1. Introduction
In recent years, due to an improvement in the quality of life, people's life expectancy has increased significantly in developed countries, but this has also led to the fact that previously rare diseases characteristic of older people, including neurodegenerative diseases, began to take on the character of an epidemic. In this regard, research aimed at developing approaches to reduce the negative effects of pathological aging and slow down the processes of natural aging have become highly relevant.
As is known, the molecular aspects of aging are a distortion of proteostasis, deregulation of intracellular signaling pathways, and an abundance of incorrectly folded proteins in the cell, which, in turn, provokes a permanent cellular stress response. To combat stress, any cell has special compartments whose task is to preserve hereditary material and a large number of proteins during stress.
Compartmentalization is primarily necessary for the cell to concentrate certain biomolecules in a limited volume of intracellular space and ensure the effective occurrence of biochemical reactions [
1]. According to modern concepts, the compartmentalization of the intracellular space is determined by the formation of two types of structures: lipid membrane-bound organelles whose contents represent specific microenvironments suited to their function which are isolated from the main contents of the cell by a membrane acting as a diffusion barrier and membrane-less organelles (MLOs); i.e., the membrane-free formations that originate in the cell as a result of the liquid-liquid phase separation (LLPS) of biopolymers, have physicochemical properties characteristic of liquids, usually contain intrinsically disordered proteins (IDPs) and RNA, and constantly exchange their contents with the intracellular environment.
Stressful conditions caused by various external factors, including changes in temperature, pH, irradiation, an increase in the intracellular concentration of reactive oxygen species, salts, and various chemical compounds activate multiple cellular signaling pathways, which is expressed in changes in the composition and characteristics of the intracellular environment, in particular, inhibition of protein synthesis, expressed under normal conditions and activation of the expression of stress response proteins. This creates conditions for changing the critical concentration of proteins necessary for the formation of a number MLOs both in the cytoplasm and in the nucleoplasm of the cell. The most studied MLOs formed in response to stress in eukaryotic cells are stress granules (SGs) and processing bodies (P-bodies).
The formation of SGs and P-bodies can be caused by both exogenous and endogenous factors, such as temperature, oxidative and osmotic stress, UV radiation, impaired proteostasis, viral infection, and other reasons [
2,
3,
4]. Stress impact on the cell causes translation arrest and dissociation of the polyribosomal complex. According to modern concepts, the dissociation of the polyribosome and the mRNA that has not yet undergone translation is accompanied in parallel by the transition of the core IDPs of SGs into the liquid-droplet phase, followed by the recruitment of free mRNA and other components of these MLOs [
3,
5,
6,
7].
SGs and P-bodies are directly involved in the regulation of a large number of signaling pathways. Furthermore, the course of a number of neurodegenerative diseases is accompanied by the transformation of proteins found in these MLOs into amyloid fibrils. SGs can act as a target in the treatment of oncological diseases. In aging
C. elegans cells, P-bodies regulate proteostasis by recruiting the IFE-2 isoform of the transcription initiation factor eIF4E into these organelles, which contributes to the blocking of protein biosynthesis and increases the lifespan of cells [
8]. Mutant forms of several proteins, such as T-cell intracellular antigen-1 (TIA-1), TIA-1-related (TIAR), RNA-binding protein fused in sarcoma (FUS), heterogeneous nuclear ribonucleoprotein A1 (hNRNPA1), transactive response DNA binding protein 43 kDa (TDP-43), and polyadenylate-binding protein 1 (PABP1) promote the transformation of SGs and P-bodies into aggregates of amyloid fibrils. A number of neurodegenerative diseases, including amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD), are directly associated with the degradation of SGs due to the inclusion in their composition of mutant forms of the FUS, TDP-43, hNRNPA1, and TIA-1 proteins with the subsequent transformation of these proteins into the amyloid-like fibrils [
9,
10,
11,
12,
13,
14,
15].
The transformation of SGs into MLOs containing amyloid fibrils in neurodegenerative diseases may be due not only to the inclusion of mutant forms of the FUS, TDP-43, hNRNPA1, and TIA-1 proteins in their composition. Impaired SG biogenesis (i.e., the timely disassembly of these MLOs) can also lead to the formation of ordered amyloid fibrils in the cell [
16,
17,
18,
19]. The SG breakdown can occur via the autophagosomal mechanism, despite the fact that SGs contain virtually no ubiquitinated proteins [
17]. The interaction of the autophagy regulator p62 with hexanucleotide repeats of the
C9orf72 gene contributes to the attraction of the formed complexes to the methylated arginine residues of intrinsically disordered stress granule proteins and the subsequent binding of SGs to the LC3-II autophagosomal membrane protein [
17,
20].
C9orf72 mutations inhibit this process and promote the transformation of liquid-like SGs into biomolecular condensates (BMCs) containing amyloid-like fibrils and the development of neurodegenerative diseases [
17]. Therefore, the permanent presence of SGs in the cell cytoplasm can provoke the development of neurodegenerative diseases, which is observed in aging cells [
21]. In other words, distortions in the time-dependent MLO degradation/disassembly during cell or organism aging increase probability of solid aggregate formation from liquid MLOs (i.e., promote a kind of liquid-solid phase transition). However, other types of aberrant phase separation besides liquid-to-solid transitions can add to proteostasis disruptions [
22].
Accordingly, changes in the material states of these MLOs may correlate with toxic effects in the cell observed during pathological and natural aging. In this work, we analyzed the proteins included in the proteome of SGs and P-bodies for their tendency to spontaneous and induced phase separation, as well as their tendency to aggregate and be engaged in nonspecific interactions with nucleic acids. Particular attention was paid to the proteins whose gene expression, according to transcriptomic data, changes significantly during the transition of human cells to the senescent state.
2. Materials and Methods
2.1. Design of the analysis
A dataset of proteins included in the proteomes of SGs and P-bodies was assembled using results reported in [
17,
23,
24,
25,
26,
27,
28,
29,
30,
31,
32,
33,
34,
35,
36,
37,
38,
39,
40,
41,
42,
43,
44,
45,
46,
47,
48,
49,
50,
51,
52,
53,
54,
55,
56,
57,
58,
59,
60,
61]. Proteins whose gene expression changes during the transition of cells to the senescent state were selected from the GenAge database [
62,
63].
We analyzed the amino acid sequences of collected proteins for the potential propensity to spontaneous and induced liquid–liquid phase separation (LLPS) and the tendency of these proteins to gelation and amyloid fibrillation. We evaluated the a broad set of sequence-based parameters, which are summarized in
Figure 1.
2.2. Disorder Prediction
The disorder analysis of the studied proteins was carried out using the RIDAO online service [
64] to predict disordered regions in proteins based on their amino acid sequences. This package combines the forecasts of several well-known disorder predictors, such as PONDR
® VLXT [
65], PONDR
® VL3 [
66], PONDR
® VLS2B [
67], PONDR
® FIT [
68], IUPred2 (Short) and IUPred2 (Long) [
69,
70]. In the case of a significant discrepancy between the results obtained by different predictors, the correctness of the protein structure disorder prediction was determined by the protein structure prediction using the AlphaFold2 algorithm [
71].
The most consistent predictions of disorder for a large dataset were generated by the PONDR® VSL2B algorithm. As a measure of the disorder of the protein structure, we used the percentage of predicted disordered residues (PPDRVSL2); i.e., the percentage of residues in the protein amino acid sequence, for which the predicted disorder score is higher than the threshold of 0.5. Such regions are defined by the algorithm as "probably disordered".
Next, we carried out a classification of proteins according to their degree of disorder. At this stage, we used very stringent criteria, and proteins with a PPDR
VSL2 value < 50% were classified as "Ordered". Proteins with 50 ≤ PPDR
VSL2 < 85% were classified as Partially Disordered (IDPRs), whereas proteins with the PPDR
VSL2 ≥ 85% were classified as Disordered (IDPs). Note that this is a more conservative classification in comparison with accepted in the field criteria, where proteins with PPDR < 10% are considered as ordered or mostly ordered; proteins with 10% ≤ PPDR < 30% are considered as moderately disordered; whereas proteins with the PPDR ≥ 30% are considered as highly disordered [
72]. Furthermore, proteins can be also classified based on their mean disorder scores (MDS) as highly ordered (MDS < 0.15), moderately disordered or flexible (MDS between 0.15 and 0.5) and highly disordered (MDS ≥ 0.5).
Next, the outputs of two binary predictors, the charge-hydropathy (CH) plot [
73,
74] and the cumulative distribution function (CDF) plot [
74,
75,
76] were combined to conduct a CH-CDF analysis [
76,
77,
78,
79] that allows classification of proteins based on their position within the CH-CDF phase space as ordered (proteins predicted to be ordered by both binary predictors), putative native “molten globules” or hybrid proteins (proteins determined to be ordered/compact by CH, but disordered by CDF), putative native coils and native pre-molten globules (proteins predicted to be disordered by both methods), and proteins predicted to be disordered by CH-plot, but ordered by CDF.
Complementary disorder evaluations together with important disorder-related functional information were retrieved from the D
2P
2 database (
http://d2p2.pro/) [
80], which is a database of predicted disorder for a large library of proteins from completely sequenced genomes [
80]. D
2P
2 database uses outputs of IUPred [
69], PONDR
® VLXT [
65], PrDOS [
81], PONDR
® VSL2B [
66,
82], PV2 [
80], and ESpritz [
83]. The visual console of D2P2 displays 9 colored bars representing the location of disordered regions as predicted by these different disorder predictors. In the middle of the D2P2 plots, the blue-green-white bar shows the predicted disorder agreement between nine disorder predictors (IUPred, PONDR
® VLXT, PONDR
® VSL2, PrDOS, PV2, and ESpritz), with blue and green parts corresponding to disordered regions by consensus. Above the disorder consensus bar are two lines with colored and numbered bars that show the positions of the predicted (mostly structured) SCOP domains [
84,
85] using the SUPERFAMILY predictor [
86]. Yellow zigzagged bar shows the location of the predicted disorder-based binding sites (MoRF regions) identified by the ANCHOR algorithm [
87], whereas differently colored circles at the bottom of the plot show location of various PTMs assigned using the outputs of the PhosphoSitePlus platform [
88], which is a comprehensive resource of the experimentally determined post-translational modifications.
The standard deviation calculated using the bootstrap was used as an error ID estimate. The subsample size is equal to the size of the set of proteins of the corresponding compartment, the number of generated subsamples is 10,000.
2.3. LLPS prediction
The studied datasets were further analyzed for the propensity of their proteins to undergo liquid–liquid phase separation (LLPS) using FuzDrop [
89] and PSPredictor [
90] predictors. The propensity of the analyzed protein to LLPS was determined based on PSPredcitor score > 0.5 and FuzDrop score > 0.6. In the event of a discrepancy between the results of predicting the propensity to phase separation obtained using both predictors, the analyzed proteins were assigned to the “controversial LLPS” group.
The FuzDrop predictor also predicts the ability of a query protein to undergo LLPS spontaneously (droplet-driving proteins) or requiring additional interactions with LLPS partners to form droplets (droplet-clients). Proteins with the pLLPS ≥ 0.60 likely drive liquid-liquid phase separation. Proteins with the propensity for liquid-liquid phase separation (pLLPS) below the threshold of 0.6 but containing the droplet-promoting regions (DPRs), defined as consecutive residues with pDP ≥ 0.60, will likely serve as drop-let-clients.
2.4. Aggregation propensity prediction
Analysis of the propensity of the studied proteins to aggregation and the formation of amyloid fibrils was performed using the AggreScan package [
91], which makes it possible to determine the so-called aggregation hot spots; i.e., areas of the protein that promote its aggregation. After preliminary analysis, it turned out that each protein in the studied datasets contained at least one such region. In this regard, the results of the performed analysis were supplemented using Normalized a4v Sequence Sum for 100 residues (Na4vSS) values, which reflect the average protein aggregation propensities of the sequences (Na4vSS>0) once corrected for their size.
In addition, the ability of the analyzed proteins to form amyloid fibrils was verified by searching the CPAD database, composed of experimentally confirmed proteins prone to aggregation [
92].
2.5. Evaluation of protein charge and hydrophobicity
The charge of the studied proteins was determined at pH = 7 according to [
73] as the average protein charge over the sequence, taking into account the charge of amino acids: R = 1, K = 1, H = 0.5, D = -1, E = -1. The hydrophobicity of the studied proteins was calculated and normalized according to the Kyte and Doolittle scale.
2.6. Analysis of the interctability of proteins
Information on the interactability of 10 human senescence-related proteins shared by SGs and P-bodies was retrieved by Search Tool for the Retrieval of Interacting Genes; STRING,
http://string-db.org/. STRING generates a network of protein-protein interactions based on predicted and experimentally-validated information on the interaction partners [
93]. In the corresponding network, the nodes correspond to proteins, whereas the edges show predicted or known functional associations. Seven types of evidence are used to build the corresponding network, where they are indicated by the differently colored lines: a green line represents neighborhood evidence; a red line – the presence of fusion evidence; a purple line – experimental evidence; a blue line – co-occurrence evidence; a light blue line – database evidence; a yellow line – text mining evidence; and a black line – co-expression evidence [
93].
In this study, STRING was utilized in two different modes: to generate the internal network protein-protein interactions (PPIs) between the 10 query proteins and to produce the global PPI network centered at these 10 proteins. Resulting PPI networks were further analyzed using STRING-embedded routines in order to retrieve the network-related statistics, such as: the number of nodes (proteins); the number of edges (interactions); average node degree (average number of interactions per protein); average local clustering coefficient (which defines how close the neighbors are to being a complete clique – if a local clustering coefficient is equal to 1, then every neighbor connected to a given node Ni is also connected to every other node within the neighborhood, and if it is equal to 0, then no node that is connected to a given node Ni connects to any other node that is connected to Ni); expected number of edges (which is a number of interactions among the proteins in a random set of proteins of similar size); and a PPI enrichment p-value (which is a reflection of the fact that query proteins in the analyzed PPI network have more interactions among themselves than what would be expected for a random set of proteins of similar size, drawn from the genome. It was pointed out that such an enrichment indicates that the proteins are at least partially biologically connected, as a group). The MCL algorithm was used to cluster the proteins that are displayed in the network.
2.7. Definition of Biological Processes and Molecular Functions of Proteins
The analysis of biological processes and molecular functions of query proteins was conducted using the Enrichr resource (
https://maayanlab.cloud/Enrichr/, accessed on 20 April 2023), and 10 terms with the lowest
p-values were selected. The
p-values were computed from the Fisher exact test, which is used to determine whether or not there are significant nonrandom associations between two categorical variables.
3. Results and discussion
3.1. Analysis of the proteomes of stress granules and P-bodies
Stress granules (SGs) and P-bodies are known to be MLOs with the overlapping compositions and functions. Almost all analyzed parameters of the studied datasets are the same for SGs and P-bodies (
Table 1,
Table 2 and
Table S1). This indicates that SGs and P-bodies are structures formed by a single mechanism. This section may be divided by subheadings. It should provide a concise and precise description of the experimental results, their interpretation, as well as the experimental conclusions that can be drawn.
