Introduction
Ubiquitin ligases and deubiquitylases form a large family that catalyze the bonding between the carboxy terminus of ubiquitin and lysine residues and the hydrolysis of the isopeptide bond at the amino terminus, respectively (1). Among the deubiquitylase family, ubiquitin-specific peptidase 21 (USP21), a cysteine protease, is highly conserved among species (2) and is known to play a role in intracellular processes and diseases such as bladder carcinoma (3) and pancreatic ductal adenocarcinoma (4). In addition, USP21 is known to be involved in epigenetic regulation as well (2). Nakagawa et al. found that USP21 increases after partial hepatectomy and catalyzes the hydrolysis of nucleosome ubiquitylated H2A, which aids in the di- and trimethylation process of H3K4 and initiates transcription of several genes associated with liver regeneration (2). In addition, overexpression of USP21 in the liver has been shown to upregulate the Serpina6 gene, which is downregulated during hepatocyte regeneration (2). In addition to the full-length USP21 consisting of 565 amino acids in humans and Usp21 consisting of 566 amino acids in mice, Okuda et al. found a short variant of Usp21 in mice. This variant is caused by alternative splicing of exon 2, resulting in the deletion of 87 amino acids from the amino terminus of the long isoform of Usp21. Usp21 short variant lacks nuclear export signal and thus localizes to the nucleus better than the Usp21 long variant (5). Despite these findings, the impact of USP21 dysfunction on disease remains to be elucidated.
Hypercholesterolemia, also referred to as dyslipidemia, results due to elevated levels of cholesterol in the blood. One of the genetic factors is the low-density lipoprotein (LDL) receptor, which is involved in lipid metabolism and whose mutations are implicated in the development of atherosclerotic cardiovascular diseases (ASCVD) (6). However, it is a rare disorder and does not explain most of the hypercholesterolemia.
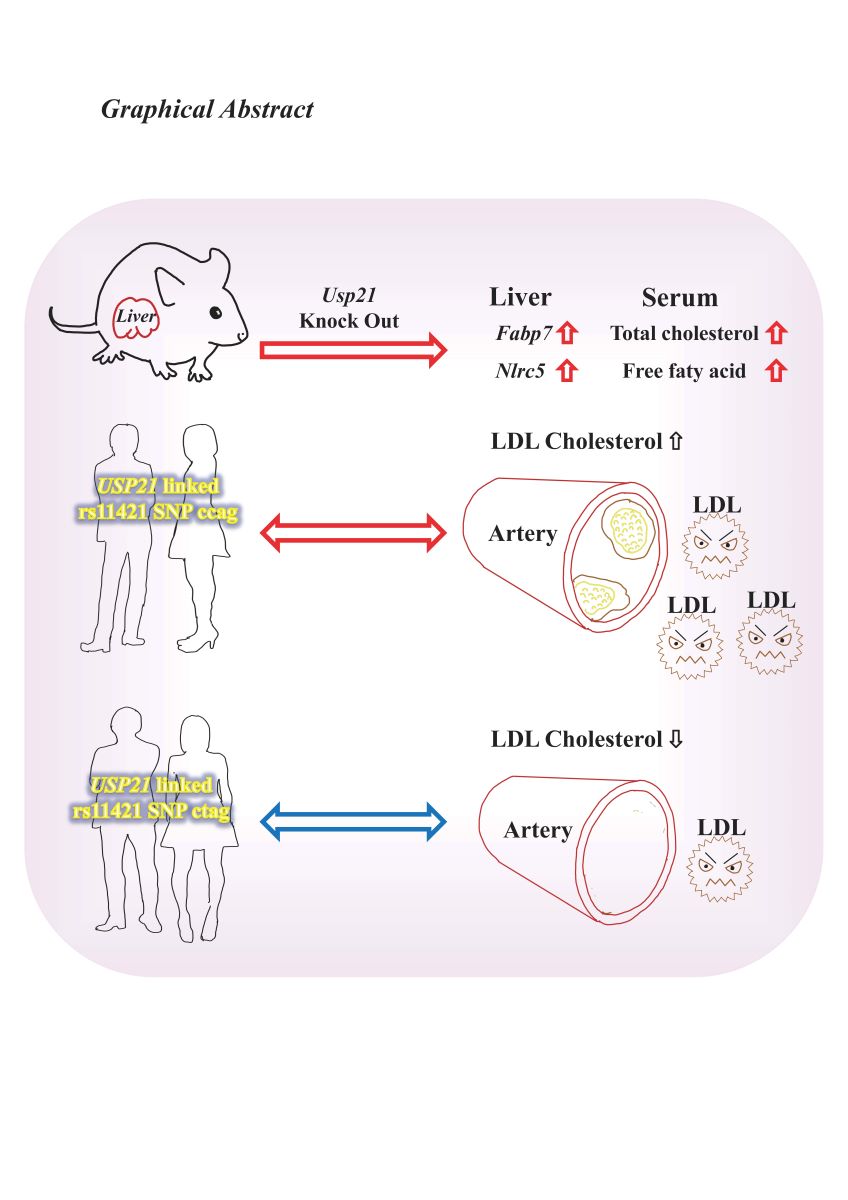
In this report, we generated Usp21 knockout (KO) mice and identified genes regulated by Usp21. The genes Fabp7 and Nlrc5 showed significant elevation in KO mice compared to wild-type mice. Blood tests revealed that KO mice exhibited elevated total cholesterol levels and free fatty acids. In humans, we examined SNPs around USP21 in non-hypercholesterolemic and hypercholesterolemic outpatients and found an association between hypercholesterolemia and the rs11421 SNP downstream of USP21. These data suggest that USP21 plays a role in lipid metabolism and that its polymorphism could serve as a diagnostic marker for hypercholesterolemia and a potential target for therapy.
Materials and Methods
Generation of USP21 KO mice
Usp21-/- conditional KO mice are created in the ERT-Cre/loxP system by using tamoxifen (TAM). Upon administration of TAM the ERT2 promoter is activated and the Cre recombinase enzyme is produced. This enzyme recognizes loxP short sequences and the sequence is excised (7). Administering TAM to genetically modified mice with loxP in the middle of the Usp21 sequence causes Usp21 to be knocked out and hence Usp21 KO mice are created. All the mice were fed a normal diet (ND) and maintained under a 12h/12h light and dark cycle.
DNA electrophoresis
The RNA was isolated from the wild-type (WT) and KO mice and amplified using Usp21-specific primers by RT-PCR. RNA was extracted using the RNeasy Plus kit (Qiagen, Germany). The Usp21 KO was confirmed with agarose gel electrophoresis. mGapdh was used as a reference gene. The primers are listed in supplementary table 1.
