 2007, 4, 27–29.
- Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. J Struct Biol. 2012, 180, 519–530. [CrossRef] [PubMed]
- Lyumkis D, Brilot AF, Theobald DL, Grigorieff N. Likelihood-based classification of cryo-EM images using FREALIGN. J Struct Biol. 2013, 183, 377–388.
- Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nature methods. 2017, 14, 290–296.
- Kimanius D, Dong L, Sharov G, Nakane T, Scheres SHW. New tools for automated cryo-EM single-particle analysis in RELION-4.0. Biochem J. 2021, 478, 4169–4185.
- Scheres SHW, Melero R, Valle M, Carazo J-M. Averaging of Electron Subtomograms and Random Conical Tilt Reconstructions through Likelihood Optimization. Structure. 2009, 17, 1563–1572.
- Stölken M, Beck F, Haller T, Hegerl R, Gutsche I, Carazo J-M, et al. Maximum likelihood based classification of electron tomographic data. Journal of Structural Biology. 2011, 173, 77–85.
- Bharat TAM, Scheres SHW. Resolving macromolecular structures from electron cryo-tomography data using subtomogram averaging in RELION. Nature Protocols. 2016, 11, 2054–2065.
- Jin Q, Sorzano COS, De La Rosa-Trevín JM, Bilbao-Castro JR, Núñez-Ramírez R, Llorca O, et al. Iterative elastic 3D-to-2D alignment method using normal modes for studying structural dynamics of large macromolecular complexes. Structure. 2014, 22, 496–506.
- Dashti A, Schwander P, Langlois R, Fung R, Li W, Hosseinizadeh A, et al. Trajectories of the ribosome as a Brownian nanomachine. Proc Natl Acad Sci U S A. 2014, 111, 17492–17497.
- Moscovich A, Halevi A, Andén J, Singer A. Cryo-EM reconstruction of continuous heterogeneity by Laplacian spectral volumes. Inverse Problems. 2020, 36, 024003.
- Lederman RR, Andén J, Singer A. Hyper-molecules: on the representation and recovery of dynamical structures for applications in flexible macro-molecules in cryo-EM. Inverse Problems. 2020, 36, 044005.
- Chen M, Ludtke SJ. Deep learning-based mixed-dimensional Gaussian mixture model for characterizing variability in cryo-EM. Nature Methods. 2021, 18, 930–936.
- Punjani A, Fleet DJ. 3D variability analysis: Resolving continuous flexibility and discrete heterogeneity from single particle cryo-EM. Journal of Structural Biology. 2021, 213, 107702.
- Zhong ED, Bepler T, Berger B, Davis JH. CryoDRGN: reconstruction of heterogeneous cryo-EM structures using neural networks. Nature Methods. 2021, 18, 176–185.
- Zhong ED, Lerer A, Davis JH, Berger B. CryoDRGN2, Ab initio neural reconstruction of 3D protein structures from real cryo-EM images. 2021 IEEE/CVF International Conference on Computer Vision (ICCV)2021. p. 4046-55.
- Levy A, Wetzstein G, Martel J, Poitevin F, Zhong ED. Amortized Inference for Heterogeneous Reconstruction in Cryo-EM. Adv Neural Inf Process Syst. 2022, 35, 13038–13049.
- Hamitouche I, Jonic S. DeepHEMNMA: ResNet-based hybrid analysis of continuous conformational heterogeneity in cryo-EM single particle images. Front Mol Biosci. 2022, 9, 965645.
- Punjani A, Fleet DJ. 3DFlex: determining structure and motion of flexible proteins from cryo-EM. Nature Methods. 2023, 20, 860–870.
- Herreros D, Lederman RR, Krieger JM, Jiménez-Moreno A, Martínez M, Myška D, et al. Estimating conformational landscapes from Cryo-EM particles by 3D Zernike polynomials. Nature Communications. 2023, 14, 154.
- Vuillemot R, Mirzaei A, Harastani M, Hamitouche I, Fréchin L, Klaholz BP, et al. MDSPACE: Extracting Continuous Conformational Landscapes from Cryo-EM Single Particle Datasets Using 3D-to-2D Flexible Fitting based on Molecular Dynamics Simulation. J Mol Biol. 2023, 435, 167951.
- Harastani M, Eltsov M, Leforestier A, Jonic S. HEMNMA-3D: Cryo Electron Tomography Method Based on Normal Mode Analysis to Study Continuous Conformational Variability of Macromolecular Complexes. Frontiers in molecular biosciences. 2021, 8, 663121.
- Vuillemot R, Rouiller I, Jonić S. MDTOMO method for continuous conformational variability analysis in cryo electron subtomograms based on molecular dynamics simulations. Scientific Reports. 2023, 13, 10596.
- Harastani M, Eltsov M, Leforestier A, Jonic S. TomoFlow: Analysis of continuous conformational variability of macromolecules in cryogenic subtomograms based on 3D dense optical flow. Journal of molecular biology. 2022, 434, 167381.
- Powell BM, Davis JH. Learning structural heterogeneity from cryo-electron sub-tomograms with tomoDRGN. bioRxiv. 2023.
