1. Introduction
The Sarcidano Horse is an autochthonous, semi-feral breed of small horses native to Sarcidano, a region in the heart of Sardinia (Italy). It belongs to the 26 minor Italian breeds of limited diffusion collected in the Stud Book, managed, since 2018, by ANAREAI, the national association for local horse and donkey breeds [
1]. These horses are locally known for their sturdy build and were traditionally used for rural work and riding. They live in conditions of geographical isolation [
2,
3,
4], due to insularity and to the wild living environment, and are recognized for their endurance and adaptability to the local terrain. According to the breed standard, Sarcidano Horses typically come only in basic coat colors, Chestnut, Bay, Black and sometimes Gray, without spotting or color dilution. Gray horses have a basic coat color at birth, but as they age, the graying of their hair eventually causes the entire coat to lose its pigmentation [
5,
6,
7]. The trait of graying with age is inherited in an autosomal dominant pattern, so it can be difficult to visually identify the real color of a Gray horse. Only two loci are involved in Chestnut, Bay and Black coat coloration: Extension locus, coding melanocortin-1-receptor (
MC1R) gene, and Agouti locus, coding the agouti-signaling protein (
ASIP) gene, which is an antagonist of the
MC1R gene. The correct functioning of the
MC1R gene produces the MC1R protein, driving eumelanin (black pigment) synthesis by melanocytes, producing Black coats [
8,
9,
10]. Contrarily, alteration in the
MC1R gene Exon 1, produced by a transition from C to T, leads to phaeomelanin (reddish pigment) synthesis, producing Chestnut coats [
11,
12]. The
ASIP gene produces the ASIP protein, which has the power to antagonize
MC1R gene functioning in the melanocytes located in the body but not in the extremities, thus producing Bay coats, resulting in a reddish-brown body color with black point coloration on the mane, tail, ear edges and lower legs [
13]. Antagonism of
MC1R by the
ASIP gene is efficient only in dominant/non-mutant variants of the
MC1R gene, whereas when the receptor is already defective, because of the C > T mutation explained above, the antagonism cannot occur, so a Chestnut coat is still produced [
8].
In a previous study on this breed, we focused on the genetic distribution of coat color as a first step aimed at contributing to the general knowledge of the Sarcidano Horse from a perspective of protection of this local genetic resource [
14]. The study highlighted the distribution of the genetic coat colors, pointing out that the absence of spotting or color dilution could be suggestive of low phenotypic variability and the absence of crossbreeding with other domestic breeds. Just thanks to that investigation, carried out on 70 horses, we became aware of a certain inconsistency in the color assigned by visual evaluation compared to that detected by genetic investigation. By analyzing this phenomenon in more detail and expanding the number of samples, these discrepancies reached a substantial number, suggesting that the phenotype of these horses is not always adequately recorded, thus raising awareness that this local resource should be better safeguarded to prevent its decline and disappearance. Since the genetic color is the only one that can be passed on to offspring, correct assignment of the parental coat color is essential to ensure both correct individual identification and the prediction of foal color, as stated in other studies conducted on other local breeds [
15,
16,
17]. In a wild population, coat color provides camouflage and protection in natural environments, helping horses to blend into their surroundings and evade predators. Domestication and human selection introduced a variety of coat colors and patterns that are currently typical of specific breeds [
18,
19]. Diluted or spotted coats, for example, allowed easier recognition of domestic from wild animals, and were, therefore, probably sought in the early stages of domestication [
20]. Thus, the study of coat color distribution could also be suggestive of possible crossing with domestic breeds, thus contributing to the observation of genetic isolation of local breeds [
21]. Correct coat color registration in individual documents avoids recognition errors and allows for appropriate archiving of data, therefore granting the correct assignment of relationships to recognize family units and to predict offspring’s coat color. Although some color assignments can be correctly determined based on phenotype alone, genetic testing may be necessary to define phenotypes that are visually ambiguous and can help to determine color possibilities for foals.
Therefore, the aims of the present research were to evaluate the discrepancies between genetic and phenotypic coat color assignment in the Sarcidano Horse and to elucidate potential reasons, in order to finally propose an effective and low-cost system for correctly recording individual phenotypes.
