Preprint
Review
Revolutionizing Vaccine Development for COVID-19: A Review of AI-Based Approaches
Altmetrics
Downloads
168
Views
54
Comments
0
A peer-reviewed article of this preprint also exists.
This version is not peer-reviewed
Preprints on COVID-19 and SARS-CoV-2
Submitted:
08 November 2023
Posted:
09 November 2023
You are already at the latest version
Alerts
Abstract
AI-based approaches are increasingly being used to revolutionize vaccine development for COVID-19. Small molecules, peptides, and epitopes are being collected for therapy discovery, which could also direct AI-based models, screening, or generation [1]. AI-based models are being used for drug discovery and vaccine development, and pre-existing data is being leveraged through machine learning approaches for COVID-19 drug discovery and vaccine development [1]. The best candidate targets for future treatment development are being identified and evaluated using AI-based models [1]. AI-based approaches can be used to overcome challenges in manufacturing, storage, logistics, and safety and efficacy issues related to different vaccine candidates for COVID-19 [2]. AI algorithms can help identify the best vaccine candidates for COVID-19 while considering the efficiency of antigenic peptides for immune response generation [2]. The presentation of antigenic peptides by major histocompatibility complex (MHC) class II molecules is the first step after COVID-19 vaccine administration for any vaccine-induced immune response [2]. Thus, AI-based models are being used to identify the best vaccine candidates for COVID-19 and to ensure that the vaccine-induced immune response is efficient and safe. The utilization of AI-based methods to address logistical, manufacturing, storage, safety, and efficacy issues regarding several COVID-19 vaccine candidates will be examined in this study. Additionally, while considering the effectiveness of antigenic peptides for the induction of an immune response, we will determine the best potential targets for the next treatment development and assess how AI-based models can help discover the best vaccine candidates for COVID-19. This research ultimately intends to offer insights into how AI-based techniques can transform COVID-19 vaccine development and how they can be applied to address vaccine development issues. In this paper, we focus on recent advances in using artificial intelligence to develop COVID-19 drugs and vaccines, as well as the potential of intelligent training in discovering COVID-19 therapeutics, highlighting potential challenges and solutions.
Keywords:
Subject: Computer Science and Mathematics - Artificial Intelligence and Machine Learning
1. Introduction
Millions of people have died because of the COVID-19 epidemic, and economies have been severely damaged. The creation of a secure and reliable vaccination is essential to stopping the virus's spread and preserving human life. Artificial intelligence (AI) has shown great promise as a tool for streamlining the process and improving vaccination efficacy in the field of vaccine development. In this review, we will look at the methods that are being used, the future research prospects, and the potential benefits and drawbacks of AI-based methodologies for the development of the COVID-19 vaccine.
Artificial intelligence (AI) is the application of computer algorithms to tasks that normally require human intelligence, such as pattern recognition, learning, and decision-making. AI can be used in vaccine development to evaluate large datasets, identify potential vaccine targets, and forecast the efficacy of vaccine candidates. The potential benefits of AI-based approaches include faster development times, improved accuracy, and scalability. AI can also help to reduce costs and improve the safety of vaccines by identifying potential adverse effects before clinical trials.
AI-based strategies for COVID-19 vaccine development include in silico modeling for vaccine design and optimization, machine learning algorithms for predicting antigenic epitopes, and AI-based genetic sequencing and analysis. Machine learning algorithms can analyze large viral protein datasets to determine which ones are most likely to elicit an immune response. In-silico modeling can be used to design and optimize vaccine candidates based on their predicted efficacy and safety profiles. Genetic sequencing and analysis using AI-based tools can help to identify mutations in the virus that may affect vaccine efficacy and inform the design of new vaccines.
AI-based methods can help with the development of COVID-19 vaccines because they are scalable, accurate, and fast. AI can analyze large datasets much faster than humans, accurately identifying potential vaccine targets and efficiently scaling up production. AI-based methods do have certain drawbacks, though, like the requirement for high-quality data and the possibility of biases in AI algorithms. For example, AI algorithms may be biased towards certain populations or may not account for all potential variables in vaccine development. Future research should focus on addressing these limitations and optimizing the use of AI-based approaches for vaccine development.
In conclusion, AI-based approaches offer new opportunities for revolutionizing vaccine development for COVID-19. These approaches can accelerate the process, improve accuracy, and reduce costs. To guarantee the security and effectiveness of vaccines, there are, nevertheless, certain restrictions that must be met. To mature safe and efficient vaccines for COVID-19 and other infectious diseases, future research should concentrate on maximizing the application of AI-based techniques and combining them with conventional vaccine development methodologies.
2. Different AI-Based Algorithms for COVID-19 Vaccine Development
2.1. Network-based algorithms
Network-based algorithms have become increasingly essential in vaccine development. To predict possible epitopes, these algorithms employ methods like deep neural networks, gradient boosting decision trees, and artificial neural networks [3]. MARIA and NetMHCPan4 are a couple of these vaccine development algorithms [3]. SARS-CoV-2 T-cell and B-cell epitopes have been identified using network-based algorithms [3]. Vaxign-ML is a machine learning tool that uses network-based algorithms to rank non-structural proteins as potential SARS-CoV-2 vaccine candidates [3]. Machine learning models for the development of the COVID-19 vaccine have also used network-based algorithms [3]. These algorithms were used to identify stable binding SARS-CoV-2 epitopes with high prediction binding scores that can bind to HLA allotypes [3]. Using these techniques, researchers can gain a better understanding of the virus and create vaccines that are more effective at preventing its spread.
Network-based algorithms are a promising avenue for COVID-19 vaccine development. This is due to stochastic computational simulations demonstrating that network structure has a significant impact on disease spread [4]. As a result of using network-based algorithms, researchers can gain a better understanding of the COVID-19 spread process as well as prevention strategies. Simulations show that the "Ring of Vaccination" approach, for example, would vaccinate a large proportion of the population while preventing new COVID-19 waves [4]. Furthermore, the dynamics of the COVID-19 pandemic in various countries can be recapitulated using various network models [4]. Through the application of network-based algorithms in vaccine development, scientists can learn more about the transmission of COVID-19 and create vaccines that are more successful in halting the disease's spread. Given the current pandemic, which has harmed millions of people globally and continues to pose a serious threat to public health, this is especially crucial.
One of the major challenges in developing a COVID-19 vaccine is the complex network structure of the virus and the human body. Several network-based algorithms have been proposed for drug and vaccine development [5], such as the novel network-based algorithm used in this study for drug discovery and development [6]. A multilayer network-based approach allows for simple, unambiguous modeling of complex systems, but it can be computationally intensive [7]. In addition, there is a global race to develop appropriate vaccines due to a dearth of potent medications and vaccines [4,5]. Traditional machine learning-based image classification algorithms can also be limited in their ability to aid in vaccine development [8]. Although several COVID-19 vaccines are now available, there is still a need for further research and clinical trials to develop a searchable drug repository and a better understanding of the disease. The development of new vaccines usually takes ten years; however, by enhancing our mechanistic and network-based understanding of disease, network-based algorithms may be able to shorten this time [9]. To investigate the efficacy of various vaccination approaches and stop fresh waves of COVID-19 infections, the epidemiological network model utilized in this work may prove to be a useful instrument [10]. However, there are still many issues that need to be resolved before network-based algorithms can significantly contribute to the creation of COVID-19 vaccines and treatments.
2.2. Expression-based Algorithms
Expression-based algorithms are vital in vaccine development, as they optimize the immunogenicity and expression of the vaccine by enhancing the sequence to increase its efficacy and safety [11]. One expression-based technique that is frequently used in the development of mRNA vaccines to increase stability, safety, and efficacy is codon optimization [12]. The balance between host tRNA availability and codon usage frequency is adjusted to achieve this optimization, which improves translation efficiency and target antigen in vivo expression [12]. Additionally, if codon optimization is modified to account for the preference of skeletal muscle, intramuscular injections of mRNA vaccines can improve immune response [12]. With just a few clicks, scientists can create a better sequence using a variety of online codon optimization tools and algorithms that are available for various research applications [12,13]. However, some of these optimization algorithms may have shortcomings, as seen with BioNTech and Moderna's algorithms [12]. Expression-based algorithms can also be used to optimize mRNA sequences for production and expression in vaccine development [13]. For example, a baculovirus expression system and insect cells can be used to produce recombinant vaccines. Using the Sf9 insect cell expression system, this system was utilized to create the modified S protein found in the NVX-CoV2373 vaccine [11]. Recombinant S protein and RBD vaccines are made using expression-based algorithms, while protein subunit vaccines are likewise based on systemically expressed viral proteins or peptides using diverse cell-expressing systems [11]. Moreover, two residues (K986 and V987) can be changed to proline to stabilize the pre-fusion conformation of S protein [11]. Expression-based algorithms can also be used in quality control steps of vaccine development to ensure optimal expression and efficacy.
