Submitted:
18 November 2023
Posted:
20 November 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Immunometabolism
a. Immunometabolism and cancer
b. A case of metabolic-dependent activation in immune cells: monocyte and macrophage
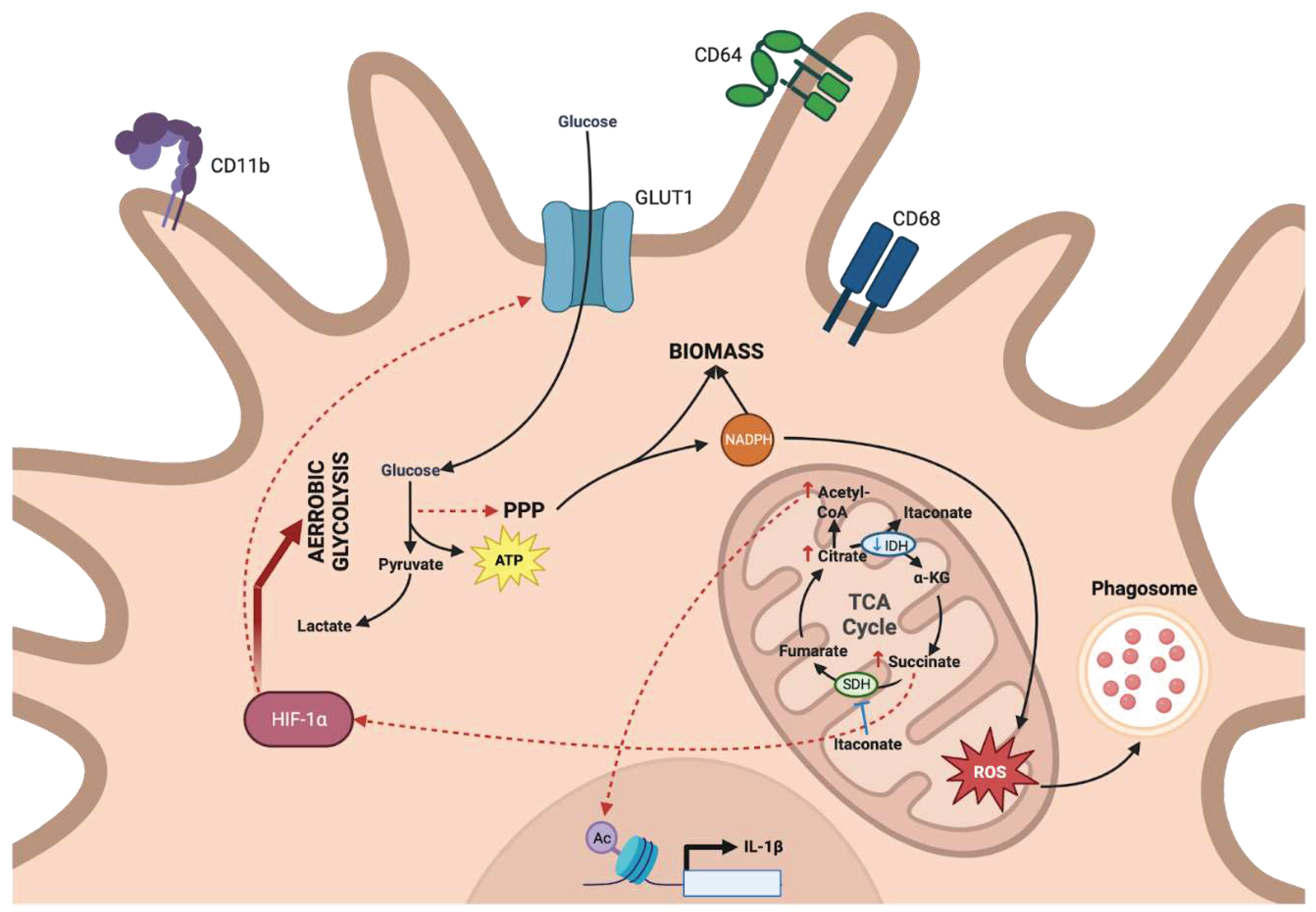
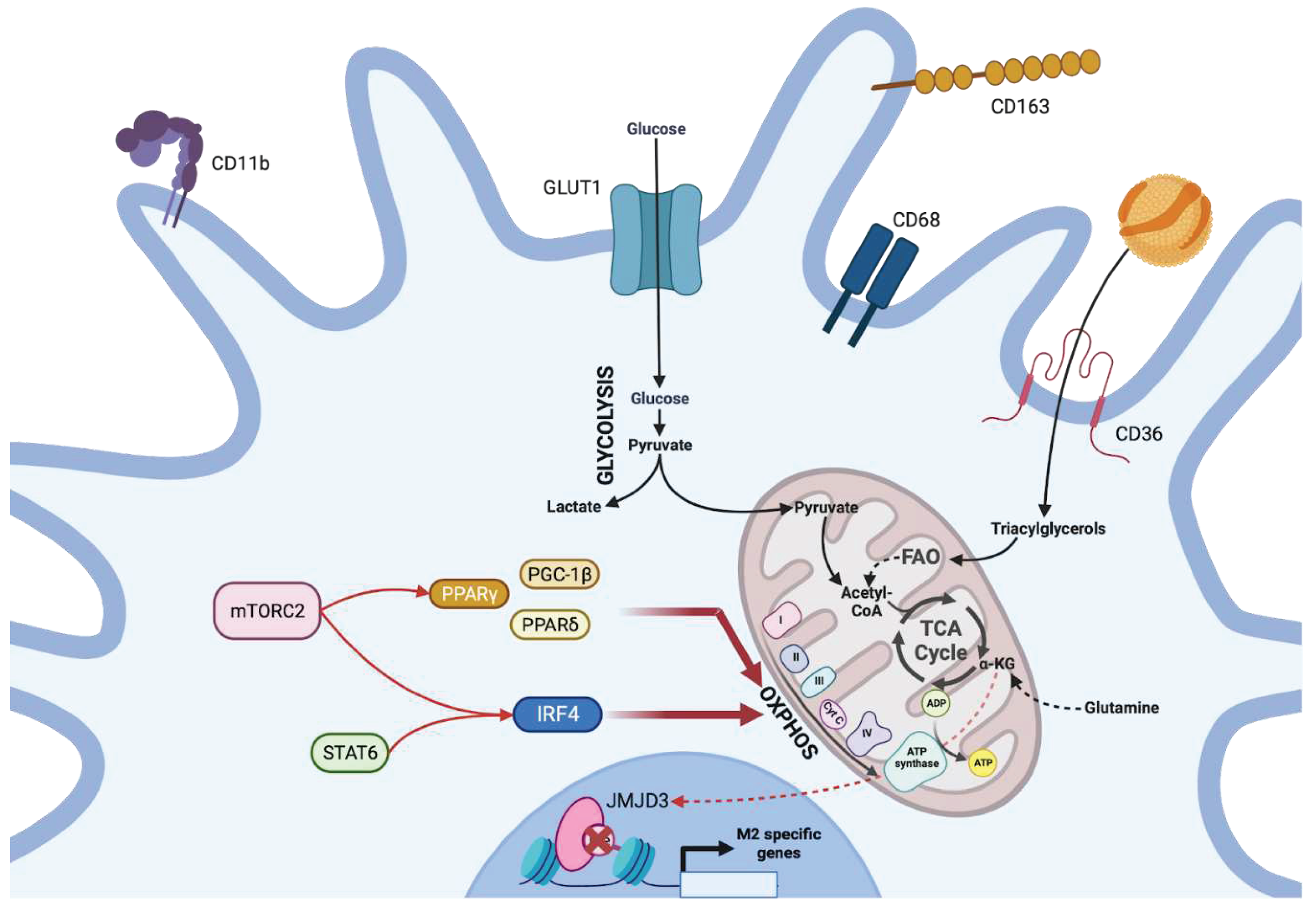
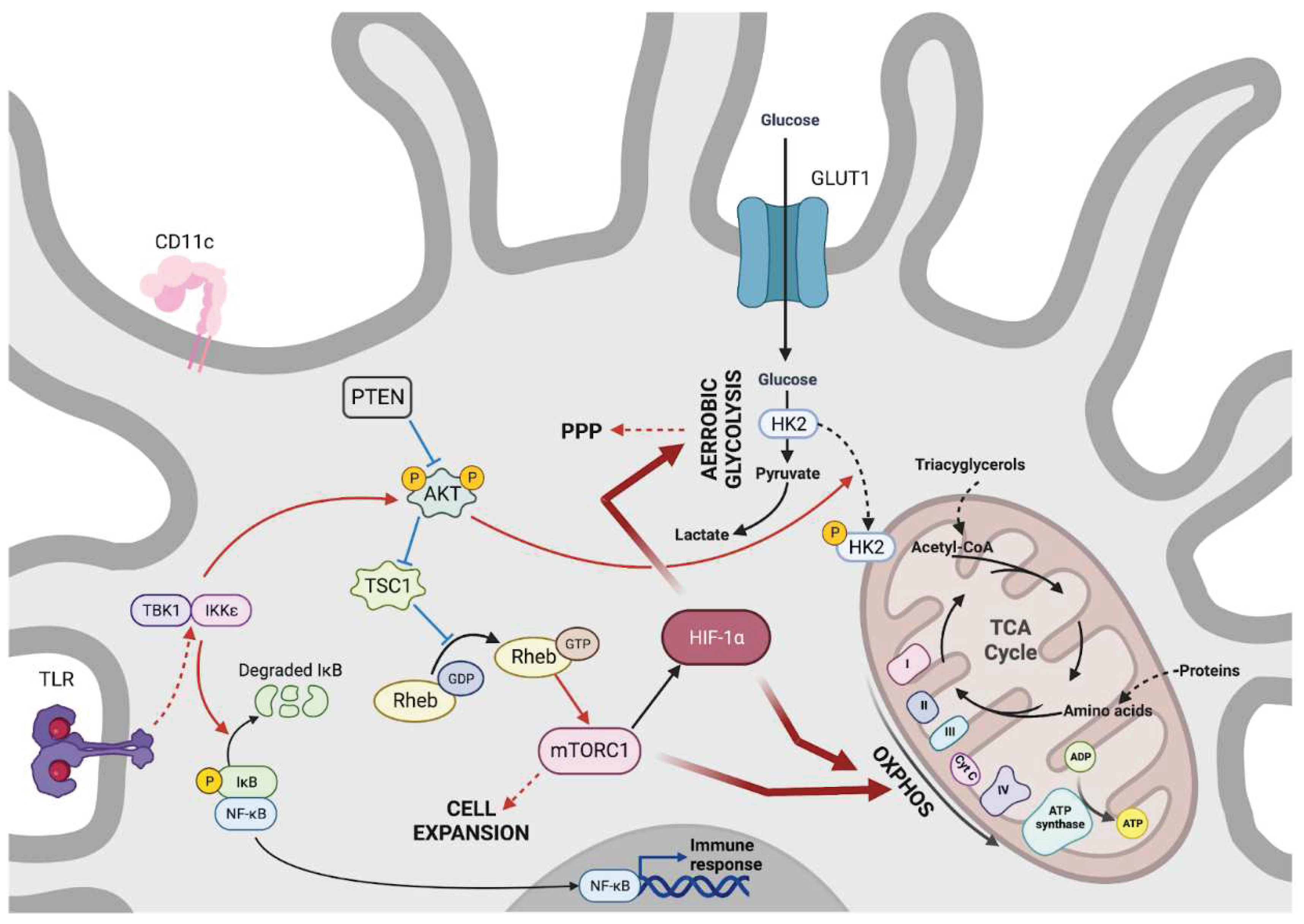
c. The metabolic-dependent activation of tissue sentinels: dendritic cells
3. At the systemic level: Interplays between host metabolic status and infectious diseases
a. Diabetes, obesity and other metabolic disorders
i. Diabetes
ii. Obesity
b. Exercise

4. Concluding remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Berbudi, A.; Rahmadika, N.; Tjahjadi, A.I.; Ruslami, R. Type 2 Diabetes and Its Impact on the Immune System. CDR 2020, 16, 442–449. [Google Scholar] [CrossRef] [PubMed]
- Palmer, Clovis. S. Innate Metabolic Responses against Viral Infections. Nat Metab 2022, 4, 1245–1259. [Google Scholar] [CrossRef] [PubMed]
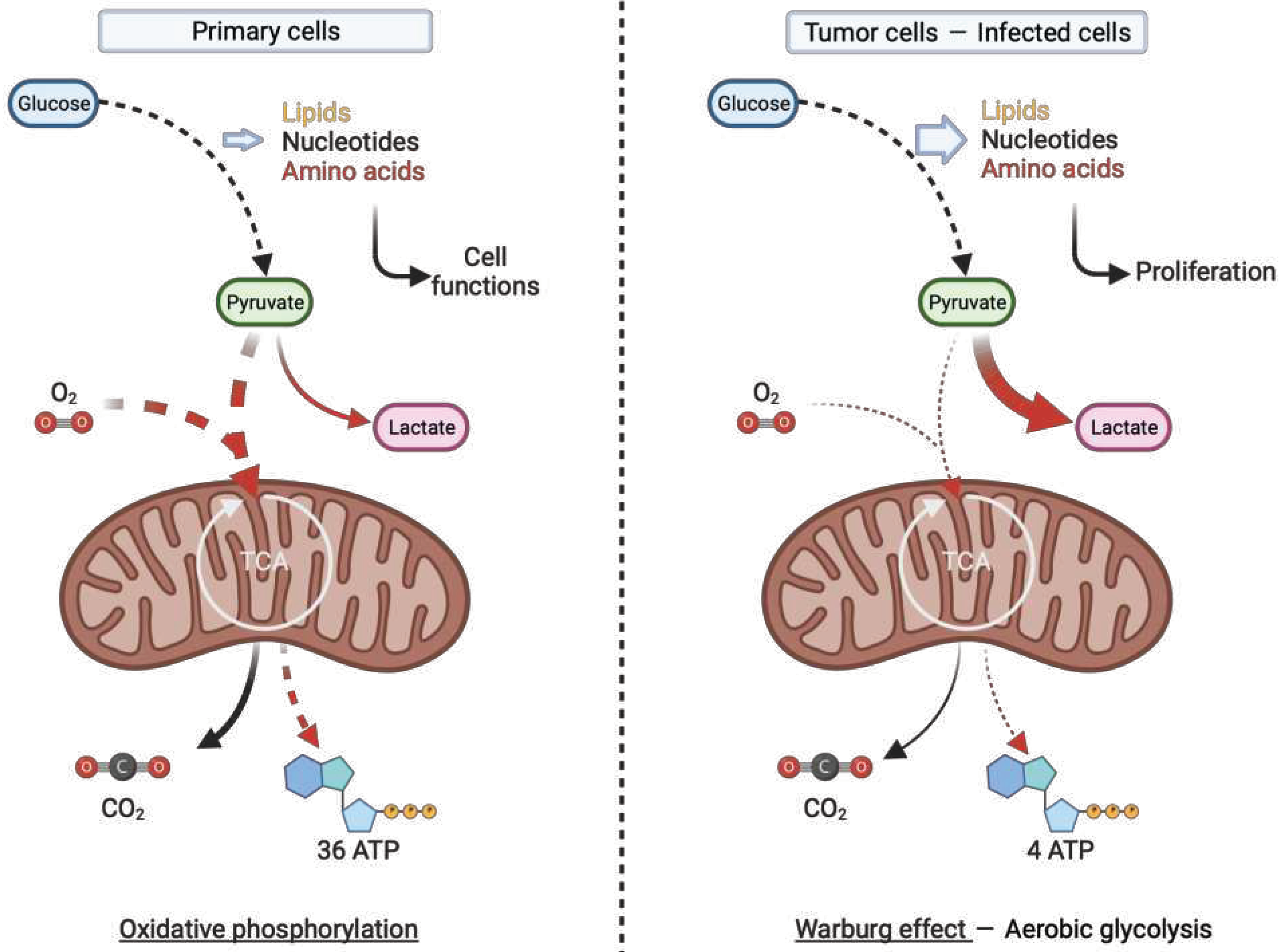
- Zhu, J.; Thompson, C.B. Metabolic Regulation of Cell Growth and Proliferation. Nat Rev Mol Cell Biol 2019, 20, 436–450. [Google Scholar] [CrossRef] [PubMed]
- Thaker, S.K.; Ch’ng, J.; Christofk, H.R. Viral Hijacking of Cellular Metabolism. BMC Biol 2019, 17, 59. [Google Scholar] [CrossRef] [PubMed]
- Warburg, O. On the Origin of Cancer Cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Allen, C.N.S.; Arjona, S.P.; Santerre, M.; Sawaya, B.E. Hallmarks of Metabolic Reprogramming and Their Role in Viral Pathogenesis. Viruses 2022, 14, 602. [Google Scholar] [CrossRef] [PubMed]
- Alivernini, S.; Tolusso, B.; Gigante, M.R.; Petricca, L.; Bui, L.; Fedele, A.L.; Di Mario, C.; Benvenuto, R.; Federico, F.; Ferraccioli, G.; et al. Overweight/Obesity Affects Histological Features and Inflammatory Gene Signature of Synovial Membrane of Rheumatoid Arthritis. Sci Rep 2019, 9, 10420. [Google Scholar] [CrossRef] [PubMed]
- Beck, S.; Zhu, Z.; Oliveira, M.F.; Smith, D.M.; Rich, J.N.; Bernatchez, J.A.; Siqueira-Neto, J.L. Mechanism of Action of Methotrexate Against Zika Virus. Viruses 2019, 11, 338. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.-Y.; Aliyari, R.; Chikere, K.; Li, G.; Marsden, M.D.; Smith, J.K.; Pernet, O.; Guo, H.; Nusbaum, R.; Zack, J.A.; et al. Interferon-Inducible Cholesterol-25-Hydroxylase Broadly Inhibits Viral Entry by Production of 25-Hydroxycholesterol. Immunity 2013, 38, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Leone, R.D.; Powell, J.D. Metabolism of Immune Cells in Cancer. Nat Rev Cancer 2020, 20, 516–531. [Google Scholar] [CrossRef]
- Pagès, F.; Galon, J.; Dieu-Nosjean, M.-C.; Tartour, E.; Sautès-Fridman, C.; Fridman, W.-H. Immune Infiltration in Human Tumors: A Prognostic Factor That Should Not Be Ignored. Oncogene 2010, 29, 1093–1102. [Google Scholar] [CrossRef]
- Paijens, S.T.; Vledder, A.; De Bruyn, M.; Nijman, H.W. Tumor-Infiltrating Lymphocytes in the Immunotherapy Era. Cell Mol Immunol 2021, 18, 842–859. [Google Scholar] [CrossRef]
- Voelxen, N.F.; Blatt, S.; Knopf, P.; Henkel, M.; Appelhans, C.; Righesso, L.A.R.; Pabst, A.; Goldschmitt, J.; Walenta, S.; Neff, A.; et al. Comparative Metabolic Analysis in Head and Neck Cancer and the Normal Gingiva. Clin Oral Invest 2018, 22, 1033–1043. [Google Scholar] [CrossRef]
- Ottensmeier, C.H.; Perry, K.L.; Harden, E.L.; Stasakova, J.; Jenei, V.; Fleming, J.; Wood, O.; Woo, J.; Woelk, C.H.; Thomas, G.J.; et al. Upregulated Glucose Metabolism Correlates Inversely with CD8+ T-Cell Infiltration and Survival in Squamous Cell Carcinoma. Cancer Research 2016, 76, 4136–4148. [Google Scholar] [CrossRef]
- Chang, C.-H.; Qiu, J.; O’Sullivan, D.; Buck, M.D.; Noguchi, T.; Curtis, J.D.; Chen, Q.; Gindin, M.; Gubin, M.M.; van der Windt, G.J.W.; et al. Metabolic Competition in the Tumor Microenvironment Is a Driver of Cancer Progression. Cell 2015, 162, 1229–1241. [Google Scholar] [CrossRef] [PubMed]
- Ho, P.-C.; Bihuniak, J.D.; Macintyre, A.N.; Staron, M.; Liu, X.; Amezquita, R.; Tsui, Y.-C.; Cui, G.; Micevic, G.; Perales, J.C.; et al. Phosphoenolpyruvate Is a Metabolic Checkpoint of Anti-Tumor T Cell Responses. Cell 2015, 162, 1217–1228. [Google Scholar] [CrossRef]
- Murray, P.J.; Wynn, T.A. Protective and Pathogenic Functions of Macrophage Subsets. Nat Rev Immunol 2011, 11, 723–737. [Google Scholar] [CrossRef] [PubMed]
- Wculek, S.K.; Dunphy, G.; Heras-Murillo, I.; Mastrangelo, A.; Sancho, D. Metabolism of Tissue Macrophages in Homeostasis and Pathology. Cell Mol Immunol 2022, 19, 384–408. [Google Scholar] [CrossRef] [PubMed]
- Freemerman, A.J.; Johnson, A.R.; Sacks, G.N.; Milner, J.J.; Kirk, E.L.; Troester, M.A.; Macintyre, A.N.; Goraksha-Hicks, P.; Rathmell, J.C.; Makowski, L. Metabolic Reprogramming of Macrophages. Journal of Biological Chemistry 2014, 289, 7884–7896. [Google Scholar] [CrossRef]