Microarray and Enrichment Analysis
RNA microarray was carried out using Sure Print G3 Mouse GE 8x60K chip (Agilent Technologies, CA) according to the manufacturer’s protocol. The RNA was extracted from the WT and KO mice. Total RNA was extracted using RNeasy Plus kit (Qiagen, Germany) and was quantified using Agilent 2100 Bioanalyzer (Agilent Technologies, CA) before performing microarray. The expression of the target genes was normalized to mGapdh expression. The Usp21 KO and WT were considered for the study. The genes that were differentially regulated as a result of Usp21 KO were analyzed using the Partek ® Genomics Suite ® (Partek, MO). All the microarray data can be found online in the NCBI GEO submission (GSE244360).
qPCR
The total RNA was extracted using the ISOGEN Ⅱ reagent (NIPPON GENE, Japan). cDNA was synthesized using 0.5 μg total RNA as a template with oligo (dT) primer (Life Technologies, CA), random hexamers (Takara Bio, Japan), and M-MuLV reverse transcriptase (New England Biolabs, MA). Real-time RT-qPCR was performed with an ABI PRISM 7900HT (Applied Biosystems, MA) with SYBR green as a reporter, cDNA as a template and gene-specific primers. The target gene expression levels were standardized to mGapdh expression levels. All the primers designed for the experiment are listed in supplementary table 1.
Mice serum analysis
Serum samples were collected from sacrificing 10-11 weeks of WT mice, Usp21 KO mice, and Usp21 heterozygous (HE) mice fed with ND. 0.5-1.0ml of blood was taken from the inferior vena cava. The collected blood was allowed to coagulate by keeping it at room temperature for 30 minutes to 2 hours. The coagulated blood is then centrifuged at 3000rpm for 10-30 mins at room temperature. The supernatant separated as a result of centrifugation is the serum which is then cryopreserved to analyze the levels of triglycerides (TG), free fatty acids (FFA), total cholesterol (T-CHOL), aspartate aminotransferase (AST), and alanine transaminase (ALT). All the mice data are represented in supplementary table 2.
SNP selection and genotyping
All the individual samples are from participants who attended an annual health check-up. This annual check-up program is conducted by the local government and directed by the Ministry of Health, Labor and Welfare in Japan. And this study was approved by the Ethics Committee of Nagasaki University Graduate School of Biomedical Sciences (project registration number 14051404). Written consent forms were available in Japanese to ensure comprehensive understanding of the study objectives, and informed consent was provided by the participants. (Supplementary table 3). A 100kb sequence upstream and downstream of USP21 was identified from the NCBI database GENBANK (GRCh37.p9) and input to GENETYX-MAC software to identify the restriction enzyme sites. All the SNPs within the restriction enzyme site around the USP21 region were identified. A total of 22 SNPs were selected. A total of 67 patient samples were collected, of which 41 had lower concentrations of LDL cholesterol (<50mg/dL), and the remaining 26 displayed higher LDL cholesterol levels (>160mg/dL). All the patient's cDNA was amplified by PCR with primers specific for each SNP. The primers for all 22 SNP are mentioned in supplementary table 4. The PCR was performed using the GoTaq DNA polymerase (Promega Cat.No.M3008, WI). The PCR master mix included 5xGoTaq buffer 2μl, 2.5mM dNTP 0.8μl, 10μM forward primer 0.5μl, 10μM reverse primer 0.5μl, Go Taq enzyme 0.05μl, cDNA (about 100ng/μl) 0.1μl and distilled water to make up the total reaction volume to 10μl. The fragments were amplified at 95℃ for 1min for the melting step, followed by 35 cycles of amplification (95℃ for 1min, 55℃ 1min, 72℃ 30sec) and terminal extension at 72℃ for 5min. Next, the PCR products were cut with specific restriction enzymes (PCR product 5μl, 100x BSA 0.1μl, 10x enzyme buffer 1μl, restriction enzyme 0.3μl and distilled water to make the volume up to 10μl). The samples were incubated for 1 hour at 37℃ and 3μl of the restriction enzyme treated sample was run on 3% agarose gel electrophoresis at 170V for 30mins. For one of the SNPs (rs2301286), sequencing was done to confirm the results or restriction enzyme cut. as the gel electrophoresis results were not clear. Sequencing was performed using the Big DyeTM Terminator v3.1 (Thermo Fisher Scientific Inc, MA) according to manufactural protocol, and analyzed with ABI PRISM 3130xl genetic analyzer (Applied Biosystems, MA). The characteristics of the SNPs and the restriction enzymes used are mentioned in supplementary table 5. The frequency of the SNP alleles in patient’s is mentioned in supplementary table 6.
Statistical analysis
Data are shown as means ± SD. Student’s t-test was used to determine the statistical significance for the target genes subjected to RT-qPCR. One-way analysis of variance (ANOVA) and Tukey’s post hoc test were used to determine the statistical significance between the mice genotypes and blood test parameters. The chi-square test was used to determine the statistical significance between the phenotypes and SNPs. P <0.05 was considered significant for all the test data. All the statistical analyses were performed using JMP Pro 17 software (SAS Institute, Cary, NC).
Ethical Approval
All animal handling and experiments were conducted following the Guidelines for Animal Experimentation of Nagasaki University and with the approval of the Institutional Animal Care and Use Committee (approval number: 1204160979).
The experiments on participants who attended in an annual health check-up in this paper were approved by the Ethics Committee of Nagasaki University Graduate School of Biomedical Sciences (project registration number 14051404).
Results
Generation of Usp21 KO mice.
The KO models serve as a valuable tool in genetics to decipher a gene's biological role. Cre/loxP technology has generated conditional KO mice (7). For this study, the deletion of exon three and exon four generated the
Usp21 KO mice (
Figure 1A). Exons 3 and 4 are known to encode essential parts of the USP21 USP domain (8). The
Usp21 with excised exons 3 and 4 is bred to Cre expressing transgenic mice strain to obtain
Usp21 KO mice (
Figure 1A, B). To further confirm, sequencing analysis of
Usp21 KO mice confirmed the successful deletion of exons 3 and 4. Exons 3 and 4 are an 181bp long sequence, and the stop codon appeared immediately after entering exon 5. The DNA gel electrophoresis confirmed the
Usp21 KO (
Figure 1C).
Figure 1D depicts the generation of
Usp21 littermates. The littermates are generated by crossing B6 mice strains harboring WT
Usp21 and B6 & SW129 mix strains.