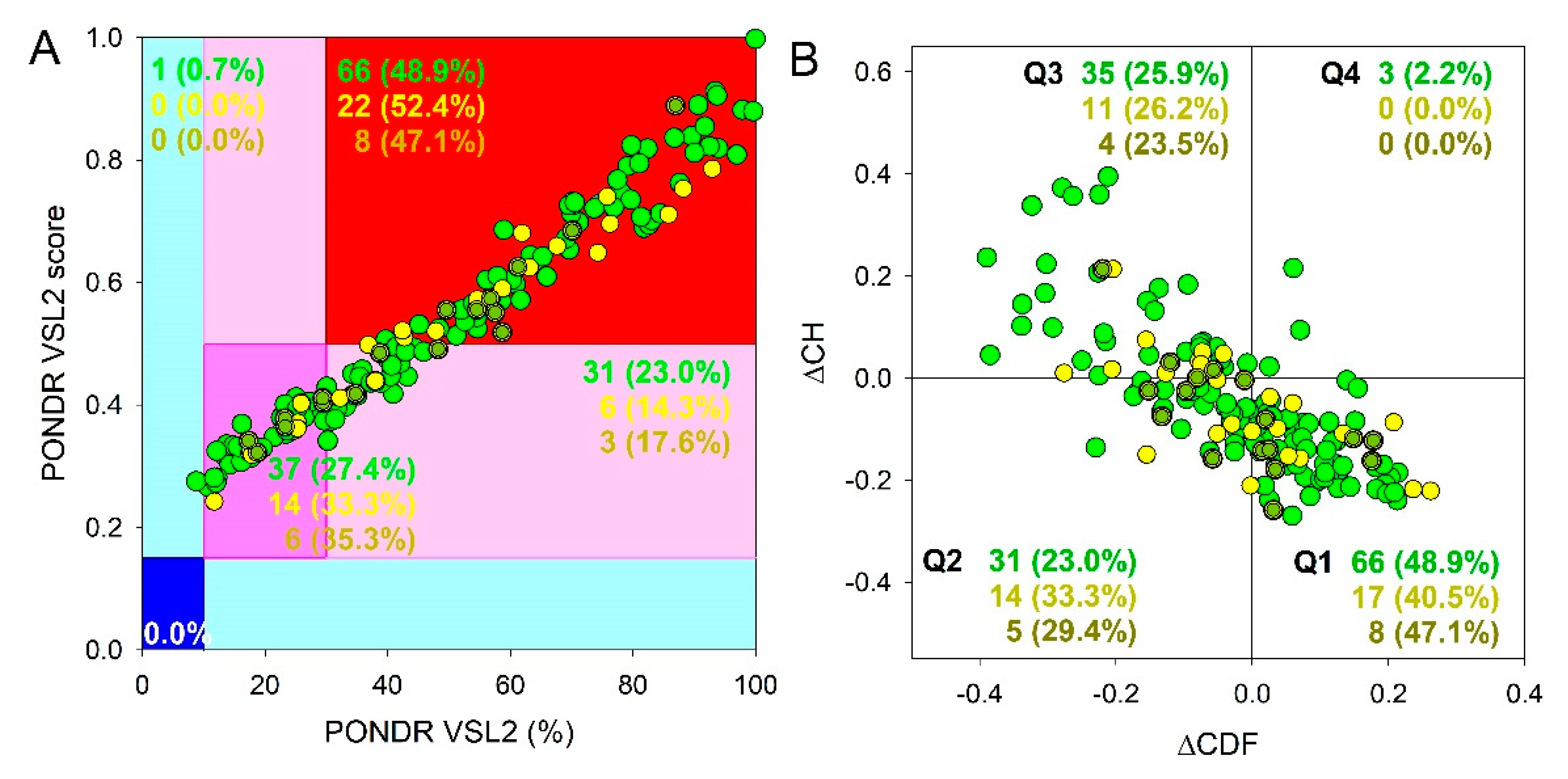
Results of global intrinsic disorder analysis of the proteomes of SGs and P-bodies as well as common proteins that are simultaneously included in SGs and P-bodies are summarized in
Figure 2. This global disorder analysis revealed that all these proteins contain noticeable levels of intrinsic disorder. For example, this is evidenced by the results of the classification of the disorder status of these proteins based on the outputs of the per-residue disorder predictor PONDR
® VSL2 in a form of the mean disorder score (MDS)
vs. percent of predicted disordered residues (PPIDR) dependence. This plot shows distribution of the proteins within MDS-PPIDR blocks containing mostly ordered (blue and light blue), moderately disordered (pink and light pink), or mostly disordered (red) proteins. Based on these classifications, none of the proteins analyzed in this study was predicted as ordered by MDS and PPIDR simultaneously (dark blue segment is empty), and only 9 SG proteins, 4 P-body proteins, and 1 common protein were predicted as mostly ordered by MDS. Therefore, the remaining proteins in these datasets are expected to be either moderately or highly disordered. In fact, according to the
Figure 2A 23.9%, 23.7%, and 21.8% of the SG, P-body and common proteins were predicted as moderately disordered and containing noticeable levels of flexible or disordered residues/regions based on both their MDS and PPIDR values (dark pink segment). Additional 20.6%/19.8%/19.% proteins were expected to be moderately disordered based on their MDS values (light pink segment), indicating that altogether 44.5%/43.5%/41.5% of the human SG/P-body/common proteins are predicated as moderately disordered. Finally, 54.3%/54.2%/57.5% of the SG/P-body/common proteins are expected to be highly disordered (red segment). These global disorder contents in the proteomes of SGs and P-bodies as well as common proteins that are simultaneously found in SGs and P-bodies are somewhat higher than that of the whole human proteome, where the moderately and highly disordered proteins account for 51.7% and 39.8%, respectively [
94].
Further support for high disorder status of human proteins in the proteomes of the SGs and P-bodies as well as common proteins that are simultaneously found in SGs and P-bodies was provided by analyzing the combined outputs of two binary disorder predictors charge-hydropathy (CH) plot and cumulative distribution function (CDF) analysis that classify proteins as mostly ordered or mostly disordered (see
Figure 2B). The result of the ΔCH-ΔCDF plot provides useful means for more detailed characterization of the global disorder status of proteins, classifying them as mostly ordered, molten globule-like or hybrid, or highly disordered (see Materials and Methods section).
Figure 2B shows that 44.7% (SGs), 44.6% (P-bodies), and 40.2% common proteins (cf. 59.1% in entire human proteome [
94]) are located within the quadrant Q1 (bottom right corner) that includes proteins predicted to be ordered by both predictors, with remaining proteins in these datasets being located outside the quadrant Q1 and can be considered as proteins with high disorder levels.
Figure 2B shows that the quadrant Q2 (bottom left corner) corresponding to molten globular or hybrid proteins predicted to be ordered/compact by the CH-plot and disordered by the CDF analysis includes 32.1%/29.4%/39.1% of SG/P-body/common proteins (21.5% in human proteome [
94]). Furthermore, 21.3%/24.3%/18.4% of SG/P-body/common proteins (12.3% in human proteome [
94]), being located in the quadrant Q3 (top left corner), are predicted as highly disordered by both predictors. Finally, the quadrant Q4 (top right corner) contains 1.9%1.7%/2.3% of SG/P-body/common proteins predicted as disordered by CH-plot and ordered by CDF analysis (3.1% in human proteome [
94]). It seems that the disorder levels of common proteins (PPIDR
VSL2 = 51.2±24.0%; MDS = 0.542±0.158) are somewhat higher than the disorderedness of the proteins in the SG (PPIDR
VSL2 = 50.8±25.1%; MDS = 0.543±0.168) and P-body proteomes (PPIDR