- Rodríguez-Prados, J.-C.; Través, P.G.; Cuenca, J.; Rico, D.; Aragonés, J.; Martín-Sanz, P.; Cascante, M.; Boscá, L. Substrate Fate in Activated Macrophages: A Comparison between Innate, Classic, and Alternative Activation. The Journal of Immunology 2010, 185, 605–614. [Google Scholar] [CrossRef]
- Palsson-McDermott, E.M.; Curtis, A.M.; Goel, G.; Lauterbach, M.A.R.; Sheedy, F.J.; Gleeson, L.E.; van den Bosch, M.W.M.; Quinn, S.R.; Domingo-Fernandez, R.; Johnston, D.G.W.; et al. Pyruvate Kinase M2 Regulates Hif-1α Activity and IL-1β Induction and Is a Critical Determinant of the Warburg Effect in LPS-Activated Macrophages. Cell Metabolism 2015, 21, 65–80. [Google Scholar] [CrossRef] [PubMed]
- Tannahill, G.M.; Curtis, A.M.; Adamik, J.; Palsson-McDermott, E.M.; McGettrick, A.F.; Goel, G.; Frezza, C.; Bernard, N.J.; Kelly, B.; Foley, N.H.; et al. Succinate Is an Inflammatory Signal That Induces IL-1β through HIF-1α. Nature 2013, 496, 238–242. [Google Scholar] [CrossRef] [PubMed]
- Meiser, J.; Krämer, L.; Sapcariu, S.C.; Battello, N.; Ghelfi, J.; D’Herouel, A.F.; Skupin, A.; Hiller, K. Pro-Inflammatory Macrophages Sustain Pyruvate Oxidation through Pyruvate Dehydrogenase for the Synthesis of Itaconate and to Enable Cytokine Expression. Journal of Biological Chemistry 2016, 291, 3932–3946. [Google Scholar] [CrossRef] [PubMed]
- Lauterbach, M.A.; Hanke, J.E.; Serefidou, M.; Mangan, M.S.J.; Kolbe, C.-C.; Hess, T.; Rothe, M.; Kaiser, R.; Hoss, F.; Gehlen, J.; et al. Toll-like Receptor Signaling Rewires Macrophage Metabolism and Promotes Histone Acetylation via ATP-Citrate Lyase. Immunity 2019, 51, 997–1011.e7. [Google Scholar] [CrossRef] [PubMed]
- Cordes, T.; Wallace, M.; Michelucci, A.; Divakaruni, A.S.; Sapcariu, S.C.; Sousa, C.; Koseki, H.; Cabrales, P.; Murphy, A.N.; Hiller, K.; et al. Immunoresponsive Gene 1 and Itaconate Inhibit Succinate Dehydrogenase to Modulate Intracellular Succinate Levels. Journal of Biological Chemistry 2016, 291, 14274–14284. [Google Scholar] [CrossRef] [PubMed]
- Mills, E.L.; Kelly, B.; Logan, A.; Costa, A.S.H.; Varma, M.; Bryant, C.E.; Tourlomousis, P.; Däbritz, J.H.M.; Gottlieb, E.; Latorre, I.; et al. Succinate Dehydrogenase Supports Metabolic Repurposing of Mitochondria to Drive Inflammatory Macrophages. Cell 2016, 167, 457–470.e13. [Google Scholar] [CrossRef]
- Baardman, J.; Verberk, S.G.S.; Prange, K.H.M.; Van Weeghel, M.; Van Der Velden, S.; Ryan, D.G.; Wüst, R.C.I.; Neele, A.E.; Speijer, D.; Denis, S.W.; et al. A Defective Pentose Phosphate Pathway Reduces Inflammatory Macrophage Responses during Hypercholesterolemia. Cell Reports 2018, 25, 2044–2052.e5. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.C.-C.; Smith, A.M.; Everts, B.; Colonna, M.; Pearce, E.L.; Schilling, J.D.; Pearce, E.J. Metabolic Reprogramming Mediated by the mTORC2-IRF4 Signaling Axis Is Essential for Macrophage Alternative Activation. Immunity 2016, 45, 817–830. [Google Scholar] [CrossRef]
- Vats, D.; Mukundan, L.; Odegaard, J.I.; Zhang, L.; Smith, K.L.; Morel, C.R.; Greaves, D.R.; Murray, P.J.; Chawla, A. Oxidative Metabolism and PGC-1β Attenuate Macrophage-Mediated Inflammation. Cell Metabolism 2006, 4, 13–24. [Google Scholar] [CrossRef]
- Huang, S.C.-C.; Everts, B.; Ivanova, Y.; O’Sullivan, D.; Nascimento, M.; Smith, A.M.; Beatty, W.; Love-Gregory, L.; Lam, W.Y.; O’Neill, C.M.; et al. Cell-Intrinsic Lysosomal Lipolysis Is Essential for Alternative Activation of Macrophages. Nat Immunol 2014, 15, 846–855. [Google Scholar] [CrossRef]
- Namgaladze, D.; Brüne, B. Fatty Acid Oxidation Is Dispensable for Human Macrophage IL-4-Induced Polarization. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids 2014, 1841, 1329–1335. [Google Scholar] [CrossRef] [PubMed]
- Nomura, M.; Liu, J.; Rovira, I.I.; Gonzalez-Hurtado, E.; Lee, J.; Wolfgang, M.J.; Finkel, T. Fatty Acid Oxidation in Macrophage Polarization. Nat Immunol 2016, 17, 216–217. [Google Scholar] [CrossRef]
- Jha, A.K.; Huang, S.C.-C.; Sergushichev, A.; Lampropoulou, V.; Ivanova, Y.; Loginicheva, E.; Chmielewski, K.; Stewart, K.M.; Ashall, J.; Everts, B.; et al. Network Integration of Parallel Metabolic and Transcriptional Data Reveals Metabolic Modules That Regulate Macrophage Polarization. Immunity 2015, 42, 419–430. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.-S.; Wang, H.; Li, X.; Chao, T.; Teav, T.; Christen, S.; Di Conza, G.; Cheng, W.-C.; Chou, C.-H.; Vavakova, M.; et al. α-Ketoglutarate Orchestrates Macrophage Activation through Metabolic and Epigenetic Reprogramming. Nat Immunol 2017, 18, 985–994. [Google Scholar] [CrossRef] [PubMed]
- Cabeza-Cabrerizo, M.; Cardoso, A.; Minutti, C.M.; Pereira Da Costa, M.; Reis E Sousa, C. Dendritic Cells Revisited. Annu. Rev. Immunol. 2021, 39, 131–166. [Google Scholar] [CrossRef] [PubMed]
- Sathaliyawala, T.; O’Gorman, W.E.; Greter, M.; Bogunovic, M.; Konjufca, V.; Hou, Z.E.; Nolan, G.P.; Miller, M.J.; Merad, M.; Reizis, B. Mammalian Target of Rapamycin Controls Dendritic Cell Development Downstream of Flt3 Ligand Signaling. Immunity 2010, 33, 597–606. [Google Scholar] [CrossRef] [PubMed]
