Submitted:
22 December 2023
Posted:
22 December 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Cell lines
2.2. Antibodies
2.3. Production of hybridomas
2.4. Flow cytometry
2.5. Determination of the binding affinity by flow cytometry
2.6. Western blot analysis
3. Results
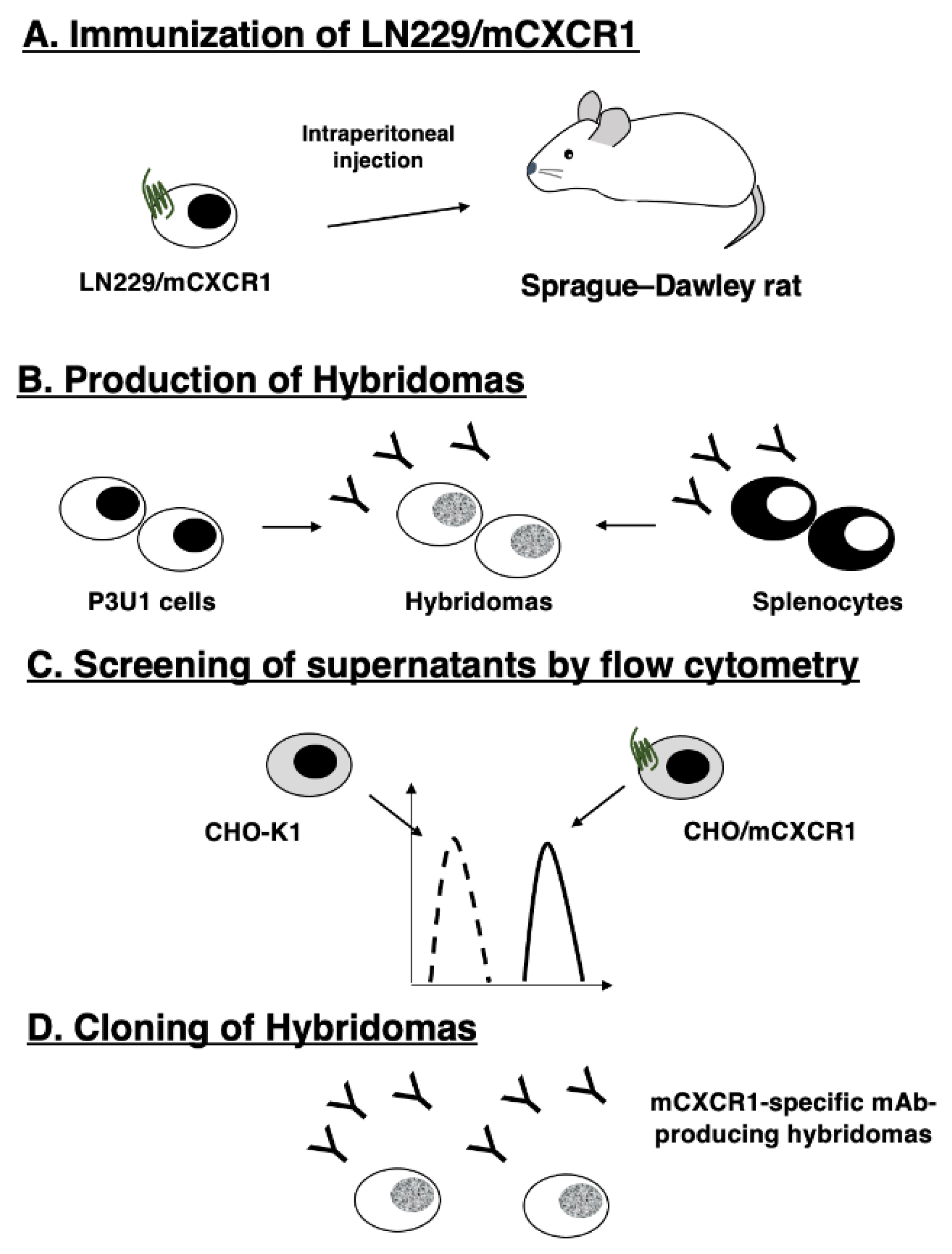
3.1. Establishment of anti-mCXCR1 mAbs
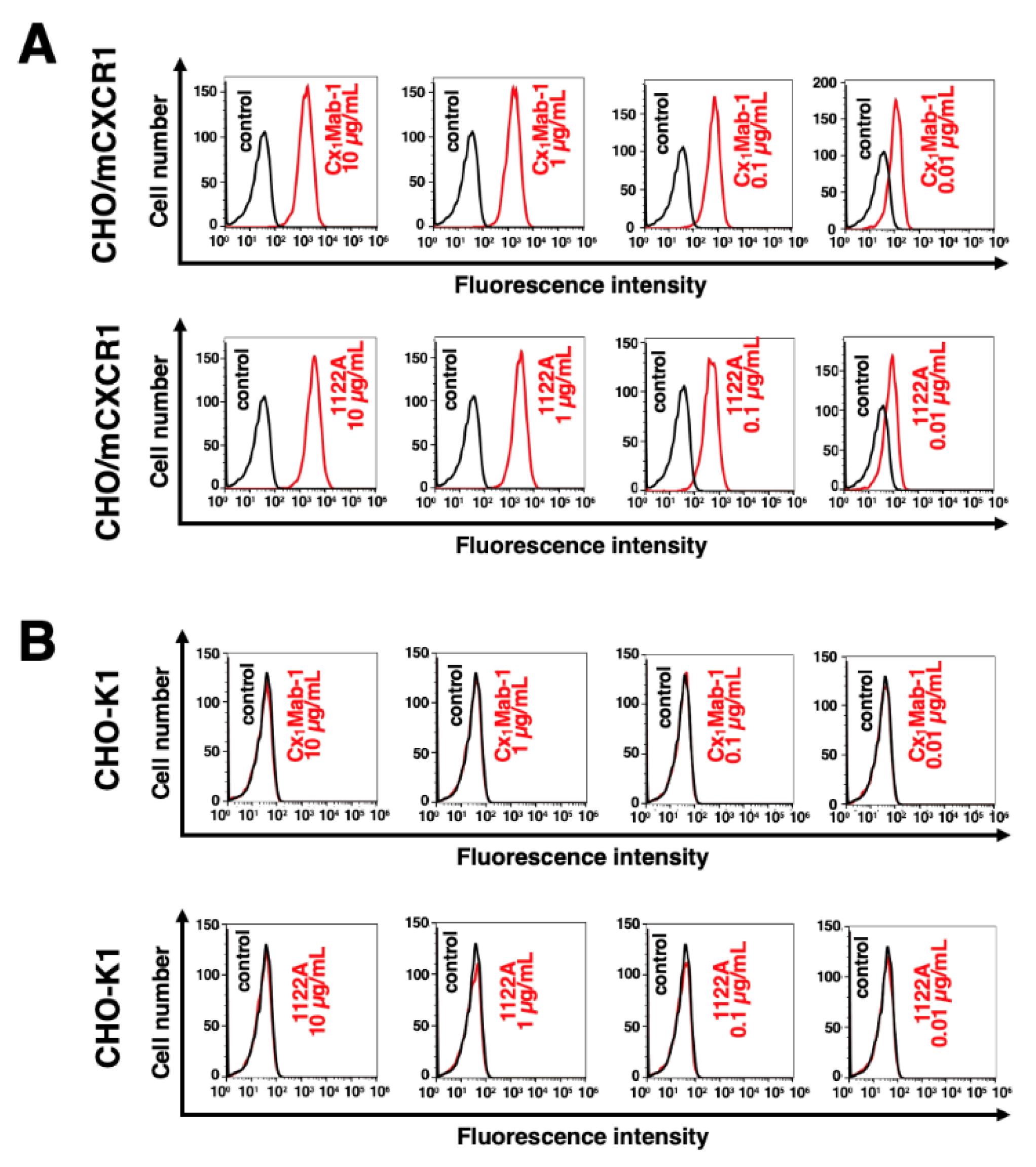
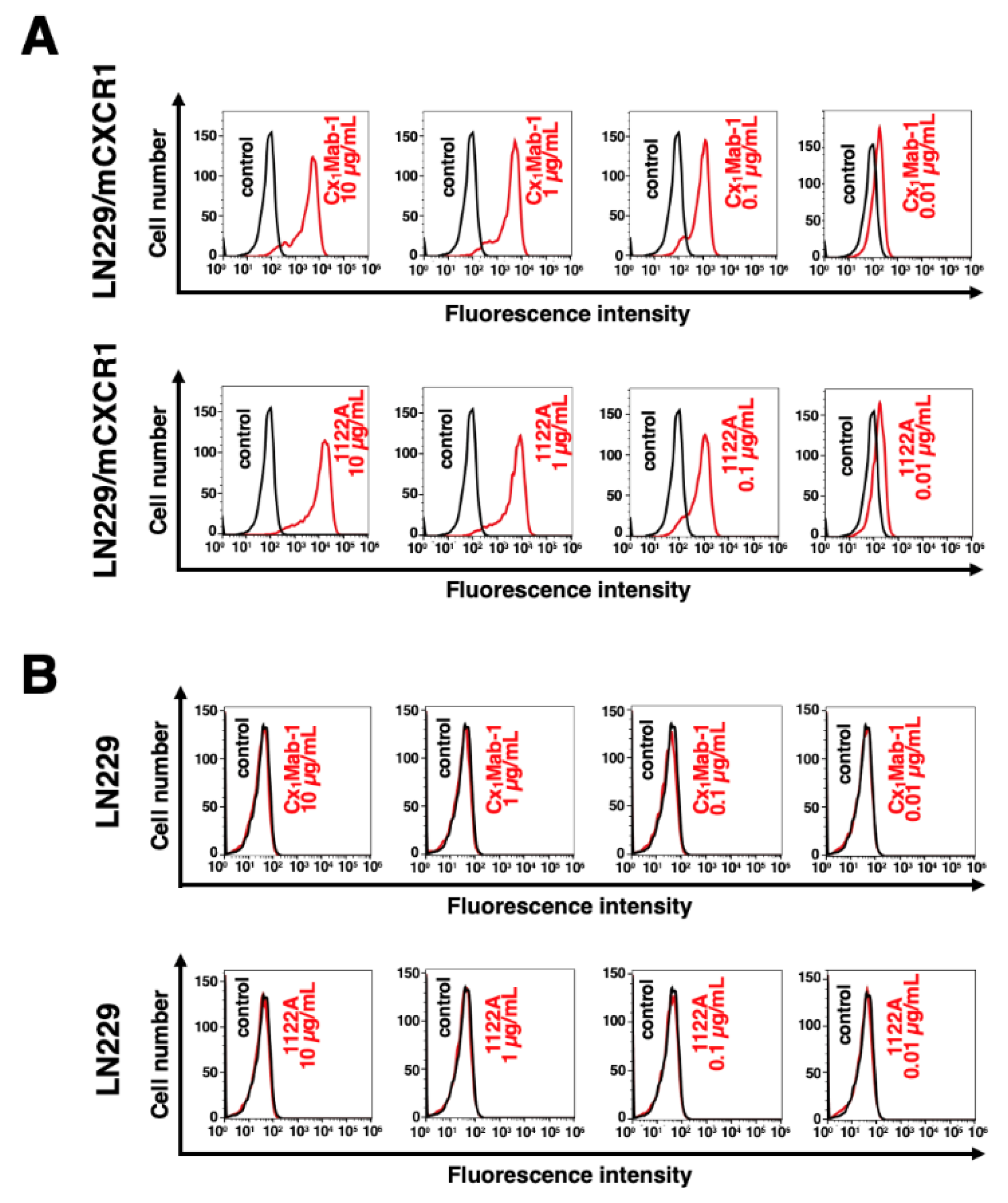
3.2. Flow cytometry
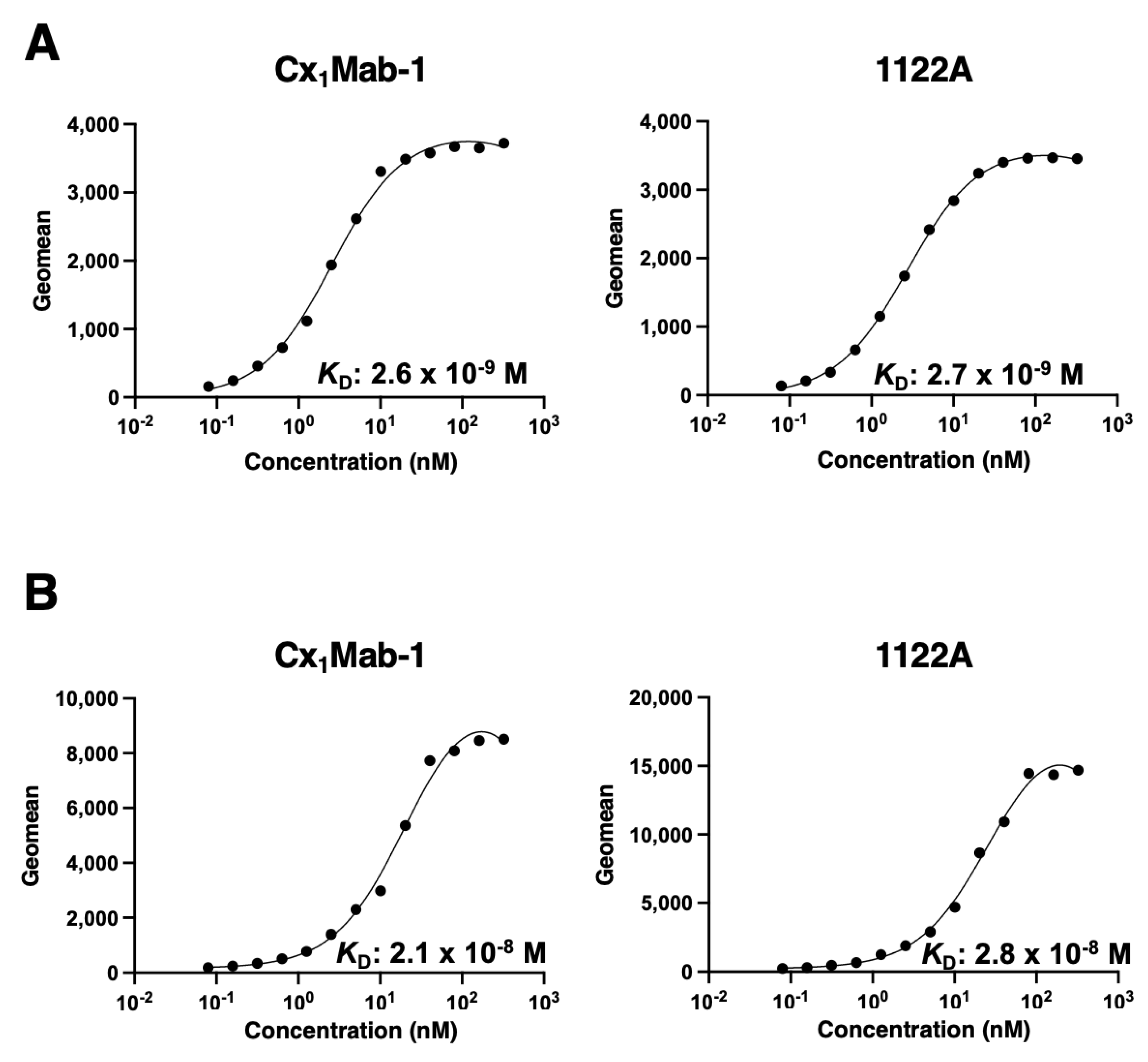
3.3. Determination of the binding affinity of Cx1Mab-1
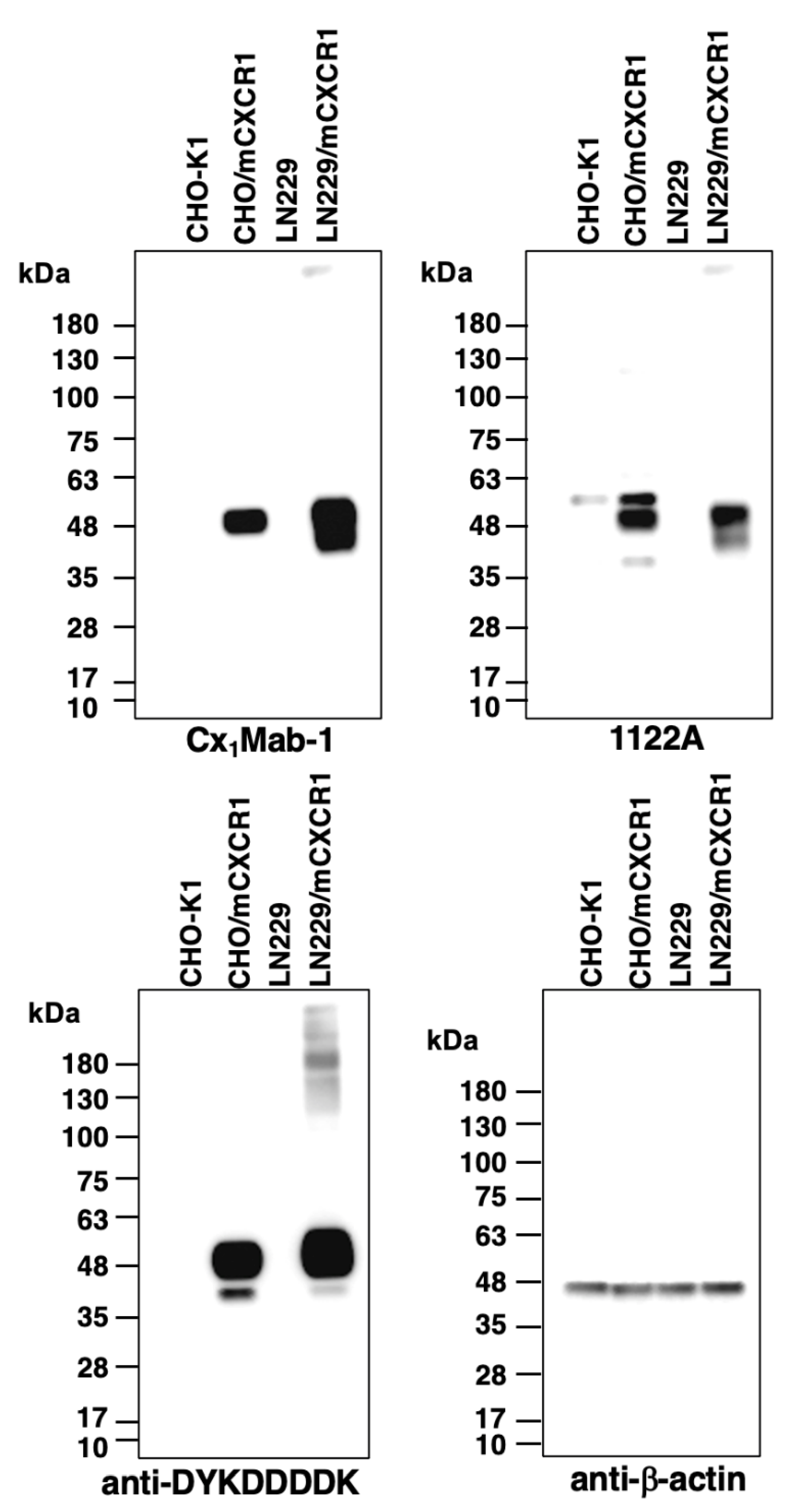
3.4. Western blot analysis
4. Discussion
References
- Hughes, C.E.; Nibbs, R.J.B. A guide to chemokines and their receptors. Febs j 2018, 285, 2944–2971. [Google Scholar] [CrossRef] [PubMed]
- Nagarsheth, N.; Wicha, M.S.; Zou, W. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat Rev Immunol 2017, 17, 559–572. [Google Scholar] [CrossRef]
- White, G.E.; Iqbal, A.J.; Greaves, D.R. CC chemokine receptors and chronic inflammation--therapeutic opportunities and pharmacological challenges. Pharmacol Rev 2013, 65, 47–89. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.J.; Li, Y.B.; Yang, L.X.; Cheng, H.J.; Liu, X.; Chen, H. Roles of the CXCL8-CXCR1/2 Axis in the Tumor Microenvironment and Immunotherapy. Molecules 2021, 27. [Google Scholar] [CrossRef] [PubMed]
- Russo, R.C.; Garcia, C.C.; Teixeira, M.M.; Amaral, F.A. The CXCL8/IL-8 chemokine family and its receptors in inflammatory diseases. Expert Rev Clin Immunol 2014, 10, 593–619. [Google Scholar] [CrossRef]