VSL2 = 48.4±25.7%; MDS = 0.523±0.157). However, significance of this difference is not clear due to the small size of the analyzed datasets. Therefore, results these analyses show that the levels of intrinsic disorder in human proteins from the proteomes of the SGs and P-bodies as well as common proteins simultaneously found in SGs and P-bodies are noticeably higher than those of entire human proteome, suggesting that intrinsic disorder might play important roles in functions of these proteins and is likely to be related to the biogenesis of these MLOs.
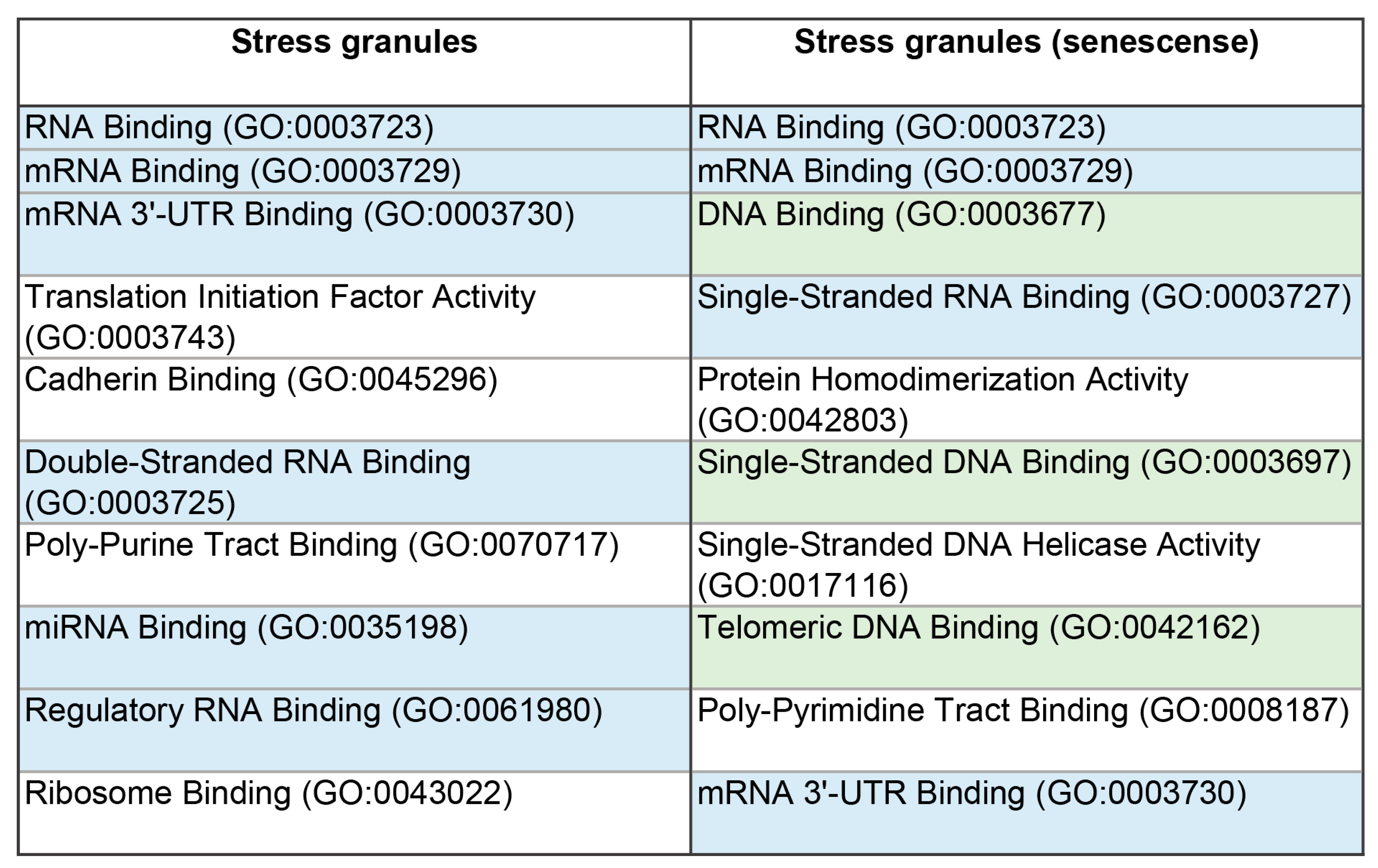
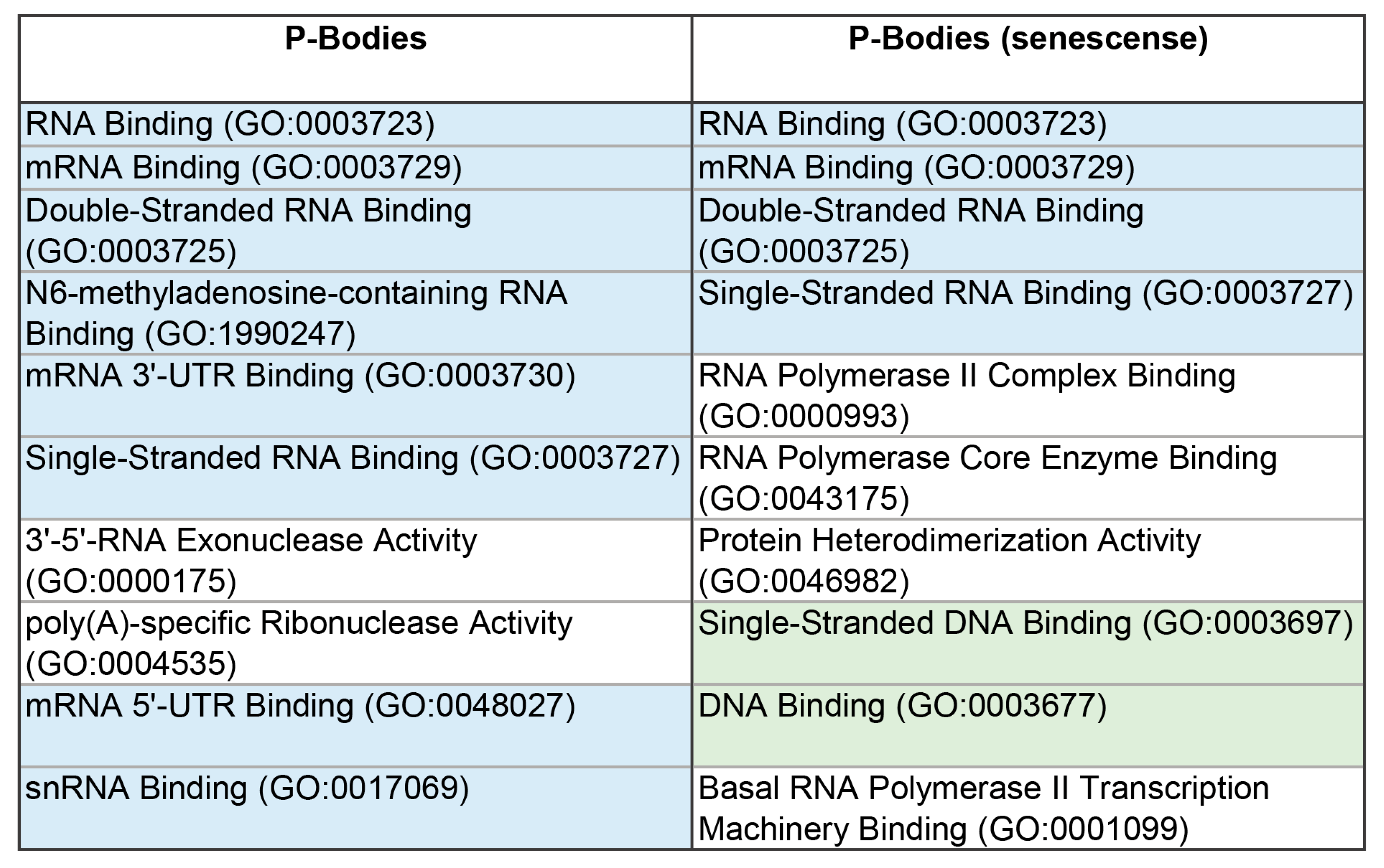
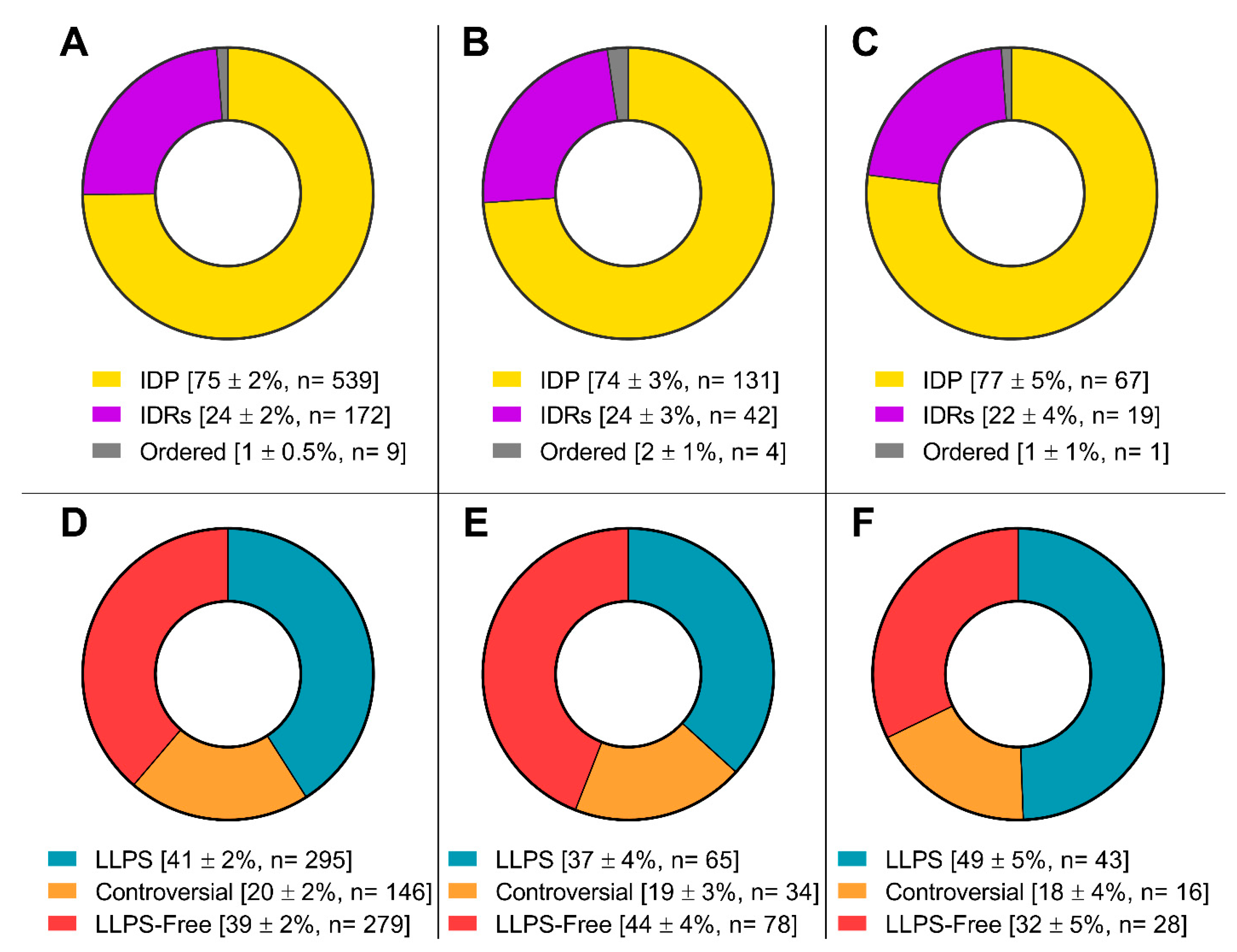
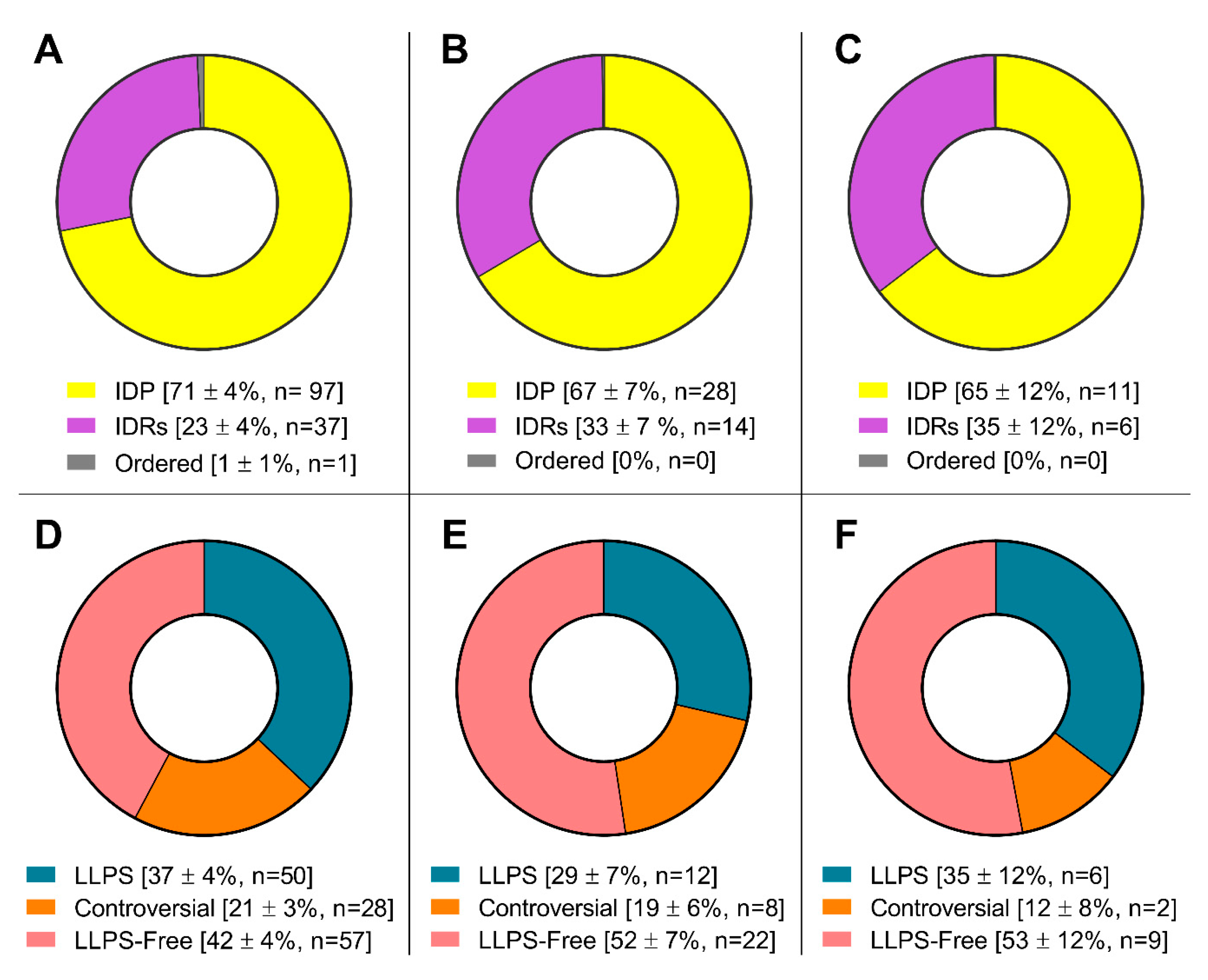
The proteomes of these MLOs are represented by proteins involved in the processes of translation, regulation, and RNA processing (
Figure 3 and
Figure 4) and is almost 100% composed of completely or partially disordered proteins, 30 - 40% of which are potentially capable of phase separation (
Figure 5).
Furthermore, the proportion of proteins capable of spontaneous LLPS is four times lower than the proportion of proteins potentially capable of LLPS induced by interactions with partners (
Table 1 and
Table 2). These data correlate with the modern concepts of the biogenesis of these MLOs, according to which the formation of SGs and P-bodies is initiated by the coacervation of their scaffold proteins and RNA molecules [
3]. This is further confirmed by the high content of RNA- and DNA-binding proteins in the proteome of the studied MLOs (
Table 1 and
Table 2,
Figure 3 and
Figure 4). The proportion of amyloidogenic proteins – proteins that potentially contribute to the transformation of these compartments into BMCs containing insoluble aggregates of amyloid fibrils – is relatively small in the proteomes of SGs and P-bodies (8 and 11%, respectively).
The only analyzed parameter in which the datasets for SGs and P-bodies differ significantly from each other is the average charge of the proteome (
Table 1 and
Table 2). SG proteins generally carry a higher positive charge than the P-body proteins. This may indirectly indicate a relatively higher content in the SG proteome of proteins capable of nonspecific interactions with RNA compared to P-bodies.
3.2. Analysis of proteins from the proteomes of SGs and P-bodies that are potentially involved in the senescence-related processes
Proteins whose gene expression changes during the transition of human cells to a senescent state account for about 20% of the studied proteomes of SGs and P-bodies.
Figure 6 represents the outputs of a global intrinsic disorder analysis in these proteins and shows that they are somewhat less disordered (SGs: PPIDR
VSL2 = 49.3±25.7%; MDS = 0.539±0.168; P-bodies: PPIDR
VSL2 = 46.9±22.7%; MDS = 0.516±0.152; common proteins: PPIDR
VSL2 = 44.6±19.8%; MDS = 0.504±0.144) than proteins in the complete proteomes of SGs and P-bodies. However, these differences are not significant.
More detailed analysis of the datasets consisting of proteins potentially involved in senescent-related processes showed that within these arrays, there is no significant change in the content of intrinsically disordered proteins and proteins prone to phase separation compared to the complete proteomes of SGs and P-bodies (
Figure 7).
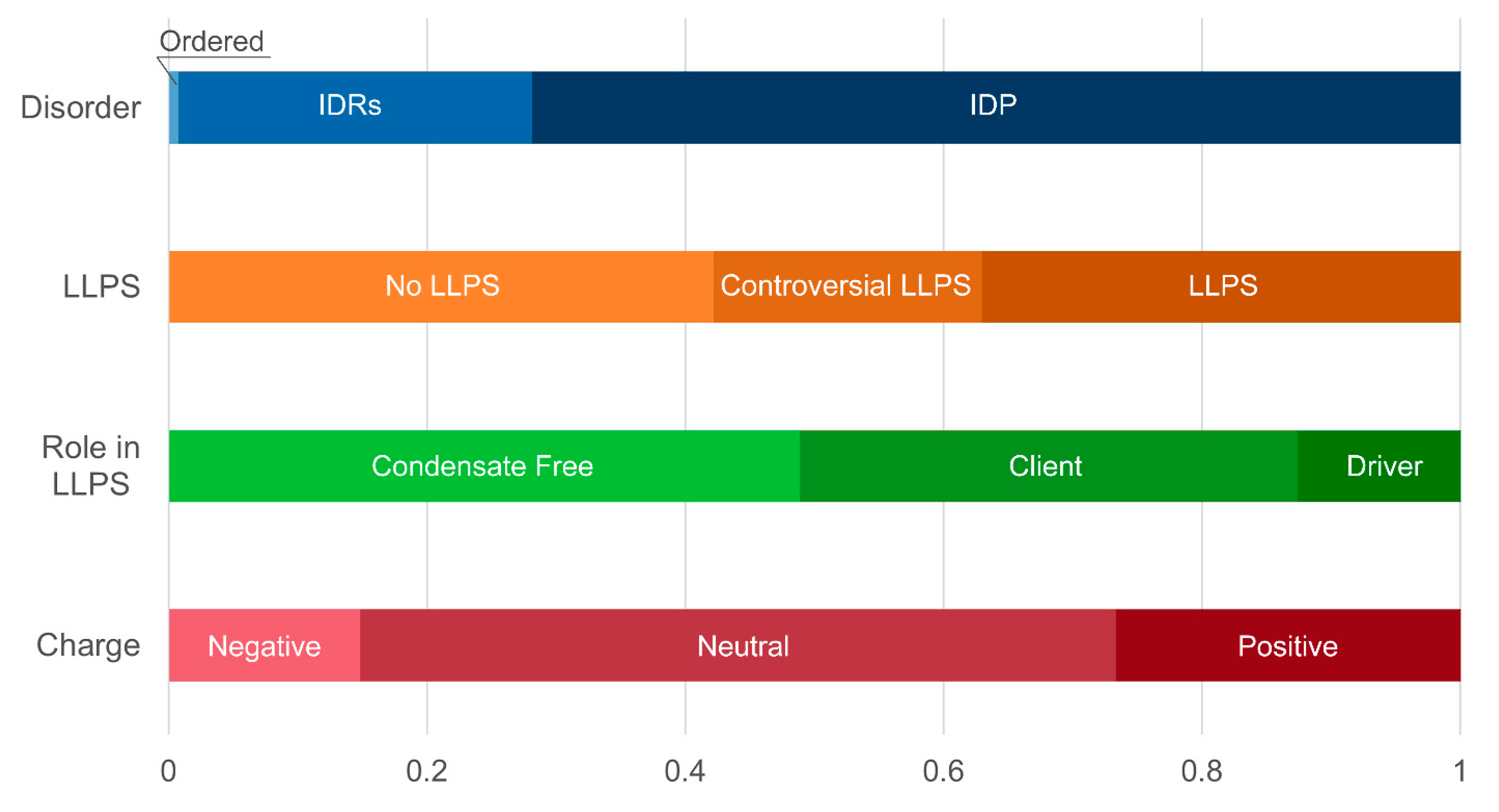
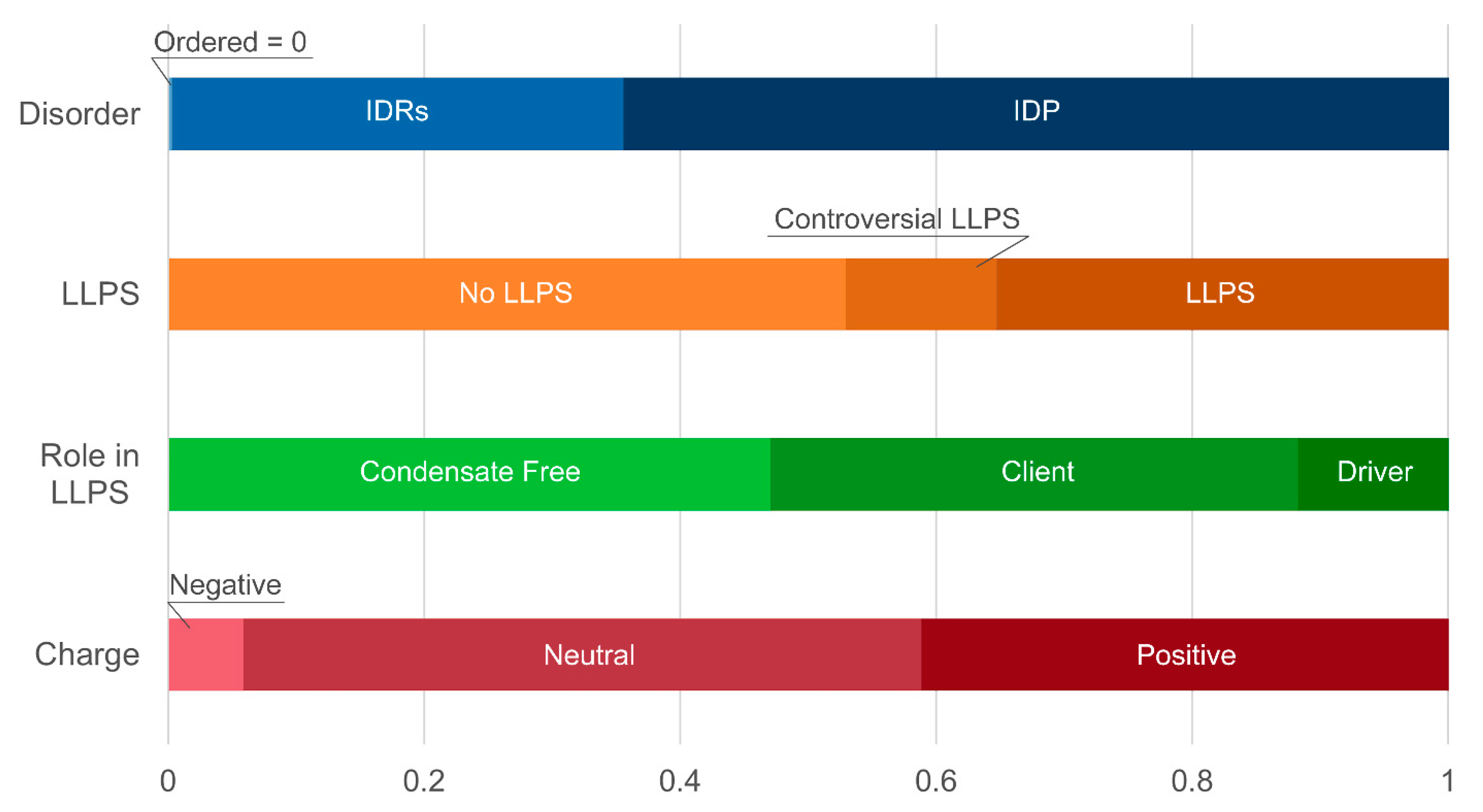
At the same time, a statistically significant increase in the number of positively charged proteins is observed in both studied datasets (
Table 1 and
Table 2,
Figure 8 and
Figure 9,
Table S2). This may indicate an increase in the relative content of proteins prone to nonspecific interaction with negatively charged nucleic acids. An increase in the relative content of DNA-, but not RNA-binding proteins, was also found in the SG dataset of proteins associated with the senescence-related processes (
Table 1 and
Table 2,
Figure 3 and
Figure 4). As is known, the SG scaffold protein G3BP1 is required for the activation of the cytosolic DNA sensor, cyclic GMP-AMP synthase (cGAS) [
95], which is involved in the regulation of the transition of cells to the senescent state when telomeres are disrupted [
96].