- Woltman, A.M.; Van Der Kooij, S.W.; Coffer, P.J.; Offringa, R.; Daha, M.R.; Van Kooten, C. Rapamycin Specifically Interferes with GM-CSF Signaling in Human Dendritic Cells, Leading to Apoptosis via Increased p27KIP1 Expression. Blood 2003, 101, 1439–1445. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.; O’Brien, T.F.; Wright, G.; Yang, J.; Shin, J.; Wright, K.L.; Zhong, X.-P. Critical Role of the Tumor Suppressor Tuberous Sclerosis Complex 1 in Dendritic Cell Activation of CD4 T Cells by Promoting MHC Class II Expression via IRF4 and CIITA. The Journal of Immunology 2013, 191, 699–707. [Google Scholar] [CrossRef] [PubMed]
- Pearce, E.J.; Everts, B. Dendritic Cell Metabolism. Nat Rev Immunol 2015, 15, 18–29. [Google Scholar] [CrossRef]
- Everts, B.; Amiel, E.; Huang, S.C.-C.; Smith, A.M.; Chang, C.-H.; Lam, W.Y.; Redmann, V.; Freitas, T.C.; Blagih, J.; Van Der Windt, G.J.W.; et al. TLR-Driven Early Glycolytic Reprogramming via the Kinases TBK1-IKKɛ Supports the Anabolic Demands of Dendritic Cell Activation. Nat Immunol 2014, 15, 323–332. [Google Scholar] [CrossRef]
- Jantsch, J.; Chakravortty, D.; Turza, N.; Prechtel, A.T.; Buchholz, B.; Gerlach, R.G.; Volke, M.; Gläsner, J.; Warnecke, C.; Wiesener, M.S.; et al. Hypoxia and Hypoxia-Inducible Factor-1α Modulate Lipopolysaccharide-Induced Dendritic Cell Activation and Function. The Journal of Immunology 2008, 180, 4697–4705. [Google Scholar] [CrossRef] [PubMed]
- Carroll, K.C.; Viollet, B.; Suttles, J. AMPKα1 Deficiency Amplifies Proinflammatory Myeloid APC Activity and CD40 Signaling. Journal of Leukocyte Biology 2013, 94, 1113–1121. [Google Scholar] [CrossRef] [PubMed]
- Saeedi, P.; Petersohn, I.; Salpea, P.; Malanda, B.; Karuranga, S.; Unwin, N.; Colagiuri, S.; Guariguata, L.; Motala, A.A.; Ogurtsova, K.; et al. Global and Regional Diabetes Prevalence Estimates for 2019 and Projections for 2030 and 2045: Results from the International Diabetes Federation Diabetes Atlas, 9th Edition. Diabetes Research and Clinical Practice 2019, 157, 107843. [Google Scholar] [CrossRef] [PubMed]
- Hodgson, K.; Morris, J.; Bridson, T.; Govan, B.; Rush, C.; Ketheesan, N. Immunological Mechanisms Contributing to the Double Burden of Diabetes and Intracellular Bacterial Infections. Immunology 2015, 144, 171–185. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Barnes, G.T.; Yang, Q.; Tan, G.; Yang, D.; Chou, C.J.; Sole, J.; Nichols, A.; Ross, J.S.; Tartaglia, L.A.; et al. Chronic Inflammation in Fat Plays a Crucial Role in the Development of Obesity-Related Insulin Resistance. J. Clin. Invest. 2003, 112, 1821–1830. [Google Scholar] [CrossRef] [PubMed]
- Mooradian, A.D.; Reed, R.L.; Meredith, K.E.; Scuderi, P. Serum Levels of Tumor Necrosis Factor and IL-1α and IL-1β in Diabetic Patients. Diabetes Care 1991, 14, 63–65. [Google Scholar] [CrossRef] [PubMed]
- Ohno, Y.; Aoki, N.; Nishimura, A. In Vitro Production of Interleukin-1, Interleukin-6, and Tumor Necrosis Factor-Alpha in Insulin-Dependent Diabetes Mellitus. The Journal of Clinical Endocrinology & Metabolism 1993, 77, 1072–1077. [Google Scholar] [CrossRef]
- Reinhold, D.; Ansorge, S. Elevated Glucose Levels Stimulate Transforming Growth Factor-Β1 (TGF-Β1), Suppress Interleukin IL-2, IL-6 and IL-10 Production and DNA Synthesis in Peripheral Blood Mononuclear Cells. Horm Metab Res 1996, 28, 267–270. [Google Scholar] [CrossRef]
- Okada, M.; Kitahara, M.; Kishimoto, S.; Matsuda, T.; Hirano, T.; Kishimoto, T. IL-6/BSF-2 Functions as a Killer Helper Factor in the in Vitro Induction of Cytotoxic T Cells. J Immunol 1988, 141, 1543–1549. [Google Scholar] [CrossRef]
- Hu, R.; Xia, C.-Q.; Butfiloski, E.; Clare-Salzler, M. Effect of High Glucose on Cytokine Production by Human Peripheral Blood Immune Cells and Type I Interferon Signaling in Monocytes: Implications for the Role of Hyperglycemia in the Diabetes Inflammatory Process and Host Defense against Infection. Clinical Immunology 2018, 195, 139–148. [Google Scholar] [CrossRef]
- Kumar, M.; Roe, K.; Nerurkar, P.V.; Orillo, B.; Thompson, K.S.; Verma, S.; Nerurkar, V.R. Reduced Immune Cell Infiltration and Increased Pro-Inflammatory Mediators in the Brain of Type 2 Diabetic Mouse Model Infected with West Nile Virus. J Neuroinflammation 2014, 11, 80. [Google Scholar] [CrossRef] [PubMed]
- Lecube, A.; Pachón, G.; Petriz, J.; Hernández, C.; Simó, R. Phagocytic Activity Is Impaired in Type 2 Diabetes Mellitus and Increases after Metabolic Improvement. PLoS ONE 2011, 6, e23366. [Google Scholar] [CrossRef] [PubMed]
- Restrepo, B.I.; Twahirwa, M.; Rahbar, M.H.; Schlesinger, L.S. Phagocytosis via Complement or Fc-Gamma Receptors Is Compromised in Monocytes from Type 2 Diabetes Patients with Chronic Hyperglycemia. PLoS ONE 2014, 9, e92977. [Google Scholar] [CrossRef] [PubMed]