Fabp7 and Nlrc5 are upregulated in Usp21 KO mice
Genes up- and down-regulated more than 1.3-fold by KO of
Usp21 by microarray were analyzed by ANOVA in the Partek Genomics Suite, followed by Gene Ontology analysis (
Figure 2A, B). A functional group with an enrichment score of more than three corresponds to an over-representation with a P<0.05. In upregulated genes, the functional group of Homeostatic process, Biological regulation, Developmental process, Biological process involved in interspecies interaction between organisms, Response to stimulus, Multicellular organismal process, locomotion, and Immune system process has an enrichment score of over three. In contrast, in downregulated genes, only the Cellular, Localization, and Metabolic processes have an enrichment score of more than three. The previous report that USP21 activates transcription by deubiquitinating ubiquitylated H2A can well explain the significant increase of upregulated genes by KO of
Usp21 in this report. Because we previously identified
USP21 as playing a role in liver regeneration (2), we try to find regulated genes related to the liver, such as liver regeneration, lipid metabolisms, and drug metabolisms. We focused on twenty-one genes described in
Figure 2C. Among the genes that were analyzed, Fatty acid binding protein 7 (
Fabp7), Cytochrome P450 family two subfamily b polypeptide 9 (
Cyp2b9), Amyloid P Component, Serum (
Apcs), C-X-C Motif Chemokine Ligand 13 (
Cxcl3), Lipocalin 2 (
Lcn2), Procollagen C-Endopeptidase Enhancer 2 (
Pcolce2), Serum Amyloid A2 (
Saa2), and Lysozyme 2, (
Lyz2) were upregulated in the microarray (
Figure 2C). To validate the microarray results, RT-qPCR confirmed their expression as described (
Figure 2D, E). Among these genes,
Fabp7 and
Nlrc5 expressed more in KO mice compared to WT mice with
P <0.05 (
Figure 2D). Fatty acid binding proteins (FABPs), members of the intracellular lipid binding protein family are known to regulate lipid metabolism by increasing the fatty acid uptake (9), fatty acid oxidation (10), and by lipolysis (11). The astrocytes and oligodendrocytes in the brain mainly expressed FABP7(12) (13). Kupffer cells express FABP7 in the liver (14). A Study has shown that FABP7 might be involved in the fatty acid metabolism in sheep liver, thereby controlling the composition of fatty acid in the muscle (15). As for
NLRC5, it is known to belong to the human Nod-like receptor (NLR) family. Of the 22 currently known human NLRs (16), obesity, inflammation, and immunity are associated with
NLRC5 (17, 18). SNPs in the
NLRC5 gene and its promoter regions link to alterations in TG, T-CHOL, and high-density lipoprotein cholesterol levels. (19). Additionally,
Nlrc5-/- mice on an HFD exhibit more significant weight gain, waist circumference, large adipose tissues, and adipocyte size than female WT mice (20). These data show that
Usp21 might play a role in lipid metabolism associated with
Fabp7 and
Nlrc5.
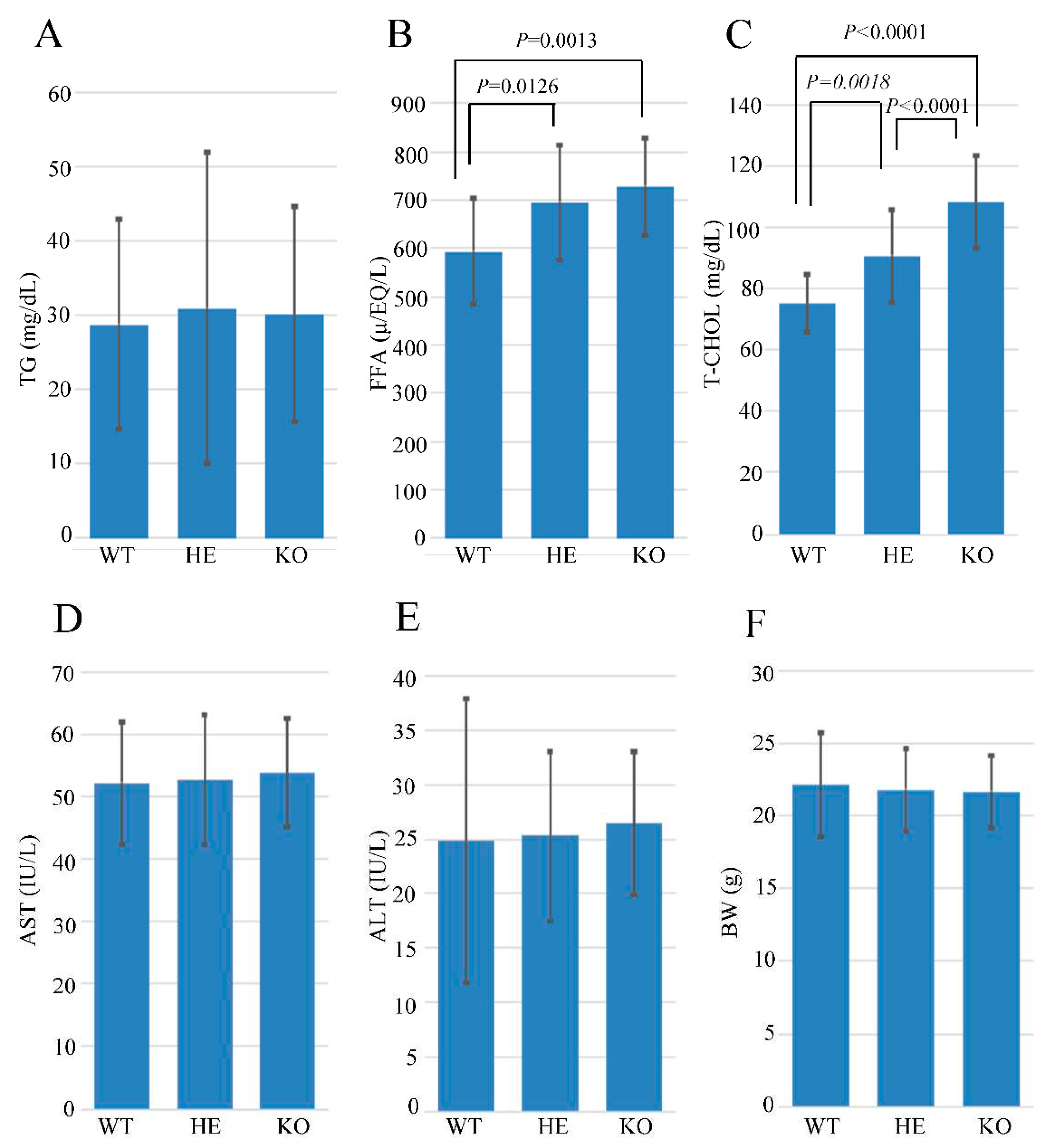
Usp21 KO mice showed an elevation of serum FFA and T-CHOL.
A blood test determined TG, FFA, AST, ALT, and T-CHOL serum levels in the littermate of 10-11 weeks WT, KO, and HE mice (
Figure 3A-E). The body weight (BW) was measured simultaneously (
Figure 3F). The results showed no significant changes in the liver enzymes AST and ALT, confirming that liver function was not impaired. Although, the levels of FFA and T-CHOL were significantly elevated in KO and HE mice compared to WT with
P-values (HE vs. WT;
P=0.0126, KO vs. WT;
P= 0.0013) and (HE vs. WT;
P=0.0018, KO vs. HE;
P<0.0001, and KO vs. WT;
P<0.0001), respectively.