- Murphy, P.M.; Tiffany, H.L. Cloning of complementary DNA encoding a functional human interleukin-8 receptor. Science 1991, 253, 1280–1283. [Google Scholar] [CrossRef]
- Holmes, W.E.; Lee, J.; Kuang, W.J.; Rice, G.C.; Wood, W.I. Structure and functional expression of a human interleukin-8 receptor. Science 1991, 253, 1278–1280. [Google Scholar] [CrossRef]
- Wang, W.; Deng, Z.; Liu, G.; Yang, J.; Zhou, W.; Zhang, C.; Shen, W.; Zhang, Y. Platelet-derived extracellular vesicles promote the migration and invasion of rheumatoid arthritis fibroblast-like synoviocytes via CXCR2 signaling. Exp Ther Med 2021, 22, 1120. [Google Scholar] [CrossRef]
- Bi, H.; Zhang, Y.; Wang, S.; Fang, W.; He, W.; Yin, L.; Xue, Y.; Cheng, Z.; Yang, M.; Shen, J. Interleukin-8 promotes cell migration via CXCR1 and CXCR2 in liver cancer. Oncol Lett 2019, 18, 4176–4184. [Google Scholar] [CrossRef]
- Chuntharapai, A.; Lee, J.; Hébert, C.A.; Kim, K.J. Monoclonal antibodies detect different distribution patterns of IL-8 receptor A and IL-8 receptor B on human peripheral blood leukocytes. J Immunol 1994, 153, 5682–5688. [Google Scholar] [CrossRef] [PubMed]
- Tavares, L.P.; Garcia, C.C.; Machado, M.G.; Queiroz-Junior, C.M.; Barthelemy, A.; Trottein, F.; Siqueira, M.M.; Brandolini, L.; Allegretti, M.; Machado, A.M.; et al. CXCR1/2 Antagonism Is Protective during Influenza and Post-Influenza Pneumococcal Infection. Front Immunol 2017, 8, 1799. [Google Scholar] [CrossRef]
- Sitaru, S.; Budke, A.; Bertini, R.; Sperandio, M. Therapeutic inhibition of CXCR1/2: Where do we stand? Intern Emerg Med 2023, 18, 1647–1664. [Google Scholar] [CrossRef]