- Chao, W.-C.; Yen, C.-L.; Wu, Y.-H.; Chen, S.-Y.; Hsieh, C.-Y.; Chang, T.-C.; Ou, H.-Y.; Shieh, C.-C. Increased Resistin May Suppress Reactive Oxygen Species Production and Inflammasome Activation in Type 2 Diabetic Patients with Pulmonary Tuberculosis Infection. Microbes and Infection 2015, 17, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Perner, A.; Nielsen, S.E.; Rask-Madsen, J. High Glucose Impairs Superoxide Production from Isolated Blood Neutrophils. Intensive Care Med 2003, 29, 642–645. [Google Scholar] [CrossRef] [PubMed]
- Stegenga, M.E.; van der Crabben, S.N.; Blümer, R.M.E.; Levi, M.; Meijers, J.C.M.; Serlie, M.J.; Tanck, M.W.T.; Sauerwein, H.P.; van der Poll, T. Hyperglycemia Enhances Coagulation and Reduces Neutrophil Degranulation, Whereas Hyperinsulinemia Inhibits Fibrinolysis during Human Endotoxemia. Blood 2008, 112, 82–89. [Google Scholar] [CrossRef] [PubMed]
- Joshi, M.B.; Lad, A.; Bharath Prasad, A.S.; Balakrishnan, A.; Ramachandra, L.; Satyamoorthy, K. High Glucose Modulates IL-6 Mediated Immune Homeostasis through Impeding Neutrophil Extracellular Trap Formation. FEBS Letters 2013, 587, 2241–2246. [Google Scholar] [CrossRef]
- Berrou, J.; Fougeray, S.; Venot, M.; Chardiny, V.; Gautier, J.-F.; Dulphy, N.; Toubert, A.; Peraldi, M.-N. Natural Killer Cell Function, an Important Target for Infection and Tumor Protection, Is Impaired in Type 2 Diabetes. PLoS ONE 2013, 8, e62418. [Google Scholar] [CrossRef]
- Mauriello, C.T.; Hair, P.S.; Rohn, R.D.; Rister, N.S.; Krishna, N.K.; Cunnion, K.M. Hyperglycemia Inhibits Complement-Mediated Immunological Control of S. Aureus in a Rat Model of Peritonitis. Journal of Diabetes Research 2014, 2014, 1–11. [Google Scholar] [CrossRef]
- Martinez, N.; Ketheesan, N.; Martens, G.W.; West, K.; Lien, E.; Kornfeld, H. Defects in Early Cell Recruitment Contribute to the Increased Susceptibility to Respiratory Klebsiella Pneumoniae Infection in Diabetic Mice. Microbes and Infection 2016, 18, 649–655. [Google Scholar] [CrossRef]
- Gupta, S.; Maratha, A.; Siednienko, J.; Natarajan, A.; Gajanayake, T.; Hoashi, S.; Miggin, S. Analysis of Inflammatory Cytokine and TLR Expression Levels in Type 2 Diabetes with Complications. Sci Rep 2017, 7, 7633. [Google Scholar] [CrossRef] [PubMed]
- Vallerskog, T.; Martens, G.W.; Kornfeld, H. Diabetic Mice Display a Delayed Adaptive Immune Response to Mycobacterium Tuberculosis. J.I. 2010, 184, 6275–6282. [Google Scholar] [CrossRef]
- Williams, N.L.; Morris, J.L.; Rush, C.; Govan, B.L.; Ketheesan, N. Impact of Streptozotocin-Induced Diabetes on Functional Responses of Dendritic Cells and Macrophages towards Burkholderia Pseudomallei. FEMS Immunol Med Microbiol 2011, 61, 218–227. [Google Scholar] [CrossRef] [PubMed]
- Carey, I.M.; Critchley, J.A.; DeWilde, S.; Harris, T.; Hosking, F.J.; Cook, D.G. Risk of Infection in Type 1 and Type 2 Diabetes Compared With the General Population: A Matched Cohort Study. Diabetes Care 2018, 41, 513–521. [Google Scholar] [CrossRef] [PubMed]
- Critchley, J.A.; Carey, I.M.; Harris, T.; DeWilde, S.; Hosking, F.J.; Cook, D.G. Glycemic Control and Risk of Infections Among People With Type 1 or Type 2 Diabetes in a Large Primary Care Cohort Study. Diabetes Care 2018, 41, 2127–2135. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhu, X.; Ji, Y.; Li, H.; Hou, F.; Xiao, C.; Yuan, P. Increased Risk of Hepatitis B Virus Infection amongst Individuals with Diabetes Mellitus. Bioscience Reports 2019, 39, BSR20181715. [Google Scholar] [CrossRef] [PubMed]
- Kulcsar, K.A.; Coleman, C.M.; Beck, S.E.; Frieman, M.B. Comorbid Diabetes Results in Immune Dysregulation and Enhanced Disease Severity Following MERS-CoV Infection. JCI Insight 2019, 4, e131774. [Google Scholar] [CrossRef] [PubMed]
- Norouzi, M.; Norouzi, S.; Ruggiero, A.; Khan, M.S.; Myers, S.; Kavanagh, K.; Vemuri, R. Type-2 Diabetes as a Risk Factor for Severe COVID-19 Infection. Microorganisms 2021, 9, 1211. [Google Scholar] [CrossRef]
- Reiterer, M.; Rajan, M.; Gómez-Banoy, N.; Lau, J.D.; Gomez-Escobar, L.G.; Ma, L.; Gilani, A.; Alvarez-Mulett, S.; Sholle, E.T.; Chandar, V.; et al. Hyperglycemia in Acute COVID-19 Is Characterized by Insulin Resistance and Adipose Tissue Infectivity by SARS-CoV-2. Cell Metabolism 2021, 33, 2174–2188.e5. [Google Scholar] [CrossRef]
- Htun, N.S.N.; Odermatt, P.; Eze, I.C.; Boillat-Blanco, N.; D’Acremont, V.; Probst-Hensch, N. Is Diabetes a Risk Factor for a Severe Clinical Presentation of Dengue? - Review and Meta-Analysis. PLoS Negl Trop Dis 2015, 9, e0003741. [Google Scholar] [CrossRef]
- Sekaran, S.D.; Liew, Z.M.; Yam, H.C.; Raju, C.S. The Association between Diabetes and Obesity with Dengue Infections. Diabetol Metab Syndr 2022, 14, 101. [Google Scholar] [CrossRef] [PubMed]
- Després, J.-P. Body Fat Distribution and Risk of Cardiovascular Disease: An Update. Circulation 2012, 126, 1301–1313. [Google Scholar] [CrossRef]
- World Health Organisation Obesity and Overweight Available online:. Available online: https://www.who.int/news-room/fact-sheets/detail/obesity-and-overweight (accessed on 30 September 2023).
