Submitted:
24 November 2023
Posted:
24 November 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Fish Sampling
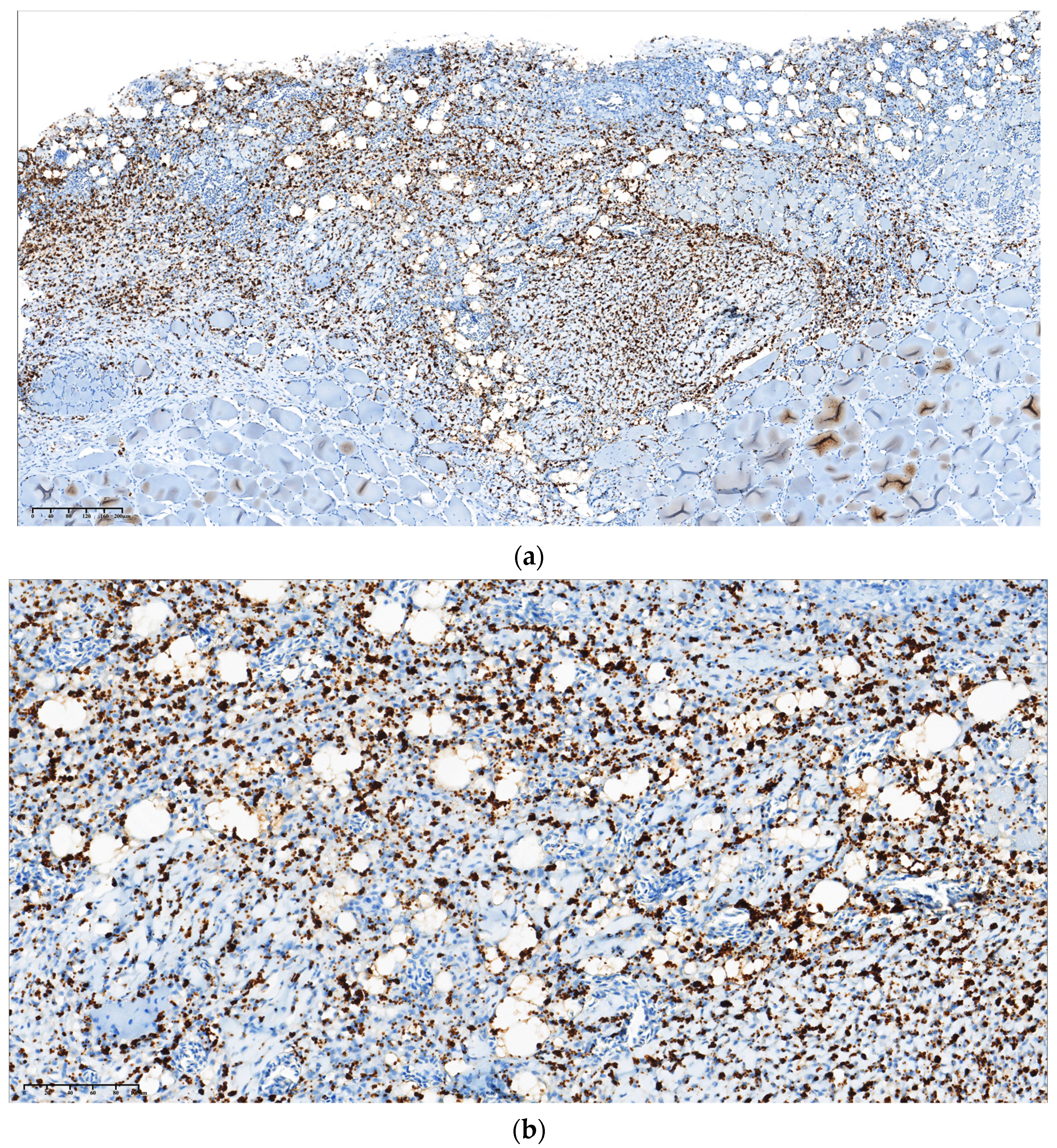
2.2. Gross Pathology and Immunohistochemistry for Detecting Piscirickettsia salmonis in Skin Tissue