- Fu, X.; Wang, Q.; Du, H.; Hao, H. CXCL8 and the peritoneal metastasis of ovarian and gastric cancer. Front Immunol 2023, 14, 1159061. [Google Scholar] [CrossRef]
- Chen, Y.; McAndrews, K.M.; Kalluri, R. Clinical and therapeutic relevance of cancer-associated fibroblasts. Nat Rev Clin Oncol 2021, 18, 792–804. [Google Scholar] [CrossRef] [PubMed]
- Naito, Y.; Yamamoto, Y.; Sakamoto, N.; Shimomura, I.; Kogure, A.; Kumazaki, M.; Yokoi, A.; Yashiro, M.; Kiyono, T.; Yanagihara, K.; et al. Cancer extracellular vesicles contribute to stromal heterogeneity by inducing chemokines in cancer-associated fibroblasts. Oncogene 2019, 38, 5566–5579. [Google Scholar] [CrossRef] [PubMed]
- Teijeira, A.; Garasa, S.; Ochoa, M.C.; Villalba, M.; Olivera, I.; Cirella, A.; Eguren-Santamaria, I.; Berraondo, P.; Schalper, K.A.; de Andrea, C.E.; et al. IL8, Neutrophils, and NETs in a Collusion against Cancer Immunity and Immunotherapy. Clin Cancer Res 2021, 27, 2383–2393. [Google Scholar] [CrossRef] [PubMed]
- Schalper, K.A.; Carleton, M.; Zhou, M.; Chen, T.; Feng, Y.; Huang, S.P.; Walsh, A.M.; Baxi, V.; Pandya, D.; Baradet, T.; et al. Elevated serum interleukin-8 is associated with enhanced intratumor neutrophils and reduced clinical benefit of immune-checkpoint inhibitors. Nat Med 2020, 26, 688–692. [Google Scholar] [CrossRef]
- Yuen, K.C.; Liu, L.F.; Gupta, V.; Madireddi, S.; Keerthivasan, S.; Li, C.; Rishipathak, D.; Williams, P.; Kadel, E.E., 3rd; Koeppen, H.; et al. High systemic and tumor-associated IL-8 correlates with reduced clinical benefit of PD-L1 blockade. Nat Med 2020, 26, 693–698. [Google Scholar] [CrossRef]
