1. Introduction
With the recent development of diagnostic and therapeutic techniques for breast cancer, the prognosis of patients with early-stage disease has greatly improved. However, approximately 20% of patients with early-stage invasive breast cancer still have a poor prognosis due to recurrence [
1]. Therefore, it is necessary to refine the prognostic and predictive factors in breast cancer.
Histological grade, which is one of the main prognostic factors in breast cancer [
2,
3], is generally classified into three grades based on three morphological features: nuclear atypia (morphological characteristics of individual cancer cells), tubule formation (morphological characteristics of the entire cancer tissue), and mitotic activity (proliferative potential of cancer cells). The Nottingham Prognostic Index is a widely used prognostic tool for patients with breast cancer, which is calculated using tumor size, histological grade, and lymph node stage. Histological grade was included in the recent staging system for breast cancer. Previous studies have shown that this histological grade classification is effective in predicting drug response and prognosis in breast cancer [
4,
5]. However, the molecular biological mechanism defining the histological grade remains unclear. Therefore, translational research is needed to elucidate the molecular mechanisms underlying such morphological features and identify genes predictive of histological grade using comprehensive gene expression data of breast cancer.
MicroRNAs (miRNAs) are single-stranded nonprotein-coding RNAs that function as critical posttranscriptional gene regulators. miRNAs negatively regulate gene expression by binding to their selective messenger RNAs, leading to translational repression [
6]. Aberrant expression of miRNAs has been reported to be associated with the development of various human diseases, including breast cancer. Although individual miRNA has been reported to be overexpressed in high-grade breast cancer, miRNA expression profiling in relation to the histological grading of breast cancer remains unclear. Discovering miRNAs related to histological grade that defines the proliferative and metastatic potentials of breast cancer can help not only discover prognostic factors in breast cancer but also develop new early diagnostic tools for patients with early-stage breast cancer and new therapeutic targets.
Thus, this study aimed to identify miRNAs differentially expressed between histological grades using comprehensive miRNA analysis data obtained by next-generation sequencing in early-stage breast cancer.
2. Results
2.1. Identification of miRNAs Associated with Breast Cancer
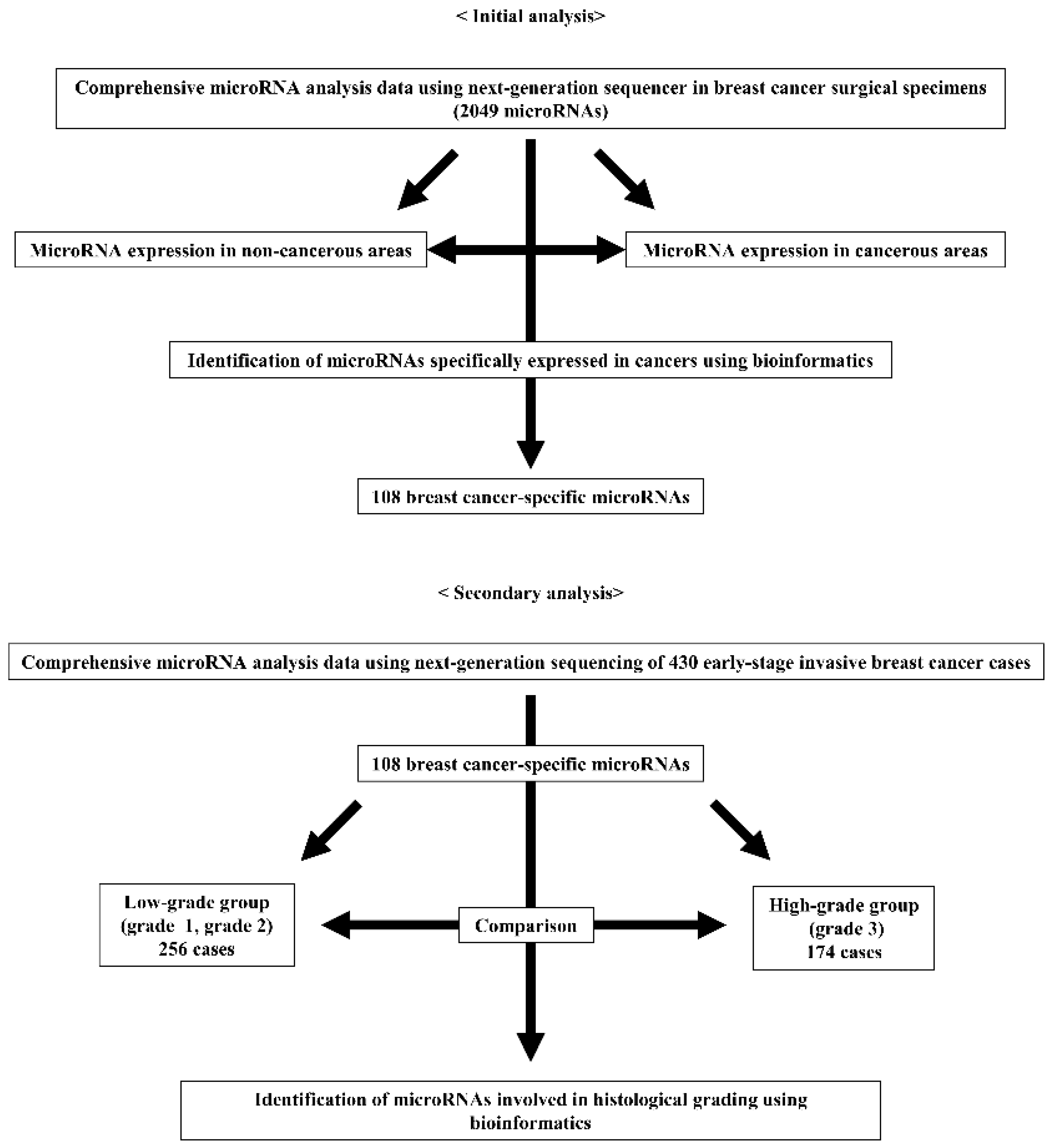
miRNAs are continuously involved in the transformation of normal cells into cancer cells. Differences in miRNAs were compared between normal tissues around the cancer and cancer cells to focus on this entire carcinogenetic process. Comparison of the RNA-seq data between normal breast and breast cancer tissues using the edgeR method showed that 108 miRNAs were significantly differentially expressed (false discovery rate [FDR] < 0.05).