2.3. DNA Isolation, 16S rRNA Gene Amplification, and Sequencing
2.4. Data Filtering, Amplicon Sequence Variants Production, and Taxonomic Assignment
2.5. Beta Diversity: Nonmetric Multidimensional Scaling (NMDS) Analysis
2.6. Alpha Diversity: Shannon Index
2.7. Linear Discriminant Analysis (LDA)
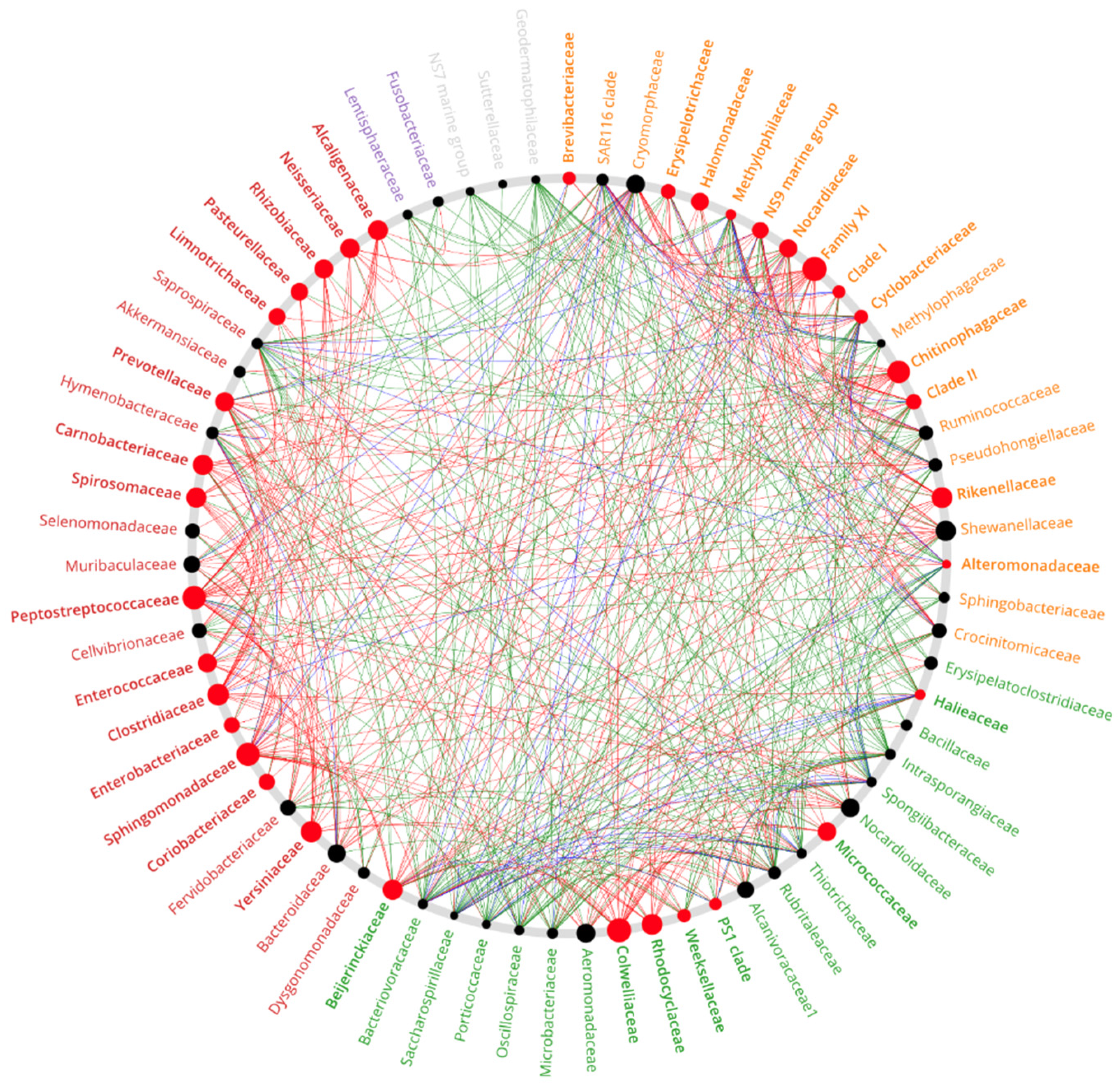
2.8. Co-Occurrence Network Analyses
3. Results
3.1. Gross Pathology and Immunohistochemistry, PCR