- Schwartz, M.W.; Seeley, R.J.; Zeltser, L.M.; Drewnowski, A.; Ravussin, E.; Redman, L.M.; Leibel, R.L. Obesity Pathogenesis: An Endocrine Society Scientific Statement. Endocrine Reviews 2017, 38, 267–296. [Google Scholar] [CrossRef] [PubMed]
- Krajmalnik-Brown, R.; Ilhan, Z.-E.; Kang, D.-W.; DiBaise, J.K. Effects of Gut Microbes on Nutrient Absorption and Energy Regulation. Nutr Clin Pract 2012, 27, 201–214. [Google Scholar] [CrossRef] [PubMed]
- Kershaw, E.E.; Flier, J.S. Adipose Tissue as an Endocrine Organ. The Journal of Clinical Endocrinology & Metabolism 2004, 89, 2548–2556. [Google Scholar] [CrossRef] [PubMed]
- Kanneganti, T.-D.; Dixit, V.D. Immunological Complications of Obesity. Nat Immunol 2012, 13, 707–712. [Google Scholar] [CrossRef] [PubMed]
- Weisberg, S.P.; McCann, D.; Desai, M.; Rosenbaum, M.; Leibel, R.L.; Ferrante, A.W. Obesity Is Associated with Macrophage Accumulation in Adipose Tissue. J. Clin. Invest. 2003, 112, 1796–1808. [Google Scholar] [CrossRef] [PubMed]
- Skurk, T.; Alberti-Huber, C.; Herder, C.; Hauner, H. Relationship between Adipocyte Size and Adipokine Expression and Secretion. The Journal of Clinical Endocrinology & Metabolism 2007, 92, 1023–1033. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, M.T.A.; Favelyukis, S.; Nguyen, A.-K.; Reichart, D.; Scott, P.A.; Jenn, A.; Liu-Bryan, R.; Glass, C.K.; Neels, J.G.; Olefsky, J.M. A Subpopulation of Macrophages Infiltrates Hypertrophic Adipose Tissue and Is Activated by Free Fatty Acids via Toll-like Receptors 2 and 4 and JNK-Dependent Pathways. Journal of Biological Chemistry 2007, 282, 35279–35292. [Google Scholar] [CrossRef]
- Shi, H.; Kokoeva, M.V.; Inouye, K.; Tzameli, I.; Yin, H.; Flier, J.S. TLR4 Links Innate Immunity and Fatty Acid–Induced Insulin Resistance. J. Clin. Invest. 2006, 116, 3015–3025. [Google Scholar] [CrossRef]
- Zheng, C.; Yang, Q.; Cao, J.; Xie, N.; Liu, K.; Shou, P.; Qian, F.; Wang, Y.; Shi, Y. Local Proliferation Initiates Macrophage Accumulation in Adipose Tissue during Obesity. Cell Death Dis 2016, 7, e2167–e2167. [Google Scholar] [CrossRef] [PubMed]
- Brotfain, E.; Hadad, N.; Shapira, Y.; Avinoah, E.; Zlotnik, A.; Raichel, L.; Levy, R. Neutrophil Functions in Morbidly Obese Subjects. Clinical and Experimental Immunology 2015, 181, 156–163. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.-C.; Kim, M.-S.; Pae, M.; Yamamoto, Y.; Eberlé, D.; Shimada, T.; Kamei, N.; Park, H.-S.; Sasorith, S.; Woo, J.R.; et al. Adipose Natural Killer Cells Regulate Adipose Tissue Macrophages to Promote Insulin Resistance in Obesity. Cell Metabolism 2016, 23, 685–698. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, S.; Manabe, I.; Nagasaki, M.; Eto, K.; Yamashita, H.; Ohsugi, M.; Otsu, M.; Hara, K.; Ueki, K.; Sugiura, S.; et al. CD8+ Effector T Cells Contribute to Macrophage Recruitment and Adipose Tissue Inflammation in Obesity. Nat Med 2009, 15, 914–920. [Google Scholar] [CrossRef] [PubMed]
- Cypess, A.M. Reassessing Human Adipose Tissue. N Engl J Med 2022, 386, 768–779. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Guerrero-Juarez, C.F.; Chen, S.X.; Zhang, X.; Yin, M.; Li, F.; Wu, S.; Chen, J.; Li, M.; Liu, Y.; et al. Diet-Induced Obesity Promotes Infection by Impairment of the Innate Antimicrobial Defense Function of Dermal Adipocyte Progenitors. Sci. Transl. Med. 2021, 13, eabb5280. [Google Scholar] [CrossRef] [PubMed]
- Shipman, A.R.; Millington, G.W.M. Obesity and the Skin: Obesity and the Skin. British Journal of Dermatology 2011, 165, 743–750. [Google Scholar] [CrossRef] [PubMed]
- Pugliese, G.; Liccardi, A.; Graziadio, C.; Barrea, L.; Muscogiuri, G.; Colao, A. Obesity and Infectious Diseases: Pathophysiology and Epidemiology of a Double Pandemic Condition. Int J Obes 2022, 46, 449–465. [Google Scholar] [CrossRef]
- Kaspersen, K.A.; Pedersen, O.B.; Petersen, M.S.; Hjalgrim, H.; Rostgaard, K.; Møller, B.K.; Juul-Sørensen, C.; Kotzé, S.; Dinh, K.M.; Erikstrup, L.T.; et al. Obesity and Risk of Infection: Results from the Danish Blood Donor Study. Epidemiology 2015, 26, 580–589. [Google Scholar] [CrossRef]
- Las Heras, V.; Clooney, A.G.; Ryan, F.J.; Cabrera-Rubio, R.; Casey, P.G.; Hueston, C.M.; Pinheiro, J.; Rudkin, J.K.; Melgar, S.; Cotter, P.D.; et al. Short-Term Consumption of a High-Fat Diet Increases Host Susceptibility to Listeria Monocytogenes Infection. Microbiome 2019, 7, 7. [Google Scholar] [CrossRef]
- Mancuso, P.; Gottschalk, A.; Phare, S.M.; Peters-Golden, M.; Lukacs, N.W.; Huffnagle, G.B. Leptin-Deficient Mice Exhibit Impaired Host Defense in Gram-Negative Pneumonia. J Immunol 2002, 168, 4018–4024. [Google Scholar] [CrossRef] [PubMed]
- Wieland, C.W.; Florquin, S.; Chan, E.D.; Leemans, J.C.; Weijer, S.; Verbon, A.; Fantuzzi, G.; van der Poll, T. Pulmonary Mycobacterium Tuberculosis Infection in Leptin-Deficient Ob/Ob Mice. International Immunology 2005, 17, 1399–1408. [Google Scholar] [CrossRef] [PubMed]
- Louie, J.K.; Acosta, M.; Samuel, M.C.; Schechter, R.; Vugia, D.J.; Harriman, K.; Matyas, B.T. the California Pandemic (H1N1) Working Group A Novel Risk Factor for a Novel Virus: Obesity and 2009 Pandemic Influenza A (H1N1). Clinical Infectious Diseases 2011, 52, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Popkin, B.M.; Du, S.; Green, W.D.; Beck, M.A.; Algaith, T.; Herbst, C.H.; Alsukait, R.F.; Alluhidan, M.; Alazemi, N.; Shekar, M. Individuals with Obesity and COVID-19: A Global Perspective on the Epidemiology and Biological Relationships. Obesity Reviews 2020, 21. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, E.A.; Sheridan, P.A.; Beck, M.A. Diet-Induced Obesity Impairs the T Cell Memory Response to Influenza Virus Infection. J.I. 2010, 184, 3127–3133. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.G.; Sheridan, P.A.; Tseng, R.J.; Sheridan, J.F.; Beck, M.A. Selective Impairment in Dendritic Cell Function and Altered Antigen-Specific CD8 + T-Cell Responses in Diet-Induced Obese Mice Infected with Influenza Virus. Immunology 2009, 126, 268–279. [Google Scholar] [CrossRef]
- Geerling, E.; Stone, E.T.; Steffen, T.L.; Hassert, M.; Brien, J.D.; Pinto, A.K. Obesity Enhances Disease Severity in Female Mice Following West Nile Virus Infection. Front. Immunol. 2021, 12, 739025. [Google Scholar] [CrossRef]
- Kim, Y.-H.; Kim, J.-K.; Kim, D.-J.; Nam, J.-H.; Shim, S.-M.; Choi, Y.-K.; Lee, C.-H.; Poo, H. Diet-Induced Obesity Dramatically Reduces the Efficacy of a 2009 Pandemic H1N1 Vaccine in a Mouse Model. The Journal of Infectious Diseases 2012, 205, 244–251. [Google Scholar] [CrossRef]