Among the SG proteins potentially involved in senescent-related processes, there is a statistically significant increase in the relative abundance of potentially amyloidogenic proteins compared to the whole proteome of these MLOs (
Table 1 and
Table 2). These are proteins involved in the regulation of the cell cycle, telomerase activity, and DNA processing (
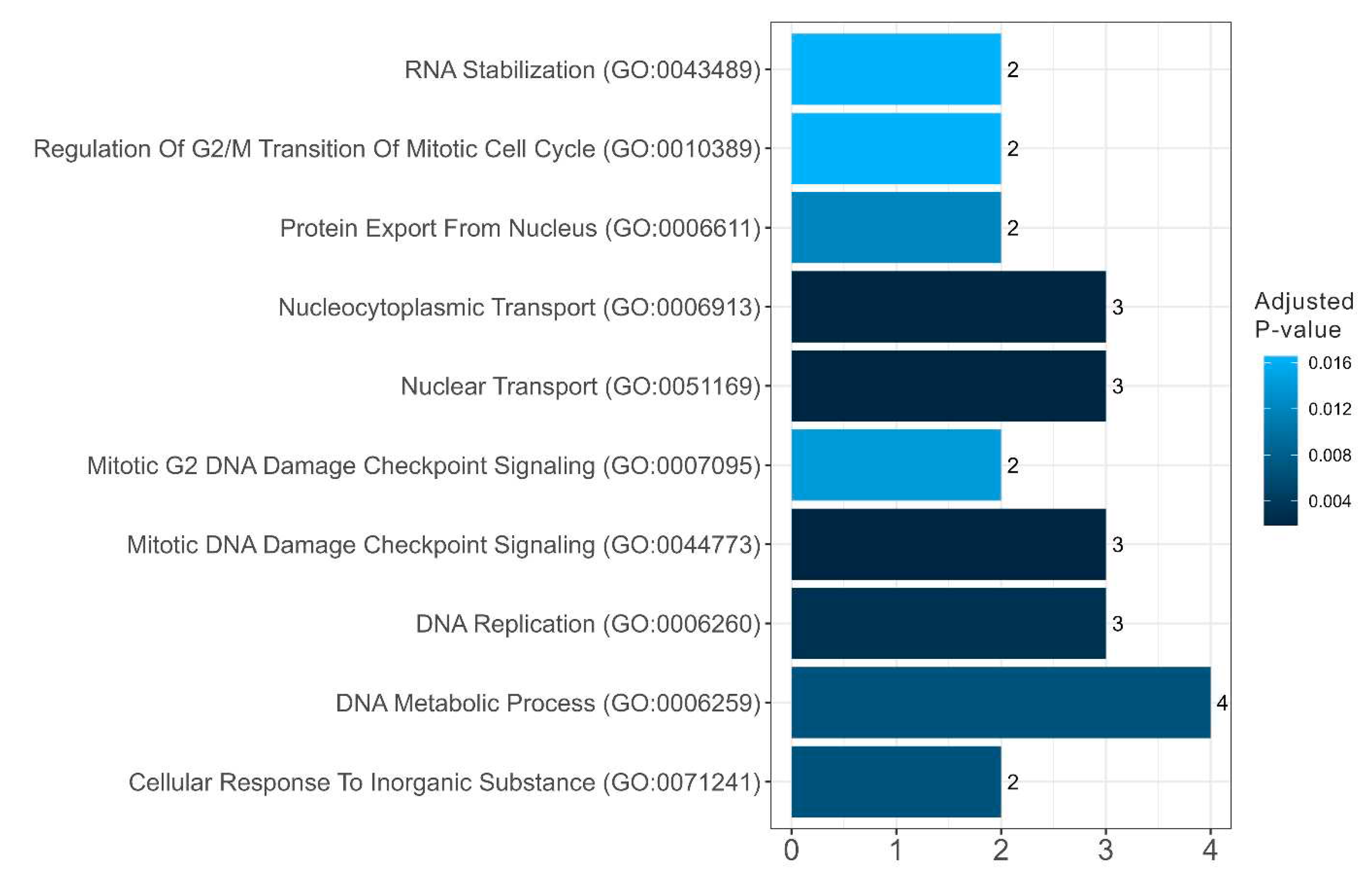
Figure 10 and
Figure 11). A significant portion of the potentially amyloidogenic SG proteins associated with senescence are components of nuclear pore complexes. It is known that the course of many neurodegenerative diseases is accompanied by disruption of nuclear-cytoplasmic transport and the inclusion of nucleoporins and other nuclear proteins in the composition of SGs.
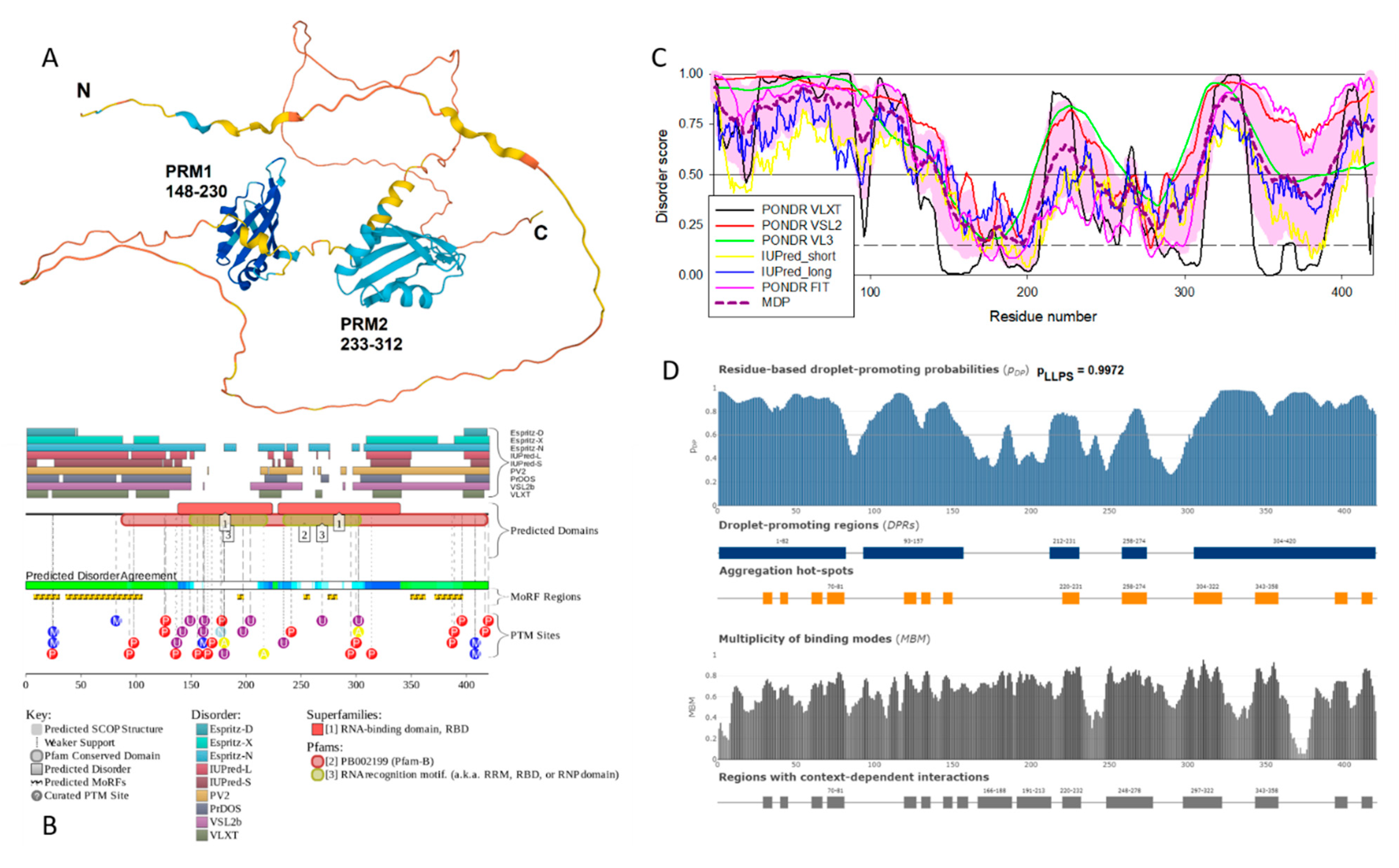
Heterogeneous nuclear ribonucleoprotein D-like protein (hnRPDL) has the highest degree of disorder and the highest propensity for LLPS among potentially amyloidogenic SG proteins associated with senescence. This nuclear protein, being a component of nuclear speckles, is directly involved in the aging process by regulating the expression of cytoskeletal and synapse genes [
97]. Mutations in the C-terminal disordered domain of this protein, characteristic of limb-girdle muscular dystrophy 1G, promote fibrillation of this protein [
98]. Taken together, these data allow us to consider this protein as “dangerous” in the context of aging and senescence-related processes.
Figure 12 represents a disorder-centric “portrait of this hero” and clearly shows that hnRPDL, being characterized by high levels of disorder, contains numerous disorder-based binding sites (molecular recognition features, which are disordered regions that undergo disorder-to-order transitions at interaction with binding partners), contain numerous sites of various post-translational modifications, shows high LLPS propensity and contains several aggregation hot spots.
3.3. Analysis of proteins simultaneously included in the proteomes of GSs and P-bodies and potentially involved in senescence-related processes
Among the proteins analyzed in this study, 87 proteins are simultaneously included in the proteomes of SGs and P-bodies. In general, the analyzed characteristics of the proteins included in this dataset correspond to the characteristics of the complete proteomes of SGs and P-bodies (
Table 3). 17 proteins from this array could potentially be involved in processes associated with the transition of cells to the senescent state. Among them are 11 RNA-binding and 3 DNA-binding proteins (
Table 3,
Figure 13).
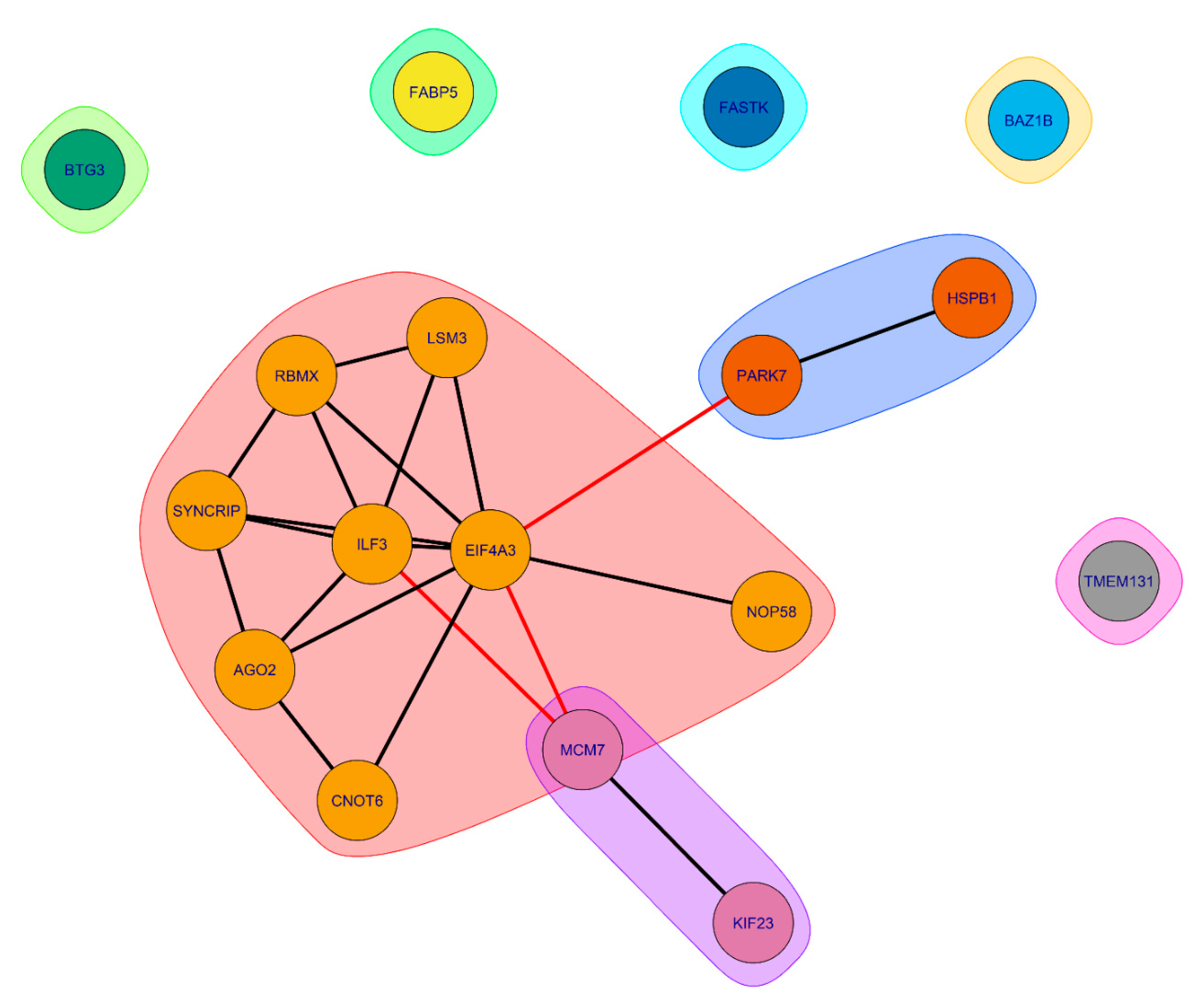
According to the STRING-based analysis (
Figure 14), proteins common to SGs and P-bodies, potentially involved in processes associated with senescence, form clusters of interacting proteins.