2. Materials and Methods
2.1. Animals and Territory
Sarcidano is the name of a plateau and district in Central-Southern Sardinia in the municipality of Laconi. The climate is influenced by the surrounding mountains, contributing to regional variation in temperature and precipitations. Summers are hot and dry, with temperatures often exceeding 30 °C, while winters are mild and relatively wet. The area has a typical Mediterranean vegetation pattern, with shrubs, olive trees and cork oaks. In this harsh environment, Sarcidano horses roam freely, grazing on natural vegetation, and are perfectly suited to the local surroundings (
Figure 1).
The present study involved 90 of the approximately 120 Sarcidano Horses recorded in 2018, corresponding to 75% of the entire population registered in that year. The current number of Sarcidano Horses in the ANAREAI Stud Book is 109 heads, but the exact number could be quite different since some subjects, although microchipped, are not yet registered. This study used blood samples provided by the veterinarians who captured the Sarcidano Horses at different times, from February 2016 to January 2018 for periodic checks or extraordinary events, such as microchip implantation or the need to move animals to safer areas based on environmental conditions (frequent bushfires in summer). These horses were anesthetized intramuscularly by a single-dart injection using a combination of tiletamine, zolazepam, detomidina, and acepromazine under the supervision of a veterinary anaesthetist. While they were under anaesthesia, they were also subjected to a general objective examination (to ascertain their health conditions, the presence of wounds and parasitic infestations) and subjected to sampling of biological material (blood, hair, faeces).
More recently, since 2021, captures and sanitary checks have occurred once a year, in late fall (November–December) to individually identify and register newborns in the ANAREAI Stud Book, and to verify the sanitary conditions of the adults. The animals are grouped in small catch paddocks and taken one by one. Each horse is equipped with an electronic transponder on the left portion of the neck, which is verified with a chip reader. The individual microchip number corresponds to the phenotypic card containing the horse’s name, date of birth, sex, biometric data, and coat color. After registration, an individual identity document (green passport) is issued.
2.2. Amplification and Mutations Dedection
DNA was extracted from the individual whole blood samples, taken from the 90 Sarcidano Horses above, using the NucleoSpin
® Blood kit (Macherey-Nagel, Düren, Germany), according to the manufacturer’s protocol. The polymerase chain reaction (PCR) technique was carried out to amplify the
MC1R gene Exon 1 and
ASIP gene Exon 3, which are known to control base coat color set in horses. A first primer set was used to amplify a 320 base pair (bp) fragment, corresponding to part of the unique Exon 1 of the
MC1R gene and one other primers’ pair was designed to delimit a 102/91-bp polymorphic fragment, corresponding to the entire Exon 3 of the
ASIP gene. The primers used were those reported by Cosso et al. [
6]. Both pairs of primers and all the obtained results refer to the latest horse genome version EquCab3.0 (GCF_002863925.1). A PCR reaction for both fragments was carried out in a 25 µL final volume, containing 150 ng of individual genomic DNA, 1X PCR Buffer (without MgCl
2); 2 mM MgCl
2; 0.2 mM dNTPs; 0.2μM primers (Bio-Fab Research srl, Roma, Italy); and 0.25 Units (U) Taq DNA polymerase (HOT FIREPol
® Polymerase, Solis BioDyne, Tartu, Estonia) and ultrapure water DNase/RNase free (Water PCR grade, Solis BioDyne, Tartu, Estonia) up to 25μL. Amplification conditions were as follows: 95 °C (5 min); 35 (
MC1R gene) or 40 (ASIP gene) cycles of 95 °C (30 s), 58 °C (
MC1R) or 60 °C (
ASIP) (30 s), 72 °C (30 s); 72 °C (10 min). The PCR reaction was performed using MAXYGENE II thermocycler (Axygen
® Tewksbury, MA, USA). Amplification results were visualized after electrophoretic run (110 V for 30 min) in a 2% ultrapure agarose gel (
w/
v) (iNtrRon Biotechnology, Sangdaewon-Dong, Republic of Korea) added with 9μL of RedSafe stain (iNtrRon Biotechnology, Sangdaewon-Dong, Republic of Korea), in 1X TAE electrophoresis buffer, and observed under ultraviolet light (UVItec, Cambridge, UK), together with a 100-bp Ladder (GeneRuler, Thermo Scientific™, Waltham, MA, USA).