Table S1 shows a list of these 108 miRNAs.
2.2. Clinicopathological and Prognostic Significance of Histological Grade in The Cancer Genome Atlas (TCGA) Cohort
Table 1 shows the clinicopathological characteristics of the TCGA cohort in this study, and
Figure 1 shows the distribution of histological grade. No patient received preoperative chemotherapy in this cohort. Of the 430 breast cancer cases in the TCGA cohort, 256 (59.5%) were of histological grades 1 and 2, and 174 (40.5%) were of histological grade 3. In this cohort, grade 3 was significantly associated with positive lymph node metastasis (p = 0.011) and estrogen receptor (ER) negativity (p < 0.0001;
Table S2). Patients with grade 3 breast cancer tended to have worse survival rates than those with grade 1 and 2 breast cancer. However, the difference was not statistically significant (hazard ratio [HR] 1.58, 95% confidence interval [CI] 0.96–2.59, p = 0.072).
2.3. miRNAs Associated with Histological Grade
Using RNA-seq data from 430 breast cancers in the TCGA cohort, the expression of the 108 miRNAs differentially expressed between normal and cancerous tissues between low-grade and high-grade breast cancer cases was compared. In high-grade cases, 28 and 18 miRNAs were significantly upregulated and downregulated, respectively (FDR < 0.05;
Table 2).
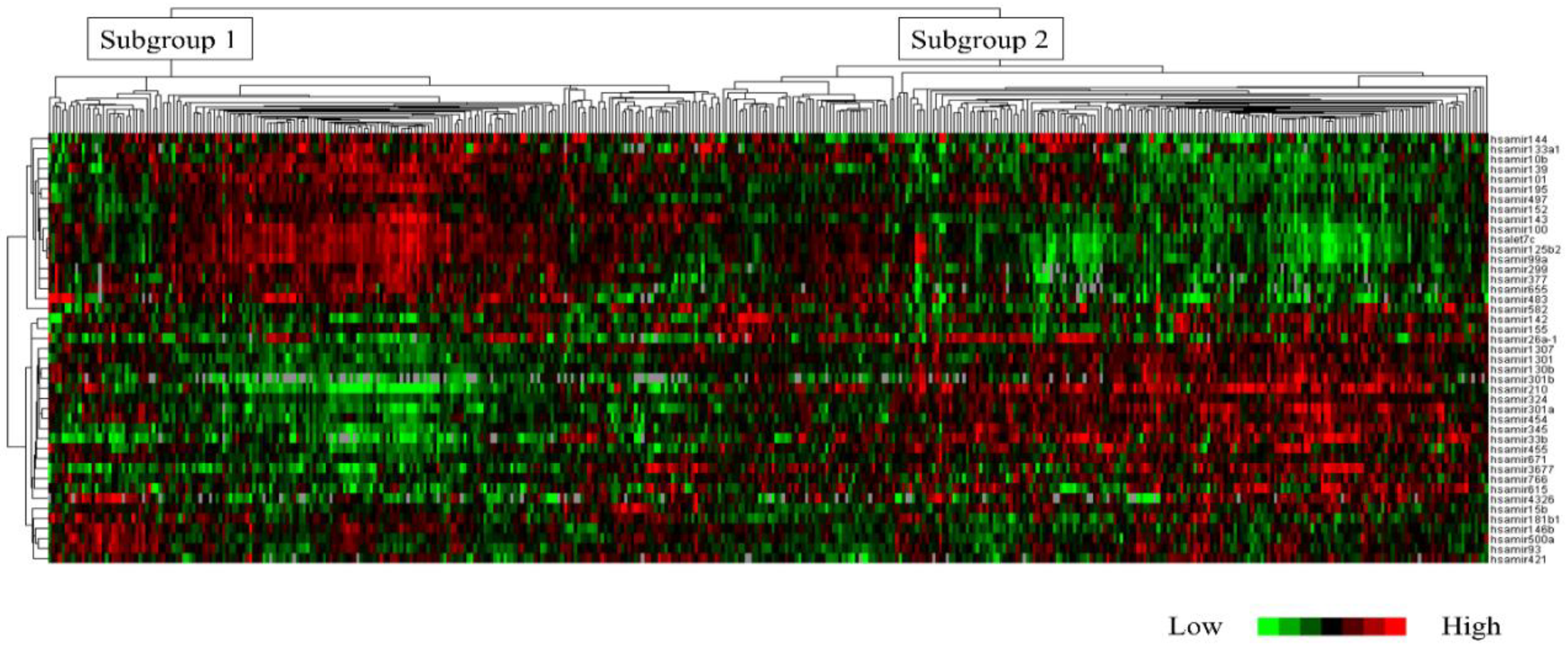
Based on clustering analysis using the TCGA cohort, the 430 cases were classified into the following two subtypes: subgroups 1 (n = 201 cases, 46.7%) and 2 (n = 229 cases, 53.3%). Hierarchical clustering was performed on the TCGA cohort using the 43 miRNAs.
Figure 2 shows a dendrogram classifying the 430 cases. Histological grade 3 was significantly more prevalent in subgroup 2 than in subgroup 1 (p < 0.0001). Additionally, subgroup 2 had a higher frequency of ER-negative cases (p < 0.0001;
Table S3). These 43 miRNAs were significantly associated with “Protein processing in endoplasmic reticulum (hsa04141)” on the KEGG pathway (p < 0.0001). Key miRNAs involved in this pathway included the highly novel miR3677. Furthermore, miR3677 was found to be involved in this pathway via a single protein, ribophorin II (RPN2).