Concerning the fact that elevated level of FFA (21) and LDL cholesterol (6, 22, 23) is known to be associated with metabolic and cardiovascular diseases, our mice serves as an excellent model to clarify the precise role that TG and T-CHOL play in the development of metabolic syndromes and cardiovascular diseases and may provide alternative treatment strategies. In addition, USP21 may serve as an essential diagnostic marker in the future.
rs11421 SNP is associated with hypercholesterolemia
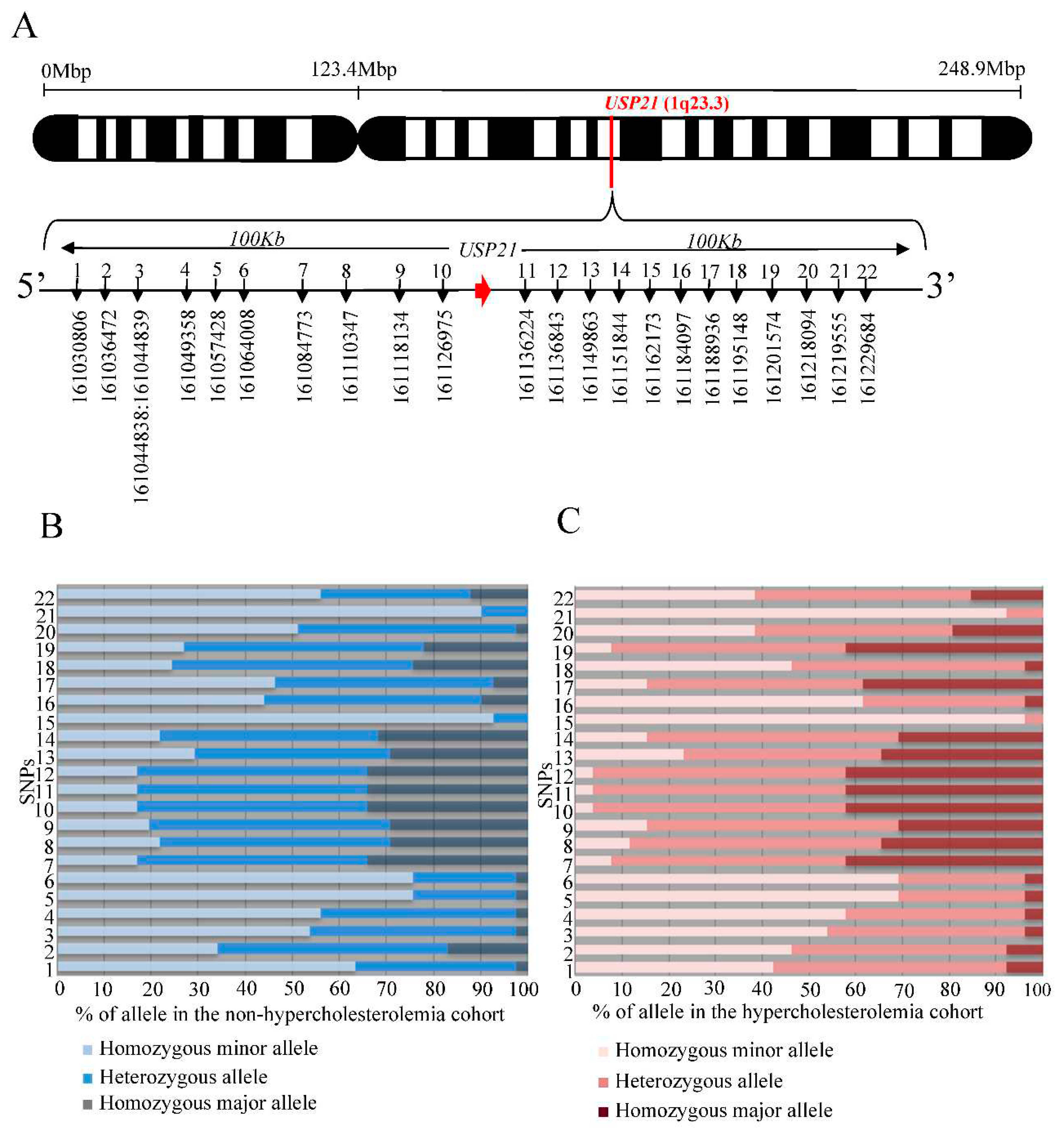
To investigate whether the human USP21 gene is involved in hypercholesterolemia, we selected SNPs around the
USP21 gene based on GRCh37 and conducted an analysis (
Figure 4A). After selecting the SNPs, we screened a sample of patients consisting of low LDL cholesterol (<50mg/dL) and high LDL cholesterol (>160mg/dL) for significant SNPs associated with serum cholesterol levels and performed a chi-square test to determine statistical significance. We found out of the 22 SNPs, the rs11421 SNP (Xsp I site ctag/ccag) showed statistical significance (
Figure 4B, C). We observed a higher frequency of the ccag allele in high LDL cholesterol patients compared to low LDL cholesterol patients with a P-value of <0.0093. Therefore, the ccag allele of the SNP rs11421 is significantly associated with hypercholesterolemia. The simplicity of PCR amplification of SNP rs11421 followed by Xsp1 restriction enzyme cleavage contributes to the practical diagnosis of hypercholesterolemia in clinical settings.
Discussion
ASCVD continues to be the leading cause of morbidity and mortality. Hypercholesterolemia, significantly increased LDL cholesterol, is a well-established risk factor for ASCVD. Hypercholesterolemia has both inherited and acquired causes. The most common genetic condition is familial hypercholesterolemia, which results from mutations in the LDL-receptor gene (6). University College London's database records two thousand two hundred twenty-one unique LDL-receptor gene variants (24, 25). However, hypercholesterolemia without apparent genetic alteration has emerged as the most common type due to the sedentary lifestyle without exercise and increased intake of an HFD, including trans fatty acids. Therefore, the prediction of hypercholesterolemia and its management, including a healthy lifestyle, reducing HFD, and intake of foods with high fiber content, is more important than drug therapy after the onset of the disease (6). We discovered that SNP rs11421 close to USP21 is significantly associated with hypercholesterolemia. Although it is unclear that USP21 causes hypercholesterolemia in humans, this SNP will help its prediction and management. Many studies have reported the association of Fabp7 and Nlrc5 in lipid metabolism. A study found that the mice fed with an HFD showed an upregulation of Fabp7 and a downregulation of miR-21. By upregulating miR-21 and subsequently downregulating Fabp7, dietary lycopene prevented hepatic steatosis in high-fat mice (26).