MC1R gene polymorphisms were detected by the restriction fragment length polymorphism (RFLP) technique, using TaqI endonuclease for the digestion of the obtained PCR products, thus allowing us to recognize a C with a T transition within the amplicons’ nucleotide sequence.
Instead, the polymorphism within the ASIP gene Exon 3 was produced by an 11 bp deletion, thanks to which it was possible to attribute the individual genotype directly through electrophoretic reading of the amplicons, producing 102 or 91 bp fragments, which identify the wild dominant or the mutant recessive allele, respectively. The results section shows how the combinations among the MC1R and ASIP genotypes determined the real genetic coat colors. To confirm the assigned genotypes, thus establishing the reliability of the used methods, 40 samples randomly chosen were sequenced in forward and reverse direction by a commercial service. Once the individual genetic color had been assigned, the genotype of each subject was compared with the color reported on the individual card after visual observation at the time of capture and identification, to verify the correspondence.
Statistical Analysis
Allele frequencies were calculated by direct count of the detected genotypes in the MC1R and ASIP genes.
The observed genotypes and the horse coat color were compared using the χ2 test by means of the R statistical software Version 4.3.2 [
22].
3. Results
Genotyping results showed the genetic coat color distribution in the examined population. Sequencing of the randomly chosen samples confirmed the assigned genotypes. Thanks to their reduced costs and simple method of execution, PCR/RFLP techniques can be a valid solution to be applied routinely for faster and cheaper analysis compared to sequencing, while ensuring correct coat color assignment.
MC1R gene genotyping started with amplification of the 320 bp fragment, corresponding to position 36,979,377–36,979,697 of the latest EquCab3.0 (GCF_002863925.1) horse genome assembly. Digestion by
TaqI endonuclease detected the C > T transition (rs68458866), which allowed us to distinguish the wildtype dominant C from the mutant recessive T allele. The resultant genotype set was homozygous wild dominant C/C, heterozygous C/T and homozygous mutant recessive T/T. The majority of the analyzed Sarcidano Horses carried the double recessive T/T genotype (58 heads, corresponding to 64% of the total studied population), 27 horses (30%) carried the heterozygous genotype C/T, and only 5 horses carried the homozygous wild dominant genotype C/C, equivalent to 6% of the studied population. The allele frequency was 79% for the T and 21% for the C allele (
Table 1).
The causative mutation falls at position chr:3: 36,979,560, producing TCC or TTC as possible codons within the protein chain, at position 83 (NCBI Reference Sequence: NP_001108006.1), resulting in a serine being replaced with a phenylalanine [
14,
23]. This single mutation in the proteins’ primary structure is expected to alter the alpha helix structure, thus producing a defective functioning of the MC1 receptor, which becomes unable to be activated by the melanocyte-stimulating hormone (MSH). This failure to function leads to pheomelanin in place of eumelanin production within melanocytes. Thereby, a Black horse exhibits at least a wildtype C allele, while a Chestnut one carries only a T/T genotype. Consequently, Agouti locus determines whether a horse carrying at least a C allele will be Bay (if it exhibits at least one recessive type 91 allele) or Black (if it carries a homozygous 91/91 genotype); there is no effect of the
ASIP gene genotype on chestnut-based horses.
ASIP gene genotyping was conducted directly through amplification by PCR of the Exon 3, producing a 102/91-bp fragment. Indeed, a polymorphic 11-bp deletion (rs396813234) allowed us to identify the three available genotype sets based on the resultant fragment size. The entire (102-bp) nucleotide sequence corresponds to the dominant wildtype allele, here named as “102”, while the 11-bp deletion identifies the mutant recessive allele, here named as “91”. Consequently, the corresponding genotype set for the ASIP gene was 102/102 for the wildtype homozygous dominant, 102/91 for the heterozygous and 91/91 for the homozygous mutant recessive genotypes.