- Weber, D.; Rutala, W.; Samsa, G.; Bradshaw, S.; Lemon, S. Impaired Immunogenicity of Hepatitis B Vaccine in Obese Persons. New England Journal of Medicine 1986, 314, 1393–1393. [Google Scholar] [CrossRef]
- Pape, K.; Ryttergaard, L.; Rotevatn, T.A.; Nielsen, B.J.; Torp-Pedersen, C.; Overgaard, C.; Bøggild, H. Leisure-Time Physical Activity and the Risk of Suspected Bacterial Infections. Medicine & Science in Sports & Exercise 2016, 48, 1737–1744. [Google Scholar] [CrossRef]
- Fondell, E.; Lagerros, Y.T.; Sundberg, C.J.; Lekander, M.; Bälter, O.; Rothman, K.J.; Bälter, K. Physical Activity, Stress, and Self-Reported Upper Respiratory Tract Infection. Medicine & Science in Sports & Exercise 2011, 43, 272–279. [Google Scholar] [CrossRef]
- Nieman, D.C.; Henson, D.A.; Austin, M.D.; Brown, V.A. Immune Response to a 30-Minute Walk: Medicine & Science in Sports & Exercise 2005, 37, 57–62. [CrossRef]
- Adams, G.R.; Zaldivar, F.P.; Nance, D.M.; Kodesh, E.; Radom-Aizik, S.; Cooper, D.M. Exercise and Leukocyte Interchange among Central Circulation, Lung, Spleen, and Muscle. Brain, Behavior, and Immunity 2011, 25, 658–666. [Google Scholar] [CrossRef]
- Fernández-Lázaro, D.; González-Bernal, J.J.; Sánchez-Serrano, N.; Navascués, L.J.; Ascaso-del-Río, A.; Mielgo-Ayuso, J. Physical Exercise as a Multimodal Tool for COVID-19: Could It Be Used as a Preventive Strategy? International Journal of Environmental Research and Public Health 2020, 17, 8496. [Google Scholar] [CrossRef] [PubMed]
- Nieman, D.C. Immune Response to Heavy Exertion. Journal of Applied Physiology 1997, 82, 1385–1394. [Google Scholar] [CrossRef] [PubMed]
- Northoff, H.; Berg, A. Immunologic Mediators as Parameters of the Reaction to Strenuous Exercise. Int J Sports Med 1991, 12, S9–S15. [Google Scholar] [CrossRef]
- Nieman, D.C.; Wentz, L.M. The Compelling Link between Physical Activity and the Body’s Defense System. Journal of Sport and Health Science 2019, 8, 201–217. [Google Scholar] [CrossRef] [PubMed]
- Clancy, R.L. Reversal in Fatigued Athletes of a Defect in Interferon Secretion after Administration of Lactobacillus Acidophilus. British Journal of Sports Medicine 2006, 40, 351–354. [Google Scholar] [CrossRef] [PubMed]
- Gleeson, M.; Pyne, D.B.; P. Austin, J.; Lynn Francis, J.; Clancy, R.L.; Mcdonald, W.A.; Fricker, P.A. Epstein-Barr Virus Reactivation and Upper-Respiratory Illness in Elite Swimmers: . Medicine & Science in Sports & Exercise 2002, 34, 411–417. [CrossRef]
- Reid, V.L. Clinical Investigation of Athletes with Persistent Fatigue and/or Recurrent Infections. British Journal of Sports Medicine 2004, 38, 42–45. [Google Scholar] [CrossRef]
- Barton, W.; Penney, N.C.; Cronin, O.; Garcia-Perez, I.; Molloy, M.G.; Holmes, E.; Shanahan, F.; Cotter, P.D.; O’Sullivan, O. The Microbiome of Professional Athletes Differs from That of More Sedentary Subjects in Composition and Particularly at the Functional Metabolic Level. Gut, 2017; gutjnl-2016-313627. [Google Scholar] [CrossRef]
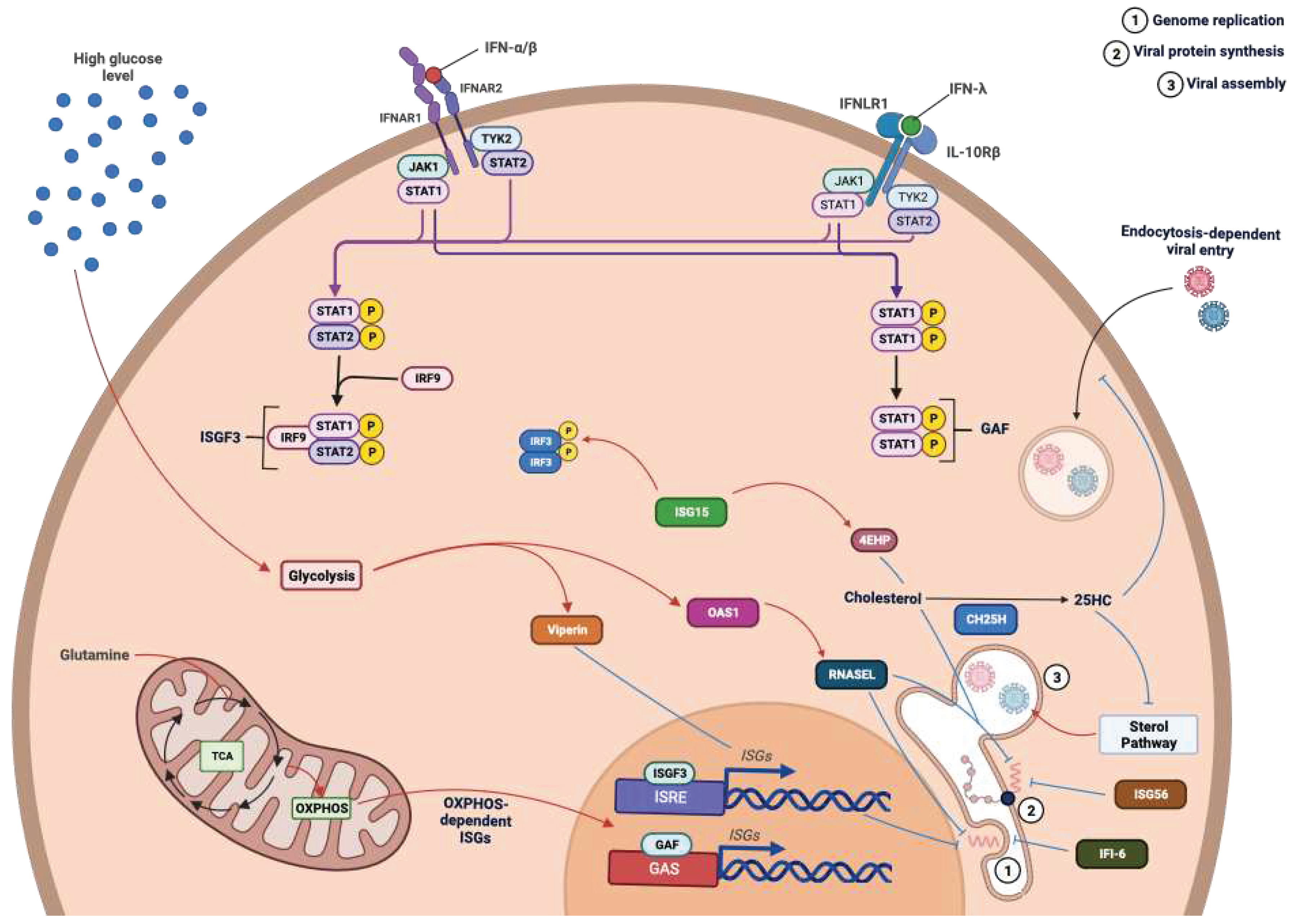
- Lazear, H.M.; Schoggins, J.W.; Diamond, M.S. Shared and Distinct Functions of Type I and Type III Interferons. Immunity 2019, 50, 907–923. [Google Scholar] [CrossRef]
- Hornung, V.; Ellegast, J.; Kim, S.; Brzózka, K.; Jung, A.; Kato, H.; Poeck, H.; Akira, S.; Conzelmann, K.-K.; Schlee, M.; et al. 5’-Triphosphate RNA Is the Ligand for RIG-I. Science 2006, 314, 994–997. [Google Scholar] [CrossRef] [PubMed]
- Pichlmair, A.; Schulz, O.; Tan, C.P.; Näslund, T.I.; Liljeström, P.; Weber, F.; Reis E Sousa, C. RIG-I-Mediated Antiviral Responses to Single-Stranded RNA Bearing 5’-Phosphates. Science 2006, 314, 997–1001. [Google Scholar] [CrossRef] [PubMed]
- Gitlin, L.; Barchet, W.; Gilfillan, S.; Cella, M.; Beutler, B.; Flavell, R.A.; Diamond, M.S.; Colonna, M. Essential Role of Mda-5 in Type I IFN Responses to Polyriboinosinic:Polyribocytidylic Acid and Encephalomyocarditis Picornavirus. Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 8459–8464. [Google Scholar] [CrossRef]
- Kato, H.; Takeuchi, O.; Sato, S.; Yoneyama, M.; Yamamoto, M.; Matsui, K.; Uematsu, S.; Jung, A.; Kawai, T.; Ishii, K.J.; et al. Differential Roles of MDA5 and RIG-I Helicases in the Recognition of RNA Viruses. Nature 2006, 441, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Kowalinski, E.; Lunardi, T.; McCarthy, A.A.; Louber, J.; Brunel, J.; Grigorov, B.; Gerlier, D.; Cusack, S. Structural Basis for the Activation of Innate Immune Pattern-Recognition Receptor RIG-I by Viral RNA. Cell 2011, 147, 423–435. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.-G.; Wang, Y.-Y.; Han, K.-J.; Li, L.-Y.; Zhai, Z.; Shu, H.-B. VISA Is an Adapter Protein Required for Virus-Triggered IFN-β Signaling. Molecular Cell 2005, 19, 727–740. [Google Scholar] [CrossRef]
- Paz, S.; Vilasco, M.; Werden, S.J.; Arguello, M.; Joseph-Pillai, D.; Zhao, T.; Nguyen, T.L.-A.; Sun, Q.; Meurs, E.F.; Lin, R.; et al. A Functional C-Terminal TRAF3-Binding Site in MAVS Participates in Positive and Negative Regulation of the IFN Antiviral Response. Cell Res 2011, 21, 895–910. [Google Scholar] [CrossRef]
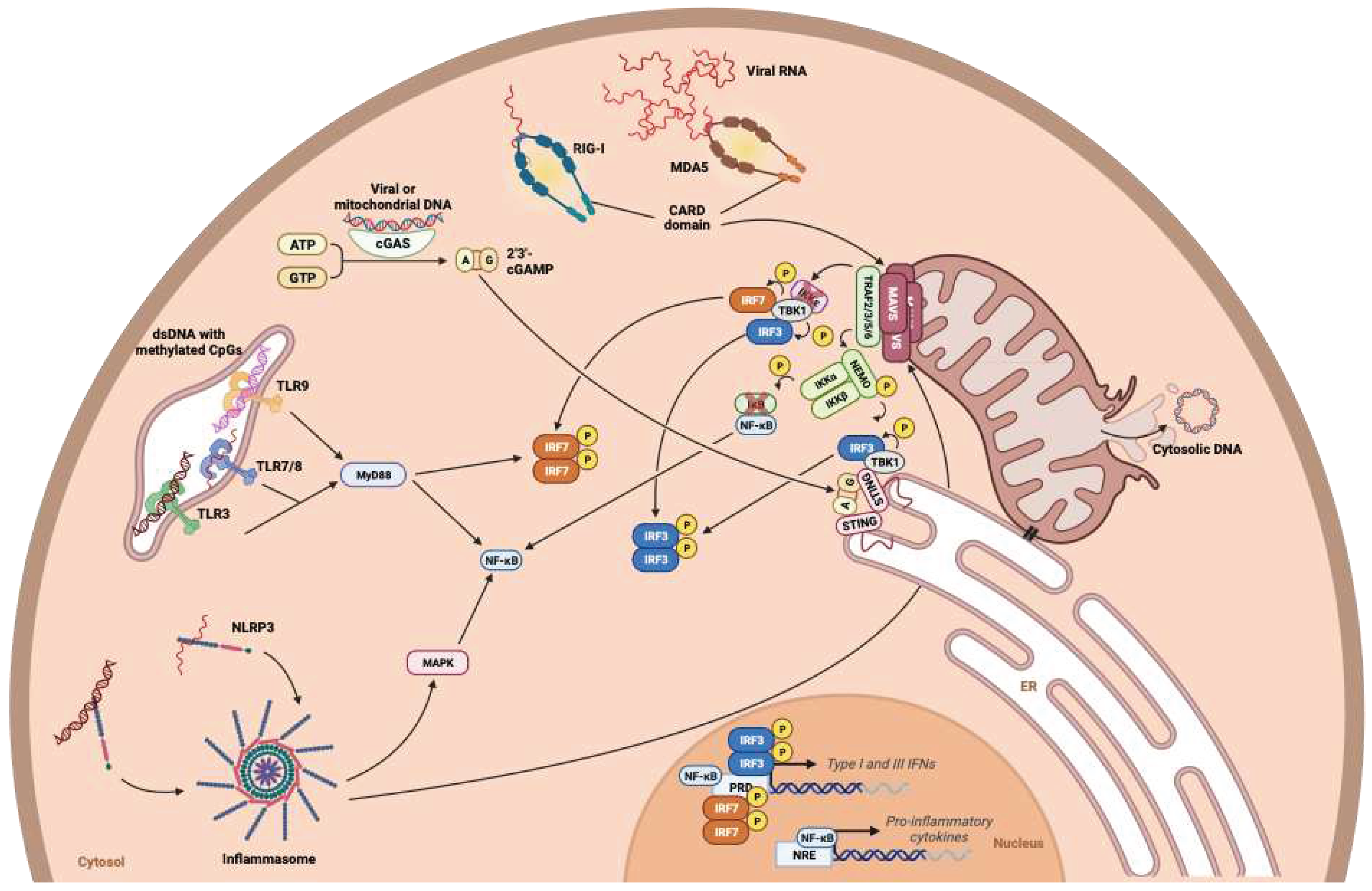
- Motwani, M.; Pesiridis, S.; Fitzgerald, K.A. DNA Sensing by the cGAS–STING Pathway in Health and Disease. Nat Rev Genet 2019, 20, 657–674. [Google Scholar] [CrossRef] [PubMed]
- Chow, K.T.; Gale, M.; Loo, Y.-M. RIG-I and Other RNA Sensors in Antiviral Immunity. Annu. Rev. Immunol. 2018, 36, 667–694. [Google Scholar] [CrossRef]
- Swanson, K.V.; Deng, M.; Ting, J.P.-Y. The NLRP3 Inflammasome: Molecular Activation and Regulation to Therapeutics. Nat Rev Immunol 2019, 19, 477–489. [Google Scholar] [CrossRef]
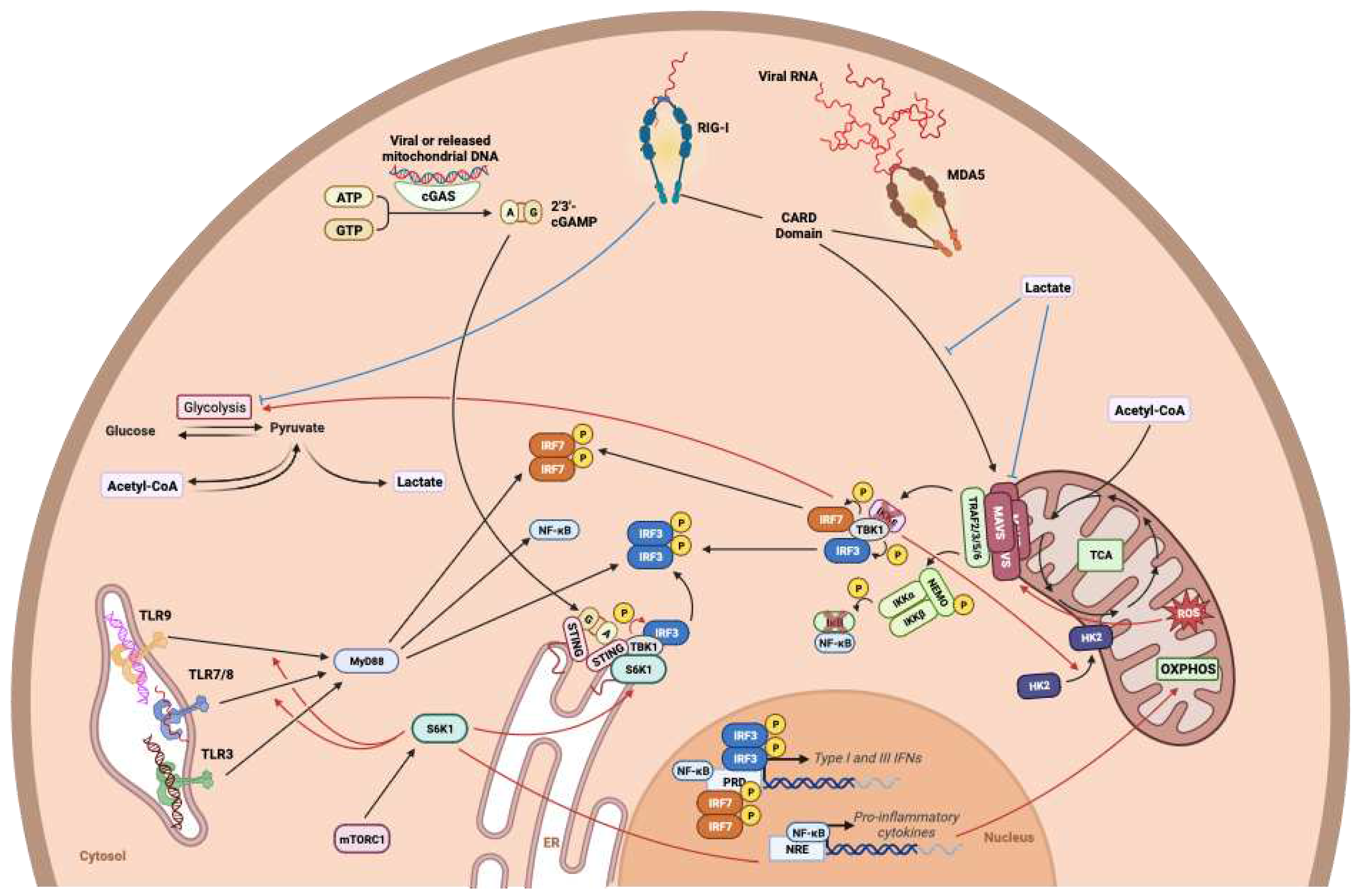
- Zhang, W.; Wang, G.; Xu, Z.-G.; Tu, H.; Hu, F.; Dai, J.; Chang, Y.; Chen, Y.; Lu, Y.; Zeng, H.; et al. Lactate Is a Natural Suppressor of RLR Signaling by Targeting MAVS. Cell 2019, 178, 176–189.e15. [Google Scholar] [CrossRef] [PubMed]
- Buskiewicz, I.A.; Montgomery, T.; Yasewicz, E.C.; Huber, S.A.; Murphy, M.P.; Hartley, R.C.; Kelly, R.; Crow, M.K.; Perl, A.; Budd, R.C.; et al. Reactive Oxygen Species Induce Virus-Independent MAVS Oligomerization in Systemic Lupus Erythematosus. Sci. Signal. 2016, 9. [Google Scholar] [CrossRef] [PubMed]