Researchers have well-documented the role of NLRC5 in liver fibrosis (27) and hepatocellular carcinoma (28). NLRC5 expression was considerably higher in mice fed ethanol with elevated TG, T-CHOL, ALT, and AST levels than in control diet-fed animals' liver tissue. NLRC5 was also increased in mouse AML-12 hepatocyte cell lines when exposed to ethanol (29). These data suggest abnormal NLRC5 expression and its involvement in hypercholesterolemia. Further experiments involving Fabp7 and Nlrc5 KO can help elucidate these genes' effects and verify any correlation between these genes and hypercholesterolemia.
We found that Usp21 KO mice showed an elevated expression of Fabp7 and Nlrc5 genes, previously reported in studies to be involved in lipid metabolism and lipid metabolism-related disorders such as obesity. In addition, the levels of FFA and T-CHOL were also significantly elevated in Usp21 KO mice and HE mice compared to WT mice, suggesting that Usp21 may contribute to lipid abnormalities. In addition, we found that the SNP rs11421 adjusting to USP21 was significantly associated with hypercholesterolemia. This clinical linkage supports the data of USP21 KO mice and may help predict and manage hypercholesterolemia.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org. Supplementary Table 1. List of primers for RT-qPCR. Supplementary Table 2. Mice characteristics and blood test data. Supplementary Table 3. Patients' characteristics. Supplementary Table 4. SNP-specific primers for PCR. Supplementary Table 5. Characteristics of SNPs. Supplementary Table 6. Frequency of which patients have SNP alleles.
Author Contributions
T. Ito, T. Nakagawa, N. Hattori, T. Maeda, and H. Koseki designed research; S. Iyer and N. Hattori analyzed data; S. Iyer, N. Hattori, and H. Okuda performed research; T. Ito, S. Iyer, N. Hattori, and H. Okuda wrote the paper; and All authors were involved in drafting and revising the manuscript.
Funding
This work was supported by the Japan Society for the Promotion of Science (JSPS) under Grant-in-Aid for Scientific Research JSPS KAKENHI Grant Numbers JP20K06489 (to T. N.), JP24118003 (to T. I.), and JP17H04044 (to T. I.) from the Ministry of Education, Culture, Sports, Science, and Technology.
Data Availability Statement
The data supporting this study's findings are available in the methods and supplementary material of this article or in GEO. All the microarray data can be found online in the NCBI GEO submission (GSE244360).
Acknowledgements
We thank Ms. Kaori Nakagawa, Ms. Hiromi Hayashida, and Dr. Mitsuhiro Yoneda for helpful discussions and technical support.
Conflicts of Interest
The authors declare that they have no conflicts of interest in this article.
Abbreviations
ALT, alanine transaminase; ANOVA, analysis of variance; Apcs, Amyloid P Component, Serum; ASCVD, atherosclerotic cardiovascular diseases; AST, aspartate aminotransferase; BW, body weight; Cxcl13, C-X-C Motif Chemokine Ligand 13; Cyp4a10, Cytochrome P450 Family 4 Subfamily A Polypeptide 10; Cyp4a14, Cytochrome P450 Family 4 Subfamily A Polypeptide 14; Cyp4a31, Cytochrome P450 Family 4 Subfamily A Polypeptide 31; Cyp2b9, Cytochrome P450 Family 2 Subfamily B Polypeptide 9; Cyp7a1, Cytochrome P450 Family 7 Subfamily A Polypeptide 1; Dusp12, Dual Specificity Phosphatase 12; Fabp7, Fatty Acid Binding Protein 7; FABPs, Fatty acid binding proteins; FFA, free fatty acids; HE, heterozygous; Hmgb1, High Mobility Group Box 1; Hmgcr, 3-Hydroxy-3-Methylglutaryl-Coenzyme A Reductase; KO, knockout; LDL, low-density lipoprotein; Lcn1, Lipocalin 1; Lyz2, Lysozyme 2; ND, normal diet; Nlrc5, Nod-like receptor Family CARD Domain Containing 5; Pparα, Peroxisome Proliferator Activated Receptor Alpha; Pcolce2, Procollagen C-Endopeptidase Enhancer 2; Rbm33, RNA Binding Motif Protein 33; Saa2, Serum Amyloid A2; Sfrs18, Splicing Factor, Arginine/Serine-rich 18; SNP, single nucleotide polymorphism; Sqle, Squalene Epoxidase; Srebf2, Sterol Regulatory Element Binding Transcription Factor 2; T-CHOL, total cholesterol; TG, triglycerides; TAM, tamoxifen; Usp21, Ubiquitin Specific Peptidase 21; WT, wild-type
References
- Dye, B. T. , and Schulman, B. A. Structural mechanisms underlying posttranslational modification by ubiquitin-like proteins. Annu Rev Biophys Biomol Struct 2007, 36, 131–150. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, T. , Kajitani, T., Togo, S., Masuko, N., Ohdan, H., Hishikawa, Y., Koji, T., Matsuyama, T., Ikura, T., Muramatsu, M., and Ito, T. Deubiquitylation of histone H2A activates transcriptional initiation via trans-histone cross-talk with H3K4 di- and trimethylation. Genes Dev 2008, 22, 37–49. [Google Scholar] [PubMed]
- Chen, Y. , Zhou, B., and Chen, D. USP21 promotes cell proliferation and metastasis through suppressing EZH2 ubiquitination in bladder carcinoma. Onco Targets Ther 2017, 10, 681–689. [Google Scholar] [CrossRef] [PubMed]
- Hou, P. , Ma, X., Zhang, Q., Wu, C. J., Liao, W., Li, J., Wang, H., Zhao, J., Zhou, X., Guan, C., Ackroyd, J., Jiang, S., Zhang, J., Spring, D. J., Wang, Y. A., and DePinho, R. A. USP21 deubiquitinase promotes pancreas cancer cell stemness via Wnt pathway activation. Genes Dev 2019, 33, 1361–1366. [Google Scholar] [PubMed]
- Okuda, H. , Ohdan, H., Nakayama, M., Koseki, H., Nakagawa, T., and Ito, T. The USP21 short variant (USP21SV) lacking NES, located mostly in the nucleus in vivo, activates transcription by deubiquitylating ubH2A in vitro. PLoS One 2013, 8, e79813. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, M. A. , Asuka, E., and Jialal, I. (2023) Hypercholesterolemia. In StatPearls, Treasure Island (FL) ineligible companies. Disclosure: Edinen Asuka declares no relevant financial relationships with ineligible companies. Disclosure: Ishwarlal Jialal declares no relevant financial relationships with ineligible companies.