Developing a COVID-19 vaccine that is both safe and effective remains a major challenge. One vaccine based on the DNA platform and two vaccines based on the mRNA platform have currently been approved for widespread use [14]. Even though many DNA and mRNA vaccines against COVID-19 are in preclinical and clinical trials, the use of expression-based algorithms in vaccine development is still limited. Despite the use of machine learning, rational COVID-19 vaccine development is still dependent on understanding the fundamental host-coronavirus interactions and protective immune mechanisms. Vaccine development outcomes may differ depending on the experimental setting [15]. Through research, six proteins were discovered, including S protein and five non-structural proteins, which are adhesins and are crucial for host invasion and virus adhesion [15]. COVID-19 vaccine candidates were predicted using the reverse vaccinology tools Vaxign and Vaxign-ML [15]. The S, nsp3 and nsp8 proteins are expected to induce highly protective antigenicity; however, vaccine candidates developed using these proteins may suffer from safety issues and incomplete induction of protection [15]. The nsp3 protein was chosen for further study because it has not been examined in any coronavirus vaccine trials [15]. The MAC1 domain of the nsp3 protein has sequence homology with the human single ADP-ribosyltransferase PARP14, even though the entire virus as well as the S protein nucleocapsid are proteins and Vaccine development against SARS and MERS tested membrane proteins [15]. Furthermore, viral adhesion proteins can activate T cells or cause autoreactions with self-antigens, reducing host reactivity to viruses [15]. Therefore, in addition to antigenicity, safety is also crucial for the development of COVID-19 vaccines [15]. Additionally, vaccination may pose safety risks that cannot be assessed by machine learning [15].
The development of COVID-19 vaccines, including those using expression-based algorithms, owes much to artificial intelligence and machine learning algorithms. Sharma noted that clinical trials developing COVID-19 vaccines and drugs benefit from the use of artificial intelligence-based models [16]. To rationally develop a COVID-19 vaccine, Ong explained how to apply reverse vaccinology and machine learning [15]. A vaccine created using an expression-based algorithm is Janssen’s COV-2-S vaccine, which uses a modified S protein of the Ad26 expression gene [11]. To develop a COVID-19 vaccine, Fang and colleagues developed a unique two-part vector system using taRNA containing a trans-replicon expression algorithm [12]. Furthermore, Magazino et al. studied the impact of vaccination on COVID-19 mortality using machine learning algorithms [17]. Amri et al. An algorithm was used to compare the immunogenicity of full-length S-type and S1-type MERS-CoV vaccines to assess vaccine effectiveness [18]. Overall, these studies suggest that the development of COVID-19 vaccines, including those produced using expression-based algorithms, benefits from the application of artificial intelligence and machine learning algorithms.
2.3. Integrated Docking Simulation Algorithms
An effective method for anticipating how ligands will interact with target proteins is to use integrated docking simulation algorithms. These algorithms obtain precise predictions of molecular interactions by utilizing virtual screening and multiple docking tools [19]. Integrated docking simulation algorithms such as PatchDock can provide the dynamic characteristics of the designed system and complexes and guess the interactions between molecules by simulating the real molecular docking environment [20]. An algorithm called PatchDock is used to undo docked molecules to form protein-protein complexes. It can accurately predict global properties such as the formation of hydrogen bonds and the conformation of molecules forming complexes [20]. Glide, AD Vina, and rDock are some other built-in docking simulation algorithms [19]. These tools were used to analyze potential ligand candidates that can inhibit SARS-CoV-2 Mpro [19]. Protein-ligand structures with low docking scores interact with amino acid residues in Mpro, and the docking score values derived from these algorithms indicate the effectiveness of the ligand [19]. Ultimately, integrated docking simulation algorithms provide a valuable means of predicting protein-ligand interactions, which can be used to design more effective drugs for a wide variety of diseases.
Integrated docking simulation algorithms are a useful tool for COVID-19 vaccine research and development. Potential vaccine targets like the spike S-protein are found using these algorithms [21]. Molecular docking and modeling are two important parts of in silico research that can help develop multi-epitope vaccines [21]. Linkers are crucial for maintaining the stability of protein structures by establishing flexible conformations, protein folding, and segregation of functional domains [21]. Epitopes removed in candidate vaccine constructs were calculated using SARS-CoV2 proteins using helper T lymphocytes, cytotoxic T lymphocytes, and B cells. Vaccine candidates are produced by fusing peptide epitopes with linkers to simplify vaccine design [21]. Molecular docking technology was used to study the immune receptor binding affinity of putative vaccine candidates [21]. Using scoring functions, molecular docking programs are crucial for early-stage drug discovery efforts as they can predict binding affinities between proteins and ligands [22,23]. Scientists around the world are using these integrated docking simulation algorithms for drug discovery in response to the COVID-19 pandemic [22]. One algorithm that can do this is the HADDOCK server, which can cluster different vaccines into different structures and clusters by docking them with the ACE-2 receptor. The top cluster score, Z-score, and water-refined models can be used to illustrate the docking interactions between the h-ACE-2 protein complex and suggested multi-epitopic vaccines. The static interaction parameters between the construct vaccines and ACE-2 are shown in a table. In the process of developing a COVID-19 vaccine, integrated docking simulation algorithms like the HADDOCK server can be used to find possible medications for the disease's treatment [21]. To improve the immune response, construct vaccines with high antigenicity were mixed with multi-epitope vaccines [21]. The broad-spectrum peptide-binding repertoires of various HLA alleles were determined using docking simulation algorithms [21]. Various vaccine constructs were then employed, combining distinct epitopes from the S1 and S2 domains of the spike SARS CoV 2 protein.
When developing COVID-19 vaccines, the use of integrated docking simulation algorithms can be very advantageous. To predict the antigenic epitopes against SARS-CoV-2 for the creation of the vaccine, these algorithms have been used in several studies [24]. The dynamics of biological macromolecules in response to different stimuli, such as interactions with other molecules, are studied through molecular dynamics simulations, which incorporate Newton's laws of motion [23]. Molecular docking and dynamics simulations can be used not only to predict antigenic epitopes but also to investigate the binding interactions between ligand and protein molecules [25]. By sifting through a sizable compound library, this approach can find drug leads and make it easier to design novel drug candidates for COVID-19 therapies [22]. Additionally, FDA-approved medications can be virtually screened for the SARS-CoV-2 virus using computational ligand-receptor binding modeling techniques, which could lead to the discovery of possible repurposing medications [19]. Furthermore, an integrative docking and simulation-based approach can also be used to develop epitope-based COVID-19 vaccines. The application of these algorithms can expedite the development of drugs and vaccines by reducing the time and cost associated with experimental studies and providing insights into molecular interactions that may not be possible to observe experimentally.
3. Exploring the Role of AI in COVID-19 Genomic Sequencing
The process of examining the virus's nucleotide base set to ascertain its genetic composition is known as COVID-19 genomic sequencing. The COVID-19 genome sequences are always changing as the virus spreads to new areas. They are stored as FASTA format files that can be combined to create a corpus of distinct sequences [26]. A crucial step in the COVID-19 genome sequencing process is acquiring the genome sequences from GenBank. These sequences list the genes' names followed by a nucleotide sequence [26]. After the genes field is removed, the entire genome sequence is a sequence of nucleotides [26]. Determining genome features can assist biomedical experts in forecasting the disease's effects on the population, even though many aspects of COVID-19-related disease behavior remain unknown [27]. Numerous organizations worldwide have undertaken the laborious and resource-intensive process of COVID-19 genomic sequencing, which calls for specialized knowledge in the field [27]. Pattern mining methods and an algorithm to examine mutations in COVID-19 genome sequences are used in the analysis process [26]. Development of drugs and vaccines is aided by the COVID-19 genomic sequencing results [28]. Furthermore, the methods described in this paper can be applied to the analysis of other human viruses in addition to the SARS-CoV-2 virus [26].
AI is being used more and more to support COVID-19 genome sequencing. Predicting the subsequent nucleotide bases in COVID-19 genome sequences is one method AI can accomplish this. To determine whether the next nucleotide bases can be predicted, cutting-edge prediction models can be trained and applied to the corpus [26]. AI can also be used to analyze the genome sequences of viruses related to COVID-19, such as SARS, MERS, and Ebola. Crucial information regarding the nucleotide frequency and composition, tri-nucleotide compositions, amino acid count, genome sequence alignment, and DNA similarity of COVID-19 and related viruses can be obtained through comparative sequence analysis [27]. Machine learning-based classifiers, like Support Vector Machines (SVM), can be used to classify different genome sequences [27]. The positions and line numbers of nucleotide variations across genome sequences can also be found and stored by AI systems [26]. AI can also determine the COVID-19 genome sequence mutation rate, analyze COVID-19 genome sequence mutations, and compute COVID-19 genome mutation rates. To contain the COVID-19 pandemic, these capabilities may be useful in the development of therapeutic and preventive agents, such as vaccinations and drugs (including drug repurposing) [26,28]. AI-based systems can help with COVID-19 genomic sequencing by analyzing a variety of factors such as genome sequence length, nucleotide frequency in the DNA, tri-nucleotide composition, GC percentage (which indicates which genome sequence has the most stable DNA), amino acid count, similarity or alignment between different genome sequences, and more [27]. Explainable AI (XAI) methods based on machine learning techniques can be used to process COVID-19 metagenomic next-generation sequencing (mNGS) samples. AI-based systems can help medical professionals diagnose COVID-19 by using genomic sequencing [29]. AI is thus critical to COVID-19 genomic sequencing, and it can assist scientists and medical professionals in understanding the virus's genetic makeup and developing effective prophylactic and treatment plans.
AI can be extremely helpful in the fight against COVID-19 by assisting researchers in developing efficient cures. Artificial intelligence's (AI) capacity to identify current medications that can be modified for COVID-19 treatment is one of the main advantages of employing AI in COVID-19 genomic sequencing [30]. Finding effective treatments and cutting down on the amount of time needed for drug discovery can be accelerated by screening billions of currently available drugs using computer simulations to see if they interact with or disrupt SARS-CoV-2 proteins [30]. Furthermore, AI is useful for determining the right drug combination to combat COVID-19 because it can link datasets faster than the human brain [30]. A list of medications that have been shortlisted can then be used to treat patients more effectively and hasten their recovery by putting them through trials to see which one works best against the viral proteins [30]. Strong medications for treating COVID-19 have been discovered using AI techniques, including salinomycin, boceprevir, homoharringtonine, tilorone, and chloroquine [30]. AI has the potential to be used in the creation of COVID-19 vaccines and drugs [28]. While the benefits of using AI for COVID-19 genomic sequencing were not discussed in the text [28], AI can be a helpful tool in the hunt for COVID-19 treatments.
