Submitted:
01 November 2023
Posted:
07 November 2023
You are already at the latest version
Abstract
Keywords:
Introduction
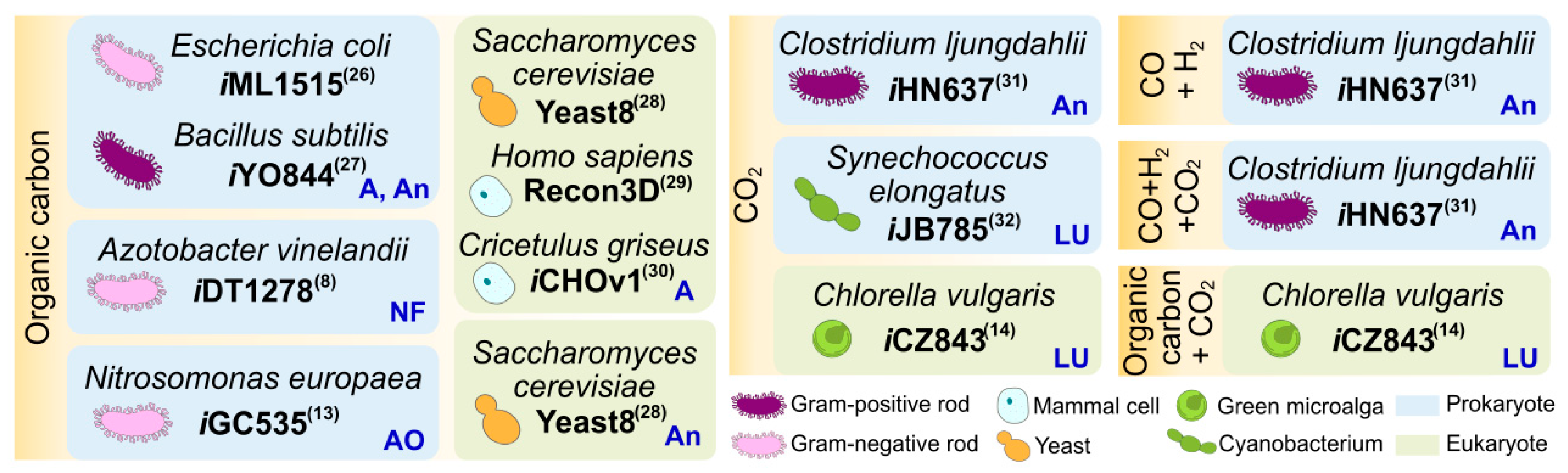
Tip 1. Retrieve the genomic and proteomic information of the target organism.
Tip 2. Identify basic metabolic your microorganism of interest.
Tip 3. Semi-automatic reconstruction of a draft model
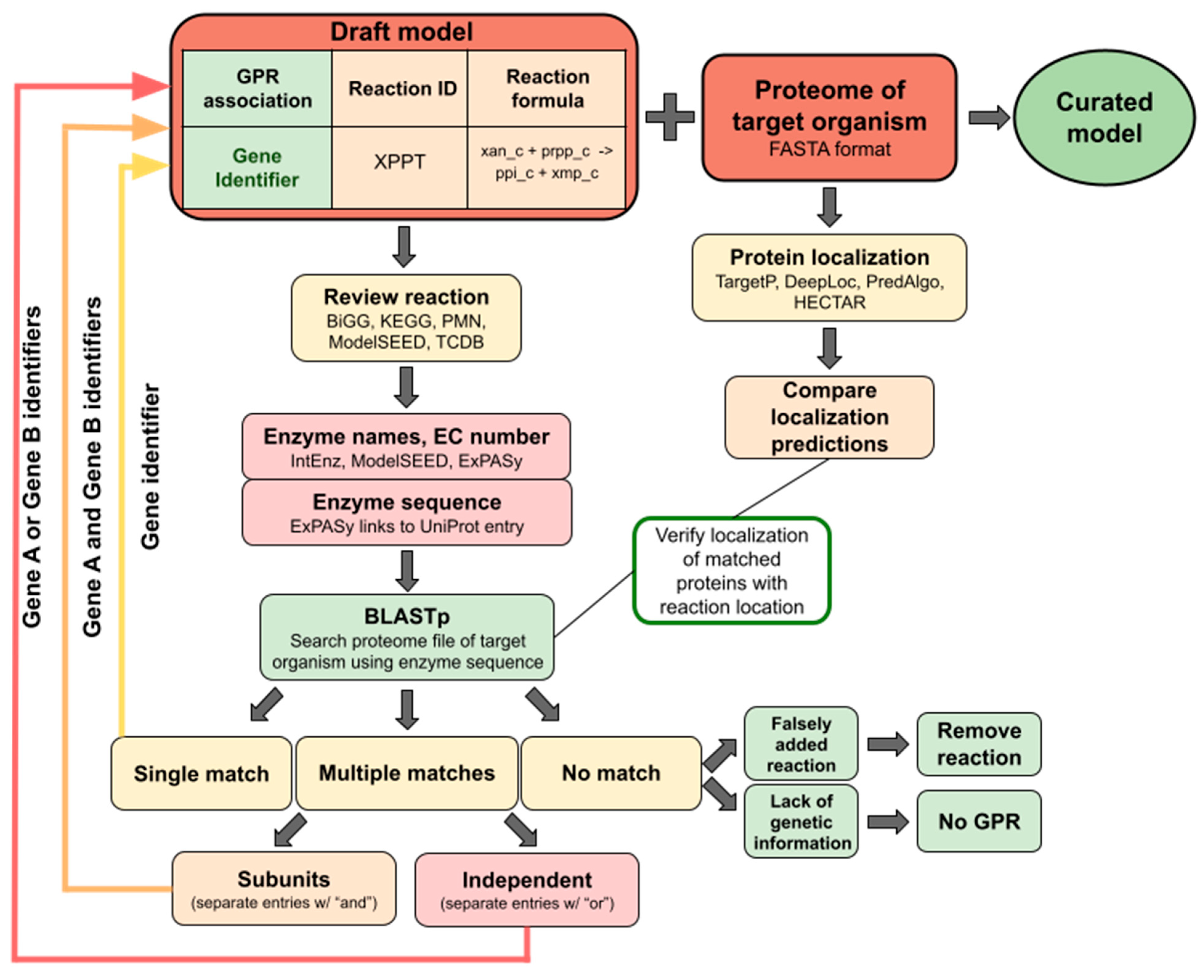
Tip 4. Manual verification of GRP associations.
Tip 5. Addition of constraints to simulate basic metabolic capabilities, generating the QC/QA script
Tip 6. Determination of the biomass objective function.
Tip 7. Addition of new metabolites and pathways based on untargeted metabolomics data
Tip 8. Gap-filling using high-throughput experimental data.
Tip 9. Addition of metadata to metabolites and reactions is critical to ensure compatibility.
Tip 10. Sharable format JSON, MAT, SBML, XML, and visualization
Conclusion
Supplementary Materials
Acknowledgements
References
- Tavassoly I, Goldfarb J, Iyengar R. Systems biology primer: The basic methods and approaches. Vol. 62, Essays in Biochemistry. Portland Press Ltd; 2018. p. 487–500. [CrossRef]
- Thomas PD, Ebert D, Muruganujan A, Mushayahama T, Albou LP, Mi H. PANTHER: Making genome-scale phylogenetics accessible to all. Vol. 31, Protein Science. John Wiley and Sons Inc; 2022. p. 8–22. [CrossRef]