- Nobre, L.; Wise, D.; Ron, D.; Volmer, R. Modulation of Innate Immune Signalling by Lipid-Mediated MAVS Transmembrane Domain Oligomerization. PLoS ONE 2015, 10, e0136883. [Google Scholar] [CrossRef]
- Imanishi, T.; Unno, M.; Kobayashi, W.; Yoneda, N.; Matsuda, S.; Ikeda, K.; Hoshii, T.; Hirao, A.; Miyake, K.; Barber, G.N.; et al. Reciprocal Regulation of STING and TCR Signaling by mTORC1 for T-Cell Activation and Function. Life Sci. Alliance 2019, 2, e201800282. [Google Scholar] [CrossRef] [PubMed]
- Shum, M.; Bellmann, K.; St-Pierre, P.; Marette, A. Pharmacological Inhibition of S6K1 Increases Glucose Metabolism and Akt Signalling in Vitro and in Diet-Induced Obese Mice. Diabetologia 2016, 59, 592–603. [Google Scholar] [CrossRef] [PubMed]
- Shum, M.; Houde, V.P.; Bellemare, V.; Junges Moreira, R.; Bellmann, K.; St-Pierre, P.; Viollet, B.; Foretz, M.; Marette, A. Inhibition of Mitochondrial Complex 1 by the S6K1 Inhibitor PF-4708671 Partly Contributes to Its Glucose Metabolic Effects in Muscle and Liver Cells. Journal of Biological Chemistry 2019, 294, 12250–12260. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Alain, T.; Szretter, K.J.; Stephenson, K.; Pol, J.G.; Atherton, M.J.; Hoang, H.-D.; Fonseca, B.D.; Zakaria, C.; Chen, L.; et al. S6K-STING Interaction Regulates Cytosolic DNA–Mediated Activation of the Transcription Factor IRF3. Nat Immunol 2016, 17, 514–522. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.; Manicassamy, S.; Tang, H.; Kasturi, S.P.; Pirani, A.; Murthy, N.; Pulendran, B. Toll-like Receptor–Mediated Induction of Type I Interferon in Plasmacytoid Dendritic Cells Requires the Rapamycin-Sensitive PI(3)K-mTOR-p70S6K Pathway. Nat Immunol 2008, 9, 1157–1164. [Google Scholar] [CrossRef]
- Schoggins, J.W. Interferon-Stimulated Genes: What Do They All Do? Annu. Rev. Virol. 2019, 6, 567–584. [Google Scholar] [CrossRef]
- Schoggins, J.W.; MacDuff, D.A.; Imanaka, N.; Gainey, M.D.; Shrestha, B.; Eitson, J.L.; Mar, K.B.; Richardson, R.B.; Ratushny, A.V.; Litvak, V.; et al. Pan-Viral Specificity of IFN-Induced Genes Reveals New Roles for cGAS in Innate Immunity. Nature 2014, 505, 691–695. [Google Scholar] [CrossRef]
- Kane, M.; Yadav, S.S.; Bitzegeio, J.; Kutluay, S.B.; Zang, T.; Wilson, S.J.; Schoggins, J.W.; Rice, C.M.; Yamashita, M.; Hatziioannou, T.; et al. MX2 Is an Interferon-Induced Inhibitor of HIV-1 Infection. Nature 2013, 502, 563–566. [Google Scholar] [CrossRef] [PubMed]
- Kochs, G.; Janzen, C.; Hohenberg, H.; Haller, O. Antivirally Active MxA Protein Sequesters La Crosse Virus Nucleocapsid Protein into Perinuclear Complexes. Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 3153–3158. [Google Scholar] [CrossRef] [PubMed]
- Feeley, E.M.; Sims, J.S.; John, S.P.; Chin, C.R.; Pertel, T.; Chen, L.-M.; Gaiha, G.D.; Ryan, B.J.; Donis, R.O.; Elledge, S.J.; et al. IFITM3 Inhibits Influenza A Virus Infection by Preventing Cytosolic Entry. PLoS Pathog 2011, 7, e1002337. [Google Scholar] [CrossRef] [PubMed]
- Huang, I.-C.; Bailey, C.C.; Weyer, J.L.; Radoshitzky, S.R.; Becker, M.M.; Chiang, J.J.; Brass, A.L.; Ahmed, A.A.; Chi, X.; Dong, L.; et al. Distinct Patterns of IFITM-Mediated Restriction of Filoviruses, SARS Coronavirus, and Influenza A Virus. PLoS Pathog 2011, 7, e1001258. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, C.; Woodward, J.; Lau, D.T.-Y.; Barnes, A.; Joyce, M.; McFarlane, N.; McKeating, J.A.; Tyrrell, D.L.; Gale, M. IFITM1 Is a Tight Junction Protein That Inhibits Hepatitis C Virus Entry. Hepatology 2013, 57, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Wee, Y.S.; Roundy, K.M.; Weis, J.J.; Weis, J.H. Interferon-Inducible Transmembrane Proteins of the Innate Immune Response Act as Membrane Organizers by Influencing Clathrin and v-ATPase Localization and Function. Innate Immun 2012, 18, 834–845. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Deng, Y.-Q.; Wang, S.; Ma, F.; Aliyari, R.; Huang, X.-Y.; Zhang, N.-N.; Watanabe, M.; Dong, H.-L.; Liu, P.; et al. 25-Hydroxycholesterol Protects Host against Zika Virus Infection and Its Associated Microcephaly in a Mouse Model. Immunity 2017, 46, 446–456. [Google Scholar] [CrossRef]
- Bordier, B.B.; Marion, P.L.; Ohashi, K.; Kay, M.A.; Greenberg, H.B.; Casey, J.L.; Glenn, J.S. A Prenylation Inhibitor Prevents Production of Infectious Hepatitis Delta Virus Particles. J Virol 2002, 76, 10465–10472. [Google Scholar] [CrossRef] [PubMed]
- Bordier, B.B.; Ohkanda, J.; Liu, P.; Lee, S.-Y.; Salazar, F.H.; Marion, P.L.; Ohashi, K.; Meuse, L.; Kay, M.A.; Casey, J.L.; et al. In Vivo Antiviral Efficacy of Prenylation Inhibitors against Hepatitis Delta Virus. J. Clin. Invest. 2003, 112, 407–414. [Google Scholar] [CrossRef]
- Chang, T.Y.; Limanek, J.S. Regulation of Cytosolic Acetoacetyl Coenzyme A Thiolase, 3-Hydroxy-3-Methylglutaryl Coenzyme A Synthase, 3-Hydroxy-3-Methylglutaryl Coenzyme A Reductase, and Mevalonate Kinase by Low Density Lipoprotein and by 25-Hydroxycholesterol in Chinese Hamster Ovary Cells. Journal of Biological Chemistry 1980, 255, 7787–7795. [Google Scholar] [CrossRef]
- Kandutsch, A.A.; Chen, H.W.; Heiniger, H.-J. Biological Activity of Some Oxygenated Sterols. Science 1978, 201, 498–501. [Google Scholar] [CrossRef] [PubMed]
- Guo, J. A New Pathway of Translational Regulation Mediated by Eukaryotic Initiation Factor 3. The EMBO Journal 2000, 19, 6891–6899. [Google Scholar] [CrossRef] [PubMed]
- Hui, D.J.; Bhasker, C.R.; Merrick, W.C.; Sen, G.C. Viral Stress-Inducible Protein P56 Inhibits Translation by Blocking the Interaction of eIF3 with the Ternary Complex eIF2·GTP·Met-tRNAi. Journal of Biological Chemistry 2003, 278, 39477–39482. [Google Scholar] [CrossRef] [PubMed]
- Pindel, A.; Sadler, A. The Role of Protein Kinase R in the Interferon Response. Journal of Interferon & Cytokine Research 2011, 31, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Terenzi, F.; Hui, D.J.; Merrick, W.C.; Sen, G.C. Distinct Induction Patterns and Functions of Two Closely Related Interferon-Inducible Human Genes, ISG54 and ISG56. Journal of Biological Chemistry 2006, 281, 34064–34071. [Google Scholar] [CrossRef] [PubMed]
- Fleith, R.C.; Mears, H.V.; Leong, X.Y.; Sanford, T.J.; Emmott, E.; Graham, S.C.; Mansur, D.S.; Sweeney, T.R. IFIT3 and IFIT2/3 Promote IFIT1-Mediated Translation Inhibition by Enhancing Binding to Non-Self RNA. Nucleic Acids Research 2018, 46, 5269–5285. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Sweeney, T.R.; Skabkin, M.A.; Skabkina, O.V.; Hellen, C.U.T.; Pestova, T.V. Inhibition of Translation by IFIT Family Members Is Determined by Their Ability to Interact Selectively with the 5′-Terminal Regions of Cap0-, Cap1- and 5′ppp- mRNAs. Nucleic Acids Research 2014, 42, 3228–3245. [Google Scholar] [CrossRef]