4. Exploring the Role of AI in COVID-19 Drug Discovery
To expedite the drug development and discovery process, machine learning techniques are being employed in the rapidly expanding field of AI-assisted drug discovery. Numerous fields in healthcare, such as pharmacology, gene therapy, regenerative medicine, and newly emerging illnesses like COVID-19, can benefit greatly from the applications of AI. AI may hasten the identification of diseases, the development of new drugs, and personalized healthcare. AI tools, such as drug repurposing and potential antiviral drug molecule prediction, are used in COVID-19 drug development. Predicting medication efficacy, adverse effects, and inhibitor molecules for pathogenic agents is done using machine learning techniques and deep learning algorithms. In drug discovery with AI support, molecules are identified from billions of compound databases. It attempts to hit several targets, which might be more useful in preventing future drug resistance. AI-assisted drug discovery reduces expenses and failures in the latter phases of drug development by employing an in-silico method. The process uses a range of AI tools, such as neural networks, homology modeling, protein fold prediction, omics analytics, SMILES, LSTM models, etc., to identify potential lead molecules among thousands of compounds. For cross-validation of hits, the Autodock Vina scoring function is utilized, and web-based toxicity evaluation tools like Deep Tox, Lim Tox, Tox21, and admetSAR are used. When everything is considered, the field of AI-assisted drug discovery is exciting and has the potential to significantly increase the effectiveness of the drug discovery process.
While AI hasn't been used extensively in COVID-19 drug discovery, scientists are constantly looking for new applications for the technology. AI has the potential to create COVID-19 medications and enhance the efficacy of the current medical and healthcare systems during the pandemic. Vaishya et al. (2020) covered several noteworthy applications of AI for COVID-19, including the creation of medications [31]. In COVID-19 research, AI can help with vaccine design and estimate dosage adjustments as well as other clinical issues like early diagnosis, risk identification, and disease evolution prediction [32]. AI can also screen substances that might be used as adjuvants in COVID-19 vaccines [33]. Ligand-based AI technologies only consider the structures of the ligands when training models for COVID-19 drug design, whereas structure-based AI technologies also take into account the protein target [32]. For COVID-19, conventional ligand-based techniques can be paired with knowledge mining methods to direct the development of experimental drugs [34]. AI is also capable of estimating drug repurposing against COVID-19 and determining the correlation between these drugs. AI methods like deep learning (DL) and generative topographic mapping (GTM) can be applied to COVID-19 drug repurposing in addition to finding new DAA agents [32,34]. In conclusion, more research should be done on the application of AI as it may be able to aid in the development of COVID-19 medication.
Even though AI has the potential to be useful in drug discovery, there are several obstacles in the way of using AI to find COVID-19 treatments. A challenge that exists is the scarcity of information and resources regarding the new virus. The identification of promising drug candidates against a variety of drug targets, including COVID-19-related targets, has been sped up with the use of computer-aided drug design [35]. Since COVID-19 is a relatively new disease, there isn't much data available for AI models to reliably predict and identify potential treatments. This is because AI-based drug discovery pipelines need a lot of data to train their models. Furthermore, many research facilities may not have easy access to high-performance computing resources, which are frequently needed for AI models. The lack of diversity in the data used to train AI models for drug discovery presents another difficulty [34]. This implies that AI models might not always generate results that are impartial or that generalize well to new data. Furthermore, a major obstacle to the application of AI in COVID-19 drug discovery is the incomplete knowledge of the virus and its mechanisms of action [31]. Additionally, although AI has been applied to drug development and discovery in other medical fields, such as radiology [36], its application to COVID-19 drug discovery is still relatively new and unproven [37]. Therefore, to fully realize the potential benefits of AI in finding COVID-19 treatments that work, it is crucial to keep researching and addressing these issues.
5. Exploring the Role of AI in COVID-19 Vaccine Discovery
The development process for COVID-19 vaccines has been sped up thanks in large part to the application of AI. AI/ML tools have been used to predict vaccine candidates' efficacy, identify potential COVID-19 vaccine targets, and support the design and optimization of vaccine candidates. Artificial neural networks, feed-forward neural networks, gradient boosting decision trees, and deep neural networks are a few examples of these tools.AI has also played a significant role in accelerating the development of vaccines by reducing the length of clinical trials and experimentation [53]. AI has a number of advantages when it comes to finding COVID-19 vaccines. These include the capacity to recognize COVID-19 genomic sequences [54], forecast putative epitopes for the development of a COVID-19 vaccine [55], and assist scientists and pharmaceutical companies in the discovery of COVID-19 medications by forecasting the SARS-CoV-2 protein structure [56]. AI/ML-based methods have also been used in drug discovery and vaccine development to combat COVID-19 [56]. Artificial intelligence (AI) tools such as MARIA, NetMHCPan4, and Vaxign-ML have been developed to identify T-cell and B-cell epitopes for the COVID-19 vaccine [56]. Using AI/ML techniques, non-structural proteins like nsp3 are found to be the most promising COVID-19 vaccine candidates [56]. Additionally, AI screening can assist in locating possible COVID-19 vaccine targets or drug candidates [56]. The AI-enabled IBM supercomputer SUMMIT can model trillions of small molecules and test them one after the other, speeding up the development of antivirals to help treat COVID-19. Additionally, AI algorithms can be trained on the CORD-19 dataset to identify generic medications that may be effective against COVID-19 [57]. However, several problems have limited the use of AI in COVID-19 diagnosis and prediction [43]. It is important to keep in mind that extensive clinical trials must be carried out prior to the authorization of medications and vaccinations developed using artificial intelligence [58]. On the other hand, AI-based technologies have significantly advanced the COVID-19 vaccine research and have the potential to be broadly embraced and employed soon [56,58].
Artificial intelligence (AI) is being used extensively in drug and vaccine development as a vital tool in the fight against COVID-19 [59]. The use of AI/ML techniques has played a significant role in the identification of promising COVID-19 vaccine targets. These technologies can also help predict the efficacy of vaccine candidates, which will ultimately lead to the development of more efficacious vaccines [60]. The largest non-structural protein in SARS-COV-2, nsp3, has been identified as a promising candidate for a vaccine [56]. AI has also been used to drive new approaches to drug discovery and public awareness because of its ability to mine data quickly and efficiently [57,61]. Artificial intelligence (AI)-based technologies such as machine learning (ML) and deep learning (DL) have been used to identify patterns in genomic sequencing, which has also aided in the development of COVID-19 vaccines and drugs [54]. These AI/ML algorithms have proven crucial in prognostication, COVID-19 treatment and vaccine development, and epidemic trajectory forecasting [58]. AI was also used to diagnose COVID-19 cases using a range of indicators and to predict the COVID-19 epidemic's spread [55]. Using AI/ML algorithms in drug discovery, plasma therapy, vaccine development, and drug repurposing are one promising approach to finding a cure for COVID-19 [62]. While there are many benefits to using AI, there are also issues that need to be resolved to ensure that it is used ethically and effectively in the research and development of vaccines.
AI has a lot of potential to help fight the ongoing COVID-19 pandemic, particularly in the areas of medication and vaccine development. In response to the urgent need to develop effective therapies, AI has been used successfully to accelerate the discovery process [59]. There are several advantages to using AI in this scenario, the most important of which is its ability to analyze massive amounts of data and identify patterns that are difficult for humans to notice. The efficient use of this technology has helped with COVID-19 surveillance, diagnosis, outcome prediction, drug discovery, and vaccine development [60]. Artificial Intelligence has already established that Nsp3 is the most promising COVID-19 vaccine candidate [56]. The search for COVID-19 drugs and vaccines has made use of several AI techniques, such as computational biology, deep learning, and machine learning (ML) [54]. Artificial intelligence (AI) can also be used to simulate human intelligence and suggest suitable COVID-19 vaccine development [61]. It could be used, for instance, to repurpose existing drugs that might be helpful in the treatment of COVID-19 and analyze genomic sequencing data to identify promising candidates for vaccines [57]. Public health officials can now target interventions in high-risk areas by using AI to forecast epidemic trends and predict the spread of COVID-19 [55,58]. Consequently, AI offers a feasible approach for the development of vaccines and is a helpful tool in the fight against COVID-19.
For COVID-19, several AI-based vaccine design initiatives are already under development. Different strategies are being used by these programs to identify possible vaccine candidates. A few programs examine the virus's internal structure. a list of the various AI-based COVID-19 vaccine design initiatives currently under development.
- EpiCC – an AI-powered platform developed by the U.S. Department of Defense (DoD) that uses machine learning to predict and design potential vaccine candidates based on the virus's genetic structure. The platform was designed to speed up the process of developing vaccines for new viruses and has already been used to design potential vaccines for viruses such as Zika and Ebola. EpiCC is open-source and available for anyone to use. The platform is constantly being updated with new data, and DoD is working on making it even more user-friendly [63].