- Montagud A, Ponce-de-Leon M, Valencia A. Systems biology at the giga-scale: Large multiscale models of complex, heterogeneous multicellular systems. Vol. 28, Current Opinion in Systems Biology. Elsevier Ltd; 2021. [CrossRef]
- Ngo RJK, Yeoh JW, Fan GHW, Loh WKS, Poh CL. BMSS2: A Unified Database-Driven Modeling Tool for Systematic Biomodel Selection. ACS Synth Biol. 2022 Aug 19;11(8):2901–6. [CrossRef]
- Erdem C, Birtwistle MR. MEMMAL: A tool for expanding large-scale mechanistic models with machine learned associations and big datasets. Frontiers in Systems Biology. 2023 Mar 9;3. [CrossRef]
- Thiele I, Palsson B. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat Protoc. 2010 Jan;5(1):93–121. [CrossRef]
- Li CT, Eng R, Zuniga C, Huang KW, Chen Y, Zengler K, et al. Optimization of nutrient utilization efficiency and productivity for algal cultures under light and dark cycles using genome-scale model process control. NPJ Syst Biol Appl. 2023 Dec 1;9(1). [CrossRef]
- Tec-Campos D, Zuñiga C, Passi A, Del Toro J, Tibocha-Bonilla JD, Zepeda A, et al. Modeling of nitrogen fixation and polymer production in the heterotrophic diazotroph Azotobacter vinelandii. Metab Eng Commun. 2020 Dec 1;11. [CrossRef]
- Passi A, Tibocha-Bonilla JD, Kumar M, Tec-Campos D, Zengler K, Zuniga C. Genome-scale metabolic modeling enables in-depth understanding of big data. Vol. 12, Metabolites. MDPI; 2022. [CrossRef]
- Gudmundsson S, Nogales J. Recent advances in model-assisted metabolic engineering. Vol. 28, Current Opinion in Systems Biology. Elsevier Ltd; 2021.