2.4. Clinicopathological and Prognostic Significance of miR-3677
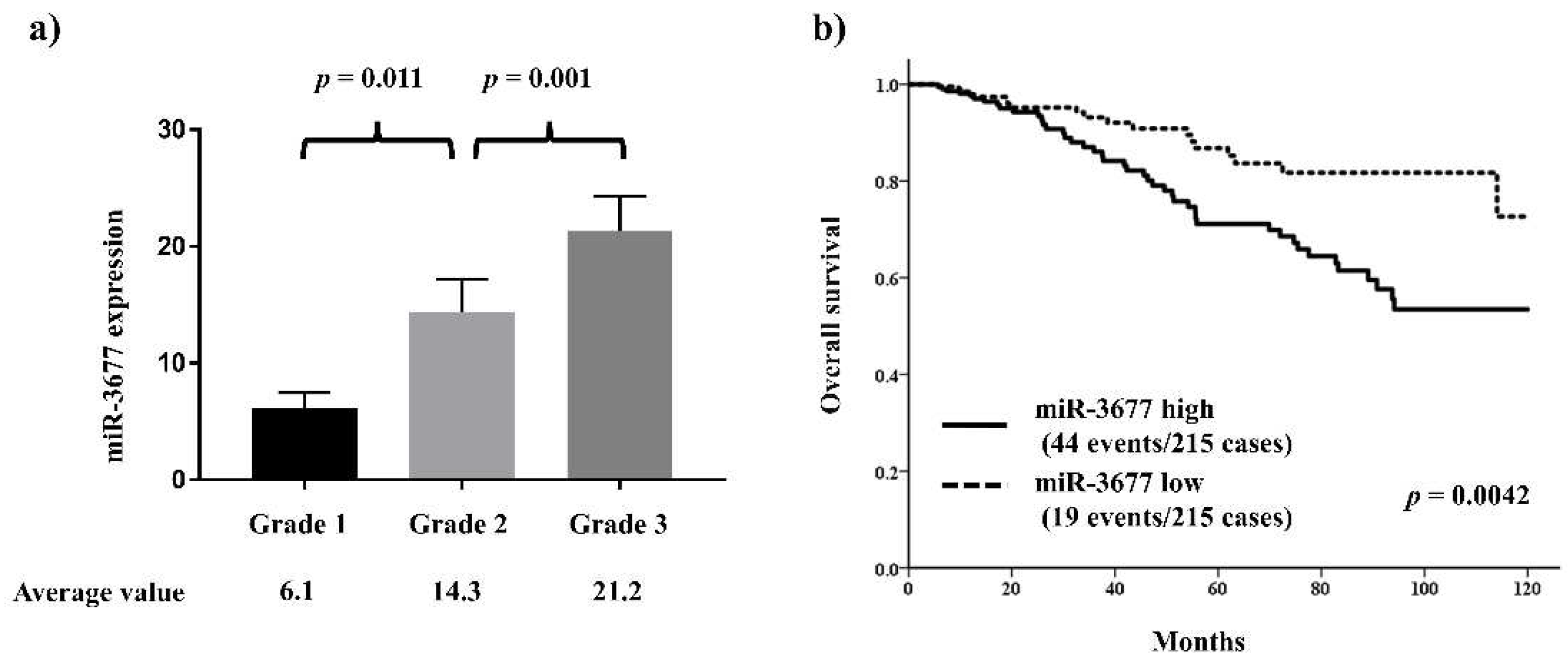
miR-3677 is one of the top miRNAs involved in histological grade and has been reported to play a key role in other cancers. Thus, the relationship between miR-3677 and clinicopathological features and histological grade was further investigated in the TCGA cohort. miR-3677 expression was significantly positively associated with histological grade (
Figure 3a). High miR-3677 expression was significantly associated with other features characteristic of aggressive behavior, including ER negativity (p = 0.00011) and HER2 positivity (p = 0.00012;
Table S4).
The outcome analysis demonstrated that patients with high miR-3677 expression had a significantly worse prognosis than those with low miR-3677 expression (HR 2.20, 95% CI 1.28–3.76, p = 0.0042;
Figure 3b). Furthermore, the multivariate analysis revealed that high miR-3677 expression was an independent predictor of poor prognosis (HR 2.45, 95% CI 1.28–4.69, p = 0.0068;
Table 3).
3. Discussion
In this study, the proprietary miRNA dataset from our facility and the TCGA miRNA database were used to identify 43 miRNAs that were significantly correlated with histological grade.
Histological grade is an extremely useful method for predicting the proliferation and metastatic potential of early-stage breast cancer. In a large-scale study conducted by Henson et al. [
7] that evaluated the survival rate of 22,616 breast cancer cases, the survival rate of histological grade 1 stage II patients was the same as that of grade 3 stage I patients. Furthermore, despite having LN positivity, grade 1 patients with a tumor size of <2 cm had a 5-year survival rate of 99% and good prognoses. These results are consistent with those of a previous study conducted by Rakha et al. [
8], which was a long-term follow-up study of 2,219 operable breast cancer cases. The study indicated that histological grade is a key determinant of breast cancer outcome. These results suggest that histological grade 1 and 3 breast cancers may have completely different molecular profiles and highlight the importance of identifying the set of genes predictive of histological grade. Sotiriou et al. [
9] developed a panel of 97 genes to differentiate between grade 1 and 3 breast cancer cases using the Nottingham grading system. Comparing the prognosis and predictive accuracy of gene signatures based on this gene panel with those of Oncotype DX®, both methods demonstrated similar differences in the distant metastasis-free survival rate between the low- and high-risk groups [
10,
11]. These studies have shed some light on the differences between the molecular profiles among tumors with different histological grades at the genomic, transcriptomic, and proteomic levels.