- COBRA – a computational method for optimizing broadly reactive epitopes for vaccines by using AI-optimization algorithms to create a cocktail of peptides that can elicit a strong immune response. The goal of COBRA is to create a vaccine that is effective against a wide range of pathogens, making it ideal for use in outbreak situations. A group of researchers from the University of Tokyo created COBRA. The group analyzed a database of peptides using machine learning to determine which ones are most likely to trigger a robust immune response. They then combined a number of these peptides to create a vaccine. So far, COBRA has been tested on mice and human cell lines. The results have been promising, and the team is now working on a clinical trial that will test the vaccine on humans [64].
- Exscalate4CoV – a supercomputing platform created by Dompé Farmaceutici with funding from the European Union those screens medications for potential COVID-19 treatment using AI and machine learning. The platform's goal is to speed up finding possible medications that might be used to treat COVID-19. It accomplishes this by searching a database of available medications for ones that might be effective against the virus using supercomputer power. Although the platform is still in its early stages of development, it has already been used to find several possible medications that might be used to treat COVID-19 [65].
- Nucleic Acid Programmed Immunity (NAPi) – a DNA vaccine platform that employs AI-based predictive algorithms to identify the optimal target antigens to elicit a strong immune response against SARS-CoV-2. The premise behind the platform is that the immune system can be trained to recognize and eradicate a pathogen by using its DNA. NAPi uses predictive algorithms to identify antigens that are the best targets for inducing an immune response against a particular pathogen. NAPi identified a set of SARS-CoV-2 antigens that should mount a strong defense against the virus. The NAPi platform is currently in its early phases [66].
- Iktos – a novel artificial intelligence technique that forecasts the ideal chemical structures of compounds that may be employed as prospective SARS-CoV-2 vaccines using machine learning. Several pharmaceutical companies, including Eli Lilly and Sanofi, have partnered with Iktos to develop new vaccine candidates. Iktos' technology could potentially speed up the development of a vaccine by months or even years, and it is hoped that it will help to get a vaccine to the market sooner rather than later [67].
6. AI-assisted Data Collection for COVID Vaccine Development
There are several obstacles associated with using traditional data collection techniques for the COVID vaccine development. The cost associated with these approaches is one of the biggest challenges. It can take several years to create an effective vaccine against a particular virus using the traditional method of vaccine development, which is expensive and time-consuming [80]. Nonetheless, some scientists are racing to develop COVID-19 vaccines due to the pressing need and the worsening global situation [80]. Verifying mistakes and anomalies that inevitably arise while gathering millions of data points presents another difficulty. For scientists, this can be an intimidating task that causes a major delay in the development process [80]. Fortunately, computational methods can expedite the creation of vaccines. By using computational techniques like artificial intelligence (AI) and machine learning to predict how a vaccine will interact with the human immune system, vaccine development can be expedited and made more accurate [80]. In conclusion, conventional data collection techniques are necessary for the development of the COVID vaccine, but they are not without difficulties. Nevertheless, some of these obstacles can be addressed and the development process accelerated with the use of computational techniques.
Following the COVID-19 pandemic, BenevolentAI, a London-based AI-enabled drug discovery company, used their AI-enhanced knowledge graph to identify possible treatment drugs [81]. This process involved integrating multiple data sources covering drugs, diseases, genes, biological mechanisms, and drug targets using machine-read literature [81]. The knowledge graph has been enhanced by AI algorithms that identify relationships between biomedical concepts to describe confidence, causality, and fill in knowledge gaps [81]. The researchers at BenevolentAI employed an interactive and iterative methodology to identify possible drugs, fusing their expertise and interactive resources with biomedical relationships produced by AI and extracted from recent coronavirus literature [81]. They focused on creating a drug to combat the "cytokine storm" that causes early COVID-19-related mortality [81]. The search was directed towards the mechanisms and proteins of the host that mediate viral infection of cells, instead of searching for viral replication [81]. They concluded that the CME pathway is most likely the way that the SARS-CoV-2 virus enters the body by using their knowledge graph and enriched proteins of the CME pathway (such as AAK1, CLTC, GAK, EPS15, and AP2M1) in the pink endocytosis cluster of PPIs in Figure 1 [81]. The human biology-focused BenevolentAI knowledge graph was employed by researchers to identify drugs that target host proteins that the virus has corrupted [81]. They also found the virus interactome and added it to the knowledge graph to find anti-inflammatory medications that could lessen the cytokine storm and have antiviral properties [81]. AI has accelerated the development of vaccines by collecting and analyzing data more quickly.
There are many benefits to using AI for data collection in the COVID vaccine development process. The guarantee of data quality with the least amount of human interaction is one such benefit. Data analysis was done using machine learning (ML) techniques, and the results were available in 22 hours [80]. Artificial intelligence (AI) has been used by the pharmaceutical industry in a variety of ways during vaccine development and trials. For example, Pfizer employed AI to find signals among millions of data points [80]. Additionally, by repurposing current medications, screening targets as vaccines, and identifying compounds that may be used as adjuvants for vaccines, AI-based methods can be used to find new drugs and vaccines [82]. Furthermore, S protein, nsp3, 3CL-pro, and nsp8-10 are examples of possible vaccine candidates that AI can help predict [80,82]. This could significantly speed up the COVID vaccine development process. By examining the whole SARS-CoV-2 proteome to find epitope hotspots for vaccine formulations, AI techniques can also investigate COVID-19 vaccine development [82]. It is well known that AI can help in the process of finding COVID vaccine candidates [80]. All things considered, there are several benefits to using AI for data collection in the COVID vaccine development process that could greatly accelerate the search for effective vaccines while upholding standards for data quality [80].
7. The challenges faced by AI in the global market for COVID vaccines.
The need for an effective vaccination and problems with the supply chain are just two of the obstacles that the global market for COVID vaccines must overcome. For some of these problems, artificial intelligence (AI) has been suggested as a possible remedy. AI systems can be used to help address COVID-19-related problems and lessen the difficulties faced by vaccine supply chains [85,86]. When it comes to creating a vaccine to combat a pathogen, however, current AI and IoT methods have drawbacks and difficulties [83]. Research has suggested combining blockchain technology with AI-specific tactics to combat COVID-19 to address these issues [87]. A significant obstacle presented by COVID-19 is ensuring equitable vaccination coverage. This will necessitate evaluating various vaccination scenarios and developing roll-out plans that consider local and national realities, particularly in Africa where there are substantial logistical challenges. It is critical to divide the difficulties related to vaccine distribution during the COVID-19 pandemic by taking data privacy, the economy, and other factors into account [88]. Additionally, to address problems related to the improvement of clinical vaccines to avoid COVID-19, current AI and IoT approaches must be improved. Developing AI systems that can forecast coronavirus infection, create medications and vaccines, and recognize probable COVID-19 symptoms are some of these challenges [89]. While the global market for COVID-19 vaccines faces many obstacles, artificial intelligence (AI) can help. However, it's critical to acknowledge these obstacles and seek to overcome them.
The COVID-19 epidemic has presented difficulties in all facets of life, including the creation and dissemination of vaccines. The use of artificial intelligence (AI) systems is one approach to overcoming these obstacles. The demand and acceptance of COVID-19 vaccines can be predicted using AI, which is important for vaccine development and distribution [90]. To reduce logistical difficulties, artificial intelligence (AI) can also be utilized to optimize the COVID-19 vaccine supply chain [85]. AI systems do, however, have certain drawbacks and difficulties, such as data privacy issues and creating AI models that are successful and efficient in identifying coronavirus infections and developing medications and vaccines [83,89]. Furthermore, distributing vaccines fairly is a significant argue during the COVID-19 endemic and must be handled while taking local and national realities into account [88]. To determine various vaccination scenarios and rollout strategies that can overcome them, logistical challenges in Africa need to be evaluated [84]. Ultimately, to support the creation and allocation of the COVID-19 vaccine, the pandemic has also exposed serious shortcomings in basic services like cleaning, delivery, and social distancing [91]. Despite these obstacles, AI has the potential to completely transform the production and distribution of vaccines. However, to meet the COVID-19 pandemic's challenges, AI systems must be designed in a way that is ethical, efficient, and operational.
8. The limitations of AI techniques in developing COVID-19 vaccines
AI has its limitations when it comes to forecasting the safety and effectiveness of vaccines, even though it can be a useful tool in vaccine development. The scarcity of data is one of the main obstacles. The COVID-19 pandemic has highlighted this issue because there is not enough data to train artificial intelligence (AI) models that can accurately diagnose and predict COVID-19 [92]. Moreover, although AI can facilitate the development of vaccines, rigorous clinical trials are still required prior to the authorization of drugs and vaccinations developed with AI [92]. This is because, to be validated, AI-generated hypotheses need to be tested in real-world situations. Furthermore, the caliber of the training data affects the caliber of AI models. Training data bias can lead to inaccurate diagnoses and predictions. Ensuring that AI models are trained on a variety of representative data sets is essential to preventing biases. In conclusion, while AI can be a helpful tool in the development of vaccines, it's important to recognize its limitations and rely on extensive clinical trials to evaluate the safety and effectiveness of vaccinations.
Since SARS-COV-2 is still a relatively new virus, and the field of study is still in its infancy, there is currently insufficient data to support the development of vaccines against it [93]. Transfer learning is a strategy that can be used to address the problem of insufficient data by leveraging previously acquired knowledge [93]. Furthermore, deep learning is crucial for the development of treatments for SARS-COV-2, enabling a timely and accurate response to the virus, despite the challenge of limited data [93]. The prediction of potential coronavirus epitopes for vaccine development has also been done using AI-based techniques. In the case of spike epitope prediction, for instance, the Long Short-Term Memory (LSTM) network has shown promising results [93]. Similarly, SARS-CoV-2 T-cell epitopes have been identified using supervised neural network-driven tools [93]. Furthermore, Crossman et al. generated spike sequences that may serve as targets for vaccine development using deep-learning RNN [93]. On the other hand, data scarcity remains a significant barrier to the creation of an AI-based COVID-19 vaccine. However, there is great potential for using AI and deep learning techniques to speed up the development of SARS-COV-2 vaccines and effective treatments.