The largest cluster is represented by RNA-binding proteins involved in RNA processing and translation regulation (
Figure 15 and
Figure 16).
Several proteins in this cluster are directly involved in the processes of aging regulation. Eukaryotic initiation factor 4A-III (eIF4A3), a core helicase component of the exon junction complex (EIF4A3), is included in P-bodies during aging, participating in the negative regulation of gene expression and ribosomal biogenesis [
8,
99].
Interleukin Enhancer Binding Factor 3 (ILF3) is involved in the regulation of gene expression during aging and chronic stress [
100,
101]
. Protein argonaute-2 (AGO-2) is an important component of the RNA-induced silencing complex (RISC) targeting microRNA (miRNA) to mRNA for translational inhibition or destruction of the target mRNA. It is known that the miRNA expression and nucleocytoplasmic transport of these molecules change significantly during aging, which correlates with changes in yjr AGO-2 expression in aging cells [
102,
103,
104]
. Associated with this cluster are two more clusters, also containing proteins directly involved in processes associated with aging. Overexpression of Small heat shock protein beta 1 (HspB1) increases lifespan in
Caenorhabiditis elegans [
105]
.
Common SG and P-body proteins that do not form clusters are also involved in aging-related processes, including collagen biogenesis, mitochondrial RNA regulation, and transcriptional control [
106,
107,
108].
4. Conclusions
The data obtained in this study suggest that SGs and P-bodies are structures formed by a common mechanism, in which RNA molecules play a leading role. These MLOs are widely involved in aging and related processes and can participate in such processes in a coordinated manner, primarily at the level of translation regulation. Stress granules are more prone to pathological transformation during aging than P-bodies, probably due to the inclusion of nucleocytoplasmic transport proteins in their content.
Supplementary Materials
The following supporting information can be downloaded at:
www.mdpi.com/xxx/s1, Table S1: Characteristics of the proteomes of the SGs and PBs their proteins involved in senescence-related processes; Table S2: Characteristics of the proteomes of the SGs and PBs proteins involved in senescence-related processes
Author Contributions
Conceptualization, A.V.F. and V.N.U; methodology, A.V.F.,V.N.U., S.V.S., Y.I.M., N.S.I., validation, Y.I.M., A.E.R., O.S.S., I.M.K., K.K.T., V.N.U., and A.V.F.; formal analysis, Y.I.M., E.Y.S., S.V.S., N.S.I.., I.M.K., K.K.T., V.N.U., and A.V.F.; investigation, S.V.S., Y.I.M., N.S.I., I.M.K., K.K.T., V.N.U., and A.V.F.; data curation, Y.I.M., N.S.I.,., I.M.K., K.K.T., V.N.U., and A.V.F.; writing—original draft preparation, Y.I.M., V.N.U., and A.V.F.; writing—review and editing, Y.I.M., S.V.S., N.S.I., I.M.K., K.K.T., V.N.U., and A.V.F.; visualization, Y.I.M., V.N.U., and A.V.F.; supervision, V.N.U and A.V.F.; project administration, A.V.F.; funding acquisition, A.V.F. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Russian Science Foundation, grant number 23-45-00041 (A.V.F.).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article or supplementary material.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Pappu, R.V. Phase Separation-A Physical Mechanism for Organizing Information and Biochemical Reactions. Developmental cell 2020, 55, 1–3. [Google Scholar] [CrossRef]
- Mahboubi, H.; Stochaj, U. Cytoplasmic stress granules: Dynamic modulators of cell signaling and disease. Biochimica et biophysica acta. Molecular basis of disease 2017, 1863, 884–895. [Google Scholar] [CrossRef] [PubMed]
- Protter, D.S.W.; Parker, R. Principles and Properties of Stress Granules. Trends in cell biology 2016, 26, 668–679. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Na, Z.; Slavoff, S.A. P-Bodies: Composition, Properties, and Functions. Biochemistry 2018, 57, 2424–2431. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Mathieu, C.; Kolaitis, R.M.; Zhang, P.; Messing, J.; Yurtsever, U.; Yang, Z.; Wu, J.; Li, Y.; Pan, Q. , et al. G3BP1 Is a Tunable Switch that Triggers Phase Separation to Assemble Stress Granules. Cell 2020, 181, 325–345 e328. [Google Scholar] [CrossRef] [PubMed]
- Guillén-Boixet, J.; Kopach, A.; Holehouse, A.S.; Wittmann, S.; Jahnel, M.; Schlüßler, R.; Kim, K.; Trussina, I.; Wang, J.; Mateju, D. , et al. RNA-Induced Conformational Switching and Clustering of G3BP Drive Stress Granule Assembly by Condensation. Cell 2020, 181, 346–361 e317. [Google Scholar] [CrossRef] [PubMed]
- Jain, S.; Wheeler, J.R.; Walters, R.W.; Agrawal, A.; Barsic, A.; Parker, R. ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure. Cell 2016, 164, 487–498. [Google Scholar] [CrossRef]
- Rieckher, M.; Markaki, M.; Princz, A.; Schumacher, B.; Tavernarakis, N. Maintenance of Proteostasis by P Body-Mediated Regulation of eIF4E Availability during Aging in Caenorhabditis elegans. Cell reports 2018, 25, 199–211 e196. [Google Scholar] [CrossRef]
- Alberti, S.; Hyman, A.A. Are aberrant phase transitions a driver of cellular aging? BioEssays : news and reviews in molecular, cellular and developmental biology 2016, 38, 959–968. [Google Scholar] [CrossRef]
- Cao, X.; Jin, X.; Liu, B. The involvement of stress granules in aging and aging-associated diseases. Aging cell 2020, 19, e13136. [Google Scholar] [CrossRef]
- Elbaum-Garfinkle, S. Matter over mind: Liquid phase separation and neurodegeneration. The Journal of biological chemistry 2019, 294, 7160–7168. [Google Scholar] [CrossRef] [PubMed]
- Lechler, M.C.; David, D.C. More stressed out with age? Check your RNA granule aggregation. Prion 2017, 11, 313–322. [Google Scholar] [CrossRef]
- Shiina, N. Liquid- and solid-like RNA granules form through specific scaffold proteins and combine into biphasic granules. The Journal of biological chemistry 2019, 294, 3532–3548. [Google Scholar] [CrossRef] [PubMed]
- Tüű-Szabó, B.; Hoffka, G.; Duro, N.; Fuxreiter, M. Altered dynamics may drift pathological fibrillization in membraneless organelles. Biochimica et biophysica acta. Proteins and proteomics 2019, 1867, 988–998. [Google Scholar] [CrossRef] [PubMed]
- Verdile, V.; De Paola, E.; Paronetto, M.P. Aberrant Phase Transitions: Side Effects and Novel Therapeutic Strategies in Human Disease. Frontiers in genetics 2019, 10, 173. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Fan, B.; Yang, P.; Temirov, J.; Messing, J.; Kim, H.J.; Taylor, J.P. Chronic optogenetic induction of stress granules is cytotoxic and reveals the evolution of ALS-FTD pathology. eLife 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Maharjan, N.; Künzli, C.; Buthey, K.; Saxena, S. C9ORF72 Regulates Stress Granule Formation and Its Deficiency Impairs Stress Granule Assembly, Hypersensitizing Cells to Stress. Molecular neurobiology 2017, 54, 3062–3077. [Google Scholar] [CrossRef]
- Zhao, Y.G.; Codogno, P.; Zhang, H. Machinery, regulation and pathophysiological implications of autophagosome maturation. Nature reviews. Molecular cell biology 2021, 22, 733–750. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, H. Phase Separation, Transition, and Autophagic Degradation of Proteins in Development and Pathogenesis. Trends in cell biology 2019, 29, 417–427. [Google Scholar] [CrossRef]
- Chitiprolu, M.; Jagow, C.; Tremblay, V.; Bondy-Chorney, E.; Paris, G.; Savard, A.; Palidwor, G.; Barry, F.A.; Zinman, L.; Keith, J. , et al. A complex of C9ORF72 and p62 uses arginine methylation to eliminate stress granules by autophagy. Nature communications 2018, 9, 2794. [Google Scholar] [CrossRef]
- Lechler, M.C.; Crawford, E.D.; Groh, N.; Widmaier, K.; Jung, R.; Kirstein, J.; Trinidad, J.C.; Burlingame, A.L.; David, D.C. Reduced Insulin/IGF-1 Signaling Restores the Dynamic Properties of Key Stress Granule Proteins during Aging. Cell reports 2017, 18, 454–467. [Google Scholar] [CrossRef] [PubMed]
- Portz, B.; Lee, B.L.; Shorter, J. FUS and TDP-43 Phases in Health and Disease. Trends in biochemical sciences 2021, 46, 550–563. [Google Scholar] [CrossRef]
- Hubstenberger, A.; Courel, M.; Bénard, M.; Souquere, S.; Ernoult-Lange, M.; Chouaib, R.; Yi, Z.; Morlot, J.B.; Munier, A.; Fradet, M. , et al. P-Body Purification Reveals the Condensation of Repressed mRNA Regulons. Molecular cell 2017, 68, 144–157 e145. [Google Scholar] [CrossRef] [PubMed]
- Youn, J.Y.; Dunham, W.H.; Hong, S.J.; Knight, J.D.R.; Bashkurov, M.; Chen, G.I.; Bagci, H.; Rathod, B.; MacLeod, G.; Eng, S.W.M. , et al. High-Density Proximity Mapping Reveals the Subcellular Organization of mRNA-Associated Granules and Bodies. Molecular cell 2018, 69, 517–532 e511. [Google Scholar] [CrossRef] [PubMed]
- Kucherenko, M.M.; Shcherbata, H.R. Stress-dependent miR-980 regulation of Rbfox1/A2bp1 promotes ribonucleoprotein granule formation and cell survival. Nature communications 2018, 9, 312. [Google Scholar] [CrossRef] [PubMed]
- Ingelfinger, D.; Arndt-Jovin, D.J.; Lührmann, R.; Achsel, T. The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. Rna 2002, 8, 1489–1501. [Google Scholar] [CrossRef] [PubMed]
- Van Treeck, B.; Protter, D.S.W.; Matheny, T.; Khong, A.; Link, C.D.; Parker, R. RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome. Proceedings of the National Academy of Sciences of the United States of America 2018, 115, 2734–2739. [Google Scholar] [CrossRef]
- Wheeler, J.R.; Jain, S.; Khong, A.; Parker, R. Isolation of yeast and mammalian stress granule cores. Methods 2017, 126, 12–17. [Google Scholar] [CrossRef]
- Markmiller, S.; Soltanieh, S.; Server, K.L.; Mak, R.; Jin, W.; Fang, M.Y.; Luo, E.C.; Krach, F.; Yang, D.; Sen, A. , et al. Context-Dependent and Disease-Specific Diversity in Protein Interactions within Stress Granules. Cell 2018, 172, 590–604 e513. [Google Scholar] [CrossRef]
- Mahboubi, H.; Moujaber, O.; Kodiha, M.; Stochaj, U. The Co-Chaperone HspBP1 Is a Novel Component of Stress Granules that Regulates Their Formation. Cells 2020, 9. [Google Scholar] [CrossRef]
- Leung, A.K.; Vyas, S.; Rood, J.E.; Bhutkar, A.; Sharp, P.A.; Chang, P. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Molecular cell 2011, 42, 489–499. [Google Scholar] [CrossRef] [PubMed]
- Shigunov, P.; Sotelo-Silveira, J.; Stimamiglio, M.A.; Kuligovski, C.; Irigoín, F.; Badano, J.L.; Munroe, D.; Correa, A.; Dallagiovanna, B. Ribonomic analysis of human DZIP1 reveals its involvement in ribonucleoprotein complexes and stress granules. BMC molecular biology 2014, 15, 12. [Google Scholar] [CrossRef]
- Cougot, N.; Babajko, S.; Séraphin, B. Cytoplasmic foci are sites of mRNA decay in human cells. The Journal of cell biology 2004, 165, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Fujimura, K.; Kano, F.; Murata, M. Identification of PCBP2, a facilitator of IRES-mediated translation, as a novel constituent of stress granules and processing bodies. Rna 2008, 14, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Wilczynska, A.; Aigueperse, C.; Kress, M.; Dautry, F.; Weil, D. The translational regulator CPEB1 provides a link between dcp1 bodies and stress granules. Journal of cell science 2005, 118, 981–992. [Google Scholar] [CrossRef]
- Reineke, L.C.; Tsai, W.C.; Jain, A.; Kaelber, J.T.; Jung, S.Y.; Lloyd, R.E. Casein Kinase 2 Is Linked to Stress Granule Dynamics through Phosphorylation of the Stress Granule Nucleating Protein G3BP1. Molecular and cellular biology 2017, 37. [Google Scholar] [CrossRef] [PubMed]
- Saito, M.; Hess, D.; Eglinger, J.; Fritsch, A.W.; Kreysing, M.; Weinert, B.T.; Choudhary, C.; Matthias, P. Acetylation of intrinsically disordered regions regulates phase separation. Nature chemical biology 2019, 15, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Salleron, L.; Magistrelli, G.; Mary, C.; Fischer, N.; Bairoch, A.; Lane, L. DERA is the human deoxyribose phosphate aldolase and is involved in stress response. Biochimica et biophysica acta 2014, 1843, 2913–2925. [Google Scholar] [CrossRef]
- Belli, V.; Matrone, N.; Sagliocchi, S.; Incarnato, R.; Conte, A.; Pizzo, E.; Turano, M.; Angrisani, A.; Furia, M. A dynamic link between H/ACA snoRNP components and cytoplasmic stress granules. Biochimica et biophysica acta. Molecular cell research 2019, 1866, 118529. [Google Scholar] [CrossRef]
- Tsai, N.P.; Tsui, Y.C.; Wei, L.N. Dynein motor contributes to stress granule dynamics in primary neurons. Neuroscience 2009, 159, 647–656. [Google Scholar] [CrossRef]
- Wippich, F.; Bodenmiller, B.; Trajkovska, M.G.; Wanka, S.; Aebersold, R.; Pelkmans, L. Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell 2013, 152, 791–805. [Google Scholar] [CrossRef] [PubMed]
- Mazroui, R.; Di Marco, S.; Kaufman, R.J.; Gallouzi, I.E. Inhibition of the ubiquitin-proteasome system induces stress granule formation. Molecular biology of the cell 2007, 18, 2603–2618. [Google Scholar] [CrossRef] [PubMed]
- Burry, R.W.; Smith, C.L. HuD distribution changes in response to heat shock but not neurotrophic stimulation. The journal of histochemistry and cytochemistry : official journal of the Histochemistry Society 2006, 54, 1129–1138. [Google Scholar] [CrossRef] [PubMed]
- Kedersha, N.; Chen, S.; Gilks, N.; Li, W.; Miller, I.J.; Stahl, J.; Anderson, P. Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Molecular biology of the cell 2002, 13, 195–210. [Google Scholar] [CrossRef] [PubMed]
- Kedersha, N.L.; Gupta, M.; Li, W.; Miller, I.; Anderson, P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. The Journal of cell biology 1999, 147, 1431–1442. [Google Scholar] [CrossRef]
- Ganassi, M.; Mateju, D.; Bigi, I.; Mediani, L.; Poser, I.; Lee, H.O.; Seguin, S.J.; Morelli, F.F.; Vinet, J.; Leo, G.; et al. A Surveillance Function of the HSPB8-BAG3-HSP70 Chaperone Complex Ensures Stress Granule Integrity and Dynamism. Molecular cell 2016, 63, 796–810. [Google Scholar] [CrossRef] [PubMed]
- Solomon, S.; Xu, Y.; Wang, B.; David, M.D.; Schubert, P.; Kennedy, D.; Schrader, J.W. Distinct structural features of caprin-1 mediate its interaction with G3BP-1 and its induction of phosphorylation of eukaryotic translation initiation factor 2alpha, entry to cytoplasmic stress granules, and selective interaction with a subset of mRNAs. Molecular and cellular biology 2007, 27, 2324–2342. [Google Scholar] [CrossRef]
- Kimball, S.R.; Horetsky, R.L.; Ron, D.; Jefferson, L.S.; Harding, H.P. Mammalian stress granules represent sites of accumulation of stalled translation initiation complexes. American journal of physiology. Cell physiology 2003, 284, C273–284. [Google Scholar] [CrossRef]