The
ASIP gene genotype distribution exhibited only two horses carrying the wildtype dominant 102/102 genotype (corresponding to 2% of the studied population), while the main part of the population exhibited the recessive 91/91 genotype (62 horses, corresponding to 69% of the analyzed Sarcidano Horses) and 26 subjects (29%) resulted heterozygous 102/91. Allele frequency resulted 83% for the recessive 91 allele and 17% for the wildtype 102 allele. Both the loci showed a very low frequency of the wild dominant allele and a very high rate of the mutant recessive type (
Table 1).
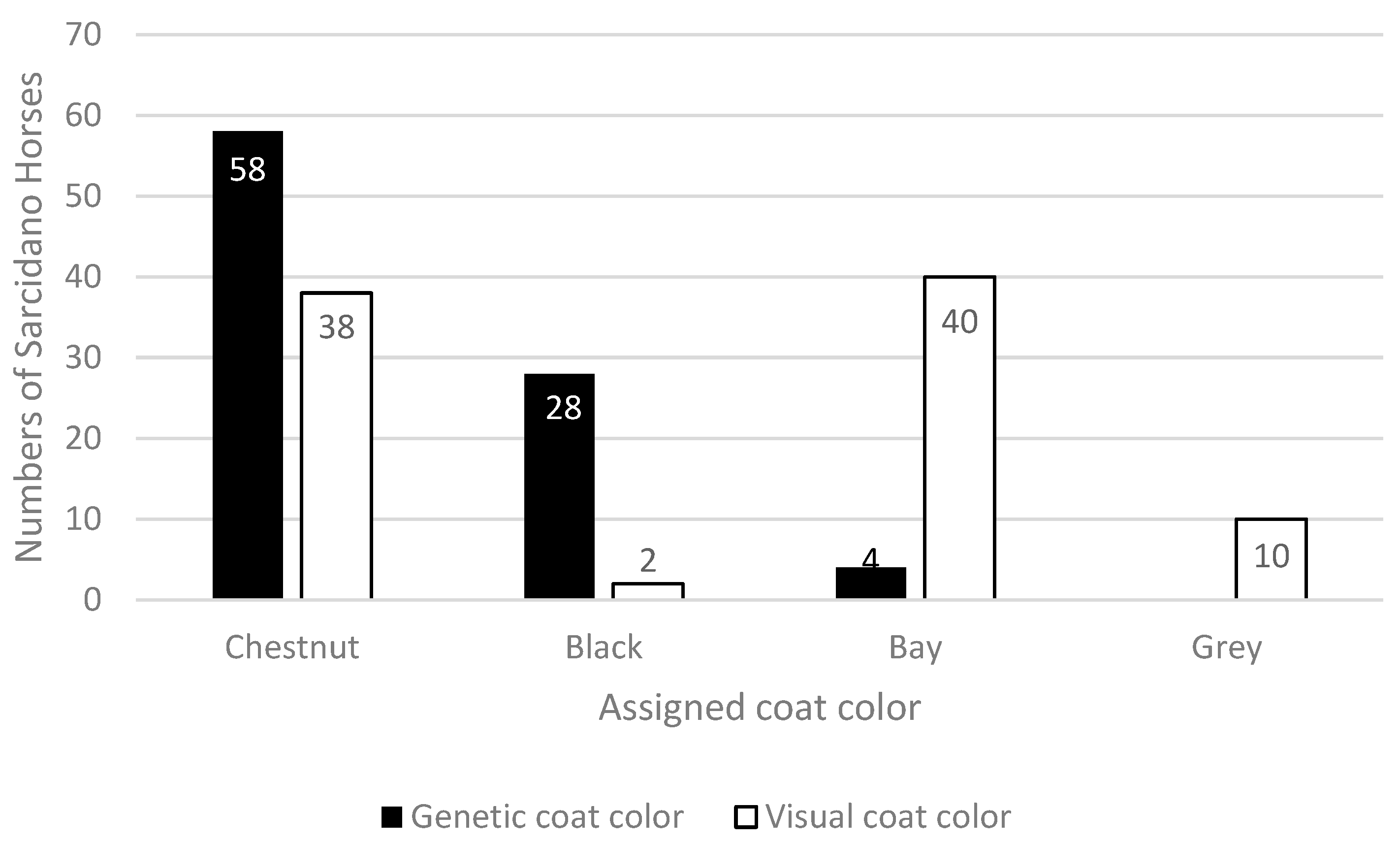
From a genetic point of view, more than half of the analyzed population resulted Chestnut, well over a quarter were Blacks and a very small portion were Bays. Inconsistencies in visual coat color registration emerged when these results were matched with the phenotypically recorded data, producing an altered distribution of coat colors in the studied population. Indeed, from a phenotypic point of view, the 90 Sarcidano Horses were registered as 38 Chestnuts, 40 Bays, only 2 Blacks, and 10 Grays. Gray horses were recorded simply as “grey”, without indication of the possible true color they carried (
Figure 2).