9. Prediction accuracy made by AI in designing COVID-19 vaccines
Artificial intelligence (AI) was used in the COVID-19 drug discovery and vaccine development process; however, it is unclear how accurate the AI predictions were at the time the COVID-19 vaccines were developed [94]. A study, however, predicted potential targets for the COVID-19 vaccine, which could lead to vaccine development [94]. Furthermore, several studies have proposed the use of artificial intelligence to predict the public's COVID-19 vaccination trend. In one study, data was trained on five different classifiers to forecast vaccination trends, and the results were evaluated using area under the curve, logarithmic loss, precision, recall, and the F1 measure [95]. The study's findings indicated that, while the possibility of a side effect is very small and infrequent, vaccinations may have some negative effects on the human body [95]. The voting classifier was used to combine the results of each classifier to provide a cumulative accuracy result [95]. The study's goal was to categorize public trends regarding COVID-19 vaccination to help policymakers make informed decisions about vaccine distribution and administration [95]. More research is needed to determine the accuracy of AI forecasts in the development of COVID-19 vaccines.
10. Conclusion and Future Directions
Artificial intelligence (AI) is being researched rapidly for use in the COVID-19 vaccination. The development of several AI/ML-based methods has accelerated the COVID-19 vaccine research process. Prachar et al., for example, used a feed-forward neural network to identify 174 SARS-CoV-2 epitopes with high prediction binding scores and the ability to stably bind to 11 HLA allotypes [96]. Non-structural protein 3 (nsp3) has been identified as the most promising COVID-19 vaccine candidate. In addition, AI/ML models use a variety of techniques to predict putative epitopes. One such tool is Vaxign-ML, an ML tool based on XGBoost that is used to rank non-structural proteins as potential SARS-CoV-2 vaccine candidates [96]. The review article discussed the advantages, disadvantages, and long-term consequences of using AI in the COVID-19 vaccine development process [97]. AI systems have proven invaluable in the COVID-19 pandemic, speeding up the development of new drugs, repurposing old ones, and developing vaccines. Using machine learning algorithms, computational analysis was able to interpret experimental data and provide structural and molecular insights into SARS-CoV2 viruses. Furthermore, it can predict viral components that may elicit protective immune responses [98]. AI has also accelerated the development of the COVID-19 vaccine, assisting in the identification of mutations that would allow the vaccine to be developed and allowing the virus to be decoded in just 48 hours [99]. However, in addition to AI, rigorous "wet" laboratory testing, animal and human trials, and regulatory approval are required to ensure vaccine safety, efficacy, and approval [98]. To summarize, the use of artificial intelligence (AI) in vaccine development speeds up the process, improves accuracy, and broadens our understanding of infectious diseases, all of which contribute to improved global health and life expectancy [98].
All things considered, it can be said that the pandemic's ultimate state is defined by regulated infection levels, viral occurrences that are like seasonal influenza in most countries, and the possibility of endemic virus presence in the populace [100]. In the meantime, image diagnostics is one task where artificial intelligence (AI) has shown promise in the fight against COVID-19 [100]. This paper, however, offers a data science viewpoint on AI's role in the pandemic response and suggests future directions for research and development. In particular, the authors suggest using a temporal step approach to examine how AI perceives and affects healthcare systems and society [100]. Even though there is still a lot of room for research to improve AI support during the current pandemic [100], the advancements made so far show how AI can benefit society and healthcare systems in the future.
Author Contributions
Aritra Ghosh collected the data and wrote the manuscript. Dr. Mirjana Pavlovic helped with some key references and information. All authors read and approved the final manuscript.
Funding
Not applicable.
Ethics Approval and Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Availability of Data and Materials
Data is available upon request.
Acknowledgments
Not applicable.
Conflicts of Interest
The authors declare that they have no competing interests.
References
- Current Artificial Intelligence (AI) Techniques, Challenges, and Approaches in Controlling and Fighting COVID-19: A Review. (n.d.) Retrieved May 27, 2023, from. www.ncbi.nlm.nih.gov/pmc/articles/PMC9140632/.
- Can artificial intelligence help us design vaccines?. (n.d.) Retrieved May 27, 2023, from. www.brookings.edu.
- Lv, H., Shi, L., Berkenpas, J., Dao, F. Application of artificial intelligence and machine learning for COVID-19 drug discovery and vaccine design (n.d.) Retrieved October 27, 2023, from. https://academic.oup.com/bib/article/22/6/bbab320/6354025?login=false.
- Tetteh, J., Hernandez-Vargas, E. Network models to evaluate vaccine strategies towards herd immunity in COVID-19. (n.d.) Retrieved October 27, 2023, from . www.sciencedirect.com/science/article/pii/S0022519321003131.
- Fiscon, G., Conte, F., Farina, L., Paci, P. [HTML][HTML] SAveRUNNER: A network-based algorithm for drug repurposing and its application to COVID-19. (n.d.) Retrieved October 27, 2023, from. https://plos.org/#journals.
- Arora, G., Joshi, J., Mandal, R., Shrivastava, N., Virmani, R. Artificial intelligence in surveillance, diagnosis, drug discovery and vaccine development against COVID-19. (n.d.) Retrieved October 27, 2023, from . www.mdpi.com/2076-0817/10/8/1048.
- Bonifazi, G., Breve, B., Cirillo, S., Corradini, EInvestigating the COVID-19 vaccine discussions on Twitter through a multilayer network-based approach. (n.d.) Retrieved October 27, 2023, from . www.sciencedirect.com/science/article/pii/S0306457322001960.
- Kibriya, H., Amin, R. [HTML][HTML] A residual network-based framework for COVID-19 detection from CXR images. (n.d.) Retrieved October 27, 2023, from. https://link.springer.com/article/10.1007/s00521-022-08127-y.
- Chen, J., Hoops, S., Marathe, A., Mortveit, H., Lewis, B. Prioritizing allocation of COVID-19 vaccines based on social contacts increases vaccination effectiveness. (n.d.) Retrieved October 27, 2023, from . www.medrxiv.org/content/10.1101/2021.02.04.21251012.abstract.
- Tetteh, J., Nguyen, V., Hernandez-Vargas, E. COVID-19 network model to evaluate vaccine strategies towards herd immunity. (n.d.) Retrieved October 27, 2023, from . www.medrxiv.org/content/10.1101/2020.12.22.20248693.abstract.
- COVID-19 vaccine development: milestones, lessons and .... (n.d.) Retrieved October 27, 2023, from . www.nature.com/articles/s41392-022-00996-y.
- Advances in COVID-19 mRNA vaccine development. (n.d.) Retrieved October 27, 2023, from . www.nature.com/articles/s41392-022-00950-y.
- AI and the COVID-19 Vaccine: Moderna's Dave Johnson. (n.d.) Retrieved October 27, 2023, from . https://sloanreview.mit.edu/.
- Biologics | Free Full-Text | Nucleic Acid Vaccines for COVID-19: A Paradigm Shift in the Vaccine Development Arena. (n.d.) Retrieved October 27, 2023, from . www.mdpi.com/2673-8449/1/3/20.
- COVID-19 Coronavirus Vaccine Design Using Reverse .... (n.d.) Retrieved October 27, 2023, from . www.ncbi.nlm.nih.gov/pmc/articles/PMC7350702/.
- Artificial Intelligence-Based Data-Driven Strategy to .... (n.d.) Retrieved October 27, 2023, from . www.ncbi.nlm.nih.gov/pmc/articles/PMC9279074/.
- A machine learning algorithm to analyse the effects of .... (n.d.) Retrieved October 27, 2023, from . www.cambridge.org.
- Coronavirus vaccine development: from SARS and MERS to .... (n.d.) Retrieved October 27, 2023, from . https://jbiomedsci.biomedcentral.com/.
- Computational molecular docking and virtual screening .... (n.d.) Retrieved October 27, 2023, from . https://academic.oup.com/pcm/article/4/1/1/6103931?login=false.
- Structural Basis for Designing Multiepitope Vaccines Against .... (n.d.) Retrieved October 27, 2023, from . https://bioinform.jmir.org/2020/1/e19371/.
- In Silico Modeling as a Perspective in Developing ... - MDPI. (n.d.) Retrieved October 27, 2023, from . www.mdpi.com/2079-6412/11/11/1273.
- AI-accelerated protein-ligand docking for SARS-CoV-2 is ....... (n.d.) Retrieved October 27, 2023, from . www.nature.com/articles/s41598-023-28785-9.
- Molecular docking and simulation studies on SARS-CoV-2 .... (n.d.) Retrieved October 27, 2023, from . www.ncbi.nlm.nih.gov/pmc/articles/PMC7484567/.
- Immunoinformatics and Molecular Docking Studies .... (n.d.) Retrieved October 27, 2023, from . www.hindawi.com/journals/bmri/2021/1596834/.
- Immunoinformatics and molecular modeling approach to .... (n.d.) Retrieved October 27, 2023, from . www.sciencedirect.com/science/article/pii/S235291482100068X.
- Nawaz, M., Fournier-Viger, P., Shojaee, A., Fujita, H. Using artificial intelligence techniques for COVID-19 genome analysis. (n.d.) Retrieved October 28, 2023, from. https://link.springer.com/article/10.1007/s10489-021-02193-w.