- Liu, H.; Zhao, Q.; Tan, L.; Wu, X.; Huang, R.; Zuo, Y.; Chen, L.; Yang, J.; Zhang, Z.X.; Ruan, W.; et al. Neutralizing IL-8 potentiates immune checkpoint blockade efficacy for glioma. Cancer Cell 2023, 41, 693–710. [Google Scholar] [CrossRef]
- Ruffini, P.A. The CXCL8-CXCR1/2 Axis as a Therapeutic Target in Breast Cancer Stem-Like Cells. Front Oncol 2019, 9, 40. [Google Scholar] [CrossRef]
- Bertini, R.; Allegretti, M.; Bizzarri, C.; Moriconi, A.; Locati, M.; Zampella, G.; Cervellera, M.N.; Di Cioccio, V.; Cesta, M.C.; Galliera, E.; et al. Noncompetitive allosteric inhibitors of the inflammatory chemokine receptors CXCR1 and CXCR2: Prevention of reperfusion injury. Proc Natl Acad Sci U S A 2004, 101, 11791–11796. [Google Scholar] [CrossRef] [PubMed]
- Chapman, R.W.; Minnicozzi, M.; Celly, C.S.; Phillips, J.E.; Kung, T.T.; Hipkin, R.W.; Fan, X.; Rindgen, D.; Deno, G.; Bond, R.; et al. A novel, orally active CXCR1/2 receptor antagonist, Sch527123, inhibits neutrophil recruitment, mucus production, and goblet cell hyperplasia in animal models of pulmonary inflammation. J Pharmacol Exp Ther 2007, 322, 486–493. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Sadanandam, A.; Nannuru, K.C.; Varney, M.L.; Mayer-Ezell, R.; Bond, R.; Singh, R.K. Small-molecule antagonists for CXCR2 and CXCR1 inhibit human melanoma growth by decreasing tumor cell proliferation, survival, and angiogenesis. Clin Cancer Res 2009, 15, 2380–2386. [Google Scholar] [CrossRef] [PubMed]