The error rate (calculated as the number of incorrectly classified color phenotypes in the total number of genetically assigned coat colors) was 53.4%, including the gray horses. This large discrepancy between phenotypic and genetic data highlighted how visual observation can greatly alter coat color assignment, leading to incorrect registration of individual data (
Table 2 and
Supplementary Table S1).
Different genotype combinations were found for each coat color, as reported in
Table 3.
These combinations produced some differences in the color shades, varying from light to dark within the basic color set, that could be responsible for the inconsistencies in visual color recognition. Chestnuts resulted in the following combinations: T/T + 91/91 (59% of the total chestnuts), T/T + 102/91 (38%) and T/T + 102/102 (3%). The most represented genotype combination in the studied Sarcidano Horses resulted T/T + 91/91, producing a liver Chestnut, seven of which were visually identified and recorded as Bay. Of the 22 horses carrying the T/T + 102/91 combination, producing a phenotypic lighter-sorrel Chestnuts, 5 were visually registered as Bays. In Black horses too, different genotype combinations were found: 23 were C/T + 91/91 (82% of the Blacks), 21 of which were visually registered as Dark Bays, while 5 were C/C + 91/91, of which 4 were visually recognized as Bays. This important discrepancy in recognizing the black coat color has different possible explanations. The non-black areas of a Bay horse can range from a light brown to near-black, whereas Black horses can range from a sun-faded brown to jet black. This range of possible shades can make it difficult to pinpoint the exact coat color and create confusion when recording [
24,
25,
26]. While all other mammals, including horses, have the genetic potential to be black, certain breed standards do not recognise this possibility and classify all dark coats as Dark Bay. Consequently, a very dark horse is registered as Dark Bay in some breeds, and many people who fill out individual forms apply this rule to all breeds. However, the genetics of a Bay coat are different to those of a Black one, thus preventing the coat color from being correctly assigned.
In this study, only 4 horses were found to be genetically Bays, while visually, as many as 40 heads were registered with this coat. All four of the Bays carried the heterozygous genotype at both the loci, C/T + 102/91. The other possible allele combinations for a Bay coat color are C/C + 102/102, C/C + 102/91 and C/T + 102/102, but none of them were found in the studied Sarcidano Horses. All these results are summarized in
Table 4.
4. Discussion
The found inconsistencies in visual vs genetic coat color recognition means that a subjective perception of shadings can alter the correct identification, making visual evaluation not completely reliable, as the same horse could be identified differently by different evaluators.
The effects of both personal and environmental elements, including age, season, housing type, and diet can make the coat color identification process challenging [
9]. As a result, a black horse kept outside all the time may show signs of “fading” in their coat, which may be mistaken for a Dark Brown horse. On the other hand, a Dark Bay horse’s coat may be confused with a black coat that has faded from sunlight because it has a black mane, tail, and legs, and very dark brown or reddish hair around the head, neck, back, and hips [
25,
26].
Instead, it is crucial to identify coat colors accurately and to correctly report them in the Stud Book, for legal and medical certification, and for the correct prediction of inheritable coat colors, which can only be achieved with genetic investigation of the
MC1R and
ASIP loci genotypes [
27].
Another source of confusion concerns Gray horses, as they are born Black, Chestnut or Bay but gradually lose color in their coat as they age [
28]. This means that an adult Gray horse masks its true color, and it is not clear what color its offspring will inherit. However, since the gray locus is epistatic to the above base coat color genes, if a horse is gray, one can be assured that it has at least one gray parent, but it is impossible to know what color its foals will be without knowing what genetic color they carry [
29]. The same can be said also for Chestnuts and Bays. Indeed, Chestnut horses, although they are surely carriers of the T/T genotype at the MC1R locus, do not externally show which genotype they have at the ASIP locus; therefore, they can generate offspring of any coat color base, depending on the partner’s genotype.