- Ahmed, I., Jeon, G. Enabling Artificial Intelligence for Genome Sequence Analysis of COVID-19 and Alike Viruses. (n.d.) Retrieved October 28, 2023, from . https://link.springer.com/article/10.1007/s12539-021-00465-0.
- Bagabir, S., Ibrahim, N., Bagabir, H. Review Covid-19 and Artificial Intelligence: Genome sequencing, drug development and vaccine discovery. (n.d.) Retrieved October 28, 2023, from . www.sciencedirect.com/science/article/pii/S1876034122000144.
- Yagin, F., Cicek, İ., Alkhateeb, A., Yagin, B. Explainable artificial intelligence model for identifying COVID-19 gene biomarkers. (n.d.) Retrieved October 28, 2023, from . www.sciencedirect.com/science/article/pii/S0010482523000847.
- Swayamsiddha, S., Prashant, K., Shaw, D. The prospective of Artificial Intelligence in COVID-19 Pandemic. (n.d.) Retrieved October 28, 2023, from . https://link.springer.com/article/10.1007/s12553-021-00601-2.
- Mikkili, I., Karlapudi, A., Venkateswarulu, T., Kodali, V. [HTML][HTML] Potential of artificial intelligence to accelerate diagnosis and drug discovery for COVID-19. (n.d.) Retrieved October 28, 2023, from . https://peerj.com/articles/12073/.
- Floresta, G., Zagni, C., Gentile, D., Patamia, V. [HTML][HTML] Artificial intelligence technologies for COVID-19 de novo drug design. (n.d.) Retrieved October 28, 2023, . from www.mdpi.com/1422-0067/23/6/3261.
- Wang, L., Zhang, Y., Wang, D., Tong, X., Liu, T. [HTML][HTML] Artificial intelligence for COVID-19: a systematic review. (n.d.) Retrieved October 28, 2023, . from www.frontiersin.org.
- Muratov, E., Amaro, R., Andrade, C., Brown, N. [HTML][HTML] A critical overview of computational approaches employed for COVID-19 drug discovery. (n.d.) Retrieved October 28, 2023, . from https://pubs.rsc.org/en/content/articlehtml/2021/cs/d0cs01065k.
- Gurung, A., Ali, M., Lee, J., Farah, M. [HTML][HTML] An updated review of computer-aided drug design and its application to COVID-19. (n.d.) Retrieved October 28, 2023, . from www.hindawi.com/journals/bmri/2021/8853056/.
- Kumar, A., Gupta, P., Srivastava, A. A review of modern technologies for tackling COVID-19 pandemic. (n.d.) Retrieved October 28, 2023, from . www.sciencedirect.com/science/article/pii/S1871402120301272.
- Monteleone, S., Kellici, T., Southey, M., Bodkin, M. Fighting COVID-19 with artificial intelligence. (n.d.) Retrieved October 28, 2023, . from https://link.springer.com/protocol/10.1007/978-1-0716-1787-8_3.
- Abubaker Bagabir, N. K. Ibrahim, H. Abubaker Bagabir, and R. Hashem Ateeq, “Covid-19 and Artificial Intelligence: Genome sequencing, drug development and Vaccine Discovery,” Journal of Infection and Public Health, vol. 15, no. 2, pp. 289–296, 2022. [CrossRef]
- Zhavoronkov A, Aladinskiy V, Zhebrak A, Zagribelnyy B, Terentiev V, Bezrukov DS, et al. Potential COVID-2019 3C-like protease inhibitors designed using generative deep learning approaches. Insilico Med Hong Kong Ltd A 2020;307:E1.
- Tang B, He F, Liu D, Fang M, Wu Z, Xu D. AI-aided design of novel targeted covalent inhibitors against SARS-CoV-2. BioRxiv 2020. [CrossRef]
- Gao K, Nguyen DD, Wang R, Wei GW. Machine intelligence design of 2019-nCoV drugs. bioRxiv 2020. [CrossRef]
- Hofmarcher M, Mayr A, Rumetshofer E, Ruch P, Renz P, Schimunek J, et al. Largescale ligand-based virtual screening for SARS-CoV-2 inhibitors using deep neural networks. Tech Rep 2020 [Available at: SSRN 3561442].
- Zhang H, Saravanan KM, Yang Y, Hossain MT, Li J, Ren X, et al. Deep learning based drug screening for novel coronavirus 2019-nCov. Interdiscip Sci: Comput Life Sci 2020;12:368–76.
- Beck BR, Shin B, Choi Y, Park S, Kang K. Predicting commercially available antiviral drugs that may act on the novel coronavirus (SARS-CoV-2) through a drugtarget interaction deep learning model. Comput Struct Biotechnol J 2020;18:784–90. [CrossRef]
- Ge Y, Tian T, Huang S, Wan F, Li J, Li S, et al. A data-driven drug repositioning framework discovered a potential therapeutic agent targeting COVID-19. Signal Transduct Target Ther 2021;6:165–70.
- Hu F, Jiang J, Yin P. Prediction of potential commercially inhibitors against SARSCoV-2 by multi-task deep model. arXiv Prepr arXiv 2020;200300728.
- Zhou Y, Hou Y, Shen J, Huang Y, Martin W, Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov 2020;6:14. [CrossRef]
- Zeng, X.; Song, X.; Ma, T.; Pan, X.; Zhou, Y.; Hou, Y.; et al. Repurpose open data to discover therapeutics for COVID-19 using deep learning. J Proteome Res 2020, 19, 4624–36. [Google Scholar] [CrossRef] [PubMed]
- Gysi DM, Do Valle Í, Zitnik M, Ameli A, Gan X, Varol O, et al. Network medicine framework for identifying drug repurposing opportunities for COVID-19. arXiv, 2020.
- Wang Z, Li L, Yan J, Yao Y. Evaluating the traditional Chinese medicine (TCM) officially recommended in China for COVID-19 using ontology-based side-effect prediction framework (OSPF) and deep learning; 2020.
- Abdel-Basset, M.; Hawash, H.; Elhoseny, M.; Chakrabortty, R.K.; Ryan, M. DeepH-DTA: deep learning for predicting drug-target interactions: a case study of COVID-19 drug repurposing. IEEE Access 2020, 8, 170433–170451. [Google Scholar] [CrossRef] [PubMed]
- Demirci, M.D.S.; Adan, A. Computational analysis of microRNA-mediated interactions in SARS-CoV-2 infection. PeerJ 2020, 8, e9369. [Google Scholar] [CrossRef] [PubMed]
- Kannan, S., Subbaram, K., Ali, S. [HTML][HTML] The role of artificial intelligence and machine learning techniques: Race for covid-19 vaccine. (n.d.) Retrieved October 28, 2023, . from https://brieflands.com/articles/archcid-103232.html.
- Bagabir, S., Ibrahim, N., Bagabir, H. [HTML][HTML] Covid-19 and Artificial Intelligence: Genome sequencing, drug development and vaccine discovery. (n.d.) Retrieved October 28, 2023, . from www.sciencedirect.com/science/article/pii/S1876034122000144.
- Abd-Alrazaq, A., Alajlani, M., Alhuwail, D. [HTML][HTML] Artificial intelligence in the fight against COVID-19: scoping review. (n.d.) Retrieved October 28, 2023, . from www.jmir.org/2020/12/e20756/.
- Lv, H., Shi, L., Berkenpas, J., Dao, F. Application of artificial intelligence and machine learning for COVID-19 drug discovery and vaccine design. (n.d.) Retrieved October 28, 2023, from . https://academic.oup.com/bib/article-abstract/22/6/bbab320/6354025.
- Ahuja, A., Reddy, V., Marques, O. [HTML][HTML] Artificial intelligence and COVID-19: A multidisciplinary approach. (n.d.) Retrieved October 28, 2023, . from www.ncbi.nlm.nih.gov/pmc/articles/PMC7255319/.
- Wang, L., Zhang, Y., Wang, D., Tong, X., Liu, T. [HTML][HTML] Artificial intelligence for COVID-19: a systematic review. (n.d.) Retrieved October 28, 2023, . from www.frontiersin.org.
- Webb, J., Salem, M. [HTML][HTML] Artificial intelligence for COVID-19 drug discovery and vaccine development. (n.d.) Retrieved October 28, 2023, from . www.frontiersin.org.
- Arora, G., Joshi, J., Mandal, R., Shrivastava, N., Virmani, R. Artificial intelligence in surveillance, diagnosis, drug discovery and vaccine development against COVID-19. (n.d.) Retrieved October 28, 2023, from . www.mdpi.com/2076-0817/10/8/1048.
- Vaishya, R., Javaid, M., Khan, I., Haleem, A. Artificial Intelligence (AI) applications for COVID-19 pandemic. (n.d.) Retrieved October 28, 2023, from . www.sciencedirect.com/science/article/pii/S1871402120300771.
- Kabra, R., Singh, S. [HTML][HTML] Evolutionary artificial intelligence based peptide discoveries for effective Covid-19 therapeutics. (n.d.) Retrieved October 28, 2023, from . www.sciencedirect.com/science/article/pii/S0925443920303264.
- U. States, “An EPICC study of SARS-COV-2 infection,” HJF. https://www.hjf.org/news/epicc-study-sars-cov-2-infection (accessed on 28 October 2023).