- Asano, T.; Nanamiya, R.; Takei, J.; Nakamura, T.; Yanaka, M.; Hosono, H.; Tanaka, T.; Sano, M.; Kaneko, M.K.; Kato, Y. Development of Anti-Mouse CC Chemokine Receptor 3 Monoclonal Antibodies for Flow Cytometry. Monoclon Antib Immunodiagn Immunother 2021, 40, 107–112. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Nanamiya, R.; Takei, J.; Nakamura, T.; Yanaka, M.; Hosono, H.; Sano, M.; Asano, T.; Kaneko, M.K.; Kato, Y. Development of Anti-Mouse CC Chemokine Receptor 8 Monoclonal Antibodies for Flow Cytometry. Monoclon Antib Immunodiagn Immunother 2021, 40, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Saito, M.; Asano, T.; Tanaka, T.; Kitamura, K.; Kudo, Y.; Kaneko, M.K.; Kato, Y. C(8)Mab-3: An Anti-Mouse CCR8 Monoclonal Antibody for Immunocytochemistry. Monoclon Antib Immunodiagn Immunother 2022, 41, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Saito, M.; Suzuki, H.; Tanaka, T.; Asano, T.; Kaneko, M.K.; Kato, Y. Development of an Anti-Mouse CCR8 Monoclonal Antibody (C(8)Mab-1) for Flow Cytometry and Immunocytochemistry. Monoclon Antib Immunodiagn Immunother 2022, 41, 333–338. [Google Scholar] [CrossRef]
- Nanamiya, R.; Takei, J.; Asano, T.; Tanaka, T.; Sano, M.; Nakamura, T.; Yanaka, M.; Hosono, H.; Kaneko, M.K.; Kato, Y. Development of Anti-Human CC Chemokine Receptor 9 Monoclonal Antibodies for Flow Cytometry. Monoclon Antib Immunodiagn Immunother 2021, 40, 101–106. [Google Scholar] [CrossRef]
- Ouchida, T.; Isoda, Y.; Tanaka, T.; Kaneko, M.K.; Suzuki, H.; Kato, Y. Cx<sub>3</sub>Mab-4: A Novel Anti-mouse CXCR3 Monoclonal Antibody for Flow Cytometry. Preprints 2023. [Google Scholar] [CrossRef]