- Gierut, J. J. , Jacks, T. E., and Haigis, K. M. Strategies to achieve conditional gene mutation in mice. Cold Spring Harb Protoc 2014, 2014, 339–349. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y. , Mao, R., Yu, Y., Liu, S., Shi, Z., Cheng, J., Zhang, H., An, L., Zhao, Y., Xu, X., Chen, Z., Kogiso, M., Zhang, D., Zhang, H., Zhang, P., Jung, J. U., Li, X., Xu, G., and Yang, J. USP21 negatively regulates antiviral response by acting as a RIG-I deubiquitinase. J Exp Med 2014, 211, 313–328. [Google Scholar] [PubMed]
- Spitsberg, V. L. , Matitashvili, E., and Gorewit, R. C. Association and Coexpression of Fatty-Acid-Binding Protein and Glycoprotein CD36 in the Bovine Mammary Gland. European Journal of Biochemistry 2008, 230, 872–878. [Google Scholar]
- Burrier, R. E. , Manson, C. R., and Brecher, P. Binding of acyl-CoA to liver fatty acid binding protein: effect on acyl-CoA synthesis. Biochim Biophys Acta 1987, 919, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Coe, N. R. , Simpson, M. A., and Bernlohr, D. A. Targeted disruption of the adipocyte lipid-binding protein (aP2 protein) gene impairs fat cell lipolysis and increases cellular fatty acid levels. Journal of Lipid Research 1999, 40, 967–972. [Google Scholar] [CrossRef]
- Owada, Y. , Yoshimoto, T., and Kondo, H. Spatio-temporally differential expression of genes for three members of fatty acid binding proteins in developing and mature rat brains. J Chem Neuroanat 1996, 12, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Sharifi, K. , Ebrahimi, M., Kagawa, Y., Islam, A., Tuerxun, T., Yasumoto, Y., Hara, T., Yamamoto, Y., Miyazaki, H., Tokuda, N., Yoshikawa, T., and Owada, Y. Differential expression and regulatory roles of FABP5 and FABP7 in oligodendrocyte lineage cells. Cell Tissue Res 2013, 354, 683–695. [Google Scholar] [CrossRef] [PubMed]
- Abdelwahab, S. A. , Owada, Y., Kitanaka, N., Iwasa, H., Sakagami, H., and Kondo, H. Localization of brain-type fatty acid-binding protein in Kupffer cells of mice and its transient decrease in response to lipopolysaccharide. Histochem Cell Biol 2003, 119, 469–475. [Google Scholar] [CrossRef] [PubMed]
- Gunawan, A. , Listyarini, K., Harahap, R. S., Jakaria, Roosita, K., Sumantri, C., Inounu, I., Akter, S. H., Islam, M. A., and Uddin, M. J. Hepatic transcriptome analysis identifies genes, polymorphisms and pathways involved in the fatty acids metabolism in sheep. PLoS One 2021, 16, e0260514. [Google Scholar] [CrossRef] [PubMed]
- Proell, M. , Riedl, S. J., Fritz, J. H., Rojas, A. M., and Schwarzenbacher, R. The Nod-like receptor (NLR) family: a tale of similarities and differences. PLoS One 2008, 3, e2119. [Google Scholar] [CrossRef] [PubMed]
- Cao-Lei, L. , Elgbeili, G., Szyf, M., Laplante, D. P., and King, S. Differential genome-wide DNA methylation patterns in childhood obesity. BMC Res Notes 2019, 12, 174. [Google Scholar] [CrossRef] [PubMed]
- Lin, X. , Peng, C., Greenbaum, J., Li, Z. F., Wu, K. H., Ao, Z. X., Zhang, T., Shen, J., and Deng, H. W. Identifying potentially common genes between dyslipidemia and osteoporosis using novel analytical approaches. Mol Genet Genomics 2018, 293, 711–723. [Google Scholar] [CrossRef] [PubMed]
- Hosseinzadeh, N. , Mehrabi, Y., Daneshpour, M. S., Zayeri, F., Guity, K., and Azizi, F. Identifying new associated pleiotropic SNPs with lipids by simultaneous test of multiple longitudinal traits: An Iranian family-based study. Gene 2019, 692, 156–169. [Google Scholar] [CrossRef] [PubMed]
- Bauer, S. , Aeissen, V., Bubeck, A. M., Kienes, I., Ellwanger, K., Scheurenbrand, M., Rexhepi, F., Ramanathan, S., Rosenstiel, P., Fricke, W. F., and Kufer, T. A. NLRC5 affects diet-induced adiposity in female mice and co-regulates peroxisome proliferator-activated receptor PPARgamma target genes. iScience 2023, 26, 106313. [Google Scholar]
- Pilz, S. , and Marz, W. Free fatty acids as a cardiovascular risk factor. Clin Chem Lab Med 2008, 46, 429–434. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S. M. , Choi, S., Kim, K., Kim, S. M., Lee, G., Park, S. Y., Kim, Y. Y., Son, J. S., Yun, J. M., and Park, S. M. Effect of Change in Total Cholesterol Levels on Cardiovascular Disease Among Young Adults. J Am Heart Assoc 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Baigent, C. , Keech, A., Kearney, P. M., Blackwell, L., Buck, G., Pollicino, C., Kirby, A., Sourjina, T., Peto, R., Collins, R., Simes, R., and Cholesterol Treatment Trialists, C. Efficacy and safety of cholesterol-lowering treatment: prospective meta-analysis of data from 90,056 participants in 14 randomised trials of statins. Lancet 2005, 366, 1267–1278. [Google Scholar] [PubMed]
- L., S. Global Variome shared LOVD the LDLR gene homepage.