- Garcia-Albornoz MA, Nielsen J. Application of Genome-Scale Metabolic Models in Metabolic Engineering. Industrial Biotechnology [Internet]. 2013 Aug 1;9(4):203–14. [CrossRef]
- Norena-Caro DA, Zuniga C, Pete AJ, Saemundsson SA, Donaldson MR, Adams AJ, et al. Analysis of the cyanobacterial amino acid metabolism with a precise genome-scale metabolic reconstruction of Anabaena sp. UTEX 2576. Biochem Eng J. 2021 Jul 1;171. [CrossRef]
- Canto-Encalada G, Tec-Campos D, Tibocha-Bonilla JD, Zengler K, Zepeda A, Zuñiga C. Flux balance analysis of the ammonia-oxidizing bacterium Nitrosomonas europaea ATCC19718 unravels specific metabolic activities while degrading toxic compounds. PLoS Comput Biol. 2022 Feb 1;18(2). [CrossRef]
- Zuñiga C, Li CT, Huelsman T, Levering J, Zielinski DC, McConnell BO, et al. Genome-scale metabolic model for the green alga Chlorella vulgaris UTEX 395 accurately predicts phenotypes under autotrophic, heterotrophic, and mixotrophic growth conditions. Plant Physiol. 2016 Sep 1;172(1):589–602.
- Zuñiga C, Peacock B, Liang B, McCollum G, Irigoyen SC, Tec-Campos D, et al. Linking metabolic phenotypes to pathogenic traits among “Candidatus Liberibacter asiaticus” and its hosts. NPJ Syst Biol Appl. 2020 Dec 1;6(1). [CrossRef]
- Seif Y, Monk JM, Mih N, Tsunemoto H, Poudel S, Zuniga C, et al. A computational knowledge-base elucidates the response of Staphylococcus aureus to different media types. PLoS Comput Biol. 2019;15(1). [CrossRef]
- Tibocha-Bonilla JD, Kumar M, Richelle A, Godoy-Silva RD, Zengler K, Zuñiga C. Dynamic resource allocation drives growth under nitrogen starvation in eukaryotes. NPJ Syst Biol Appl. 2020 Dec 1;6(1). [CrossRef]
- Tec-Campos D, Posadas C, Tibocha-Bonilla JD, Thiruppathy D, Glonek N, Zuñiga C, et al. The genome-scale metabolic model for the purple non-sulfur bacterium Rhodopseudomonas palustris Bis A53 accurately predicts phenotypes under chemoheterotrophic, chemoautotrophic, photoheterotrophic, and photoautotrophic growth conditions. PLoS Comput Biol [Internet]. 2023 Aug 9;19(8):e1011371-. [CrossRef]
- Machado D, Andrejev S, Tramontano M, Patil KR. Fast automated reconstruction of genome-scale metabolic models for microbial species and communities. Nucleic Acids Res. 2018 Sep 6;46(15):7542–53. [CrossRef]
- Henry CS, Dejongh M, Best AA, Frybarger PM, Linsay B, Stevens RL. High-throughput generation, optimization and analysis of genome-scale metabolic models. Nat Biotechnol. 2010 Sep;28(9):977–82. [CrossRef]
- Wang H, Marcišauskas S, Sánchez BJ, Domenzain I, Hermansson D, Agren R, et al. RAVEN 2.0: A versatile toolbox for metabolic network reconstruction and a case study on Streptomyces coelicolor. PLoS Comput Biol. 2018 Oct 1;14(10). [CrossRef]