Recently, the clinical utility of histological grade in the selection of drug therapy for early-stage breast cancer has increased. Chemotherapy targets molecular pathways related to cell proliferation and affects biomarkers associated with cell proliferation, including histological grade. Although invasive breast cancer with histological grade 3 has a poor prognosis, chemotherapy is highly efficient for treating this type of cancer. Histological grade before neoadjuvant chemotherapy has been reported to be associated with a pathological complete response rate after neoadjuvant treatment. Histological grade is a predictor of response to preoperative trastuzumab combination chemotherapy in patients with HER2-positive breast cancer [
12,
13]. According to the St. Gallen guidelines [
14], individual treatment decision is recommended based on clinicopathological factors, such as tumor size, lymph node metastasis status, histological grade, Ki67 labeling index, quantitative expression of hormone receptors, and genomic signature, in stage 2 breast cancer when selecting chemotherapy for ER-positive HER2-negative early invasive breast cancer. Cyclin-dependent kinase inhibitors, including palbociclib, ribociclib, and abemaciclib, target factors related to the cell cycle, and their utility has already been confirmed in clinical trials [
15,
16,
17]. The monarchE trial demonstrated that abemaciclib improves prognosis in patients with high-risk ER-positive and HER2-negative early-stage breast cancer. In this trial, “high risk” was defined as “patients with four or more positive pathologic axillary lymph nodes or one–three positive axillary lymph nodes and at least one of the following: tumor size > 5 cm, histological grade 3, or centrally assessed Ki-67 > 20%” [
18].
miRNAs comprise small noncoding RNA molecules that regulate gene expression posttranscriptionally and are involved in the onset, progression, and metastasis of breast cancer [
19,
20]. Recent studies have reported that a diagnostic miRNA signature can be employed to assess tumor aggressiveness, drug response, and outcomes in patients with breast cancer [
19,
21,
22]. Furthermore, miRNA expression profiles have been reported to be different for each molecular subtype of breast cancer [
6]. Liquid biopsy using a blood sample for collecting data is equivalent to the method that uses a solid tissue sample [
23]. Substances produced by tumors, such as carcinoembryonic antigen, carbohydrate antigen 19-9, circulating tumor cells, cell-free DNA/circulating tumor DNA, and miRNA, can be measured by liquid biopsy [
24]. Liquid biopsy enables the monitoring of circulating tumor cells throughout the body via the blood and their biological features, such as genes originating from such tumor cells [
25]. Since the molecular features of biological substances can change in a short period of time, this method can provide real-time identification of factors directly contributing to tumor proliferation and metastasis. However, the scientific stage where all the data can be obtained by liquid biopsy alone has not been reached, and histopathological evaluations by pathologists are still crucial in clinical practice. Few studies have confirmed the clinical utility of liquid biopsies for monitoring serum miRNA fluctuations. Newly discovered miRNAs can provide a strategically novel option for cancer diagnosis and treatment by determining the histological grade of breast cancer.
This study focused on one of the miRNAs, miR-3677, because of its novelty and significance. This miRNA has been reported to be a significant prognostic predictor for hepatocellular carcinoma [
26]. Peng et al. [
27] compared the expression levels of miR-3677 in noncancerous and breast cancer cells and showed significantly higher miR-3677 levels in breast cancer cells. Furthermore, they reported that the overexpression of miR-3677 promoted the proliferation and colony formation of breast cancer cells. Additionally, they performed a bioinformatic analysis using data obtained from the TCGA database and identified transducin-like enhancer protein 3 (TLE3) as a potential target of miR-3677 [
27]. TLE3 is a full-length human TLE family member that plays an important role in breast cancer cell differentiation and proliferation [
28,
29]. Moreover, patients with breast and ovarian cancers with high TLE3 expression have been reported to respond well to a chemotherapeutic regimen that includes taxane [
30,
31]. The expression levels of miR-3677 have been reported to be negatively correlated with TLE3 in breast cancer tissue, suggesting that miR-3677 is involved in breast cancer cell proliferation and metastasis by suppressing TLE3 expression. This study demonstrated that miR-3677 was significantly correlated with histological grade. Therefore, it may be a strong prognostic predictor. The pathway analysis revealed that miR-3677 was associated with the “protein processing in the endoplasmic reticulum (hsa04141)” KEGG pathway via a single protein, RPN2, which is a membrane protein that localizes to the rough endoplasmic reticulum. Based on gene expression profiles of breast cancer, Ochiya et al. [
32] revealed that RPN2 was involved in the treatment resistance of breast cancer and found that RPN2 was specifically expressed in the ESA+/CD44+/CD24− breast cancer cells, which is considered “breast cancer stem cell.” Further studies are needed to analyze the molecular mechanisms involved in miR-3677 expression and RPN2 using in vitro and in vivo assays.