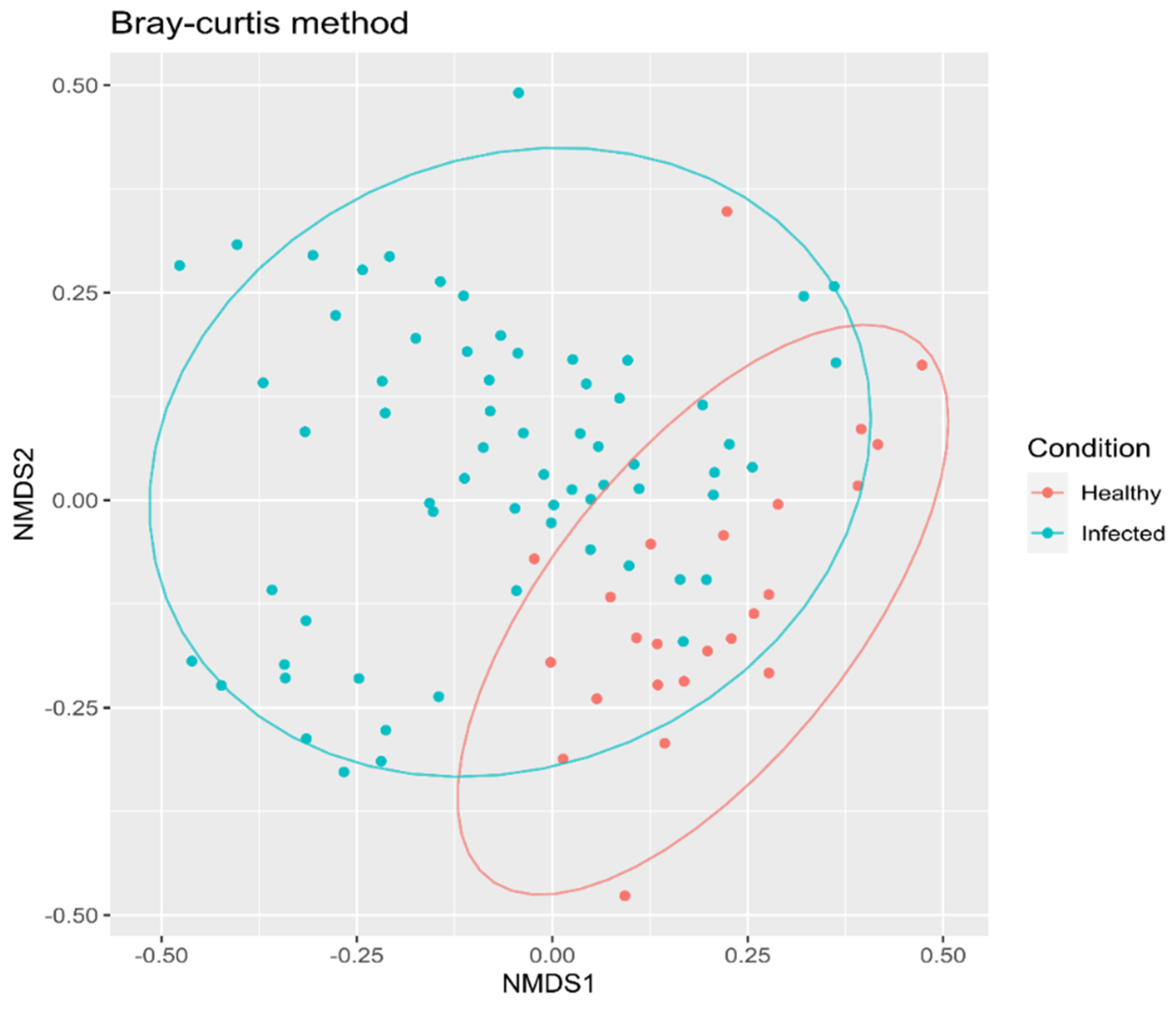
3.2. Beta Diversity
Bray–Curtis Method
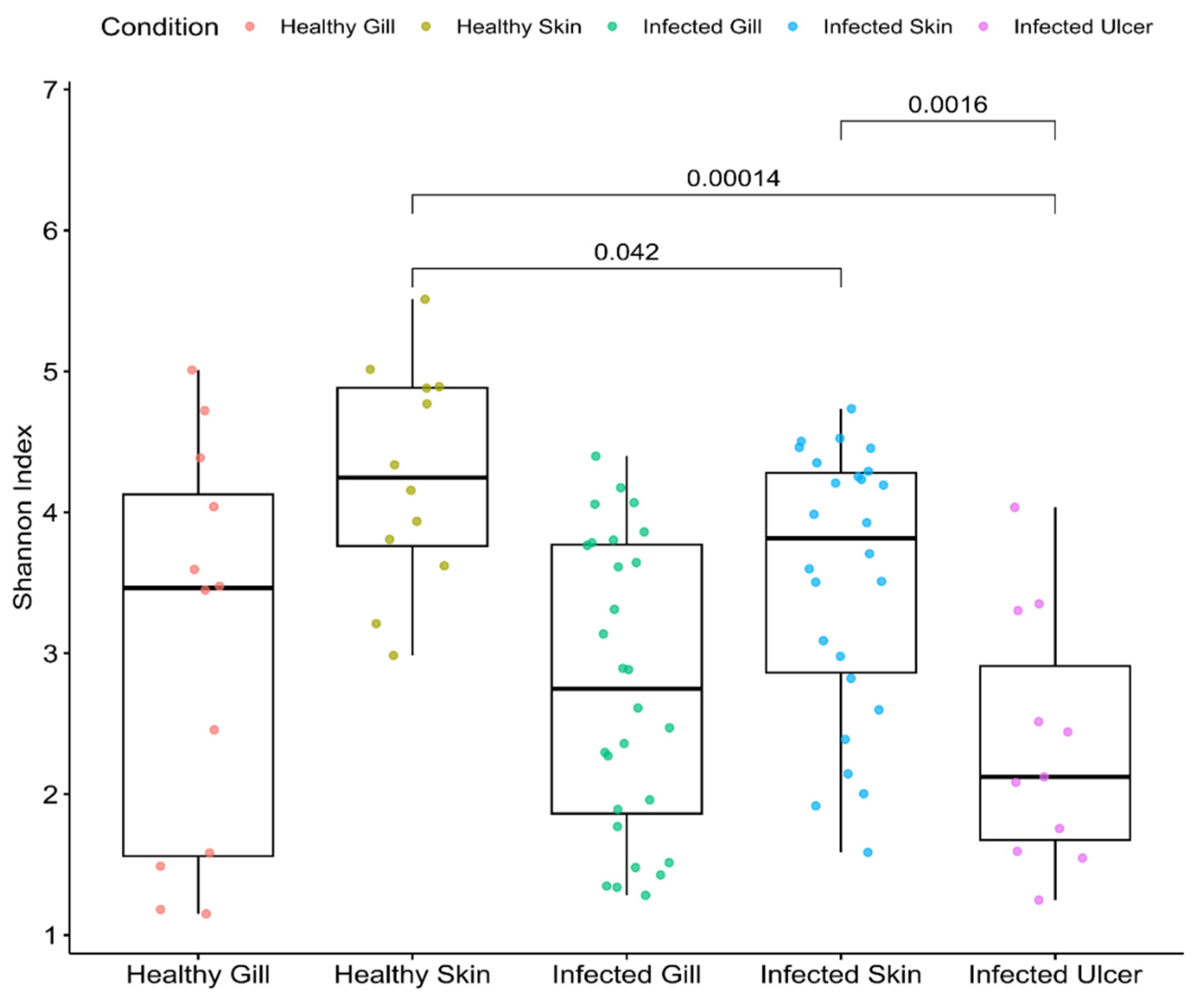
3.3. Alpha Diversity
Shannon Index
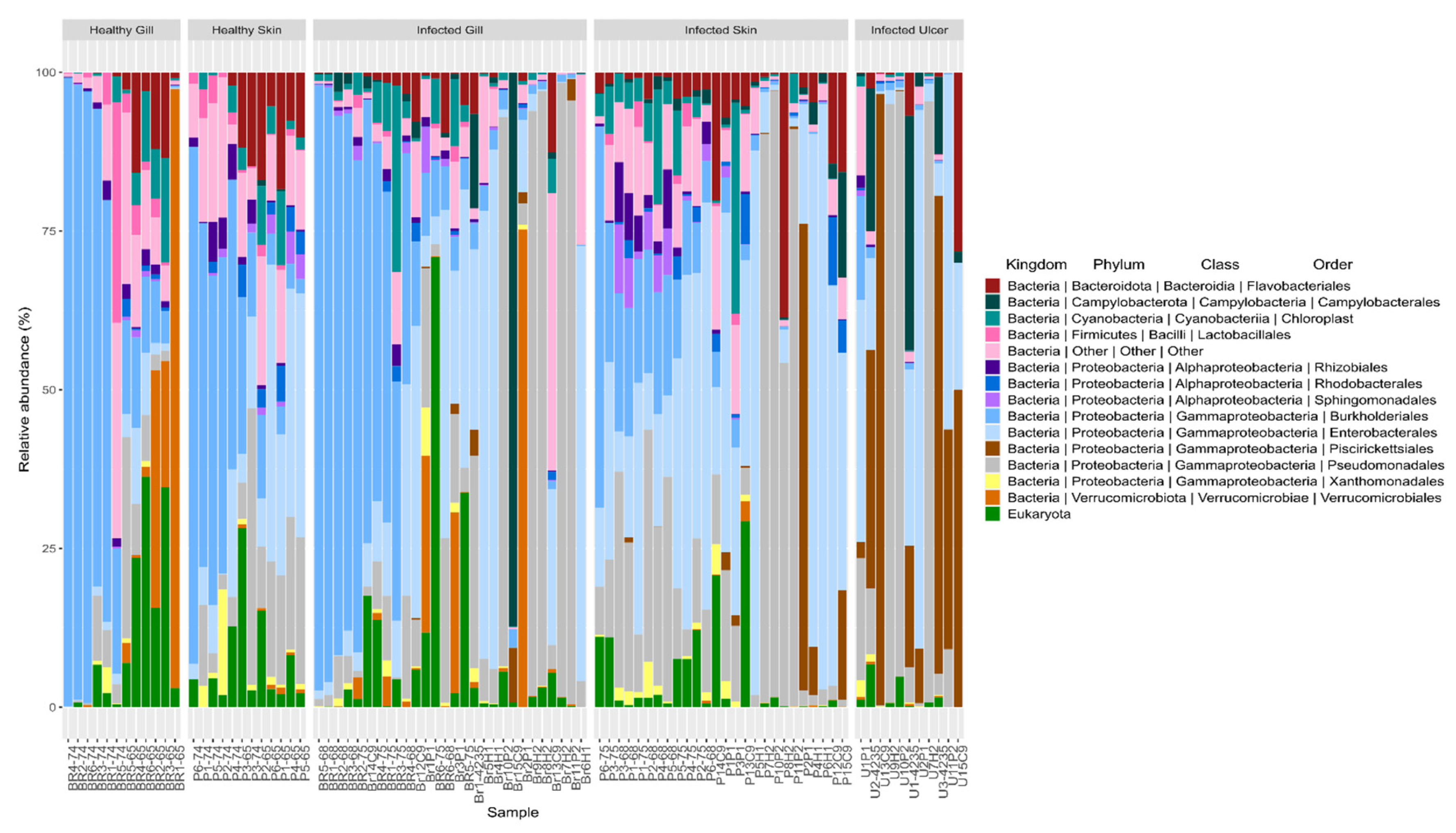
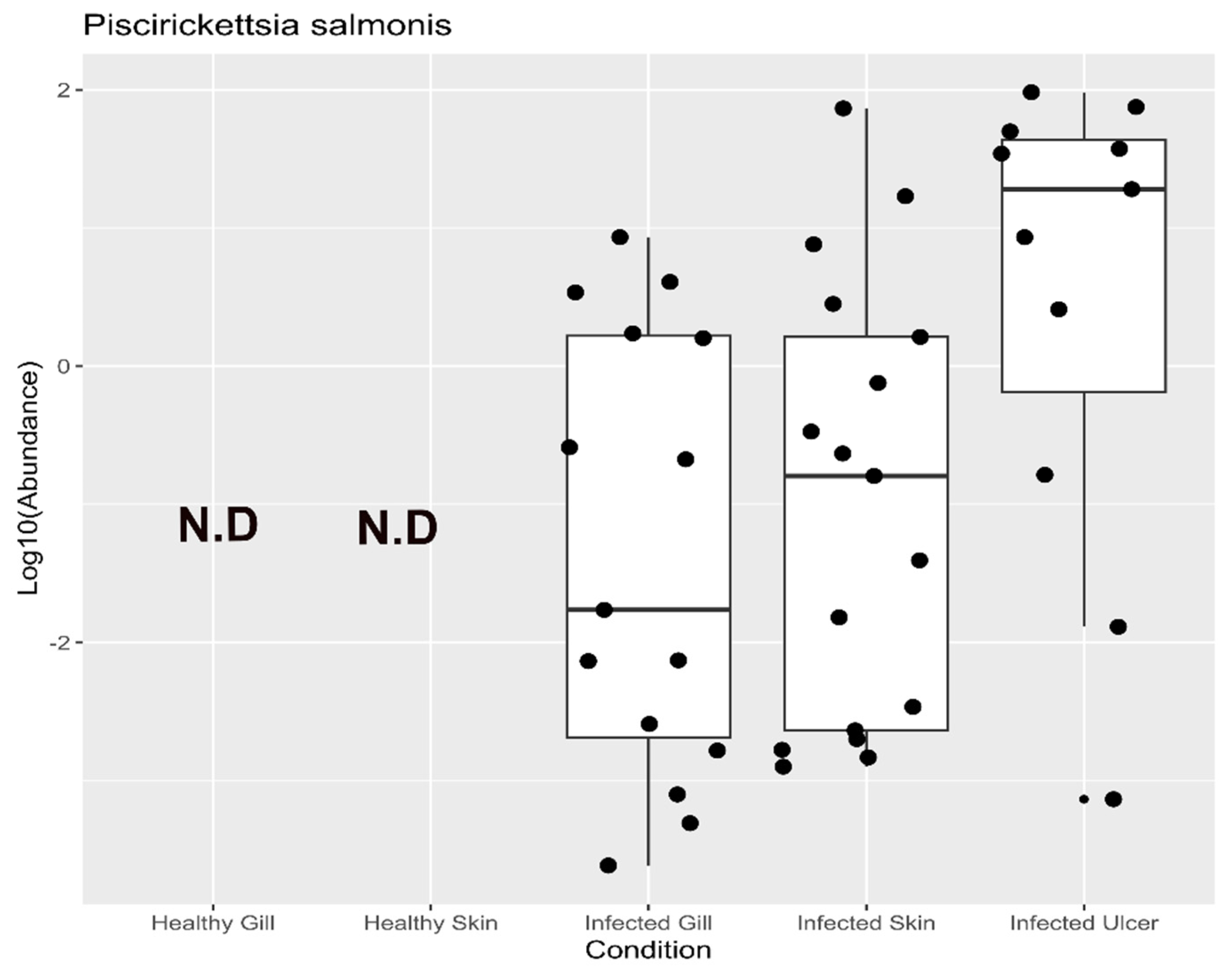