- Tibocha-Bonilla JD, Zuñiga C, Lekbua A, Lloyd C, Rychel K, Short K, et al. Predicting stress response and improved protein overproduction in Bacillus subtilis. NPJ Syst Biol Appl. 2022 Dec 1;8(1). [CrossRef]
- O’Brien EJ, Lerman JA, Chang RL, Hyduke DR, Palsson B. Genome-scale models of metabolism and gene expression extend and refine growth phenotype prediction. Mol Syst Biol. 2013;9. [CrossRef]
- Zuñiga C, Li T, Guarnieri MT, Jenkins JP, Li CT, Bingol K, et al. Synthetic microbial communities of heterotrophs and phototrophs facilitate sustainable growth. Nat Commun. 2020 Dec 1;11(1). [CrossRef]
- Zuñiga C, Li CT, Yu G, Al-Bassam MM, Li T, Jiang L, et al. Environmental stimuli drive a transition from cooperation to competition in synthetic phototrophic communities. Nat Microbiol. 2019;4(12):2184–91.
- Monk JM, Lloyd CJ, Brunk E, Mih N, Sastry A, King Z, et al. iML1515, a knowledgebase that computes Escherichia coli traits. Vol. 35, Nature Biotechnology. Nature Publishing Group; 2017. p. 904–8. [CrossRef]
- Oh YK, Palsson BO, Park SM, Schilling CH, Mahadevan R. Genome-scale reconstruction of metabolic network in Bacillus subtilis based on high-throughput phenotyping and gene essentiality data. Journal of Biological Chemistry. 2007 Sep 28;282(39):28791–9. [CrossRef]
- Lu H, Li F, Sánchez BJ, Zhu Z, Li G, Domenzain I, et al. A consensus S. cerevisiae metabolic model Yeast8 and its ecosystem for comprehensively probing cellular metabolism. Nat Commun. 2019 Dec 1;10(1).
- Brunk E, Sahoo S, Zielinski DC, Altunkaya A, Dräger A, Mih N, et al. Recon3D enables a three-dimensional view of gene variation in human metabolism. Nat Biotechnol. 2018 Mar 1;36(3):272–81. [CrossRef]
- Hefzi H, Ang KS, Hanscho M, Bordbar A, Ruckerbauer D, Lakshmanan M, et al. A Consensus Genome-scale Reconstruction of Chinese Hamster Ovary Cell Metabolism. Cell Syst. 2016 Nov 23;3(5):434-443.e8. [CrossRef]
- Nagarajan H, Sahin M, Nogales J, Latif H, Lovley DR, Ebrahim A, et al. Characterizing acetogenic metabolism using a genome-scale metabolic reconstruction of Clostridium ljungdahlii. Microb Cell Fact [Internet]. 2013 Nov 25;12(118). Available online: http://www.microbialcellfactories.
- Broddrick JT, Rubin BE, Welkie DG, Du N, Mih N, Diamond S, et al. Unique attributes of cyanobacterial metabolism revealed by improved genome-scale metabolic modeling and essential gene analysis. Proc Natl Acad Sci U S A. 2016 Dec 20;113(51):E8344–53. [CrossRef]
- Heirendt L, Arreckx S, Pfau T, Mendoza SN, Richelle A, Heinken A, et al. Creation and analysis of biochemical constraint-based models using the COBRA Toolbox v.3.0. Nature Protocols 2019 14:3 [Internet]. 2019 Feb 20 [cited 2023 Jun 4];14(3):639–702. Available online: https://www.nature.com/articles/s41596-018-0098-2.