Even Bay horses do not display the genotype they carry at the ASIP locus and are consequently able to transmit different variants of ASIP alleles; a homozygous 102/102 horse will necessarily pass on the 102 allele to its offspring, while a heterozygous horse (102/91) will have a 50% chance of passing on the 102 allele.
Therefore, when the individual coat color is genetically identified, it is possible to know without any doubt, and from birth, what type of foal each horse will be able to generate, in the absence of other color modifiers, as is the case for the Sarcidano Horse. From homozygous MC1R and ASIP loci parents (C/C + 102/102), all offspring will be Bay. From C/C + 102/91 parents, all offspring will be dark based, but could be both Dark Bay and true Black. From C/T + 102/102 parents, the offspring will be Bay as they receive homozygous wild type ASIP alleles, diluting black body color, so that they can be visually appear more red or more black depending on coat color tone and different external and individual factors, such as age, environmental conditions, and nutritional status [
30]. Finally, from heterozygous C/T + 102/91 parents, any color offspring are possible.
5. Conclusions
To conclude, this study tested the genetic coat colors of the Sarcidano Horse and revealed a high percentage of discrepancies that were assigned phenotypically. Furthermore, this research contributes to laying out a reliable, easy, and low-cost method for the identification of coat color in horses, avoiding incorrect recording of collected data and uncertain heritable color prediction. Given the wild nature of the Sarcidano Horse, visual observation of coat color is not always an easy activity, since it occurs outdoors, under any weather condition, without guaranteeing optimal observation time, due to the capture requirements. Inconsistencies in coat color registration are a common problem for many horse breeds, in which assignment only through observation can lead to inaccurate attribution of lineage. Incorrect parentage may be a consequence of the observed discrepancies, which could have a detrimental effect on the health and variability of such a small population, but the main goal should be to preserve all of these ancient native breeds. The introduction of routine paternity testing would be useful to avoid incorrect attribution of lineage, but it still entails high costs and would not solve the problem of incorrect color attributions. Enhanced training of technicians for visual coat color identification is surely recommended, but it should be highlighted that discrepancies in coat color assignment could be completely remedied if genetic testing was routinely carried out. Although further initiatives to defend this important genetic resource would be desirable, at present, only a few breeders and local authorities and passionate researchers are actively involved in increasing interest in this small population.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org, Table S1: All individual coat colors identified by genotyping and visually assigned.
Author Contributions
All authors have made substantial contributions to all of the following: the conception and design of the study, M.C.M., V.C., S.L. and G.C.; acquisition of data, N.C. and E.S.P.; analysis and interpretation of data, G.C., M.C.M., V.C. and S.L.; and drafting the article and revising it critically for important intellectual content, M.C.M., V.C., S.L., G.C., N.C. and E.S.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by grants from Sassari University research fund 2020 (FAR/2020).
Institutional Review Board Statement
All the procedures were reviewed and approved by the Organism in charge for the Animal Welfare and Experimentation (OPBSA) of the University of Sassari (Protocol number: 2018UNISSMEV 0000177).
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available upon request from the corresponding author.
Acknowledgments
We are grateful to the Laconi municipality for its availability in this research.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- ANAREAI. Available online: https://www.anareai.it/ (accessed on 15 September 2023).