4. Materials and Methods
4.1. Discovery Cohort
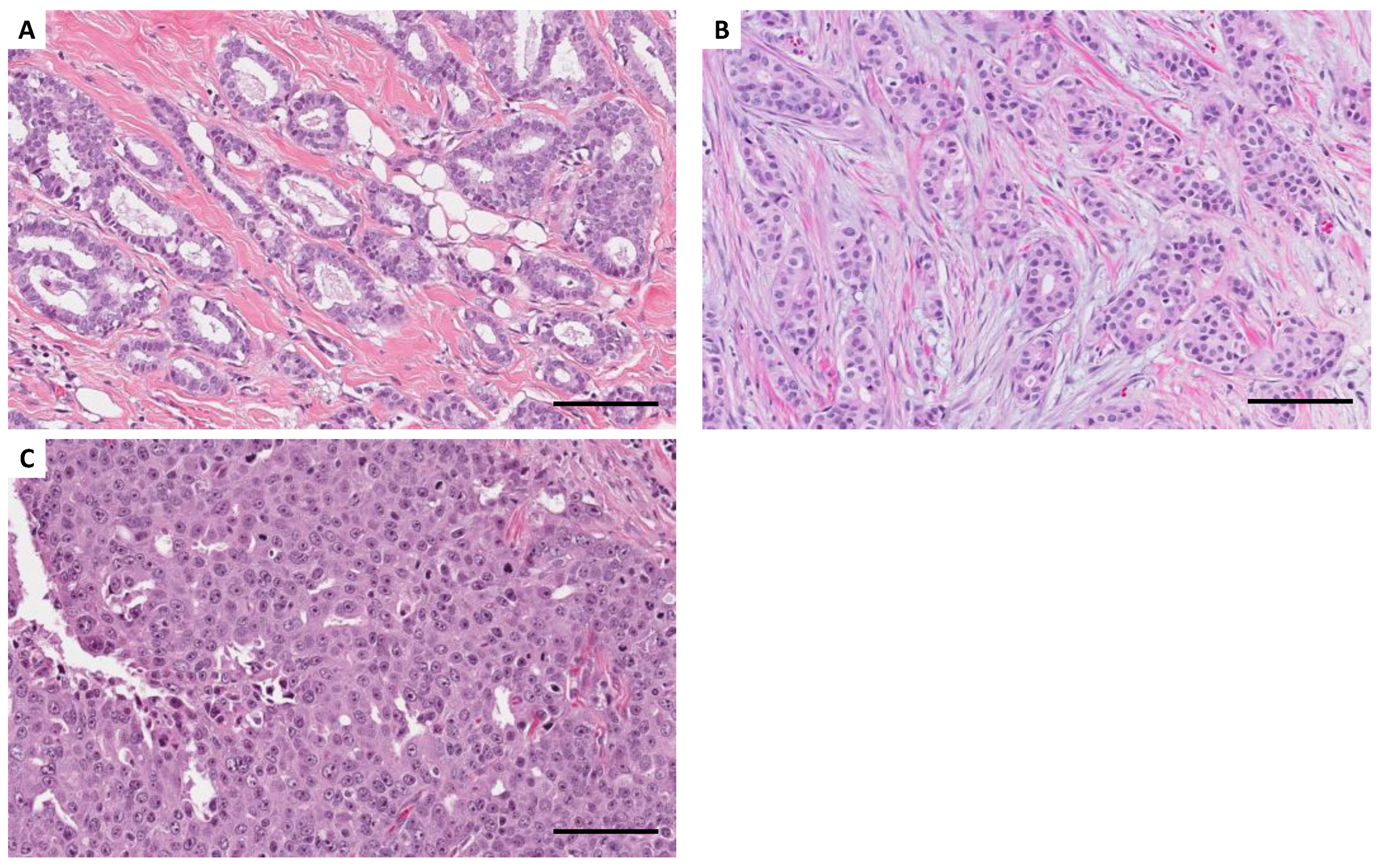
A total of 20 surgically excised tissue samples were collected at Gunma University Hospital to construct miRNA expression signatures of human breast cancer, including five normal breast, five ER-positive/HER2-negative breast cancer, 5 HER2-positive breast cancer, and 5 triple-negative breast cancer tissue samples. The clinicopathological characteristics of the 15 breast cancer cases are shown in
Table S5. Total RNA was isolated from these frozen samples using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The miRNA expression signatures of the 20 samples were generated by small RNA sequencing using HiSeq 2500 (Illumina, San Diego, CA, USA). Quality control and normalization were performed according to previous analyses [
33,
34]. Clean reads were obtained after removing adaptor sequences, reads having greater than 10% of unknown bases, and reads with low-quality bases (base with quality value ≤ 5) greater than 10% in a read. A comparison between the five noncancerous and 15 cancerous cells was performed, and miRNAs with different expressions were identified using the edgeR method (
Figure 4).
4.2. TCGA Validation Cohort
A total of 430 patients with breast cancer were enrolled from TCGA with clinicopathological information and miRNA data as measured by RNA-seq downloaded from the Genomic Data Commons Data Portal and cBioPortal [
35,
36]. Differences in miRNA expression between low-grade and high-grade breast cancers were compared using the edgeR method [
37], and FDR < 0.05 was considered significant (
Figure 1). The MA plot of miRNAs is shown in
Figure S1. TCGA data were log2 transformed and normalized to the provided miRNA expression data before cluster analysis. Cluster analysis and heatmap generation were performed using Cluster 3.0 and Java Treeview [
38]. Pathway analysis was performed using mirPath v.3 (
https://dianalab.e-ce.uth.gr/html/mirpathv3/index.php?r=mirpath) to calculate the pathways that were significantly associated with the candidate miRNAs in the KEGG analysis [
39].
4.3. Statistical Analysis
The association between histological grade and candidate miRNAs was assessed using the Mann–Whitney test, whereas the association between candidate miRNAs and other clinicopathological factors was assessed using the chi-square test. Survival analysis was performed using the Kaplan–Meier method to illustrate the survival curve, and HR and 95% CI were calculated using the Cox proportional hazards regression model. A p-value < 0.05 was considered significant. Statistical analyses were performed using SPSS Statistics version 24.0 (IBM Corp, Armonk, NY, USA) and GraphPad Prism 7.03 (GraphPad Software Inc., La Jolla, CA, USA).