- Karp PD, Midford PE, Billington R, Kothari A, Krummenacker M, Latendresse M, et al. Pathway Tools version 23.0 update: Software for pathway/genome informatics and systems biology. Vol. 22, Briefings in Bioinformatics. Oxford University Press; 2021. p. 109–26. [CrossRef]
- Magnúsdóttir S, Heinken A, Kutt L, Ravcheev DA, Bauer E, Noronha A, et al. Generation of genome-scale metabolic reconstructions for 773 members of the human gut microbiota. Nat Biotechnol. 2017 Jan 1;35(1):81–9. [CrossRef]
- Boratyn GM, Camacho C, Cooper PS, Coulouris G, Fong A, Ma N, et al. BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 2013;41(Web Server issue). [CrossRef]
- Fleischmann A, Darsow M, Degtyarenko K, Fleischmann W, Boyce S, Axelsen KB, et al. IntEnz, the integrated relational enzyme database. Nucleic Acids Res. 2004 Jan 1;32(DATABASE ISS.). [CrossRef]
- Saier MH, Tran C V., Barabote RD. TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res. 2006;34(Database issue).
- Duvaud S, Gabella C, Lisacek F, Stockinger H, Ioannidis V, Durinx C. Expasy, the Swiss Bioinformatics Resource Portal, as designed by its users. Nucleic Acids Res. 2021 Jul 2;49(W1):W216–27. [CrossRef]
- Bremer E, Calteau A, Danchin A, Harwood C, Helmann JD, Médigue C, et al. A model industrial workhorse: Bacillus subtilis strain 168 and its genome after a quarter of a century. Vol. 16, Microbial Biotechnology. John Wiley and Sons Ltd; 2023. p. 1203–31. [CrossRef]
- Emanuelsson O, Nielsen H, Brunak S, Von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000 Jul 21;300(4):1005–16. [CrossRef]
- Gschloessl B, Guermeur Y, Cock JM. HECTAR: A method to predict subcellular targeting in heterokonts. BMC Bioinformatics. 2008 Sep 23;9. [CrossRef]
- Almagro Armenteros JJ, Sønderby CK, Sønderby SK, Nielsen H, Winther O. DeepLoc: prediction of protein subcellular localization using deep learning. Bioinformatics. 2017 Nov 1;33(21):3387–95.
- Tardif M, Atteia A, Specht M, Cogne G, Rolland N, Brugière S, et al. Predalgo: A new subcellular localization prediction tool dedicated to green algae. In: Molecular Biology and Evolution. 2012. p. 3625–39. [CrossRef]
- King ZA, Lu J, Dräger A, Miller P, Federowicz S, Lerman JA, et al. BiGG Models: A platform for integrating, standardizing and sharing genome-scale models. Nucleic Acids Res. 2016;44(D1):D515–22. [CrossRef]
- Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000 Jan 1;28(1):27–30.
- Hawkins C, Ginzburg D, Zhao K, Dwyer W, Xue B, Xu A, et al. Plant Metabolic Network 15: A resource of genome-wide metabolism databases for 126 plants and algae. Vol. 63, Journal of Integrative Plant Biology. John Wiley and Sons Inc; 2021. p. 1888–905. [CrossRef]
- Seaver SMD, Liu F, Zhang Q, Jeffryes J, Faria JP, Edirisinghe JN, et al. The ModelSEED Biochemistry Database for the integration of metabolic annotations and the reconstruction, comparison and analysis of metabolic models for plants, fungi and microbes. Nucleic Acids Res. 2021 Jan 8;49(D1):D575–88.
- Bairoch, A. The ENZYME database in 2000. Nucleic Acids Res. 2000;28(1):304–5. [CrossRef]
- Bateman A, Martin MJ, Orchard S, Magrane M, Ahmad S, Alpi E, et al. UniProt: the Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023 Jan 6;51(D1):D523–31.
- Orth JD, Thiele I, Palsson BO. What is flux balance analysis? Vol. 28, Nature Biotechnology. 2010. p. 245–8.
- Gustafsson J, Anton M, Roshanzamir F, Jörnsten R, Kerkhoven EJ, Robinson JL, et al. Generation and analysis of context-specific genome-scale metabolic models derived from single-cell RNA-Seq data. Proc Natl Acad Sci U S A. 2023 Feb 7;120(6).