- Kang, J.S.; Hwang, Y.S.; Kim, L.K.; Lee, S.; Lee, W.B.; Kim-Ha, J.; Kim, Y.J. OASL1 Traps Viral RNAs in Stress Granules to Promote Antiviral Responses. Molecules and cells 2018, 41, 214–223. [Google Scholar] [CrossRef]
- Wehner, K.A.; Schütz, S.; Sarnow, P. OGFOD1, a novel modulator of eukaryotic translation initiation factor 2alpha phosphorylation and the cellular response to stress. Molecular and cellular biology 2010, 30, 2006–2016. [Google Scholar] [CrossRef]
- Lin, J.C.; Hsu, M.; Tarn, W.Y. Cell stress modulates the function of splicing regulatory protein RBM4 in translation control. Proceedings of the National Academy of Sciences of the United States of America 2007, 104, 2235–2240. [Google Scholar] [CrossRef] [PubMed]
- Bakkar, N.; Kousari, A.; Kovalik, T.; Li, Y.; Bowser, R. RBM45 Modulates the Antioxidant Response in Amyotrophic Lateral Sclerosis through Interactions with KEAP1. Molecular and cellular biology 2015, 35, 2385–2399. [Google Scholar] [CrossRef] [PubMed]
- Reineke, L.C.; Lloyd, R.E. The stress granule protein G3BP1 recruits protein kinase R to promote multiple innate immune antiviral responses. Journal of virology 2015, 89, 2575–2589. [Google Scholar] [CrossRef] [PubMed]
- Eisinger-Mathason, T.S.; Andrade, J.; Groehler, A.L.; Clark, D.E.; Muratore-Schroeder, T.L.; Pasic, L.; Smith, J.A.; Shabanowitz, J.; Hunt, D.F.; Macara, I.G.; et al. Codependent functions of RSK2 and the apoptosis-promoting factor TIA-1 in stress granule assembly and cell survival. Molecular cell 2008, 31, 722–736. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Fu, X.; Song, J.; Zhang, Y.; Cui, X.; Su, C.; Ge, L.; Shao, J.; Xin, L.; Saarikettu, J.; et al. Poly(A)(+) mRNA-binding protein Tudor-SN regulates stress granules aggregation dynamics. The FEBS journal 2015, 282, 874–890. [Google Scholar] [CrossRef] [PubMed]
- Onomoto, K.; Jogi, M.; Yoo, J.S.; Narita, R.; Morimoto, S.; Takemura, A.; Sambhara, S.; Kawaguchi, A.; Osari, S.; Nagata, K.; et al. Critical role of an antiviral stress granule containing RIG-I and PKR in viral detection and innate immunity. PloS one 2012, 7, e43031. [Google Scholar] [CrossRef]
- Zhu, C.H.; Kim, J.; Shay, J.W.; Wright, W.E. SGNP: an essential Stress Granule/Nucleolar Protein potentially involved in 5.8s rRNA processing/transport. PloS one 2008, 3, e3716. [Google Scholar] [CrossRef]
- Das, R.; Schwintzer, L.; Vinopal, S.; Aguado Roca, E.; Sylvester, M.; Oprisoreanu, A.M.; Schoch, S.; Bradke, F.; Broemer, M. New roles for the de-ubiquitylating enzyme OTUD4 in an RNA-protein network and RNA granules. Journal of cell science 2019, 132. [Google Scholar] [CrossRef]
- Ryu, H.H.; Jun, M.H.; Min, K.J.; Jang, D.J.; Lee, Y.S.; Kim, H.K.; Lee, J.A. Autophagy regulates amyotrophic lateral sclerosis-linked fused in sarcoma-positive stress granules in neurons. Neurobiology of aging 2014, 35, 2822–2831. [Google Scholar] [CrossRef]
- Kunde, S.A.; Musante, L.; Grimme, A.; Fischer, U.; Müller, E.; Wanker, E.E.; Kalscheuer, V.M. The X-chromosome-linked intellectual disability protein PQBP1 is a component of neuronal RNA granules and regulates the appearance of stress granules. Human molecular genetics 2011, 20, 4916–4931. [Google Scholar] [CrossRef]
- Aditi; Folkmann, A.W.; Wente, S.R. Cytoplasmic hGle1A regulates stress granules by modulation of translation. Molecular biology of the cell 2015, 26, 1476–1490. [CrossRef] [PubMed]
- Chatsirisupachai, K.; Palmer, D.; Ferreira, S.; de Magalhães, J.P. A human tissue-specific transcriptomic analysis reveals a complex relationship between aging, cancer, and cellular senescence. Aging cell 2019, 18, e13041. [Google Scholar] [CrossRef] [PubMed]
- Avelar, R.A.; Ortega, J.G.; Tacutu, R.; Tyler, E.J.; Bennett, D.; Binetti, P.; Budovsky, A.; Chatsirisupachai, K.; Johnson, E.; Murray, A.; et al. A multidimensional systems biology analysis of cellular senescence in aging and disease. Genome biology 2020, 21, 91. [Google Scholar] [CrossRef] [PubMed]
- Dayhoff, G.W., 2nd; Uversky, V.N. Rapid prediction and analysis of protein intrinsic disorder. Protein science : a publication of the Protein Society 2022, 31, e4496. [Google Scholar] [CrossRef] [PubMed]
- Romero, P.; Obradovic, Z.; Li, X.; Garner, E.C.; Brown, C.J.; Dunker, A.K. Sequence complexity of disordered protein. Proteins 2001, 42, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Peng, K.; Radivojac, P.; Vucetic, S.; Dunker, A.K.; Obradovic, Z. Length-dependent prediction of protein intrinsic disorder. BMC bioinformatics 2006, 7, 208. [Google Scholar] [CrossRef] [PubMed]
- Peng, K.; Vucetic, S.; Radivojac, P.; Brown, C.J.; Dunker, A.K.; Obradovic, Z. Optimizing long intrinsic disorder predictors with protein evolutionary information. Journal of bioinformatics and computational biology 2005, 3, 35–60. [Google Scholar] [CrossRef]
- Xue, B.; Dunbrack, R.L.; Williams, R.W.; Dunker, A.K.; Uversky, V.N. PONDR-FIT: a meta-predictor of intrinsically disordered amino acids. Biochimica et biophysica acta 2010, 1804, 996–1010. [Google Scholar] [CrossRef]
- Dosztányi, Z.; Csizmok, V.; Tompa, P.; Simon, I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 2005, 21, 3433–3434. [Google Scholar] [CrossRef]
- Dosztanyi, Z.; Csizmok, V.; Tompa, P.; Simon, I. The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J Mol Biol 2005, 347, 827–839. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Rajagopalan, K.; Mooney, S.M.; Parekh, N.; Getzenberg, R.H.; Kulkarni, P. A majority of the cancer/testis antigens are intrinsically disordered proteins. J Cell Biochem 2011, 112, 3256–3267. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Gillespie, J.R.; Fink, A.L. Why are "natively unfolded" proteins unstructured under physiologic conditions? Proteins 2000, 41, 415–427. [Google Scholar] [CrossRef] [PubMed]
- Oldfield, C.J.; Cheng, Y.; Cortese, M.S.; Brown, C.J.; Uversky, V.N.; Dunker, A.K. Comparing and combining predictors of mostly disordered proteins. Biochemistry 2005, 44, 1989–2000. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Wang, K.; Liu, Y.; Xue, B.; Uversky, V.N.; Dunker, A.K. Predicting intrinsic disorder in proteins: an overview. 2009, 19, 929-949, doi:10.1038/cr.2009.87. [CrossRef]
- Xue, B.; Oldfield, C.J.; Dunker, A.K.; Uversky, V.N. CDF it all: consensus prediction of intrinsically disordered proteins based on various cumulative distribution functions. FEBS Lett 2009, 583, 1469–1474. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Oldfield, C.; Meng, J.; Hsu, W.L.; Xue, B.; Uversky, V.N.; Romero, P.; Dunker, A.K. Subclassifying disordered proteins by the CH-CDF plot method. Pac Symp Biocomput 2012, 128–139. [Google Scholar]
- Mohan, A.; Sullivan, W.J., Jr.; Radivojac, P.; Dunker, A.K.; Uversky, V.N. Intrinsic disorder in pathogenic and non-pathogenic microbes: discovering and analyzing the unfoldomes of early-branching eukaryotes. Mol Biosyst 2008, 4, 328–340. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Oldfield, C.J.; Xue, B.; Hsu, W.L.; Meng, J.; Liu, X.; Shen, L.; Romero, P.; Uversky, V.N.; Dunker, A. Improving protein order-disorder classification using charge-hydropathy plots. BMC bioinformatics 2014, 15 Suppl 17, S4. [Google Scholar] [CrossRef]
- Oates, M.E.; Romero, P.; Ishida, T.; Ghalwash, M.; Mizianty, M.J.; Xue, B.; Dosztanyi, Z.; Uversky, V.N.; Obradovic, Z.; Kurgan, L.; et al. D(2)P(2): database of disordered protein predictions. Nucleic acids research 2013, 41, D508–516. [Google Scholar] [CrossRef]
- Ishida, T.; Kinoshita, K. PrDOS: prediction of disordered protein regions from amino acid sequence. Nucleic acids research 2007, 35, W460–464. [Google Scholar] [CrossRef]
- Obradovic, Z.; Peng, K.; Vucetic, S.; Radivojac, P.; Dunker, A.K. Exploiting heterogeneous sequence properties improves prediction of protein disorder. Proteins: Structure, Function, and Bioinformatics 2005, 61, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Walsh, I.; Martin, A.J.; Di Domenico, T.; Tosatto, S.C. ESpritz: accurate and fast prediction of protein disorder. Bioinformatics 2012, 28, 503–509. [Google Scholar] [CrossRef]
- Andreeva, A.; Howorth, D.; Brenner, S.E.; Hubbard, T.J.; Chothia, C.; Murzin, A.G. SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic acids research 2004, 32, D226–229. [Google Scholar] [CrossRef] [PubMed]
- Murzin, A.G.; Brenner, S.E.; Hubbard, T.; Chothia, C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol 1995, 247, 536–540. [Google Scholar] [CrossRef] [PubMed]
- de Lima Morais, D.A.; Fang, H.; Rackham, O.J.; Wilson, D.; Pethica, R.; Chothia, C.; Gough, J. SUPERFAMILY 1.75 including a domain-centric gene ontology method. Nucleic acids research 2011, 39, D427–434. [Google Scholar] [CrossRef]
- Meszaros, B.; Simon, I.; Dosztanyi, Z. Prediction of protein binding regions in disordered proteins. PLoS Comput Biol 2009, 5, e1000376. [Google Scholar] [CrossRef] [PubMed]
- Hornbeck, P.V.; Kornhauser, J.M.; Tkachev, S.; Zhang, B.; Skrzypek, E.; Murray, B.; Latham, V.; Sullivan, M. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic acids research 2012, 40, D261–270. [Google Scholar] [CrossRef]
- Hardenberg, M.; Horvath, A.; Ambrus, V.; Fuxreiter, M.; Vendruscolo, M. Widespread occurrence of the droplet state of proteins in the human proteome. Proceedings of the National Academy of Sciences of the United States of America 2020, 117, 33254–33262. [Google Scholar] [CrossRef]
- Chu, X.; Sun, T.; Li, Q.; Xu, Y.; Zhang, Z.; Lai, L.; Pei, J. Prediction of liquid-liquid phase separating proteins using machine learning. BMC bioinformatics 2022, 23, 72. [Google Scholar] [CrossRef]
- Conchillo-Solé, O.; de Groot, N.S.; Avilés, F.X.; Vendrell, J.; Daura, X.; Ventura, S. AGGRESCAN: a server for the prediction and evaluation of "hot spots" of aggregation in polypeptides. BMC bioinformatics 2007, 8, 65. [Google Scholar] [CrossRef]
- Thangakani, A.M.; Nagarajan, R.; Kumar, S.; Sakthivel, R.; Velmurugan, D.; Gromiha, M.M. CPAD, Curated Protein Aggregation Database: A Repository of Manually Curated Experimental Data on Protein and Peptide Aggregation. PloS one 2016, 11, e0152949. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Kuhn, M.; Simonovic, M.; Roth, A.; Minguez, P.; Doerks, T.; Stark, M.; Muller, J.; Bork, P.; et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic acids research 2011, 39, D561–568. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, A.S.; Uversky, V.N. Intrinsic Disorder as a Natural Preservative: High Levels of Intrinsic Disorder in Proteins Found in the 2600-Year-Old Human Brain. Biology (Basel) 2022, 11. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Xia, T.; Xing, J.Q.; Yin, L.H.; Li, X.W.; Pan, J.; Liu, J.Y.; Sun, L.M.; Wang, M.; Li, T.; et al. The stress granule protein G3BP1 promotes pre-condensation of cGAS to allow rapid responses to DNA. EMBO reports 2022, 23, e53166. [Google Scholar] [CrossRef] [PubMed]
- Abdisalaam, S.; Bhattacharya, S.; Mukherjee, S.; Sinha, D.; Srinivasan, K.; Zhu, M.; Akbay, E.A.; Sadek, H.A.; Shay, J.W.; Asaithamby, A. Dysfunctional telomeres trigger cellular senescence mediated by cyclic GMP-AMP synthase. The Journal of biological chemistry 2020, 295, 11144–11160. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhang, J.; Ye, J.; Li, X.; Liu, H.; Ma, X.; Wang, C.; He, K.; Zhang, W.; Yuan, J.; et al. Nuclear speckle specific hnRNP D-like prevents age- and AD-related cognitive decline by modulating RNA splicing. Molecular neurodegeneration 2021, 16, 66. [Google Scholar] [CrossRef] [PubMed]
- Batlle, C.; Yang, P.; Coughlin, M.; Messing, J.; Pesarrodona, M.; Szulc, E.; Salvatella, X.; Kim, H.J.; Taylor, J.P.; Ventura, S. hnRNPDL Phase Separation Is Regulated by Alternative Splicing and Disease-Causing Mutations Accelerate Its Aggregation. Cell reports 2020, 30, 1117–1128 e1115. [Google Scholar] [CrossRef] [PubMed]
- Kanellis, D.C.; Espinoza, J.A.; Zisi, A.; Sakkas, E.; Bartkova, J.; Katsori, A.M.; Boström, J.; Dyrskjøt, L.; Broholm, H.; Altun, M.; et al. The exon-junction complex helicase eIF4A3 controls cell fate via coordinated regulation of ribosome biogenesis and translational output. Science advances 2021, 7. [Google Scholar] [CrossRef]
- Chaturvedi, P.; Neelamraju, Y.; Arif, W.; Kalsotra, A.; Janga, S.C. Uncovering RNA binding proteins associated with age and gender during liver maturation. Scientific reports 2015, 5, 9512. [Google Scholar] [CrossRef]
- Yamashita, A.; Shichino, Y.; Fujii, K.; Koshidaka, Y.; Adachi, M.; Sasagawa, E.; Mito, M.; Nakagawa, S.; Iwasaki, S.; Takao, K.; et al. ILF3 prion-like domain regulates gene expression and fear memory under chronic stress. iScience 2023, 26, 106229. [Google Scholar] [CrossRef]
- Jorge, S.-R.; Cristina, M.-B.; Nekane, R.-G.; Javier, H.-A.; Mar, D.; Consuelo, B. MicroRNA biogenesis pathway alterations in aging. Extracellular Vesicles and Circulating Nucleic Acids 2023, 4, 486–501. [Google Scholar] [CrossRef]
- Min, K.W.; Zealy, R.W.; Davila, S.; Fomin, M.; Cummings, J.C.; Makowsky, D.; McDowell, C.H.; Thigpen, H.; Hafner, M.; Kwon, S.H.; et al. Profiling of m6A RNA modifications identified an age-associated regulation of AGO2 mRNA stability. Aging cell 2018, 17, e12753. [Google Scholar] [CrossRef] [PubMed]
- Guan, L.; Grigoriev, A. Age-Related Argonaute Loading of Ribosomal RNA Fragments. MicroRNA 2020, 9, 142–152. [Google Scholar] [CrossRef] [PubMed]
- Alexander, C.C.; Munkáscy, E.; Tillmon, H.; Fraker, T.; Scheirer, J.; Holstein, D.; Lozano, D.; Khan, M.; Gidalevitz, T.; Lechleiter, J.D.; et al. HspB1 Overexpression Improves Life Span and Stress Resistance in an Invertebrate Model. The journals of gerontology. Series A, Biological sciences and medical sciences 2022, 77, 268–275. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Bai, M.; Barbosa, G.O.; Chen, A.; Wei, Y.; Luo, S.; Wang, X.; Wang, B.; Tsukui, T.; Li, H.; et al. Broadly conserved roles of TMEM131 family proteins in intracellular collagen assembly and secretory cargo trafficking. Science advances 2020, 6, eaay7667. [Google Scholar] [CrossRef] [PubMed]
- Torres-Pérez, J.V.; Anagianni, S.; Mech, A.M.; Havelange, W.; García-González, J.; Fraser, S.E.; Vallortigara, G.; Brennan, C.H. baz1b loss-of-function in zebrafish produces phenotypic alterations consistent with the domestication syndrome. iScience 2023, 26, 105704. [Google Scholar] [CrossRef] [PubMed]
- Jourdain, A.A.; Popow, J.; de la Fuente, M.A.; Martinou, J.C.; Anderson, P.; Simarro, M. The FASTK family of proteins: emerging regulators of mitochondrial RNA biology. Nucleic acids research 2017, 45, 10941–10947. [Google Scholar] [CrossRef]
Figure 1.