3.4. Taxonomic Assignment
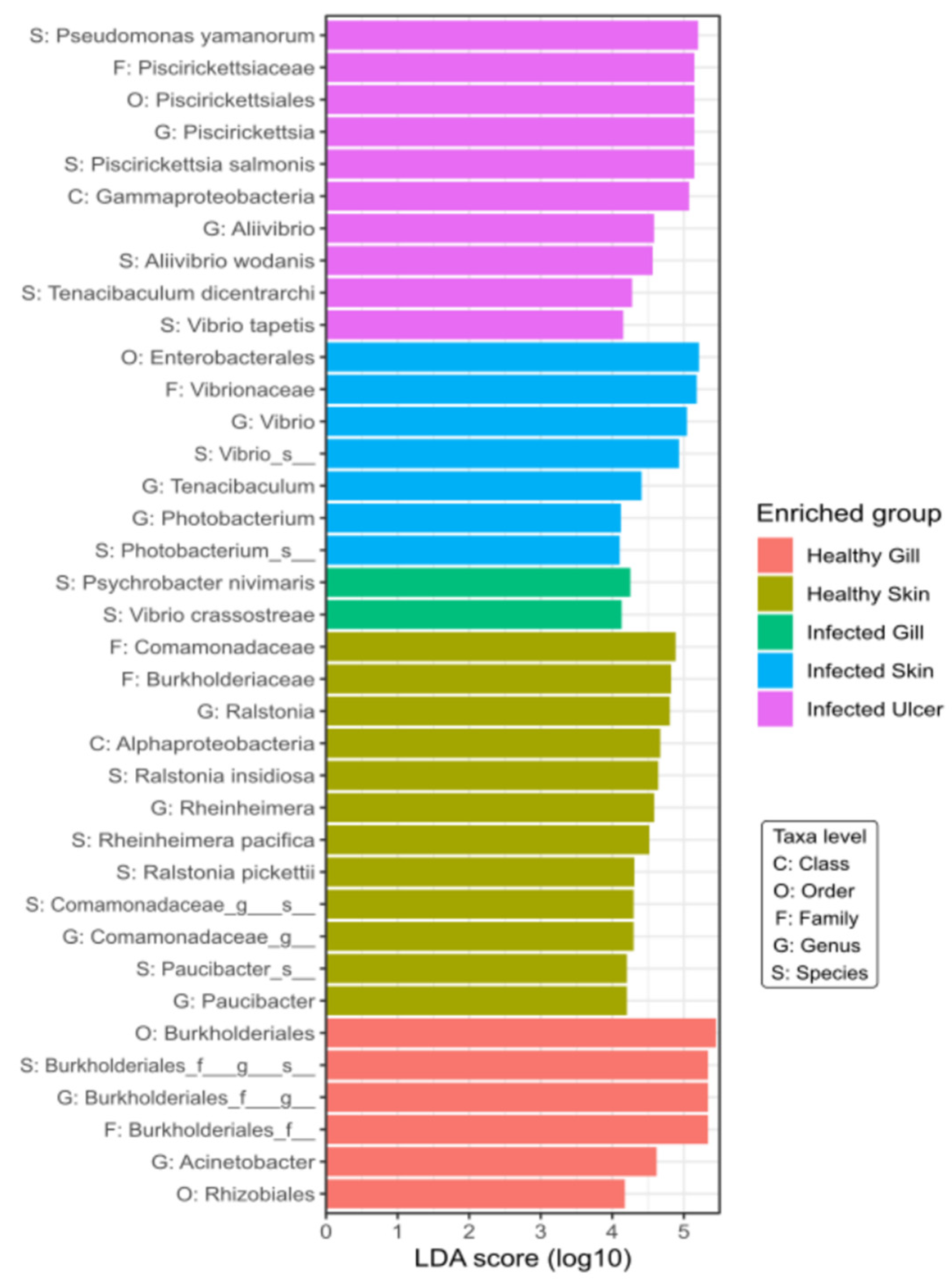
3.5. Linear Discriminant Analysis (LDA)
3.6. Co-Occurrence Network
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
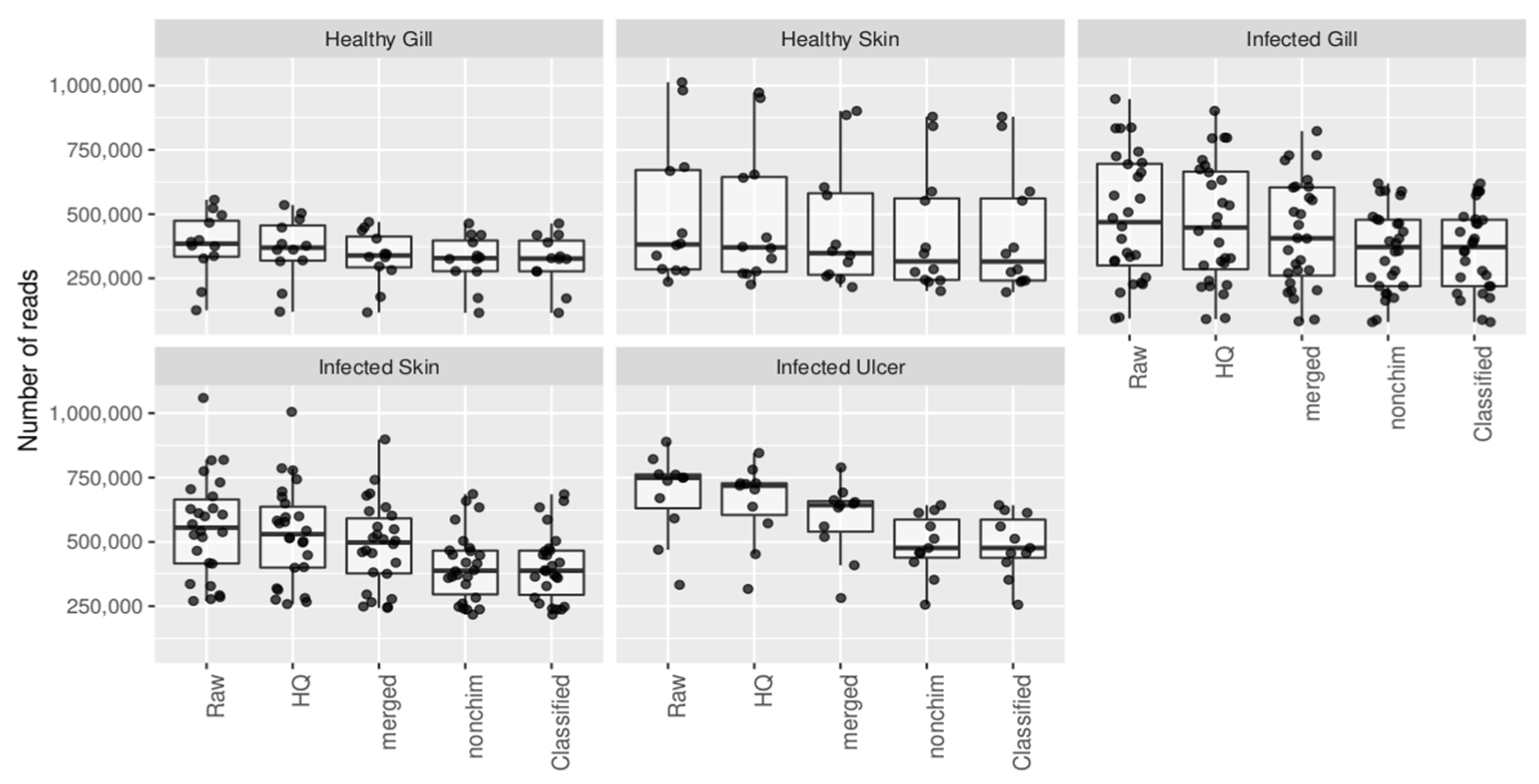