- Hamilton JJ, Dwivedi V, Reed JL. Quantitative assessment of thermodynamic constraints on the solution space of genome-scale metabolic models. Biophys J. 2013 Jul 16;105(2):512–22. [CrossRef]
- Fritzemeier CJ, Hartleb D, Szappanos B, Papp B, Lercher MJ. Erroneous energy-generating cycles in published genome scale metabolic networks: Identification and removal. PLoS Comput Biol. 2017 Apr 1;13(4). [CrossRef]
- Lieven C, Beber ME, Olivier BG, Bergmann FT, Ataman M, Babaei P, et al. MEMOTE for standardized genome-scale metabolic model testing. Nat Biotechnol [Internet]. 2020;38(3):272–6. [CrossRef]
- Chang RL, Ghamsari L, Manichaikul A, Hom EFY, Balaji S, Fu W, et al. Metabolic network reconstruction of Chlamydomonas offers insight into light-driven algal metabolism. Mol Syst Biol. 2011;7. [CrossRef]
- Tibocha-Bonilla JD, Kumar M, Zengler K, Zuniga C. Integrating Metabolic Modeling and High-Throughput Data to Characterize Diatoms Metabolism. In: The Mathematical Biology of Diatoms. Wiley; 2023. p. 165–91.
- Feist AM, Palsson BO. The biomass objective function. Vol. 13, Current Opinion in Microbiology. 2010. p. 344–9.
- Lachance JC, Lloyd CJ, Monk JM, Yang L, Sastry A V., Seif Y, et al. BOFDAT: Generating biomass objective functions for genome-scale metabolic models from experimental data. PLoS Comput Biol. 2019;15(4). [CrossRef]
- Broddrick JT, Welkie DG, Jallet D, Golden SS, Peers G, Palsson BO. Predicting the metabolic capabilities of Synechococcus elongatus PCC 7942 adapted to different light regimes. Metab Eng [Internet]. 2019;52:42–56. Available online: https://www.sciencedirect.com/science/article/pii/S1096717618303288.
- Schrimpe-Rutledge AC, Codreanu SG, Sherrod SD, McLean JA. Untargeted Metabolomics Strategies—Challenges and Emerging Directions. J Am Soc Mass Spectrom. 2016 Dec 1;27(12):1897–905. [CrossRef]
- Zhou F, Zuo J, Gao L, Sui Y, Wang Q, Jiang A, et al. An untargeted metabolomic approach reveals significant postharvest alterations in vitamin metabolism in response to LED irradiation in pak-choi (Brassica campestris L. ssp. chinensis (L.) Makino var. communis Tsen et Lee). Metabolomics. 2019 Dec 1;15(12). [CrossRef]
- Lommen A, van der Weg G, van Engelen MC, Bor G, Hoogenboom LAP, Nielen MWF. An untargeted metabolomics approach to contaminant analysis: Pinpointing potential unknown compounds. Anal Chim Acta. 2007 Feb 12;584(1):43–9. [CrossRef]
- Bernstein DB, Sulheim S, Almaas E, Segrè D. Addressing uncertainty in genome-scale metabolic model reconstruction and analysis. Genome Biol [Internet]. 2021;22(1):64. [CrossRef]
- Pan S, Reed JL. Advances in gap-filling genome-scale metabolic models and model-driven experiments lead to novel metabolic discoveries. Vol. 51, Current Opinion in Biotechnology. Elsevier Ltd; 2018. p. 103–8. [CrossRef]
- Karp PD, Weaver D, Latendresse M. How accurate is automated gap filling of metabolic models? BMC Syst Biol. 2018 Jun 19;12(1).
- Thiele I, Vlassis N, Fleming RMT. FASTGAPFILL: Efficient gap filling in metabolic networks. Bioinformatics. 2014 Sep 1;30(17):2529–31. [CrossRef]
- Hartleb D, Jarre F, Lercher MJ. Improved Metabolic Models for E. coli and Mycoplasma genitalium from GlobalFit, an Algorithm That Simultaneously Matches Growth and Non-Growth Data Sets. PLoS Comput Biol. 2016 Aug 1;12(8). [CrossRef]
- Boughorbel S, Jarray F, El-Anbari M. Optimal classifier for imbalanced data using Matthews Correlation Coefficient metric. PLoS One. 2017 Jun 1;12(6). [CrossRef]
- Leonidou N, Fritze E, Renz A, Dr¨ager AD. SBOannotator: a Python Tool for the Automated Assignment of Systems Biology Ontology Terms. 2023. Available online: https://uni-tuebingen.de/en/216529.