5. Conclusions
In this study, a miRNA profile associated with histological grade was found. The study results may lead to the discovery of new miRNAs that may become candidates for new RNA-targeted therapies. However, research clarifying the mechanisms of miRNA molecules on histological grade using comprehensive expression data obtained by next-generation sequencing for breast cancer is insufficient. Thus, further functional analysis including real-time polymerase chain reaction assay of the miRNAs related to histological grade is necessary to use them in breast cancer diagnosis and treatment.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org. Table S1: List of 108 microRNAs associated with breast cancer; Table S2: Clinicopathological significance of histological grade in the present TCGA cohort; Table S3: Clinicopathological significance of a subgroup based on microRNAs related to histological grade; Table S4: Clinicopathological significance of miR-3677 in the present TCGA cohort; Figure S1: MA plot of miRNAs.
Funding
This study was supported by KAKENHI, grant number 16K19906.
Institutional Review Board Statement
The study was conducted in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards and approved by the Institutional Review Board of Gunma University (approval nos. 2016-023 and 2017-163).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The datasets generated and/or analyzed during the current study are not publicly available due to the regulation of the Institutional Review Board of Gunma University but are available from the corresponding author upon reasonable request.
Acknowledgments
We gratefully acknowledge the work of our research technician (Kumiko Sudo). This work was supported by Gunma University and International University of Health and Welfare.
Conflicts of Interest
SK has received honoraria from Kyowa Kirin Co., Ltd., Daiichi Sankyo Co. Ltd., Taiho Pharmaceutical Co. Ltd., Eli Lilly and Company, MSD K.K., AstraZeneca K.K., CHUGAI Pharmaceutical, Ltd., Dinow Inc., Eisai Co., Ltd., Takeda Pharmaceutical Co., Ltd., and Novartis Japan. TY has received research grants from Ono Pharmaceutical Co., Ltd., CHUGAI Pharmaceutical Co., Ltd., and Memolead Co. TF has received research funding from Eisai Co., Ltd. KS has received research grants from CHUGAI Pharmaceutical Co., Ltd. and Ono Pharmaceutical Co., Ltd. The other authors declare no conflict of interest.
References
- Early Breast Cancer Trialists’ Collaborative Group (EBCTCG). Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomized trials. Lancet. 2015, 365, 1687–1717.
- Rakha, E.A.; Reis-Filho, J.S.; Baehner, F.; Dabbs, D.J.; Decker, T.; Eusebi, V.; Fox, S.B.; Ichihara, S.; Jacquemier, J.; Lakhani, S.R.; Palacios, J.; Richardson, A.L.; Schnitt, S.J.; Schmitt, F.C.; Tan, P.H.; Tse, G.M.; Badve, S.; Ellis, I.O. Breast cancer prognostic classification in the molecular era: the role of histological grade. Breast Cancer Res. 2010, 12, 207. [CrossRef]
- Elston, C.W.; Ellis, I.O. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991, 19, 403–410. [CrossRef]
- Kurozumi, S.; Matsumoto, H.; Hayashi, Y.; Tozuka, K.; Inoue, K.; Horiguchi, J.; Takeyoshi, I.; Oyama, T.; Kurosumi, M. Power of PgR expression as a prognostic factor for ER-positive/HER2-negative breast cancer patients at intermediate risk classified by the Ki67 labeling index. BMC Cancer. 2017, 17, 354. [CrossRef]
- Kurozumi, S.; Inoue, K.; Takei, H.; Matsumoto, H.; Kurosumi, M.; Horiguchi, J.; Takeyoshi, I.; Oyama, T. ER, PgR, Ki67, p27(Kip1), and histological grade as predictors of pathological complete response in patients with HER2-positive breast cancer receiving neoadjuvant chemotherapy using taxanes followed by fluorouracil, epirubicin, and cyclophosphamide concomitant with trastuzumab. BMC Cancer. 2015, 15, 622. [CrossRef]
- Kurozumi, S.; Yamaguchi, Y.; Kurosumi, M.; Ohira, M.; Matsumoto, H.; Horiguchi, J. Recent trends in microRNA research into breast cancer with particular focus on the associations between microRNAs and intrinsic subtypes. J. Hum. Genet. 2017, 62, 15–24. [CrossRef]
- Henson, D.E.; Ries, L.; Freedman, L.S.; Carriaga, M. Relationship among outcome, stage of disease, and histologic grade for 22,616 cases of breast cancer. The basis for a prognostic index. Cancer. 1991, 68, 2142–2149. [CrossRef]
- Rakha, E.A.; El-Sayed, M.E.; Lee, A.H.; Elston, C.W.; Grainge, M.J.; Hodi, Z.; Blamey, R.W.; Ellis, I.O. Prognostic significance of Nottingham histologic grade in invasive breast carcinoma. J. Clin. Oncol. 2008, 26, 3153–3158. [CrossRef]