- Morelli, L.; Useli, A.; Sanna, D.; Barbato, M.; Contu, D.; Pala, M.; Cancedda, M.; Francalacci, P. Mitochondrial DNA lineages of Italian Giara and Sarcidano horses. Genet. Mol. Res. 2014, 13, 8241–8257. [Google Scholar] [CrossRef] [PubMed]
- Cardinali, I.; Lancioni, H.; Giontella, A.; Capodiferro, M.R.; Capomaccio, S.; Buttazzoni, L.; Biggio, G.P.; Cherchi, R.; Albertini, E.; Olivieri, A.; et al. An overview of ten Italian horse breeds through mitochondrial DNA. PLoS ONE 2016, 7, e0153004. [Google Scholar] [CrossRef] [PubMed]
- Giontella, A.; Cardinali, I.; Pieramati, C.; Cherchi, R.; Biggio, G.P.; Achilli, A.; Silvestrelli, M.; Lancioni, H. A Genetic Window on Sardinian Native Horse Breeds through Uniparental Molecular Systems. Animals 2020, 10, 1544. [Google Scholar] [CrossRef]
- Teixeira, R.B.; Rendahl, A.K.; Anderson, S.M.; Mickelson, J.R.; Sigler, D.; Buchanan, B.R.; Coleman, R.J.; McCue, M.E. Coat color genotypes and risk and severity of melanoma in gray quarter horses. J. Vet. Intern. Med. 2013, 27, 1201–1208. [Google Scholar] [CrossRef] [PubMed]
- Gao, K.X.; Chen, N.B.; Liu, W.J.; Li, R.; Lan, X.Y.; Chen, H.; Lei, C.Z.; Dang, R.H. Frequency of gray coat color in native Chinese horse breeds. Genet. Mol. Res. 2015, 30, 14144–14150. [Google Scholar] [CrossRef] [PubMed]
- Nowacka-Woszuk, J.; Mackowski, M.; Stefaniuk-Szmukier, M.; Cieslak, J. The equine graying with age mutation of the STX17 gene: A copy number study using droplet digital PCR reveals a new pattern. Anim. Genet. 2021, 52, 223–227. [Google Scholar] [CrossRef]
- Li, B.; He, X.; Zhao, Y.; Bai, D.; Shiraigo, W.; Zhao, Q.; Manglai, D. Regulatory pathway analysis of coat color genes in Mongolian horses. Hereditas 2017, 29, 13. [Google Scholar] [CrossRef]
- Neves, A.P.; Schwengber, E.B.; Albrecht, F.F.; Isola, J.V.; van der Linden, L.D.S. Beyond fifty shades: The genetics of horse colors. In Trends and Advances in Veterinary Genetics; IntechOpen: London, UK, 2017. [Google Scholar]
- Sponenberg, D.P.; Bellone, R. Equine Color Genetics, 4th ed.; John Wiley & Sons: Hoboken, NJ, USA, 2017; ISBN 978-1-119-13058-1. [Google Scholar]
- Beaumont, K.A.; Shekar, S.L.; Newton, R.A.; James, M.R.; Stow, J.L.; Duffy, D.L.; Sturm, R.A. Receptor function, dominant negative activity and phenotype correlations for MC1R variant alleles. Hum. Mol. Genet. 2007, 16, 2249–2260. [Google Scholar] [CrossRef]
- Shang, S.; Yu, Y.; Zhao, Y.; Dang, W.; Zhang, J.; Qin, X.; Irwin, D.M.; Qin Wang, Q.; Liu, F.; Wang, Z.; et al. Synergy between MC1R and ASIP for coat color in horses (Equus caballus). J. Anim. Sci. 2019, 97, 1578–1585. [Google Scholar] [CrossRef]
- Thiruvenkadan, A.K.; Kandasamy, N.; Panneerselvam, S. Coat colour inheritance in horses. Livest. Sci. 2008, 117, 109–129. [Google Scholar] [CrossRef]
- Cosso, G.; Carcangiu, V.; Luridiana, S.; Fiori, S.; Columbano, N.; Masala, G.; Careddu, G.M.; Sanna Passino, E.; Mura, M.C. Characterization of the Sarcidano Horse Coat Color Genes. Animals 2022, 12, 2677. [Google Scholar] [CrossRef]
- Mackowski, M.; Wodas, L.; Brooks, S.A.; Cieslak, J. TBX3 and ASIP genotypes reveal discrepancies in officially recorded coat colors of Hucul horses. Animal 2019, 13, 1811–1816. [Google Scholar] [CrossRef] [PubMed]
- Silva, I.L.S.; Junqueira, G.S.B.; Oliveira, C.A.A.; Costa, R.B.; De Camargo, G.M.F. Inconsistencies in horse coat color registration: A case study. J. Equine Sci. 2020, 31, 57–60. [Google Scholar] [CrossRef]