- Anton M, Almaas E, Benfeitas R, Benito-Vaquerizo S, Blank LM, Dräger A, et al. standard-GEM: standardization of open-source genome-scale metabolic models. bioRxiv [Internet]. 2023. [CrossRef]
- Carey MA, Dräger A, Beber ME, Papin JA, Yurkovich JT. Community standards to facilitate development and address challenges in metabolic modeling. Mol Syst Biol. 2020 Aug;16(8). [CrossRef]
- Thiele I, Preciat G, Fleming RMT. MetaboAnnotator: an efficient toolbox to annotate metabolites in genome-scale metabolic reconstructions. Bioinformatics. 2022 Oct 14;38(20):4831–2. [CrossRef]
- Bansal P, Morgat A, Axelsen KB, Muthukrishnan V, Coudert E, Aimo L, et al. Rhea, the reaction knowledgebase in 2022. Nucleic Acids Res. 2022 Jan 7;50(D1):D693–700. [CrossRef]
- Moretti S, Tran VDT, Mehl F, Ibberson M, Pagni M. MetaNetX/MNXref: Unified namespace for metabolites and biochemical reactions in the context of metabolic models. Nucleic Acids Res. 2021 Jan 8;49(D1):D570–4. [CrossRef]
- Karp PD, Billington R, Caspi R, Fulcher CA, Latendresse M, Kothari A, et al. The BioCyc collection of microbial genomes and metabolic pathways. Brief Bioinform. 2018 Mar 27;20(4):1085–93. [CrossRef]
- Gillespie M, Jassal B, Stephan R, Milacic M, Rothfels K, Senff-Ribeiro A, et al. The reactome pathway knowledgebase 2022. Nucleic Acids Res. 2022 Jan 7;50(D1):D687–92.
- Hucka M, Bergmann FT, Hoops S, Keating SM, Sahle S, Schaff JC, et al. The Systems Biology Markup Language (SBML): Language Specification for Level 3 Version 1 Core. J Integr Bioinform. 2015;12(2):266.
- Hucka M, Bergmann FT, Dräger A, Hoops S, Keating SM, Le Novère N, et al. Systems Biology Markup Language (SBML) Level 2 Version 5: Structures and Facilities for Model Definitions. J Integr Bioinform. 2015;12(2):271.
- JSON [Internet]. [cited 2023 Jul 18]. Available online: https://www.json.org/json-en.html.
- Ebrahim A, Lerman JA, Palsson BO, Hyduke DR. COBRApy: COnstraints-Based Reconstruction and Analysis for Python. BMC Syst Biol. 2013 Aug 8;7. [CrossRef]
- Arkin AP, Cottingham RW, Henry CS, Harris NL, Stevens RL, Maslov S, et al. KBase: The United States department of energy systems biology knowledgebase. Vol. 36, Nature Biotechnology. Nature Publishing Group; 2018. p. 566–9. [CrossRef]
- Ben-Kiki O, Evans C, Ingerson B, Oren Ben-Kiki by. YAML Ain’t Markup Language (YAMLTM) Version 1.1 Working Draft 2005-01-18-CVS XSL • FO RenderX YAML Ain’t Markup Language (YAMLTM) Version 1.1 Working Draft 2005-01-18-CVS [Internet]. 2001. Available online: http://www.unicode.org/.
- Norsigian CJ, Fang X, Seif Y, Monk JM, Palsson BO. A workflow for generating multi-strain genome-scale metabolic models of prokaryotes. Nat Protoc. 2020 Jan 1;15(1):1–14. [CrossRef]
- Domenzain I, Sánchez B, Anton M, Kerkhoven EJ, Millán-Oropeza A, Henry C, et al. Reconstruction of a catalogue of genome-scale metabolic models with enzymatic constraints using GECKO 2.0. Nat Commun. 2022 Dec 1;13(1). [CrossRef]
- Heinken A, Thiele I. Microbiome Modelling Toolbox 2.0: efficient, tractable modelling of microbiome communities. Bioinformatics. 2022 Apr 15;38(8):2367–8. [CrossRef]
- Heinken A, Basile A, Thiele I. Advances in constraint-based modelling of microbial communities. Curr Opin Syst Biol [Internet]. 2021;27:100346. Available online: https://www.sciencedirect.com/science/article/pii/S2452310021000317.
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