- Tagare HD, Kucukelbir A, Sigworth FJ, Wang H, Rao M. Directly reconstructing principal components of heterogeneous particles from cryo-EM images. Journal of structural biology. 2015, 191, 245–262.
- Katsevich E, Katsevich A, Singer A. Covariance Matrix Estimation for the Cryo-EM Heterogeneity Problem. SIAM J Imaging Sci. 2015, 8, 126–185.
- Liao Hstau Y, Hashem Y, Frank J. Efficient Estimation of Three-Dimensional Covariance and its Application in the Analysis of Heterogeneous Samples in Cryo-Electron Microscopy. Structure. 2015, 23, 1129–1137.
- Marshall NF, Mickelin O, Shi Y, Singer A. Fast principal component analysis for cryo-electron microscopy images. Biological Imaging. 2023, 3, e2.
- Tama F, Miyashita O, Brooks CL. Flexible Multi-scale Fitting of Atomic Structures into Low-resolution Electron Density Maps with Elastic Network Normal Mode Analysis. Journal of Molecular Biology. 2004, 337, 985–999.
- Orzechowski M, Tama F. Flexible fitting of high-resolution x-ray structures into cryoelectron microscopy maps using biased molecular dynamics simulations. Biophysical journal. 2008, 95, 5692–5705.
- Miyashita O, Kobayashi C, Mori T, Sugita Y, Tama F. Flexible fitting to cryo-EM density map using ensemble molecular dynamics simulations. Journal of computational chemistry. 2017, 38, 1447–1461.
- Miyashita O, Tama F. Hybrid Methods for Macromolecular Modeling by Molecular Mechanics Simulations with Experimental Data. Adv Exp Med Biol. 2018, 1105, 199–217.
- Vuillemot R, Miyashita O, Tama F, Rouiller I, Jonic S. NMMD: Efficient cryo-EM flexible fitting based on simultaneous Normal Mode and Molecular Dynamics atomic displacements. Journal of Molecular Biology. 2022, 434, 167483.
- Harastani M, Vuillemot R, Hamitouche I, Barati Moghadam N, Jonic S. ContinuousFlex: Software package for analyzing continuous conformational variability of macromolecules in cryo electron microscopy and tomography data. Journal of Structural Biology. 2022, 107906.
- Conesa P, Fonseca YC, Jiménez de la Morena J, Sharov G, de la Rosa-Trevín JM, Cuervo A, et al. Scipion3, A workflow engine for cryo-electron microscopy image processing and structural biology. Biological Imaging. 2023, 3, e13.
- De la Rosa-Trevín J, Quintana A, Del Cano L, Zaldívar A, Foche I, Gutiérrez J, et al. Scipion: A software framework toward integration, reproducibility and validation in 3D electron microscopy. Journal of structural biology. 2016, 195, 93–99.
- Jiménez de la Morena J, Conesa P, Fonseca YC, de Isidro-Gómez FP, Herreros D, Fernández-Giménez E, et al. ScipionTomo: Towards cryo-electron tomography software integration, reproducibility, and validation. Journal of Structural Biology. 2022, 214, 107872.
- Herreros D, Krieger JM, Fonseca Y, Conesa P, Harastani M, Vuillemot R, et al. Scipion Flexibility Hub: an integrative framework for advanced analysis of conformational heterogeneity in cryoEM. Acta Crystallographica Section D. 2023, 79, 569–584.
- Harastani M, Sorzano COS, Jonić S. Hybrid Electron Microscopy Normal Mode Analysis with Scipion. Protein Science. 2020, 29, 223–236.
- Pearson K. LIII. On lines and planes of closest fit to systems of points in space. The London, Edinburgh, and Dublin Philosophical Magazine and Journal of Science. 1901, 2, 559–572.
- McInnes L, Healy J, Melville J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. ArXiv. 2018, abs/1802.03426.
- Kobayashi C, Jung J, Matsunaga Y, Mori T, Ando T, Tamura K, et al. GENESIS 1.1, A hybrid-parallel molecular dynamics simulator with enhanced sampling algorithms on multiple computational platforms. J Comput Chem. 2017, 38, 2193–2206.
- Huang J, MacKerell Jr AD. CHARMM36 all-atom additive protein force field: Validation based on comparison to NMR data. Journal of computational chemistry. 2013, 34, 2135–2145.
- Karanicolas J, Brooks CL. Improved Gō-like Models Demonstrate the Robustness of Protein Folding Mechanisms Towards Non-native Interactions. Journal of Molecular Biology. 2003, 334, 309–325.
- Noel JK, Levi M, Raghunathan M, Lammert H, Hayes RL, Onuchic JN, et al. SMOG 2, A Versatile Software Package for Generating Structure-Based Models. PLoS computational biology2016. p. e1004794.
- Suhre K, Sanejouand YH. ElNemo: a normal mode web server for protein movement analysis and the generation of templates for molecular replacement. Nucleic Acids Res. 2004, 32, W610–W614.
- Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Science. 2018, 27, 14–25.
- Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996, 14, 33–38.
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