The scheme illustrating analysis design
Figure 1.
The scheme illustrating analysis design
Figure 2.
Global disorder analysis of proteins in the proteomes of human SGs (720 proteins, green circles) and P-bodies (177 proteins, yellow circles), as well as proteins simultaneously found in SGs and P-bodies (87 proteins, yellow-green circles). (A) PONDR® VSL2 output. PONDR® VSL2 score is the mean disorder score (MDS) for a protein, which is calculated as a sequence-length normalized sum of its per-residue disorder scores. PONDR VSL2 (%) is a percent of predicted intrinsically disordered residues (PPIDR); i.e., the percent of residues with disorder scores above 0.5. Color blocks indicate regions in which proteins are mostly ordered (blue and light blue), moderately disordered (pink and light pink), or mostly disordered (red). If the two parameters agree, the corresponding part of the background is dark (blue or pink), whereas light blue and light pink reflect areas in which only one of these criteria applies. The boundaries of the colored regions represent arbitrary and accepted cutoffs for MDS (y-axis) and the percentage of predicted disordered residues (PPDR; x-axis). (B) Charge-hydropathy and cumulative distribution function (ΔCH-ΔCDF) plot. The Y-coordinate is calculated as the distance of the corresponding protein from the boundary in the CH plot. The X-coordinate is calculated as the average distance of the corresponding protein’s CDF curve from the CDF boundary. The quadrant where the proteins are located determines their classification. Q1, 111 proteins (67.3%) predicted to be ordered by CDF and compact/ordered by CH-plot. Q2, 30 proteins (18.2%) predicted to be ordered/compact by CH-plot and disordered by CDF. Q3, 16 proteins (9.7%) predicted to be disordered by CH-plot and CDF. Q4, 8 proteins (4.8%) predicted to be disordered by CH-plot and ordered by CDF.
Figure 2.
Global disorder analysis of proteins in the proteomes of human SGs (720 proteins, green circles) and P-bodies (177 proteins, yellow circles), as well as proteins simultaneously found in SGs and P-bodies (87 proteins, yellow-green circles). (A) PONDR® VSL2 output. PONDR® VSL2 score is the mean disorder score (MDS) for a protein, which is calculated as a sequence-length normalized sum of its per-residue disorder scores. PONDR VSL2 (%) is a percent of predicted intrinsically disordered residues (PPIDR); i.e., the percent of residues with disorder scores above 0.5. Color blocks indicate regions in which proteins are mostly ordered (blue and light blue), moderately disordered (pink and light pink), or mostly disordered (red). If the two parameters agree, the corresponding part of the background is dark (blue or pink), whereas light blue and light pink reflect areas in which only one of these criteria applies. The boundaries of the colored regions represent arbitrary and accepted cutoffs for MDS (y-axis) and the percentage of predicted disordered residues (PPDR; x-axis). (B) Charge-hydropathy and cumulative distribution function (ΔCH-ΔCDF) plot. The Y-coordinate is calculated as the distance of the corresponding protein from the boundary in the CH plot. The X-coordinate is calculated as the average distance of the corresponding protein’s CDF curve from the CDF boundary. The quadrant where the proteins are located determines their classification. Q1, 111 proteins (67.3%) predicted to be ordered by CDF and compact/ordered by CH-plot. Q2, 30 proteins (18.2%) predicted to be ordered/compact by CH-plot and disordered by CDF. Q3, 16 proteins (9.7%) predicted to be disordered by CH-plot and CDF. Q4, 8 proteins (4.8%) predicted to be disordered by CH-plot and ordered by CDF.
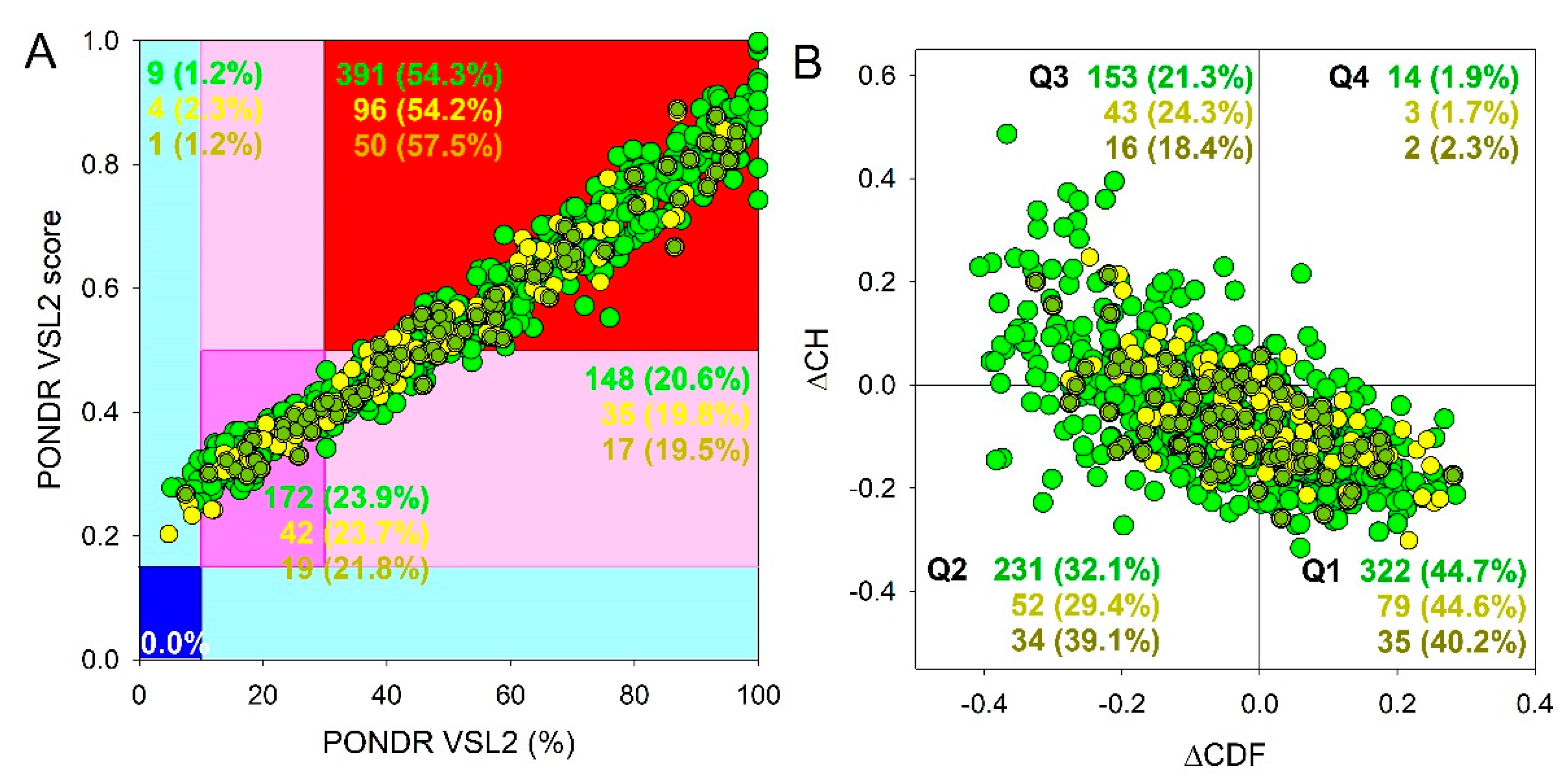
Figure 3.
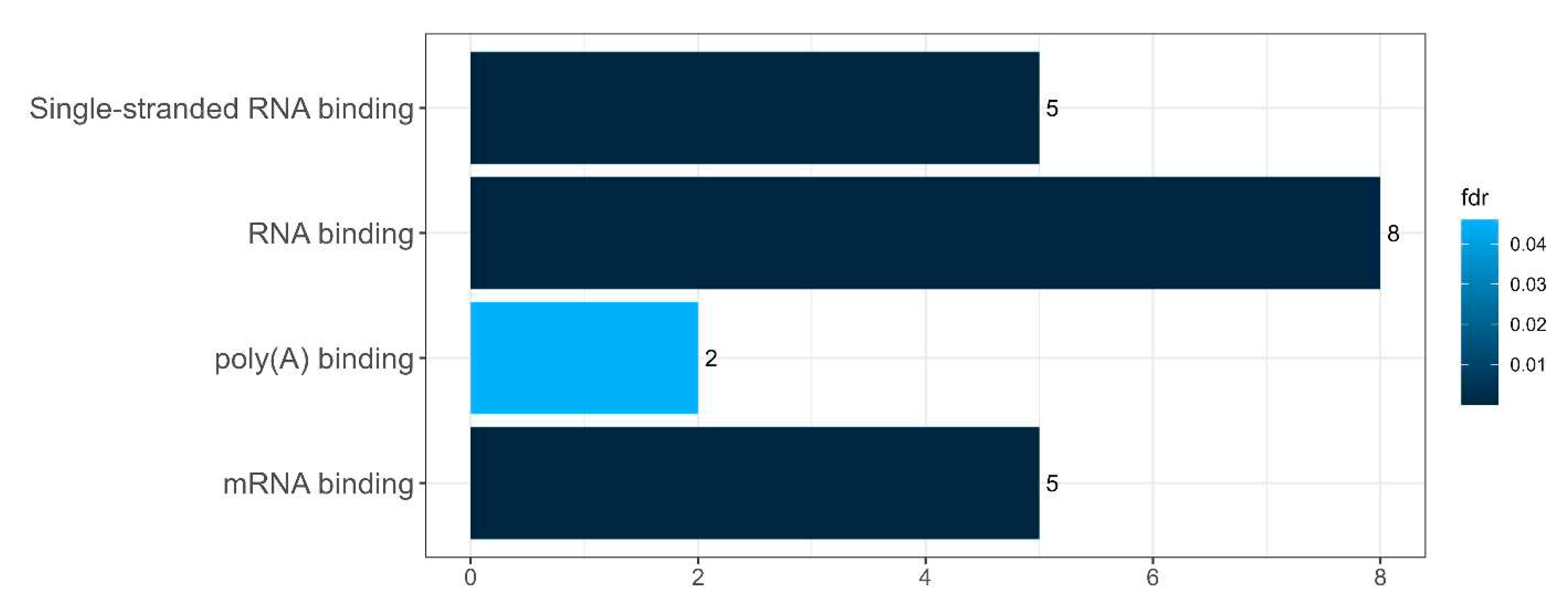
Diagram presenting the molecular functions of the proteins of the SGs and their proteins involved in senescence-related processes
Figure 3.
Diagram presenting the molecular functions of the proteins of the SGs and their proteins involved in senescence-related processes
Figure 4.
Diagram presenting the molecular functions of the proteins of the P-bodies and their proteins involved in senescence-related processes
Figure 4.
Diagram presenting the molecular functions of the proteins of the P-bodies and their proteins involved in senescence-related processes
Figure 5.