- Leigh, S. , Futema, M., Whittall, R., Taylor-Beadling, A., Williams, M., den Dunnen, J. T., and Humphries, S. E. The UCL low-density lipoprotein receptor gene variant database: pathogenicity update. J Med Genet 2017, 54, 217–223. [Google Scholar] [CrossRef]
- Ahn, J. , Lee, H., Jung, C. H., and Ha, T. Lycopene inhibits hepatic steatosis via microRNA-21-induced downregulation of fatty acid-binding protein 7 in mice fed a high-fat diet. Mol Nutr Food Res 2012, 56, 1665–1674. [Google Scholar] [CrossRef] [PubMed]
- Liu, X. , Wu, Y., Yang, Y., Li, W., Huang, C., Meng, X., and Li, J. Role of NLRC5 in progression and reversal of hepatic fibrosis. Toxicol Appl Pharmacol 2016, 294, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y. Y. , He, Y. H., Chen, C., Xu, T., Li, L., Ni, M. M., Meng, X. M., Huang, C., and Li, J. NLRC5 regulates cell proliferation, migration and invasion in hepatocellular carcinoma by targeting the Wnt/beta-catenin signaling pathway. Cancer Lett 2016, 376, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y. , Huang, C., Shi, T., and Li, J. Deficiency of NLR family member NLRC5 alleviates alcohol induced hepatic injury and steatosis by enhancing autophagy of hepatocytes. Toxicol Appl Pharmacol 2023, 461, 116406. [Google Scholar] [CrossRef] [PubMed]
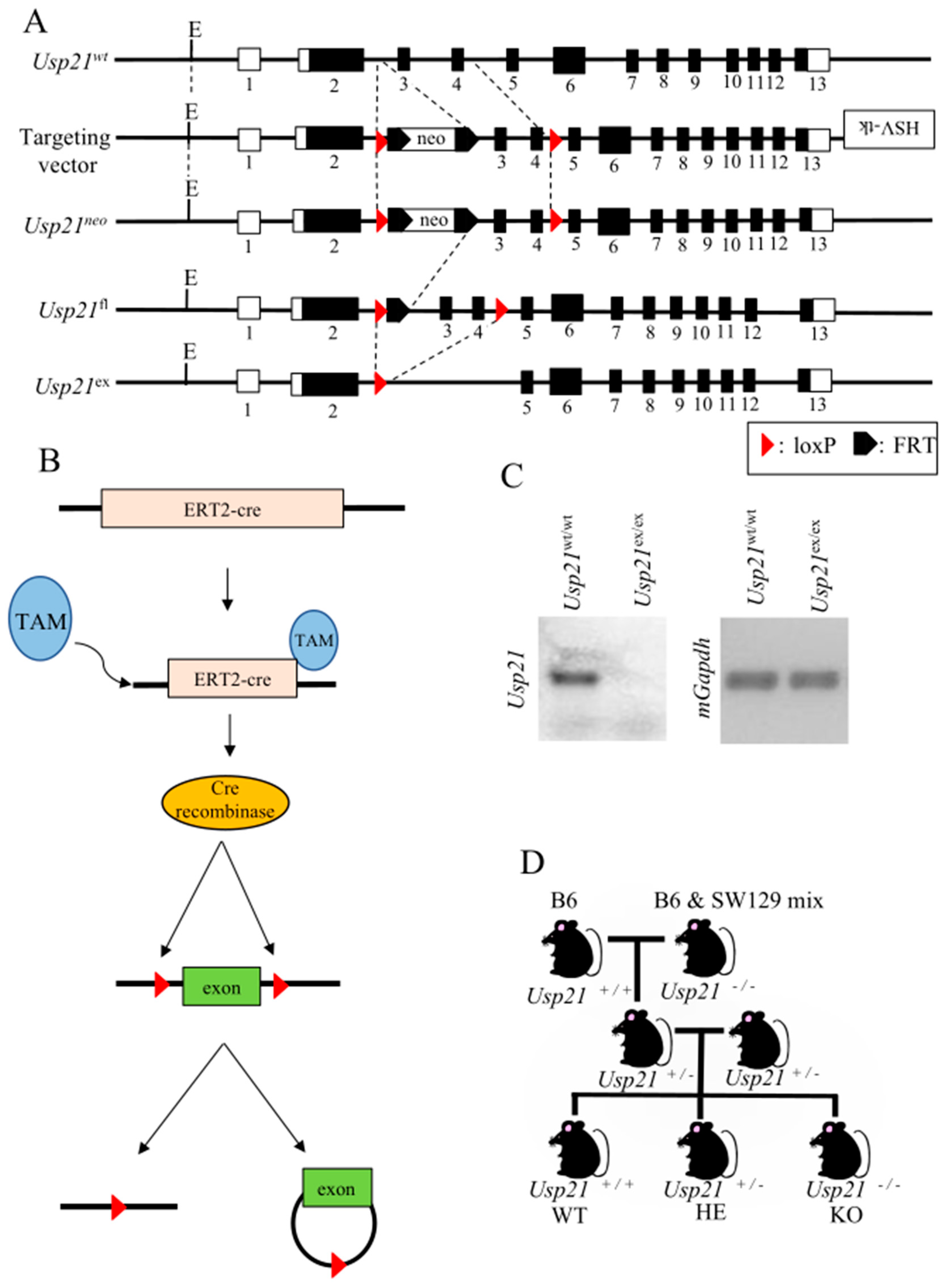
Figure 1.
Figure 1. Generation of Usp21 KO mice and its confirmation. (A-B) Pictorial representation of the generation of Usp21 conditional KO mice using the ERT2-Cre/loxP system. (C) DNA gel electrophoresis of Usp21 wt/wt mice and Usp21 ex/ex mice to check the deletion of Usp21. The mGapdh is used as the reference gene. (D) Schematic representation of the generation of Usp21 KO mice littermates by mating B6 harboring Usp21 WT gene with B6&SW129 mix having Usp21 knocked out. ex, excision; fl, floxed; FRT, flippase recognition target sites; HE, heterozygous; HSV-tk, herpes simplex virus- thymidine kinase; KO, knockout; neo, neomycin resistance gene; TAM, tamoxifen; WT, wild-type.
Figure 1.
Figure 1. Generation of Usp21 KO mice and its confirmation. (A-B) Pictorial representation of the generation of Usp21 conditional KO mice using the ERT2-Cre/loxP system. (C) DNA gel electrophoresis of Usp21 wt/wt mice and Usp21 ex/ex mice to check the deletion of Usp21. The mGapdh is used as the reference gene. (D) Schematic representation of the generation of Usp21 KO mice littermates by mating B6 harboring Usp21 WT gene with B6&SW129 mix having Usp21 knocked out. ex, excision; fl, floxed; FRT, flippase recognition target sites; HE, heterozygous; HSV-tk, herpes simplex virus- thymidine kinase; KO, knockout; neo, neomycin resistance gene; TAM, tamoxifen; WT, wild-type.
Figure 2.
Fabp7 and Nlrc5 are upregulated in Usp21 KO mice. (A, B) Gene ontology analysis of upregulated genes and downregulated genes in Usp21 KO mice. (C) Analysis of microarray results. (D, E) RT-qPCR results of (D) upregulated or (E) downregulated genes in Usp21 KO mice. The values are normalized with the mGapdh expression. BR, Biological regulation; BP, Biological phase; CP, Cellular process; D, Detoxification; DP, Developmental process; G, Growth; HP, Homeostatic process; IN, Biological process involved in interspecies interaction between organisms; INT, Biological process involved in intraspecies interaction between organisms; ISP, Immune system process; KO, knockout. L, Localization; LO, Locomotion; MOP, Multicellular organismal process; MP, Metabolic process; P, Pigmentation; R, Reproduction; REP, Reproductive process; RP, Rhythmic process; RS, Response to stimulus; V, Viral process; WT, wild-type. Data are shown as mean ± SD. P values were tested with a student t-test. P <0.05 is considered significant.
Figure 2.