- Sotiriou, C.; Wirapati, P.; Loi, S.; Harris, A.; Fox, S.; Smeds, J.; Nordgren, H.; Farmer, P.; Praz, V.; Haibe-Kains, B.; Desmedt, C.; Larsimont, D.; Cardoso, F.; Peterse, H.; Nuyten, D.; Buyse, M.; Van de Vijver, M.J.; Bergh, J.; Piccart, M.; Delorenzi, M. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J. Natl Cancer Inst. 2006, 98, 262–272. [CrossRef]
- Sparano, J.A.; Gray, R.J.; Makower, D.F.; Pritchard, K.I.; Albain, K.S.; Hayes, D.F.; Geyer, C.E. Jr; Dees, E.C.; Goetz, M.P.; Olson, J.A. Jr; Lively, T.; Badve, S.S.; Saphner, T.J.; Wagner, L.I.; Whelan, T.J.; Ellis, M.J.; Paik, S.; Wood, W.C.; Ravdin, P.M.; Keane, M.M.; Gomez Moreno, H.L.; Reddy, P.S.; Goggins, T.F.; Mayer, I.A.; Brufsky, A.M.; Toppmeyer, D.L.; Kaklamani, V.G.; Berenberg, J.L.; Abrams, J.; Sledge, G.W. Jr. Adjuvant chemotherapy guided by a 21-gene expression assay in breast cancer. N. Engl. J. Med. 2018, 379, 111–121. [CrossRef]
- Sotiriou, C.; Pusztai, L. Gene-expression signatures in breast cancer. N. Engl. J. Med. 2009, 360, 790–800. [CrossRef]
- Kurozumi, S.; Inoue, K.; Takei, H.; Matsumoto, H.; Kurosumi, M.; Horiguchi, J.; Takeyoshi, I.; Oyama, T. ER, PgR, Ki67, p27(Kip1), and histological grade as predictors of pathological complete response in patients with HER2-positive breast cancer receiving neoadjuvant chemotherapy using taxanes followed by fluorouracil, epirubicin, and cyclophosphamide concomitant with trastuzumab. BMC Cancer. 2015, 15, 622. [CrossRef]
- Katayama, A.; Miligy, I.M.; Shiino, S.; Toss, M.S.; Eldib, K.; Kurozumi, S.; Quinn, C.M.; Badr, N.; Murray, C.; Provenzano, E.; Callagy, G.; Martyn, C.; Millican-Slater, R.; Purdie, C.; Purnell, D.; Pinder, S.E.; Oyama, T.; Shaaban, A.M.; Ellis, I.; Lee, A.H.S.; Rakha, E.A. Predictors of pathological complete response to neoadjuvant treatment and changes to post-neoadjuvant HER2 status in HER2-positive invasive breast cancer. Mod. Pathol. 2021, 34, 1271–1281. [CrossRef]
- Curigliano, G.; Burstein, H.J.; Gnant, M.; Loibl, S.; Cameron, D.; Regan, M.M.; Denkert, C.; Poortmans, P.; Weber, W.P.; Thürlimann, B.; St Gallen Consensus Conference Panelists 2023. Understanding breast cancer complexity to improve patient outcomes: the St Gallen international consensus conference for the primary therapy of individuals with early breast cancer 2023. Ann. Oncol. 2023. S0923-7534(23)00835-9. [CrossRef]
- Turner, N.C.; Slamon, D.J.; Ro, J.; Bondarenko, I.; Im, S.A.; Masuda, N.; Colleoni, M.; DeMichele, A.; Loi, S.; Verma, S.; Iwata, H.; Harbeck, N.; Loibl, S.; André, F.; Puyana Theall, K.; Huang, X.; Giorgetti, C.; Huang Bartlett, C.; Cristofanilli, M. Overall survival with palbociclib and fulvestrant in advanced breast cancer. N. Engl. J. Med. 2018, 379, 1926–1936. [CrossRef]
- Hortobagyi, G.N.; Stemmer, S.M.; Burris, H.A.; Yap, Y.S.; Sonke, G.S.; Paluch-Shimon, S.; Campone, M.; Petrakova, K.; Blackwell, K.L.; Winer, E.P.; Janni, W.; Verma, S.; Conte, P.; Arteaga, C.L.; Cameron, D.A.; Mondal, S.; Su, F.; Miller, M.; Elmeliegy, M.; Germa, C.; O'Shaughnessy, J. Updated results from MONALEESA-2, a phase III trial of first-line ribociclib plus letrozole versus placebo plus letrozole in hormone receptor-positive, HER2-negative advanced breast cancer. Ann. Oncol. 2019, 30, 1842. [CrossRef]
- Johnston, S.; Martin, M.; Di Leo, A.; Im, S.A.; Awada, A.; Forrester, T.; Frenzel, M.; Hardebeck, M.C.; Cox, J.; Barriga, S.; Toi, M.; Iwata, H.; Goetz, M.P.. MONARCH 3 final PFS: a randomized study of abemaciclib as initial therapy for advanced breast cancer. npj Breast Cancer. 2019, 5, 5. [CrossRef]
- Johnston, S.R.D.; Harbeck, N.; Hegg, R.; Toi, M.; Martin, M.; Shao, Z.M.; Zhang, Q.Y.; Martinez Rodriguez, J.L.; Campone, M.; Hamilton, E.; Sohn, J.; Guarneri, V.; Okada, M.; Boyle, F.; Neven, P.; Cortés, J.; Huober, J.; Wardley, A.; Tolaney, S.M.; Cicin, I.; Smith, I.C.; Frenzel, M.; Headley, D.; Wei, R.; San Antonio, B.; Hulstijn, M.; Cox, J.; O’Shaughnessy, J.; Rastogi, P.; monarchE Committee Members and Investigators Abemaciclib Combined With Endocrine Therapy for the Adjuvant Treatment of HR+, HER2−, Node-Positive, High-Risk, Early Breast Cancer (monarchE). J. Clin. Oncol. 2020, 38, 3987–3998. [CrossRef]
- Serpico, D.; Molino, L.; Di Cosimo, S. microRNAs in breast cancer development and treatment. Cancer Treat. Rev. 2014, 40, 595–604. [CrossRef]
- Kayani, M.; Kayani, M.A.; Malik, F.A.; Faryal, R. Role of miRNAs in breast cancer. Asian Pac. J. Cancer Prev. 2011, 12, 3175–3180.