Pie charts presenting the proportion of IDPs (upper panels) and LLPS-related proteins (bottom panels) in the proteomes of stress-granules (panels A and D), P-bodies (panels B and E) and intersecting proteins (panels C and F) according to the PONDR® VSL2b and FuzDrop/PSP predictor analysis.
Figure 5.
Pie charts presenting the proportion of IDPs (upper panels) and LLPS-related proteins (bottom panels) in the proteomes of stress-granules (panels A and D), P-bodies (panels B and E) and intersecting proteins (panels C and F) according to the PONDR® VSL2b and FuzDrop/PSP predictor analysis.
Figure 6.
Global disorder analysis of human proteins that are potentially involved in the senescence-related processes and can be found in SGs (135 proteins, green circles) and P-bodies (42 proteins, yellow circles), as well as proteins simultaneously found in SGs and P-bodies (17 proteins, yellow-green circles). (A) PONDR® VSL2 output. (B) Charge-hydropathy and cumulative distribution function (ΔCH-ΔCDF) plot.
Figure 6.
Global disorder analysis of human proteins that are potentially involved in the senescence-related processes and can be found in SGs (135 proteins, green circles) and P-bodies (42 proteins, yellow circles), as well as proteins simultaneously found in SGs and P-bodies (17 proteins, yellow-green circles). (A) PONDR® VSL2 output. (B) Charge-hydropathy and cumulative distribution function (ΔCH-ΔCDF) plot.
Figure 7.
Pie charts presenting the proportion of IDPs (upper panels) and LLPS-related proteins (bottom panels) in the senescence-related proteomes of SGs (panels A and D), P-bodies (panels B and E) and intersecting proteins (panels C and F) according to the PONDR® VSL2b and FuzDrop/PSP predictor analysis.
Figure 7.
Pie charts presenting the proportion of IDPs (upper panels) and LLPS-related proteins (bottom panels) in the senescence-related proteomes of SGs (panels A and D), P-bodies (panels B and E) and intersecting proteins (panels C and F) according to the PONDR® VSL2b and FuzDrop/PSP predictor analysis.
Figure 8.
Distribution of the proteins from senescence-related proteomes of stress-granules based on protein disorder propensity, tendency to LLPS, roles in LLPS according to FuzDrop, and protein charge.
Figure 8.
Distribution of the proteins from senescence-related proteomes of stress-granules based on protein disorder propensity, tendency to LLPS, roles in LLPS according to FuzDrop, and protein charge.
Figure 9.
Distribution of the proteins from senescence-related proteomes of P-bodies based on protein disorder propensity, tendency to LLPS, roles in LLPS according to FuzDrop, and protein charge.
Figure 9.
Distribution of the proteins from senescence-related proteomes of P-bodies based on protein disorder propensity, tendency to LLPS, roles in LLPS according to FuzDrop, and protein charge.
Figure 10.
Diagram presenting the molecular functions of amyloidogenic proteins in senescence-related proteome of stress-granules
Figure 10.
Diagram presenting the molecular functions of amyloidogenic proteins in senescence-related proteome of stress-granules
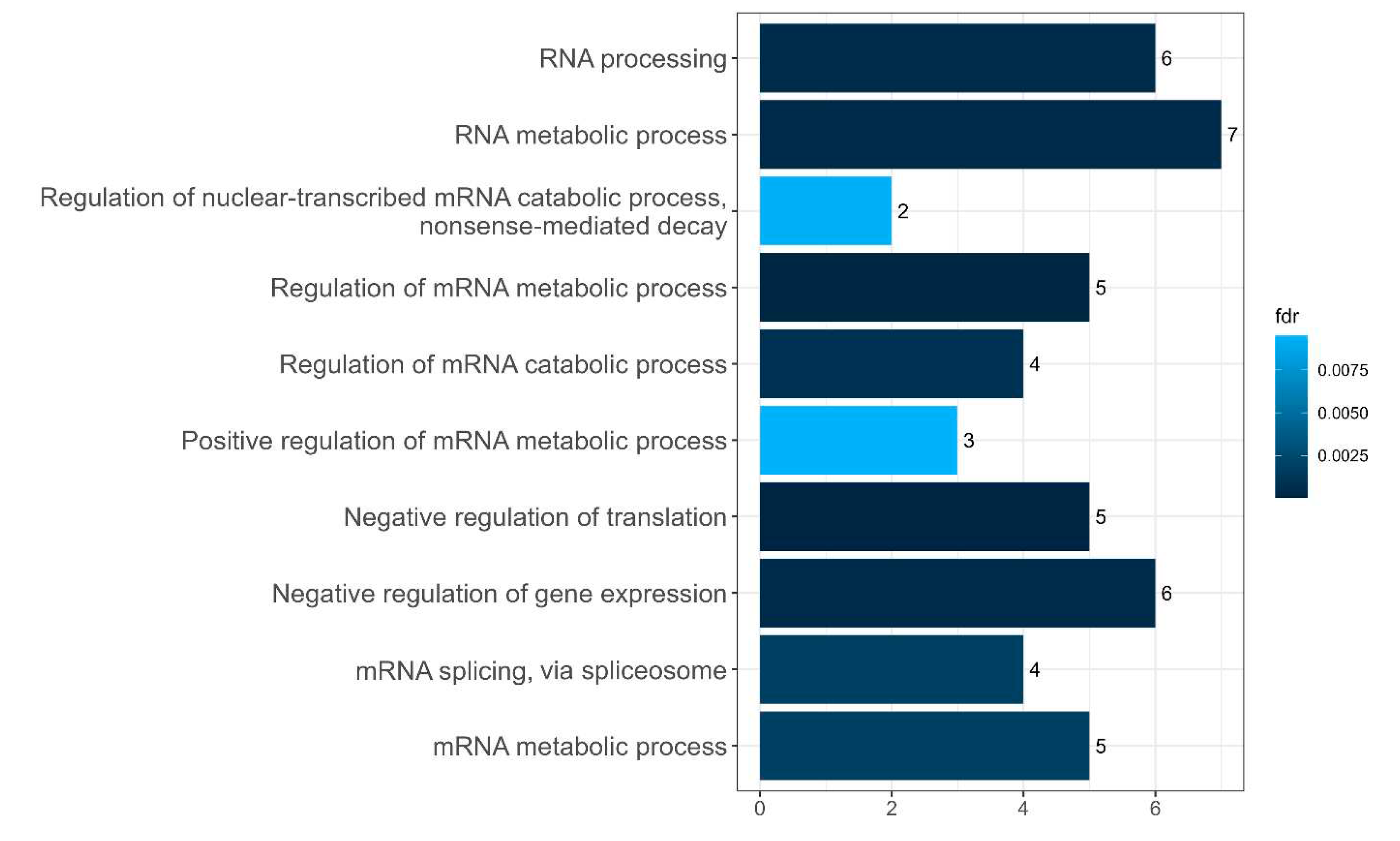
Figure 11.
Diagram presenting the biological processes of amyloidogenic proteins in senescence-related proteome of stress-granules
Figure 11.
Diagram presenting the biological processes of amyloidogenic proteins in senescence-related proteome of stress-granules
Figure 12.
Functional disorder analysis of human hnRPDL protein. A. Structural model generated by AlphaFold. B. Functional disorder profile generated by D2P2 platform. C. Per-residue intrinsic disorder profile generated using outputs of RIDAO. D. FuzDrop-generated profile of the LLPS potential of hnRPDL showing positions droplet-promoting regions, aggregation hot spots and regions with context-dependent interactions.
Figure 12.
Functional disorder analysis of human hnRPDL protein. A. Structural model generated by AlphaFold. B. Functional disorder profile generated by D2P2 platform. C. Per-residue intrinsic disorder profile generated using outputs of RIDAO. D. FuzDrop-generated profile of the LLPS potential of hnRPDL showing positions droplet-promoting regions, aggregation hot spots and regions with context-dependent interactions.
Figure 13.
Diagram presenting the molecular functions of the intersecting proteins between proteomes of the P-bodies and SGs and their proteins involved in senescence-related processes
Figure 13.
Diagram presenting the molecular functions of the intersecting proteins between proteomes of the P-bodies and SGs and their proteins involved in senescence-related processes
Figure 14.
Interactome of intersecting senescence-related proteins between SGs and P-bodies according to STRING. Colors presenting proteins clusters.
Figure 14.
Interactome of intersecting senescence-related proteins between SGs and P-bodies according to STRING. Colors presenting proteins clusters.
Figure 15.
Diagram presenting the molecular functions of the EIF4A3-related cluster in
interactome of intersecting senescence-related proteins between SGs and P-bodies
Figure 15.
Diagram presenting the molecular functions of the EIF4A3-related cluster in
interactome of intersecting senescence-related proteins between SGs and P-bodies
Figure 16.
Diagram presenting the biological processes of the EIF4A3-related cluster in interactome of intersecting senescence-related proteins between SGs and P-bodies
Figure 16.
Diagram presenting the biological processes of the EIF4A3-related cluster in interactome of intersecting senescence-related proteins between SGs and P-bodies
Table 1.
Comparative characteristics of the proteomes of the SGs and their proteins involved in senescence-related processes
Table 1.
Comparative characteristics of the proteomes of the SGs and their proteins involved in senescence-related processes
| Parameter |
SGs |
SGs senescence-related |
Are the differences significant? |
Statistical test |
| Size of dataset |
720 |
135 |
- |
- |
| Median molecular weight, kDa |
60.44 |
57.67 |
No |
t-test |
| Disorder |
Median Disorder |
48.5 |
44.3 |
No |
t-test |
| Ordered Proteins |
9 (1 %) |
1 (1%) |
No |
exact fisher |
| IDRs |
172 (24%) |
37 (27%) |
No |
exact fisher |
| IDPs |
539 (75%) |
97 (72%) |
No |
exact fisher |
| LLPS |
LLPS-free |
279 (39%) |
57 (42%) |
No |
exact fisher |
| Controversial |
146 (20%) |
28 (21%) |
No |
exact fisher |
| LLPS |
295 (41%) |
50 (37%) |
No |
exact fisher |
| Role in LLPS (FuzDrop) |
LLPS-free |
391 (54%) |
66 (49%) |
No |
exact fisher |
| Client |
261 (36%) |
52 (39%) |
No |
exact fisher |
| Driver |
68 (9%) |
17 (13%) |
No |
exact fisher |
| Charge |
Median charge of IDRs |
0.08 |
0.06 |
No |
t-test |
| Negative charge |
118 (16%) |
20 (15%) |
No |
exact fisher |
| Near-zero charge |
490 (68%) |
79 (59%) |
Yes |
exact fisher |
| Positive charge |
112 (16%) |
36 (27%) |
Yes |
exact fisher |
| Aggregation |
LAHS per length (Summary Length of AHS per protein length) |
0.089 |
0.08 |
No |
t-test |
| Amyloidogenic proteins |
57 (8%) |
18 (13%) |
Yes |
exact fisher |
| Nucleic acid-binding |
RNA-binding |
389 (54 %) |
67 (50%) |
No |
exact fisher |
| DNA-binding |
110 (15 %) |
31 (23%) |
Yes |
exact fisher |
Table 2.
Comparative characteristics of the proteomes of the P-bodies and their proteins involved in senescence-related processes
Table 2.
Comparative characteristics of the proteomes of the P-bodies and their proteins involved in senescence-related processes
| Parameter |
PBs |
PBs senescence-related |
Are the differences significant? |
Statistical test |
| Size of data |
177 |
42 |
- |
- |
| Median molecular weight, kDa |
69.8 |
66.2 |
No |
t-test |
| Disorder |
Median Disorder |
46.76 |
45.3 |
No |
t-test |
| Ordered Proteins |
4 (2%) |
0 (0%) |
No |
exact fisher |
| IDRs |
42 (24%) |
14 (33%) |
No |
exact fisher |
| IDP |
131 (74 %) |
28 (67%) |
No |
exact fisher |
| LLPS |
LLPS-free |
78 (44 %) |
22 (52%) |
No |
exact fisher |
| Controversial |
34 (19%) |
8 (19%) |
No |
exact fisher |
| LLPS |
65 (36%) |
12 (29%) |
No |
exact fisher |
| Role in LLPS (FuzDrop) |
LLPS-free |
87 (49%) |
17 (40%) |
No |
exact fisher |
| Client |
71 (40 %) |
19 (45%) |
No |
exact fisher |
| Driver |
19 (11%) |
6 (14%) |
No |
exact fisher |
| Charge |
Median charge of IDRs |
0.003 |
0.031 |
No |
t-test |
| Negative charge |
17 (10%) |
4 (10%) |
No |
exact fisher |
| Near-zero charge |
120 (68%) |
21 (50%) |
Yes |
exact fisher |
| Positive charge |
40 (23%) |
17 (40%) |
Yes |
exact fisher |
| Aggregation |
LAHS per length (Summary Length of AHS per protein length) |
0.091 |
0.084 |
No |
t-test |
| Amyloidogenic proteins |
19 (11%) |
3 (7%) |
No |
exact fisher |
| Nucleic acid-binding |
RNA-binding |
116 (66%) |
31 (74%) |
No |
exact fisher |
| DNA-binding |
31 (18 %) |
10 (24%) |
No |
exact fisher |
Table 3.
Comparative characteristics of the intersecting proteins between proteomes of the P-bodies and SGs and their proteins involved in senescence-related processes
Table 3.
Comparative characteristics of the intersecting proteins between proteomes of the P-bodies and SGs and their proteins involved in senescence-related processes
| Parameter |
SG&PBs |
SG&PBs senescence-related |
Are the differences significant? |
Statistical test |
| Size of data |
87 |
17 |
- |
- |
| Median molecular weight, kDa |
67.7 |
61.1 |
No |
t-test |
| Disorder |
Median Disorder |
48.4 |
48.3 |
No |
t-test |
| Ordered Proteins |
1 (1%) |
0 (0%) |
No |
exact fisher |
| IDRs |
19 (22%) |
6 (35%) |
No |
exact fisher |
| IDP |
67 (77%) |
11 (65%) |
No |
exact fisher |
| LLPS |
LLPS-free |
28 (32%) |
9 (53 %) |
No |
exact fisher |
| Controversial |
16 (18%) |
2 (12 %) |
No |
exact fisher |
| LLPS |
43 (49%) |
6 (35%) |
No |
exact fisher |
| Role in LLPS (FuzDrop) |
LLPS-free |
54 (62%) |
8 (47%) |
No |
exact fisher |
| Client |
26 (30%) |
7 (41%) |
No |
exact fisher |
| Driver |
7 (8%) |
2 (12%) |
No |
exact fisher |
| Charge |
Median charge of IDRs |
0.014 |
0.035 |
No |
t-test |
| Negative charge |
7 (8%) |
1 (6%) |
No |
exact fisher |
| Near-zero charge |
66 (76%) |
9 (53%) |
No |
exact fisher |
| Positive charge |
14 (16%) |
7 (41%) |
Yes |
exact fisher |
| Aggregation |
LAHS per length (Summary Length of AHS per protein length) |
0.11 |
0.095 |
No |
t-test |
| Amyloidogenic proteins |
8 (9%) |
0 (0%) |
No |
exact fisher |
| Nucleic acid-binding |
RNA-binding |
63 |
11 |
No |
exact fisher |
| DNA-binding |
13 |
3 |
No |
exact fisher |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).