- A. Nuñez, Y. Huang, and T. M. Ross, “Next-generation computationally designed influenza hemagglutinin vaccines protect against H5NX virus infections,” Pathogens, vol. 10, no. 11, p. 1352, 2021. [CrossRef]
- S. Coletti and G. Bernardi, in Exscalate4CoV high-performance computing for Covid Drug Discovery, Cham: Springer International Publishing, 2023.
- R. Baghban, A. Ghasemian, and S. Mahmoodi, “Nucleic acid-based vaccine platforms against the coronavirus disease 19 (covid-19),” Archives of Microbiology, vol. 205, no. 4, 202. [CrossRef]
- D. Tirumalaraju, “Covid-19 drug development to include AI by Iktos and Sri.,” Pharmaceutical Technology. https://www.pharmaceutical-technology.com/news/iktos-sri-covid-19-drug-development/ (accessed on 28 October 2023).
- Abdelmageed MI, Abdelmoneim AH, Mustafa MI, Elfadol NM, Murshed NS, Shantier SW, et al. Design of multi epitope-based peptide vaccine against E protein of human 2019-nCoV: an immunoinformatics approach. bioRxiv 2020:11. [Preprint posted online February].
- Sarkar B, Ullah MA, Johora FT, Taniya MA, Araf Y. The essential facts of Wuhan novel coronavirus outbreak in China and epitope-based vaccine designing against 2019-nCoV. BioRxiv 2020. [CrossRef]
- Fast E, Altman RB, Chen B. Potential T-cell and B-cell epitopes of 2019-nCoV. bioRxiv 2020. [CrossRef]
- Ong E, Wong MU, Huffman A, He Y. COVID-19 coronavirus vaccine design using reverse vaccinology and machine learning. Front Immunol 2020;11:1581.
- Rahman MS, Hoque MN, Islam MR, Akter S, Rubayet-Ul-Alam ASM, Siddique MA, et al. Epitope-based chimeric peptide vaccine design against S, M and E proteins of SARS-CoV-2 etiologic agent of global pandemic COVID-19: an in-silico approach. BioRxiv 2020. [03. 30.015164. 2020].
- Susithra Priyadarshni M, Isaac Kirubakaran S, Harish MC. In silico approach to design a multi-epitopic vaccine candidate targeting the non-mutational immunogenic regions in envelope protein and surface glycoprotein of SARS-CoV-2. J Biomol Struct Dyn 2021:1–16. [CrossRef]
- Russo G, Di Salvatore V, Sgroi G, Parasiliti Palumbo GA, Reche PA, Pappalardo F. A multi-step and multi-scale bioinformatic protocol to investigate potential SARSCoV-2 vaccine targets. Brief Bioinform 2021. [CrossRef]
- G. Liu et al., “Computationally optimized SARS-COV-2 MHC class I and II vaccine formulations predicted to target human haplotype distributions,” Cell Systems, vol. 11, no. 2, 2020. [CrossRef]
- Baruah, “TCS Partners with CSIR to find cure for covid-19,” mint,. https://www.livemint.com/companies/news/tcs-partners-with-csir-to-find-cure-for-covid-19-11585561862046.html (accessed on 29 October 2023).
- Bali and N. Bali, “Role of artificial intelligence in fast-track drug discovery and vaccine development for covid-19,” Novel AI and Data Science Advancements for Sustainability in the Era of COVID-19, pp. 201–229, 2022. [CrossRef]
- “Press release details,” Exscientia. https://investors.exscientia.ai/press-releases/press-release-details/2020/Exscientia-announces-joint-initiative-to-identify-COVID-19-drugs-with-Diamond-Light-Source-and-Scripps-Research/default.aspx (accessed on 29 October 2023).
- Chandra Kaushik and, U. Raj, “Ai-driven drug discovery: A Boon against covid-19?,” AI Open, vol. 1, pp. 1–4, 2020. [CrossRef]
- Sharma, A., Virmani, T., Pathak, V., Sharma, A. BioMed Research International. (n.d.) Retrieved October 28, 2023, from . www.hindawi.com/journals/bmri/2022/7205241/.
- Richardson, P., Robinson, B., Smith, D., Stebbing, J. Vaccines | Free Full-Text | The AI-Assisted Identification and Clinical Efficacy of Baricitinib in the Treatment of COVID-19. (n.d.) Retrieved October 28, 2023, from . www.mdpi.com/2076-393X/10/6/951.
- Wang, L., Zhang, Y., Wang, D., Tong, X., Liu, T. Artificial Intelligence for COVID-19: A Systematic Review. (n.d.) Retrieved October 28, 2023, from . www.frontiersin.org.
- Arora, G., Joshi, J., Mandal, R., Shrivastava, N., Virmani, R. Artificial intelligence in surveillance, diagnosis, drug discovery and vaccine development against COVID-19. (n.d.) Retrieved October 29, 2023, from . www.mdpi.com/2076-0817/10/8/1048.
- Mellado, B., Wu, J., Kong, J., Bragazzi, N. Leveraging artificial intelligence and big data to optimize COVID-19 clinical public health and vaccination roll-out strategies in Africa. (n.d.) Retrieved October 29, 2023, from . www.mdpi.com/1660-4601/18/15/7890.
- Cano-Marin, E., Ribeiro-Soriano, D., Mardani, A. [HTML][HTML] Exploring the challenges of the COVID-19 vaccine supply chain using social media analytics: a global perspective. (n.d.) Retrieved October 29, 2023, from . www.sciencedirect.com/science/article/pii/S277303282300010X.
- Velu, A., Reddy, R., Sharma, P. Impact of Covid Vaccination on the Globe using data analytics. (n.d.) Retrieved October 29, 2023, from . www.taylorfrancis.com.
- Meghla, T., Rahman, M., Biswas, A. Supply chain management with demand forecasting of covid-19 vaccine using blockchain and machine learning. (n.d.) Retrieved October 29, 2023, from . https://ieeexplore.ieee.org/abstract/document/9580006/.
- Kamran, M., Kia, R., Goodarzian, F., Ghasemi, P. A new vaccine supply chain network under COVID-19 conditions considering system dynamic: Artificial intelligence algorithms. (n.d.) Retrieved October 29, 2023, from . www.sciencedirect.com/science/article/pii/S0038012122001732.
- Pham, Q., Nguyen, D., Huynh-The, T., Hwang, W. Artificial intelligence (AI) and big data for coronavirus (COVID-19) pandemic: a survey on the state-of-the-arts. (n.d.) Retrieved October 29, 2023, from . https://ieeexplore.ieee.org/abstract/document/9141265/.
- Almars, A., Gad, I., Atlam, E. Applications of AI and IoT in COVID-19 vaccine and its impact on social life. (n.d.) Retrieved October 29, 2023, from . https://link.springer.com/chapter/10.1007/978-3-030-91103-4_7.
- Arora, N., Banerjee, A., Narasu, M. The role of artificial intelligence in tackling COVID-19. (n.d.) Retrieved October 29, 2023, from . www.futuremedicine.com/doi/abs/10.2217/fvl-2020-0130.
- Wang, L., Zhang, Y., Wang, D., Tong, X., Liu, T. Artificial Intelligence for COVID-19: A Systematic Review. (n.d.) Retrieved October 29, 2023, from . www.frontiersin.org.
- Webb, J., Salem, M. Artificial Intelligence for COVID-19 Drug Discovery and Vaccine Development. (n.d.) Retrieved October 29, 2023, from . www.frontiersin.org.
- Artificial Intelligence for COVID-19: A Systematic Review. (n.d.) Retrieved October 29, 2023, from . www.frontiersin.org/articles/10.3389/fmed.2021.704256.
- Data | Free Full-Text | Future Prediction of COVID-19 Vaccine Trends Using a Voting Classifier. (n.d.) Retrieved October 29, 2023, from . www.mdpi.com/2306-5729/6/11/112.
- Twitter. (n.d.) Retrieved October 29, 2023, from . www.ncbi.nlm.nih.gov/pmc/articles/PMC8511807/.
- Mathematics | Free Full-Text | Role of Artificial Intelligence for Analysis of COVID-19 Vaccination-Related Tweets: Opportunities, Challenges, and Future Trends. (n.d.) Retrieved October 29, 2023, from . www.mdpi.com/2227-7390/10/17/3199.
- Artificial intelligence in accelerating vaccine development - current and future perspectives. (n.d.) Retrieved October 29, 2023, from. www.frontiersin.org/articles/10.3389/fbrio.2023.1258159.
- How leveraging the power of AI is changing the way Moderna vaccines are made and distributed | Euronews. (n.d.) Retrieved October 29, 2023. from. www.euronews.com.
- The Role of Artificial Intelligence in Fighting the COVID-19 Pandemic. (n.d.) Retrieved October 29, 2023. from . https://link.springer.com/article/10.1007/s10796-021-10131-x.
Figure 1.
The PPI network was chosen based on the knowledge graph query "SARS-CoV-2 AND Endocytosis". Differently colored clusters represent different pathways and processes (e.g., cytokine signaling in green and orange, and endocytosis in pink). Every node represents a protein, and the edges show which protein-protein interactions are enhancing (green) or inhibiting (red). Pink modules make up CME. [81].
Figure 1.
The PPI network was chosen based on the knowledge graph query "SARS-CoV-2 AND Endocytosis". Differently colored clusters represent different pathways and processes (e.g., cytokine signaling in green and orange, and endocytosis in pink). Every node represents a protein, and the edges show which protein-protein interactions are enhancing (green) or inhibiting (red). Pink modules make up CME. [81].