- Jayasingam, S.D.; Citartan, M.; Mat Zin, A.A.; Rozhdestvensky, T.S.; Tang, T.H.; Ch’ng, E.S.An Eleven-microRNA Signature Related to Tumor-Associated Macrophages Predicts Prognosis of Breast Cancer. Int. J. Mol. Sci. 2022, 23. [CrossRef]
- van Schooneveld, E.; Wildiers, H.; Vergote, I.; Vermeulen, P.B.; Dirix, L.Y.; Van Laere, S.J. Dysregulation of microRNAs in breast cancer and their potential role as prognostic and predictive biomarkers in patient management. Breast Cancer Res. 2015, 17, 21. [CrossRef]
- Pantel, K.; Alix-Panabières, C. Circulating tumor cells in cancer patients: challenges and perspectives. Trends Mol. Med. 2010, 16, 398–406. [CrossRef]
- Domínguez-Vigil, I.G.; Moreno-Martínez, A.K.; Wang, J.Y.; Roehrl, M.H.A.; Barrera-Saldaña, H.A. The dawn of the liquid biopsy in the fight against cancer. Oncotarget. 2018, 9, 2912–2922. [CrossRef]
- Merker, J.D.; Oxnard, G.R.; Compton, C.; Diehn, M.; Hurley, P.; Lazar, A.J.; Lindeman, N.; Lockwood, C.M.; Rai, A.J.; Schilsky, R.L.; Tsimberidou, A.M.; Vasalos, P.; Billman, B.L.; Oliver, T.K.; Bruinooge, S.S.; Hayes, D.F.; Turner, N.C. Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists joint review. J. Clin. Oncol. 2018, 36, 1631–1641. [CrossRef]
- Yao, B.; Li, Y.; Niu, Y.; Wang, L.; Chen, T.; Guo, C.; Liu, Q. Hypoxia-induced miR-3677-3p promotes the proliferation, migration, and invasion of hepatocellular carcinoma cells by suppressing SIRT5. J. Cell. Mol. Med. 2020, 24, 8718–8731. [CrossRef]
- Peng, L.N.; Deng, X.Y.; Gan, X.X.; Zhang, J.H.; Ren, G.H.; Shen, F.; Feng, J.H.; Cai, W.S.; Xu, B. Targeting of TLE3 by miR-3677 in human breast cancer promotes cell proliferation, migration, and invasion. Oncol. Lett. 2020, 19, 1409–1417. [CrossRef]
- van Agthoven, T.; Sieuwerts, A.M.; Meijer-van Gelder, M.E.; Look, M.P.; Smid, M.; Veldscholte, J.; Sleijfer, S.; Foekens, J.A.; Dorssers, L.C. Relevance of breast cancer antiestrogen resistance genes in human breast cancer progression and tamoxifen resistance. J. Clin. Oncol. 2009, 27, 542–549. [CrossRef]
- Jangal, M.; Couture, J.P.; Bianco, S.; Magnani, L.; Mohammed, H.; Gévry, N. The transcriptional co-repressor TLE3 suppresses basal signaling on a subset of estrogen receptor α target genes. Nucleic Acids Res. 2014, 42, 11339–11348. [CrossRef]
- Kulkarni, S.A.; Hicks, D.G.; Watroba, N.L.; Murekeyisoni, C.; Hwang, H.; Khoury, T.; Beck, R.A.; Ring, B.Z.; Estopinal, N.C.; Schreeder, M.T.; Seitz, R.S.; Ross, D.T. TLE3 as a candidate biomarker of response to taxane therapy. Breast Cancer Res. 2009, 11, R17. [CrossRef]
- Samimi, G.; Ring, B.Z.; Ross, D.T.; Seitz, R.S.; Sutherland, R.L.; O'Brien, P.M.; Hacker, N.F.; Huh, W.K. TLE3 expression is associated with sensitivity to taxane treatment in ovarian carcinoma. Cancer Epidemiol. Biomarkers Prev. 2012, 21, 273–279. [CrossRef]
- Honma, K.; Iwao-Koizumi, K.; Takeshita, F.; Yamamoto, Y.; Yoshida, T.; Nishio, K.; Nagahara, S.; Kato, K.; Ochiya, T. RPN2 gene confers docetaxel resistance in breast cancer. Nat. Med. 2008, 14, 939–948. [CrossRef]
- Toda, H.; Kurozumi, S.; Kijima, Y.; Idichi, T.; Shinden, Y.; Yamada, Y.; Arai, T.; Maemura, K.; Fujii, T.; Horiguchi, J.; Natsugoe, S.; Seki, N. Molecular pathogenesis of triple-negative breast cancer based on microRNA expression signatures: antitumor miR-204-5p targets AP1S3. J. Hum. Genet. 2018, 63, 1197–1210. [CrossRef]
- Toda, H.; Seki, N.; Kurozumi, S.; Shinden, Y.; Yamada, Y.; Nohata, N.; Moriya, S.; Idichi, T.; Maemura, K.; Fujii, T.; Horiguchi, J.; Kijima, Y.; Natsugoe, S. RNA-sequence-based microRNA expression signature in breast cancer: tumor-suppressive miR-101-5p regulates molecular pathogenesis. Mol. Oncol. 2020, 14, 426–446. [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; Antipin, Y.; Reva, B.; Goldberg, A.P.; Sander, C.; Schultz, N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; Cerami, E.; Sander, C.; Schultz, N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [CrossRef]
- Lohse, M.; Bolger, A.M.; Nagel, A.; Fernie, A.R.; Lunn, J.E.; Stitt, M.; Usadel, B. RobiNA: a user-friendly, integrated software solution for RNA-Seq-based transcriptomics. Nucleic Acids Res. 2012, 40, W622–W627. [CrossRef]
- de Hoon, M.J.; Imoto, S.; Nolan, J.; Miyano, S. Open source clustering software. Bioinformatics. 2004, 20, 1453–1454. [CrossRef]
- Vlachos, I.S.; Paraskevopoulou, M.D.; Karagkouni, D.; Georgakilas, G.; Vergoulis, T.; Kanellos, I.; Anastasopoulos, I.L.; Maniou, S.; Karathanou, K.; Kalfakakou, D.; Fevgas, A.; Dalamagas, T.; Hatzigeorgiou, A.G. Diana-TarBase v7.0: indexing more than half a million experimentally supported miRNA:mRNA interactions. Nucleic Acids Res. 2015, 43, D153–D159. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions, and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions, or products referred to in the content. |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).