- Cieslak, J.; Brooks, S.A.; Wodas, L.; Mantaj, W.; Borowska, A.; Sliwowska, J.H.; Ziarniak, K.; Mackowski, M. Genetic Background of the Polish Primitive Horse (Konik) Coat Color Variation-New Insight into Dun Dilution Phenotypic Effect. J. Hered. 2021, 112, 436–442. [Google Scholar] [CrossRef]
- Cieslak, M.; Reissmann, M.; Hofreiter, M.; Lydwig, A. Colours of domestication. Biol. Rev. 2011, 86, 885–899. [Google Scholar] [CrossRef]
- Ahmad, H.I.; Ahmad, M.J.; Jabbir, F.; Ahmar, S.; Ahmad, N.; Elokil, A.A.; Chen, J. The Domestication Makeup: Evolution, Survival, and Challenges. Front. Ecol. Evol. 2020, 8, 103. [Google Scholar] [CrossRef]
- Ludwig, A.; Pruvost, M.; Reissmann, M.; Benecke, N.; Brockmann, G.A.; Castaños, P.; Cieslak, M.; Lippold, S.; Llorente, L.; Malaspinas, A.S.; et al. Coat color variation at the beginning of horse domestication. Science 2009, 324, 485. [Google Scholar] [CrossRef] [PubMed]
- Reissmann, M.; Musa, L.; Zakizadeh, S.; Ludwig, A. Distribution of coat-color-associated alleles in the domestic horse population and Przewalski’s horse. J. Appl. Genet. 2016, 57, 519–525. [Google Scholar] [CrossRef] [PubMed]
- R Core Team 2023-Version 4.3.2 R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. Available online: https://www.R-project.org/ (accessed on 30 October 2023).
- Marklund, L.; Moller, M.J.; Sandberg, K.; Andersson, L. A missense mutation in the gene for melanocyte-stimulating hormone receptor (MC1R) is associated with the chestnut coat color in horses. Mamm. Genome 1996, 7, 895–899. [Google Scholar] [CrossRef]
- Rieder, S.; Taourit, S.; Mariat, D.; Langlois, B.; Guerin, G. Mutations in the agouti (ASIP), the extension (MC1R), and the brown (TYRP1) loci and their association to coat color phenotypes in horses (Equus caballus). Mamm. Genome 2001, 12, 450–455. [Google Scholar] [CrossRef]
- Druml, T.; Grilz-Seger, G.; Horna, M.; Brem, G. Discriminant Analysis of Colour Measurements Reveals Allele Dosage Effect of ASIP/MC1R in Bay Horses. Czech J. Anim. Sci. 2018, 63, 347–355. [Google Scholar] [CrossRef]
- Corbin, L.J.; Pope, J.; Sanson, J.; Antczak, D.F.; Miller, D.; Sadeghi, R.; Brooks, S.A. An Independent Locus Upstream of ASIP Controls Variation in the Shade of the Bay Coat Colour in Horses. Genes 2020, 11, 606. [Google Scholar] [CrossRef] [PubMed]
- Avila, F.; Hughes, S.S.; Magdesian, K.G.; Penedo, M.C.T.; Bellone, R.R. Breed Distribution and Allele Frequencies of Base Coat Color, Dilution, and White Patterning Variants across 28 Horse Breeds. Genes 2022, 13, 1641. [Google Scholar] [CrossRef] [PubMed]
- Kavar, T.; Čeh, E.; Dovč, P. A simplified PCR-based method for detection of gray coat color allele in horse. Mol. Cell Probes. 2012, 26, 256–258. [Google Scholar] [CrossRef] [PubMed]
- Pielberg, R.G.; Golovko, A.; Sundström, E.; Curik, I.; Lennartsson, J.; Seltenhammer, M.H.; Druml, T.; Binns, M.; Fitzsimmons, C.; Lindgren, G.; et al. A cis-acting regulatory mutation causes premature hair graying and susceptibility to melanoma in the horse. Nat. Genet. 2008, 40, 1004–1009. [Google Scholar] [CrossRef]
- Zhao, R.Y.; Zhao, Y.P.; Li, B.; Bou, G.; Zhang, X.Z.; Mongke, T.; Bao, T.; Gereliin, S.; Gereltuuin, T.; Li, C.; et al. Overview of the genetic control of horse coat color patterns. Yi Chuan 2018, 40, 357–368. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions, or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).