- Durfee, L.A.; Lyon, N.; Seo, K.; Huibregtse, J.M. The ISG15 Conjugation System Broadly Targets Newly Synthesized Proteins: Implications for the Antiviral Function of ISG15. Molecular Cell 2010, 38, 722–732. [Google Scholar] [CrossRef]
- Okumura, F.; Zou, W.; Zhang, D.-E. ISG15 Modification of the eIF4E Cognate 4EHP Enhances Cap Structure-Binding Activity of 4EHP. Genes Dev. 2007, 21, 255–260. [Google Scholar] [CrossRef]
- Shi, H.-X.; Yang, K.; Liu, X.; Liu, X.-Y.; Wei, B.; Shan, Y.-F.; Zhu, L.-H.; Wang, C. Positive Regulation of Interferon Regulatory Factor 3 Activation by Herc5 via ISG15 Modification. Molecular and Cellular Biology 2010, 30, 2424–2436. [Google Scholar] [CrossRef]
- Richardson, R.B.; Ohlson, M.B.; Eitson, J.L.; Kumar, A.; McDougal, M.B.; Boys, I.N.; Mar, K.B.; De La Cruz-Rivera, P.C.; Douglas, C.; Konopka, G.; et al. A CRISPR Screen Identifies IFI6 as an ER-Resident Interferon Effector That Blocks Flavivirus Replication. Nat Microbiol 2018, 3, 1214–1223. [Google Scholar] [CrossRef] [PubMed]
- Lebeau, G.; El Safadi, D.; Paulo-Ramos, A.; Hoareau, M.; Desprès, P.; Krejbich-Trotot, P.; Chouchou, F.; Roche, M.; Viranaicken, W. The Efficient Antiviral Response of A549 Cells Is Enhanced When Mitochondrial Respiration Is Promoted. Pathogens 2022, 11, 1168. [Google Scholar] [CrossRef] [PubMed]
- Lebeau, G.; Paulo-Ramos, A.; Hoarau, M.; El Safadi, D.; Meilhac, O.; Krejbich-Trotot, P.; Roche, M.; Viranaicken, W. Metabolic Dependency Shapes Bivalent Antiviral Response in Host Cells: The Role of Glutamine, Immunology. 2023.
- Reslan, A.; Haddad, J.G.; Desprès, P.; Bascands, J.-L.; Gadea, G. High Glucose Induces in HK2 Kidney Cells an IFN–Dependent ZIKV Antiviral Status Fueled by Viperin. Biomedicines 2022, 10, 1577. [Google Scholar] [CrossRef] [PubMed]
- Koufaris, C.; Nicolaidou, V. Glutamine Addiction in Virus-Infected Mammalian Cells: A Target of the Innate Immune System? Medical Hypotheses 2021, 153, 110620. [Google Scholar] [CrossRef]
- Allonso, D.; Andrade, I.S.; Conde, J.N.; Coelho, D.R.; Rocha, D.C.P.; Da Silva, M.L.; Ventura, G.T.; Silva, E.M.; Mohana-Borges, R. Dengue Virus NS1 Protein Modulates Cellular Energy Metabolism by Increasing Glyceraldehyde-3-Phosphate Dehydrogenase Activity. J Virol 2015, 89, 11871–11883. [Google Scholar] [CrossRef] [PubMed]
- Fontaine, K.A.; Sanchez, E.L.; Camarda, R.; Lagunoff, M. Dengue Virus Induces and Requires Glycolysis for Optimal Replication. J Virol 2015, 89, 2358–2366. [Google Scholar] [CrossRef]
- Heaton, N.S.; Perera, R.; Berger, K.L.; Khadka, S.; LaCount, D.J.; Kuhn, R.J.; Randall, G. Dengue Virus Nonstructural Protein 3 Redistributes Fatty Acid Synthase to Sites of Viral Replication and Increases Cellular Fatty Acid Synthesis. Proc. Natl. Acad. Sci. U.S.A. 2010, 107, 17345–17350. [Google Scholar] [CrossRef] [PubMed]
- Ledur, P.F.; Karmirian, K.; Pedrosa, C.D.S.G.; Souza, L.R.Q.; Assis-de-Lemos, G.; Martins, T.M.; Ferreira, J.D.C.C.G.; De Azevedo Reis, G.F.; Silva, E.S.; Silva, D.; et al. Zika Virus Infection Leads to Mitochondrial Failure, Oxidative Stress and DNA Damage in Human iPSC-Derived Astrocytes. Sci Rep 2020, 10, 1218. [Google Scholar] [CrossRef] [PubMed]
- Yau, C.; Low, J.Z.H.; Gan, E.S.; Kwek, S.S.; Cui, L.; Tan, H.C.; Mok, D.Z.L.; Chan, C.Y.Y.; Sessions, O.M.; Watanabe, S.; et al. Dysregulated Metabolism Underpins Zika-Virus-Infection-Associated Impairment in Fetal Development. Cell Reports 2021, 37, 110118. [Google Scholar] [CrossRef]
- Sahoo, B.R.; Crook, A.A.; Pattnaik, A.; Torres-Gerena, A.D.; Khalimonchuk, O.; Powers, R.; Franco, R.; Pattnaik, A.K. Redox Regulation and Metabolic Dependency of Zika Virus Replication: Inhibition by Nrf2-Antioxidant Response and NAD(H) Antimetabolites. J Virol 2023, 97, e01363–22. [Google Scholar] [CrossRef]
- Anzinger, J.J.; Butterfield, T.R.; Gouillou, M.; McCune, J.M.; Crowe, S.M.; Palmer, C.S. Glut1 Expression Level on Inflammatory Monocytes Is Associated With Markers of Cardiovascular Disease Risk in HIV-Infected Individuals. JAIDS Journal of Acquired Immune Deficiency Syndromes 2018, 77, e28–e30. [Google Scholar] [CrossRef] [PubMed]
- Loisel-Meyer, S.; Swainson, L.; Craveiro, M.; Oburoglu, L.; Mongellaz, C.; Costa, C.; Martinez, M.; Cosset, F.-L.; Battini, J.-L.; Herzenberg, L.A.; et al. Glut1-Mediated Glucose Transport Regulates HIV Infection. Proc. Natl. Acad. Sci. U.S.A. 2012, 109, 2549–2554. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Jia, L.; Lin, W.; Yip, Y.L.; Lo, K.W.; Lau, V.M.Y.; Zhu, D.; Tsang, C.M.; Zhou, Y.; Deng, W.; et al. Epstein-Barr Virus-Encoded Latent Membrane Protein 1 Upregulates Glucose Transporter 1 Transcription via the mTORC1/NF-κB Signaling Pathways. J Virol 2017, 91, e02168–16. [Google Scholar] [CrossRef] [PubMed]
- Meade, N.; Furey, C.; Li, H.; Verma, R.; Chai, Q.; Rollins, M.G.; DiGiuseppe, S.; Naghavi, M.H.; Walsh, D. Poxviruses Evade Cytosolic Sensing through Disruption of an mTORC1-mTORC2 Regulatory Circuit. Cell 2018, 174, 1143–1157.e17. [Google Scholar] [CrossRef] [PubMed]
- Liang, Q.; Luo, Z.; Zeng, J.; Chen, W.; Foo, S.-S.; Lee, S.-A.; Ge, J.; Wang, S.; Goldman, S.A.; Zlokovic, B.V.; et al. Zika Virus NS4A and NS4B Proteins Deregulate Akt-mTOR Signaling in Human Fetal Neural Stem Cells to Inhibit Neurogenesis and Induce Autophagy. Cell Stem Cell 2016, 19, 663–671. [Google Scholar] [CrossRef] [PubMed]
- Angin, M.; Volant, S.; Passaes, C.; Lecuroux, C.; Monceaux, V.; Dillies, M.-A.; Valle-Casuso, J.C.; Pancino, G.; Vaslin, B.; Le Grand, R.; et al. Metabolic Plasticity of HIV-Specific CD8+ T Cells Is Associated with Enhanced Antiviral Potential and Natural Control of HIV-1 Infection. Nat Metab 2019, 1, 704–716. [Google Scholar] [CrossRef] [PubMed]
- Chatel-Chaix, L.; Cortese, M.; Romero-Brey, I.; Bender, S.; Neufeldt, C.J.; Fischl, W.; Scaturro, P.; Schieber, N.; Schwab, Y.; Fischer, B.; et al. Dengue Virus Perturbs Mitochondrial Morphodynamics to Dampen Innate Immune Responses. Cell Host & Microbe 2016, 20, 342–356. [Google Scholar] [CrossRef]
- Van Valen, L. A New Evolutionary Law. EVOL. THEORY 1973, 1, 1–30. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).