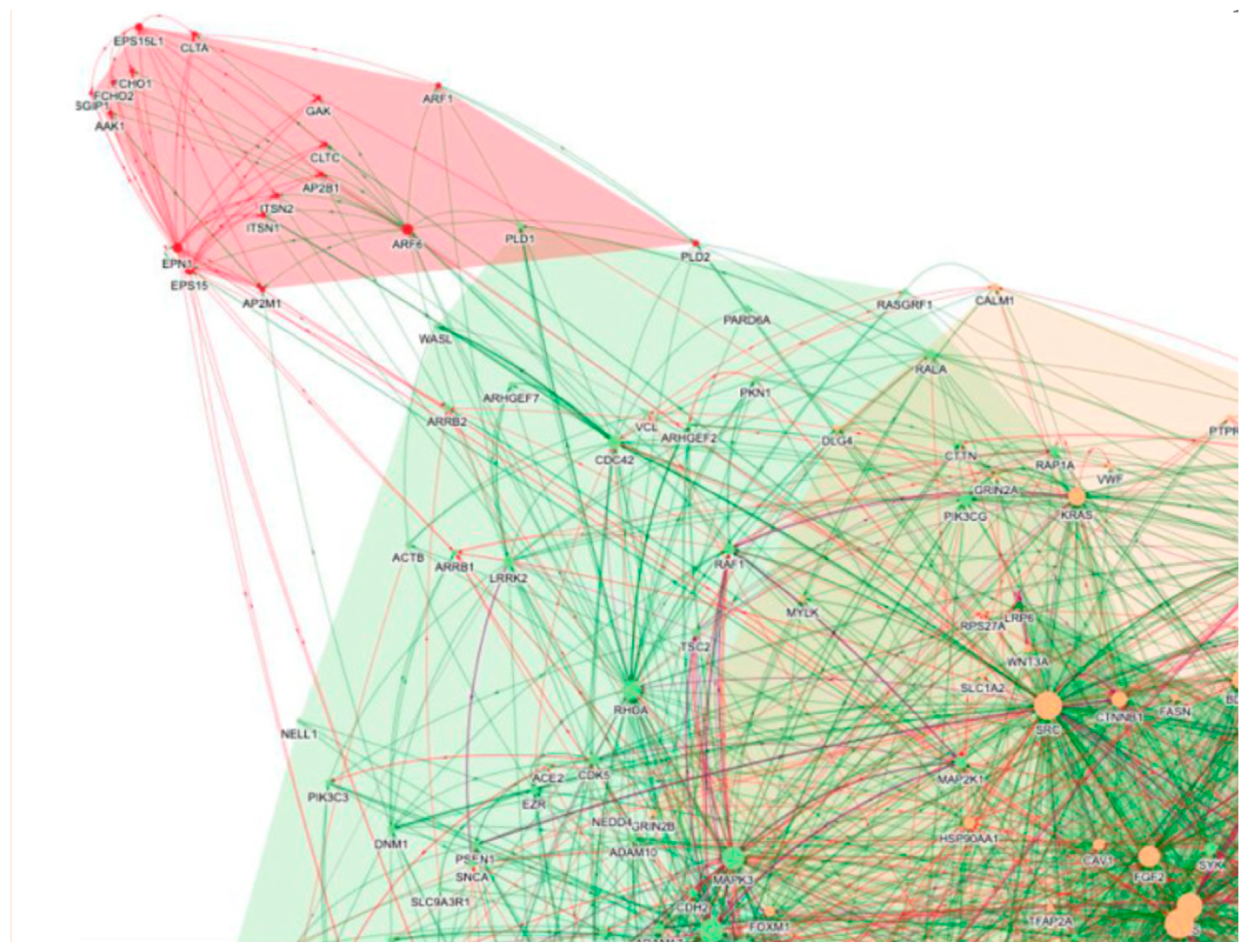
Table 1.
COVID-19 drug discovery using several AI techniques.
| Research | AI Methodology | Clinical Benefits |
|---|---|---|
| Zhavoronkov et al. (2020) [39] | Generative Deep Learning | Obtain cost and time effective drug compound |
| Tang et al. (2020) [40] | Advanced Deep Q-Learning Network (ADQN-FBDD) | There are 47 lead compounds known to target the SARS-CoV2 3C-like main protease. |
| Gao et al. (2020) [41] | Generative network complex (GNC) powered by AI | Create 15 possible medications. |
| Hofmarcher et al. (2020) Hofmarcher et al. (2020) [42] | Deep neural network protocol; ChemAI | Make minuscule components that inhibit SARS-CoV-2. |
| Zhang et al. (2020) [43] | DFCNN, or Dense Fully Convolutional Neural Network | A list of available chemical ligands and peptide medications was provided. |
| Beck et al. (2020) [44] | Molecule Transformer-Drug Target Interaction (MT-DTI) | Ascertain the drug's relationship to the target. A list of antiviral drugs was present. |
| Ge et al. (2020) [45] | Algorithms for network-based knowledge mining that combine statistical analysis and machine learning within an integrated framework and have experimental validation | PARP1 inhibitors have been shown to have antiviral effects against SARS-CoV-2. |
| Hu et al. (2020) [46] | Pre-trained multi-task deep model | Cut down on the time and money spent looking for a cure. |
| Zhou et al. (2020) [47] | Repurposing an integrative antiviral drug via a pharmacologically based network medicine platform. | A total of sixteen possible medications were identified as candidate drugs. |
| Zeng et al. (2020) [48] | A network-DL methodology with a graph named CoV-KGE. | It has been discovered that a cloud provider has 41 repurposing medications. |
| Gysi et al. (2020) [49] | A comprehensive graph neural network. | Defining 81 drug repurposing candidates through the use of in vitro data. |
| Wang et al. (2020) [50] | Ontology-based side-effect prediction framework (OSPF). | Seven TCM have high safety indicators (SI of greater than 0.8). |
| Abdel-Basset et al. (2020) [51] | DeepH-DTA: Developed an HGAT model to learn topological information from compound molecules and a bidirectional ConvLSTM layer to model spatial sequence information in SMILES sequences of drug data. | Determine a medication's affinity score with respect to the amino acid sequences of SARS-CoV-2. |
| Demirci et al. (2020) [52] | ML-based miRNA prediction analysis. | Explore the SARS-CoV-2 infection mechanism. |
Table 2.
COVID-19 vaccine discovery using several AI techniques.
| Research | AI Methodology | Clinical Benefits |
|---|---|---|
| Abdelmageed et al. (2020) [68] | Bioinformatics databases and tools (ACT, VaxiJen Software, and the comparative genomic method). | Epitope vaccines were developed using protein E as an antigenic site. |
| Sarkar et al. (2020) [69] | Molecular docking analysis, reverse vaccinology, and immune informatics. | There are now three known epitope-based subunit vaccines. |
| Fast et al. (2020) [70] | Computational methodology. | Locating multiple SARS-CoV-2 epitopes in order to develop potential vaccinations. |
| Ong et al. (2020) [71] | ML and reverse vaccinology | Notably, there is a good chance that numerous nonstructural proteins will one day be turned into vaccines. |
| Rahman et al. (2020) [72] | Combined with comparative genomics and immuno-informatics methodology. | To target the S, M, and E proteins, a chimeric peptide vaccine called CoV-RMEN was developed. It is based on multiple epitopes. |
| Susithra Priyadarshni et al. (2021) [73] | In silico approach. Molecular docking analysis. | Development of multi-epitope vaccine candidates that specifically target the non-mutated immunogenic regions of the surface glycoprotein and coat protein of SARS-CoV-2. |
| Russo et al. (2021) [74] | A combined bioinformatics pipeline that brings together different programs' predictive capabilities. | Calculate the expected effectiveness of established immunization programs against SARS-CoV-2. |
| Liu et al. (2020) [75] | OptiVax searches a variety of viral or tumor proteins for all peptide fragments that might be suitable candidates for vaccination. | This method increases the presentation likelihood of a diverse set of vaccine peptides based on the target human population's HLA haplotype distribution and predicted epitope drift. |
| A supplementary tool called EvalVax was created at MIT and predicts vaccination coverage while enabling others to assess various vaccine formulations. | ||
| Report by Ayushman Baruah [76] | TCS used generative and predictive models based on deep neural networks to design small compounds that can block the 3CL protease from the ground up. The resultant tiny components were sorted and tested against the SARS-3CL CoV-2 binding site's protease structure. | They came to the conclusion that 31 possible components were strong candidates for production and testing against SARS-CoV-2 based on the screening results and further investigation. |
| Bali et al. (2022) [77] | Instead of concentrating mostly on drugs that might directly impact the virus, Benevolent AI investigated ways to prevent the virus from infecting human cells via biological mechanisms. | Look for approved drugs that may slow the progression of COVID-19, reduce the "cytokine storm," and lessen the inflammation the illness causes. |
| Exscientia press release (2020) [78] | Exscientia, a UK-based company, and Diamond Light Source and Calibr, a US division of Scripps Research, screened a set of 15,000 clinically ready molecules using new biosensor platforms. | Finding any existing drugs that can be modified to protect people is the first goal. |
| Kaushik et al. (2020) [79] | The seven compounds that exhibit potential as COVID-19 inhibitors were discovered by the Hong Kong-based pharmaceutical research company Insilco Medicine. For testing purposes, two of the compounds have already been synthesized. | Utilized VR to Optimize Novel AI-Created COVID-19 medications. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Copyright: This open access article is published under a Creative Commons CC BY 4.0 license, which permit the free download, distribution, and reuse, provided that the author and preprint are cited in any reuse.
MDPI Initiatives
Important Links
© 2024 MDPI (Basel, Switzerland) unless otherwise stated