Fabp7 and Nlrc5 are upregulated in Usp21 KO mice. (A, B) Gene ontology analysis of upregulated genes and downregulated genes in Usp21 KO mice. (C) Analysis of microarray results. (D, E) RT-qPCR results of (D) upregulated or (E) downregulated genes in Usp21 KO mice. The values are normalized with the mGapdh expression. BR, Biological regulation; BP, Biological phase; CP, Cellular process; D, Detoxification; DP, Developmental process; G, Growth; HP, Homeostatic process; IN, Biological process involved in interspecies interaction between organisms; INT, Biological process involved in intraspecies interaction between organisms; ISP, Immune system process; KO, knockout. L, Localization; LO, Locomotion; MOP, Multicellular organismal process; MP, Metabolic process; P, Pigmentation; R, Reproduction; REP, Reproductive process; RP, Rhythmic process; RS, Response to stimulus; V, Viral process; WT, wild-type. Data are shown as mean ± SD. P values were tested with a student t-test. P <0.05 is considered significant.
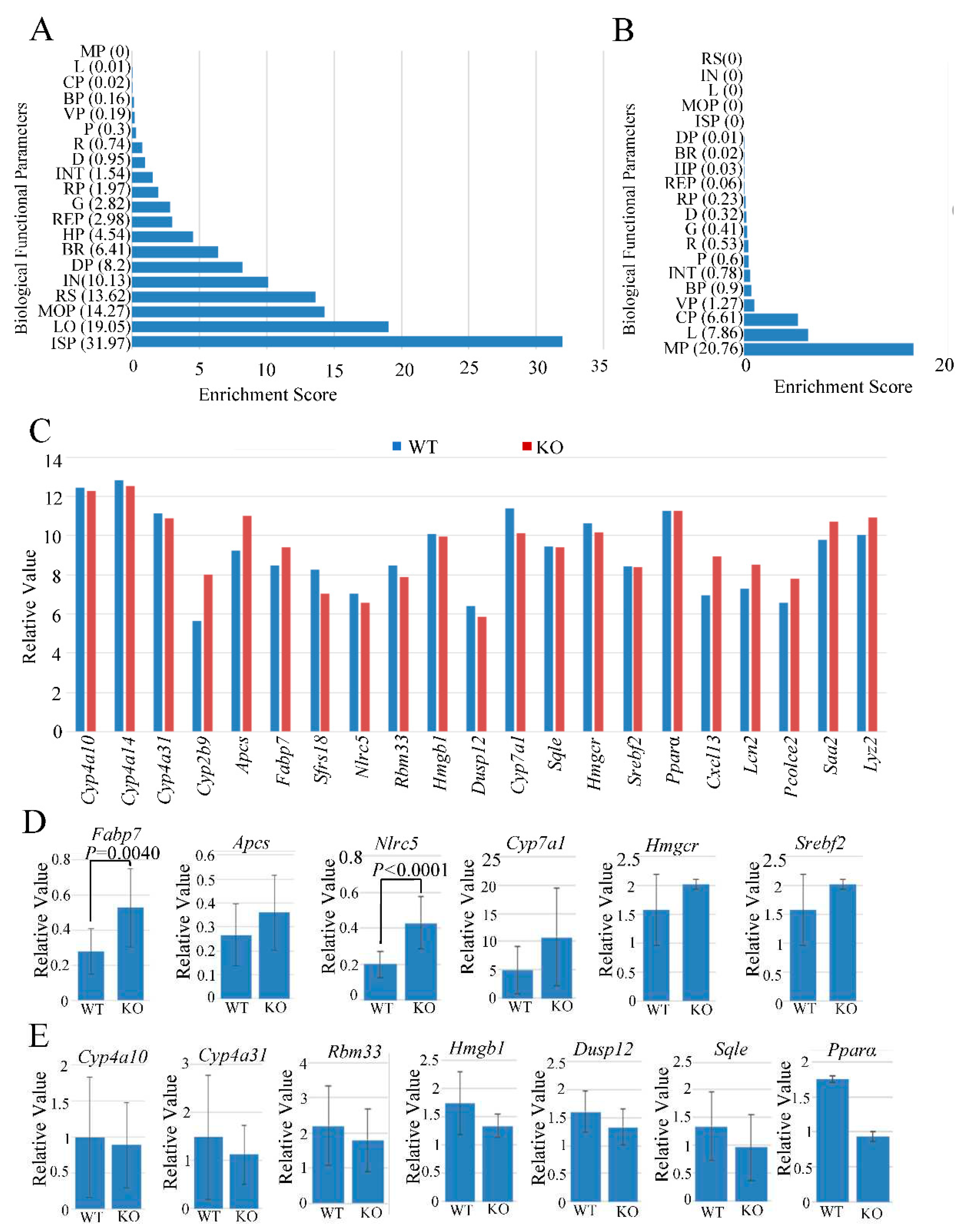
Figure 3.
FFA and T-CHOL were elevated in Usp21 KO mice. (A- E) The blood test data of (A) TG, (B) FFA, (C) T-CHOL, (D) AST, and (E) ALT 10-11 weeks WT, HE, and KO mice. (F) BW in 10-11 weeks WT, HE, and KO mice. ALT, Alanine transaminase; AST, Aspartate aminotransferase; BW, Body weight; FFA, Free fatty acid; HE, heterozygous; KO, knockout; TG, Triglyceride; T-CHOL, Total cholesterol; WT, wild-type. Data are shown as mean ± SD. P values were tested with one-way ANOVA, followed by Tukey’s test. P <0.05 is considered significant.
Figure 3.
FFA and T-CHOL were elevated in Usp21 KO mice. (A- E) The blood test data of (A) TG, (B) FFA, (C) T-CHOL, (D) AST, and (E) ALT 10-11 weeks WT, HE, and KO mice. (F) BW in 10-11 weeks WT, HE, and KO mice. ALT, Alanine transaminase; AST, Aspartate aminotransferase; BW, Body weight; FFA, Free fatty acid; HE, heterozygous; KO, knockout; TG, Triglyceride; T-CHOL, Total cholesterol; WT, wild-type. Data are shown as mean ± SD. P values were tested with one-way ANOVA, followed by Tukey’s test. P <0.05 is considered significant.
Figure 4.
rs11421 is associated with hypercholesterolemia. (A) Schema of SNPs surrounding the human USP21 gene. (B) Genotypes at each SNP locus in non-hypercholesterolemic outpatients. (C) Genotypes at each SNP locus in hypercholesterolemic outpatients. SNP, single nucleotide polymorphism.
Figure 4.
rs11421 is associated with hypercholesterolemia. (A) Schema of SNPs surrounding the human USP21 gene. (B) Genotypes at each SNP locus in non-hypercholesterolemic outpatients. (C) Genotypes at each SNP locus in hypercholesterolemic outpatients. SNP, single nucleotide polymorphism.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).