- Ouchida, T.S., H.; Tanaka, T.; Kaneko, M.K.; Kato, Y. Cx4Mab-1: A Novel Anti-mouse CXCR4 Monoclonal Antibody for Flow Cytometry. Preprints 2023 2023, 2023110501. [Google Scholar] [CrossRef]
- Kitamura, K.; Suzuki, H.; Kaneko, M.K.; Kato, Y. Cx(6)Mab-1: A Novel Anti-Mouse CXCR6 Monoclonal Antibody Established by N-Terminal Peptide Immunization. Monoclon Antib Immunodiagn Immunother 2022, 41, 133–141. [Google Scholar] [CrossRef]
- Borlongan, M.C.; Saha, D.; Wang, H. Tumor Microenvironment: A Niche for Cancer Stem Cell Immunotherapy. Stem Cell Rev Rep 2023. [Google Scholar] [CrossRef]
- Hammond, M.E.; Lapointe, G.R.; Feucht, P.H.; Hilt, S.; Gallegos, C.A.; Gordon, C.A.; Giedlin, M.A.; Mullenbach, G.; Tekamp-Olson, P. IL-8 induces neutrophil chemotaxis predominantly via type I IL-8 receptors. J Immunol 1995, 155, 1428–1433. [Google Scholar] [CrossRef]
- Thai, T.D.; Chuenchom, C.; Donsa, W.; Faksri, K.; Sripa, B.; Edwards, S.W.; Salao, K. Helicobacter pylori extract induces purified neutrophils to produce reactive oxygen species only in the presence of plasma. Biomed Rep 2023, 19, 89. [Google Scholar] [CrossRef] [PubMed]
- Qin, R.; Ren, W.; Ya, G.; Wang, B.; He, J.; Ren, S.; Jiang, L.; Zhao, S. Role of chemokines in the crosstalk between tumor and tumor-associated macrophages. Clin Exp Med 2023, 23, 1359–1373. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; He, H.; Liu, H.; Li, R.; Chen, Y.; Qi, Y.; Jiang, Q.; Chen, L.; Zhang, P.; Zhang, H.; et al. Tumour-associated macrophages-derived CXCL8 determines immune evasion through autonomous PD-L1 expression in gastric cancer. Gut 2019, 68, 1764–1773. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Ma, Z.; Wang, R.; Wang, Y.; Wang, S.; Cheng, Z.; Xu, H.; Jin, X.; Li, W.; Wang, X. Thrombin facilitates invasion of ovarian cancer along peritoneum by inducing monocyte differentiation toward tumor-associated macrophage-like cells. Cancer Immunol Immunother 2010, 59, 1097–1108. [Google Scholar] [CrossRef] [PubMed]
- Gonsiorek, W.; Fan, X.; Hesk, D.; Fossetta, J.; Qiu, H.; Jakway, J.; Billah, M.; Dwyer, M.; Chao, J.; Deno, G.; et al. Pharmacological characterization of Sch527123, a potent allosteric CXCR1/CXCR2 antagonist. J Pharmacol Exp Ther 2007, 322, 477–485. [Google Scholar] [CrossRef]
- Ginestier, C.; Liu, S.; Diebel, M.E.; Korkaya, H.; Luo, M.; Brown, M.; Wicinski, J.; Cabaud, O.; Charafe-Jauffret, E.; Birnbaum, D.; et al. CXCR1 blockade selectively targets human breast cancer stem cells in vitro and in xenografts. J Clin Invest 2010, 120, 485–497. [Google Scholar] [CrossRef]
- Brandolini, L.; Cristiano, L.; Fidoamore, A.; De Pizzol, M.; Di Giacomo, E.; Florio, T.M.; Confalone, G.; Galante, A.; Cinque, B.; Benedetti, E.; et al. Targeting CXCR1 on breast cancer stem cells: Signaling pathways and clinical application modelling. Oncotarget 2015, 6, 43375–43394. [Google Scholar] [CrossRef]
- Gulhati, P.; Schalck, A.; Jiang, S.; Shang, X.; Wu, C.J.; Hou, P.; Ruiz, S.H.; Soto, L.S.; Parra, E.; Ying, H.; et al. Targeting T cell checkpoints 41BB and LAG3 and myeloid cell CXCR1/CXCR2 results in antitumor immunity and durable response in pancreatic cancer. Nat Cancer 2023, 4, 62–80. [Google Scholar] [CrossRef]
- Teijeira, Á.; Garasa, S.; Gato, M.; Alfaro, C.; Migueliz, I.; Cirella, A.; de Andrea, C.; Ochoa, M.C.; Otano, I.; Etxeberria, I.; et al. CXCR1 and CXCR2 Chemokine Receptor Agonists Produced by Tumors Induce Neutrophil Extracellular Traps that Interfere with Immune Cytotoxicity. Immunity 2020, 52, 856–871. [Google Scholar] [CrossRef] [PubMed]
- Kargl, J.; Zhu, X.; Zhang, H.; Yang, G.H.Y.; Friesen, T.J.; Shipley, M.; Maeda, D.Y.; Zebala, J.A.; McKay-Fleisch, J.; Meredith, G.; et al. Neutrophil content predicts lymphocyte depletion and anti-PD1 treatment failure in NSCLC. JCI Insight 2019, 4. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Suzuki, H.; Ohishi, T.; Asano, T.; Tanaka, T.; Yanaka, M.; Nakamura, T.; Yoshikawa, T.; Kawada, M.; Kaneko, M.K.; et al. Antitumor activities of a defucosylated anti-EpCAM monoclonal antibody in colorectal carcinoma xenograft models. Int J Mol Med 2023, 51. [Google Scholar] [CrossRef] [PubMed]
- Tateyama, N.; Nanamiya, R.; Ohishi, T.; Takei, J.; Nakamura, T.; Yanaka, M.; Hosono, H.; Saito, M.; Asano, T.; Tanaka, T.; et al. Defucosylated Anti-Epidermal Growth Factor Receptor Monoclonal Antibody 134-mG(2a)-f Exerts Antitumor Activities in Mouse Xenograft Models of Dog Epidermal Growth Factor Receptor-Overexpressed Cells. Monoclon Antib Immunodiagn Immunother 2021, 40, 177–183. [Google Scholar] [CrossRef]
- Tanaka, T.; Suzuki, H.; Ohishi, T.; Kaneko, M.K.; Kato, Y. Antitumor activities against breast cancers by an afucosylated anti-HER2 monoclonal antibody H(2) Mab-77-mG(2a) -f. Cancer Sci 2023. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