Appendix B
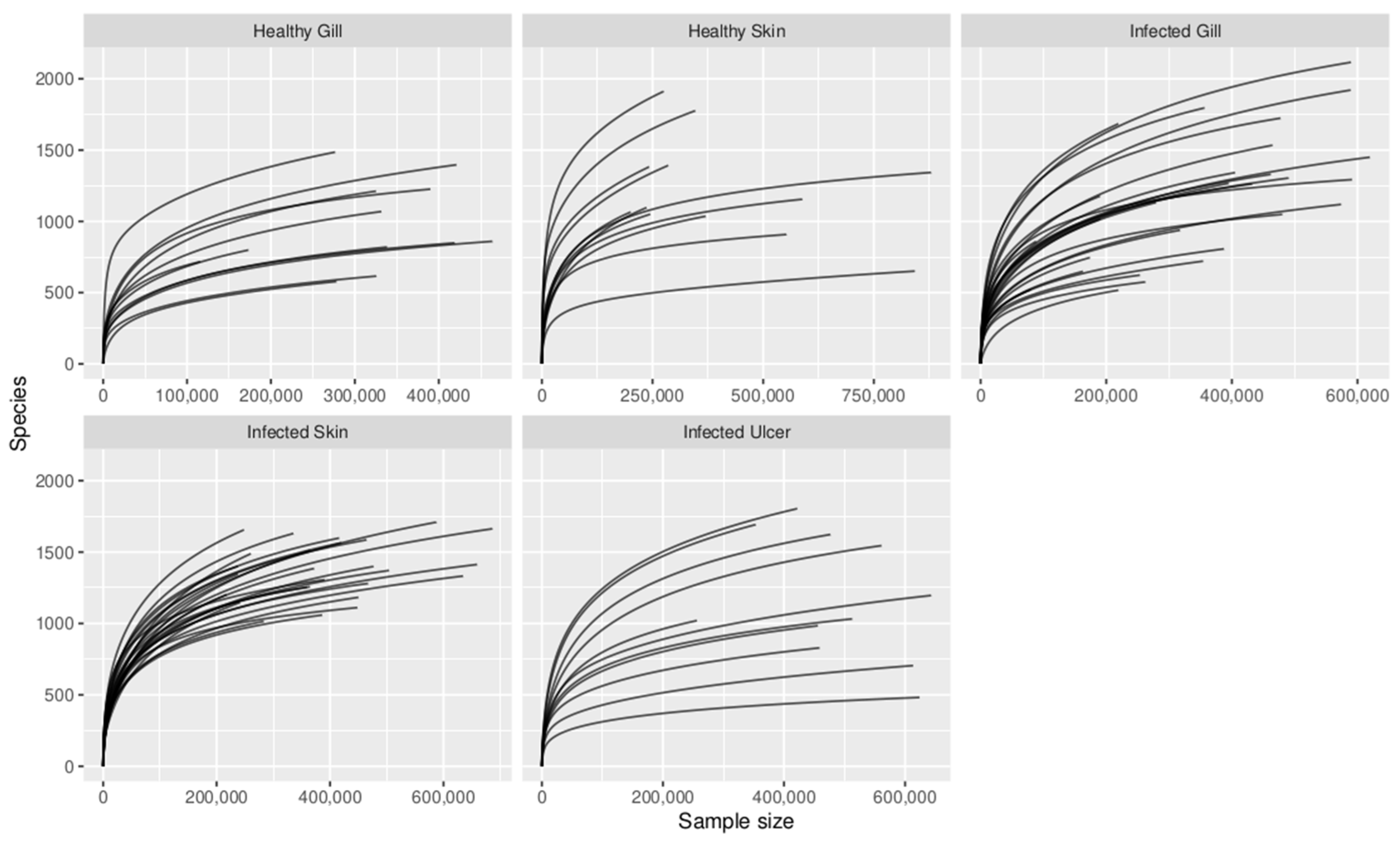
Appendix C
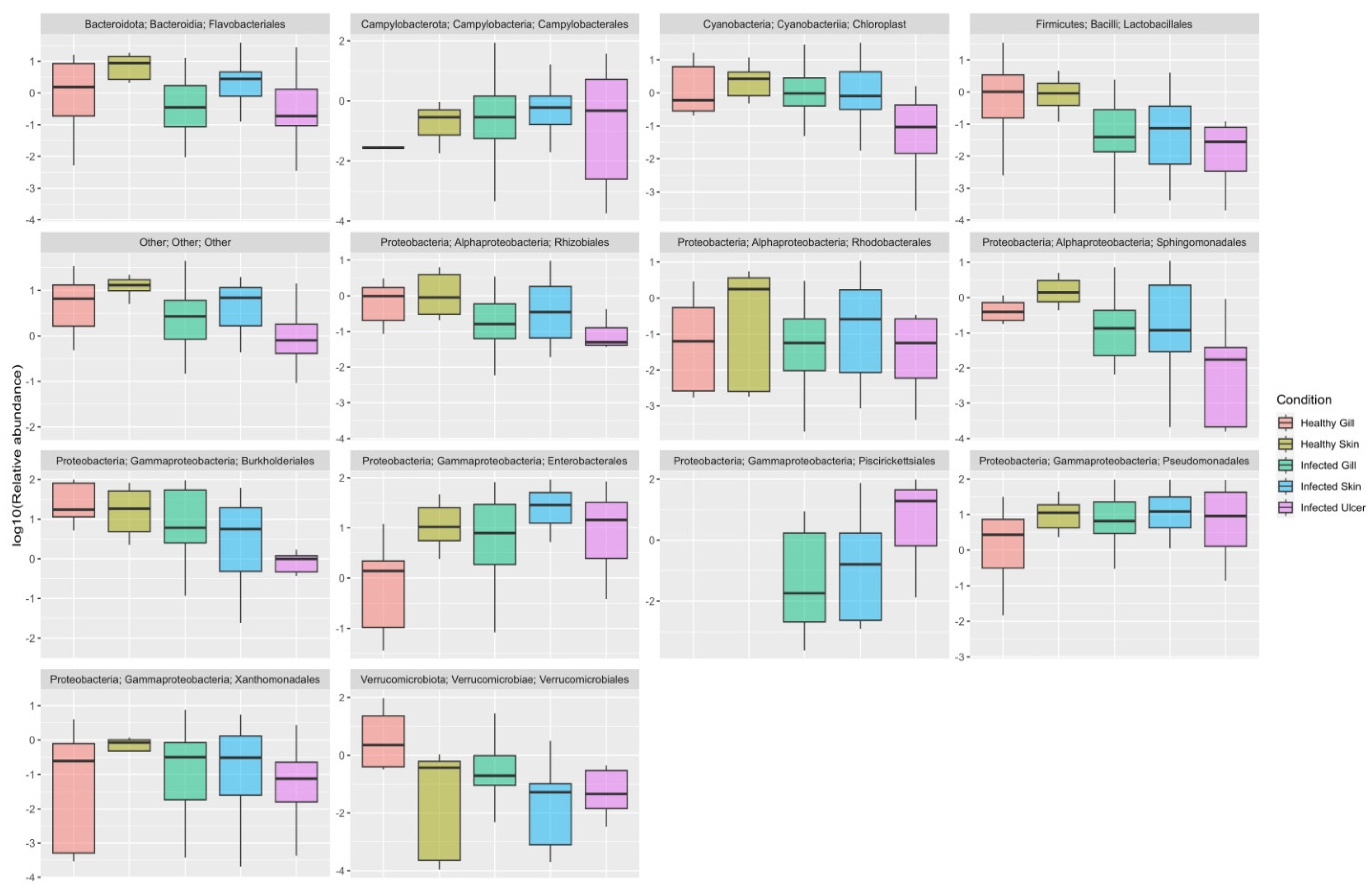
Appendix D
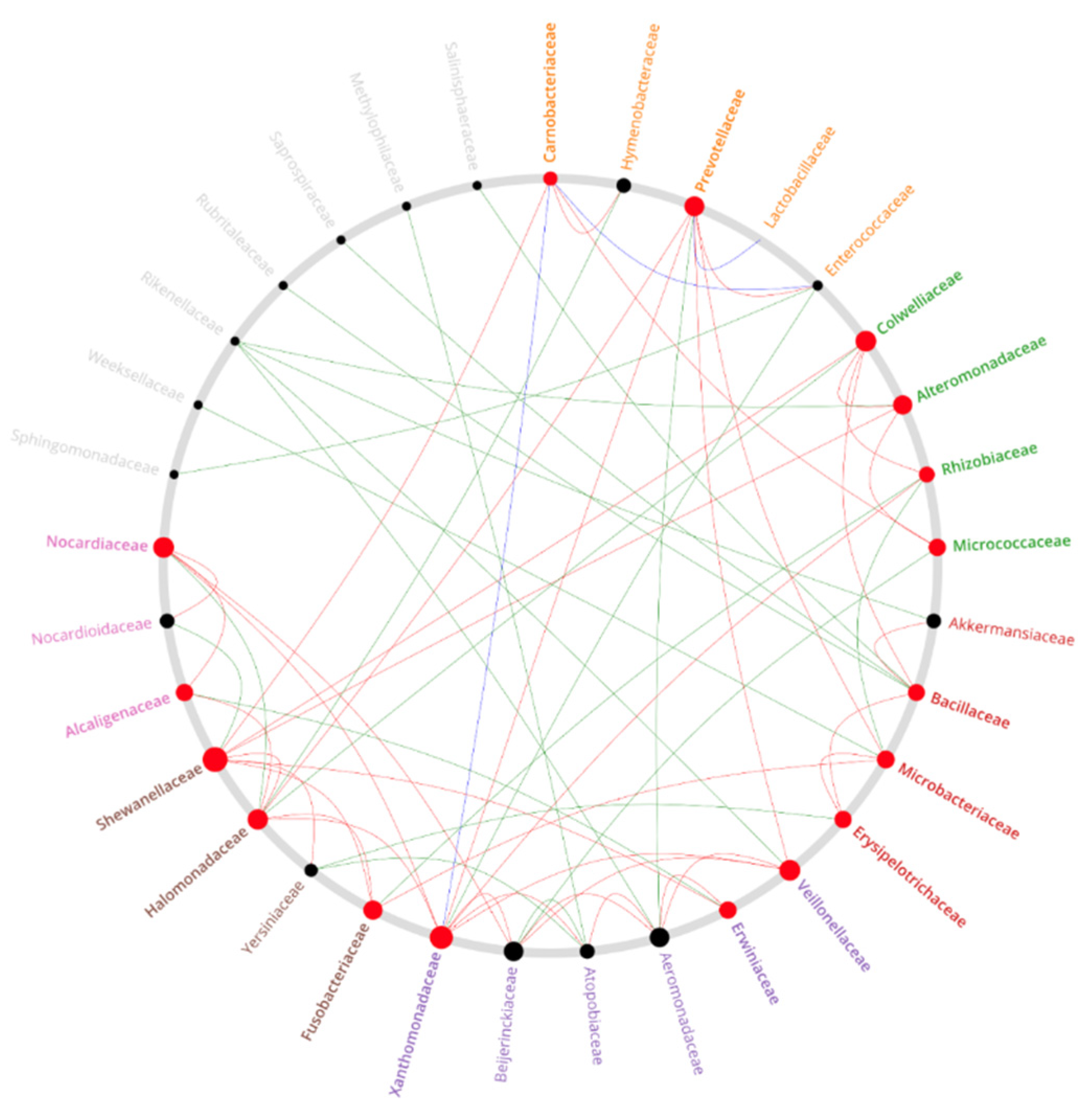
Appendix E
References
- Garcés, L.H.; Larenas, J.J.; Smith, P.A.; Sandino, S.; Lannan, C.N.; Fryer, J.L. Infectivity of a rickettsia isolated from coho salmon Oncorhynchus kisutch. Dis. Aquat. Organ. 1991, 11, 93–97. [Google Scholar] [CrossRef]
- Chen, S.C.; Wang, P.C.; Tung, M.C.; Thompson, K.D.; Adams, A. A Piscirickettsia salmonis-like organism in grouper, Epinephelus melanostigma, in Taiwan. J. Fish Dis. 2000, 23, 415–418. [Google Scholar] [CrossRef]
- Arkush, K.D.; McBride, A.M.; Mendonca, H.L.; Okihiro, M.S.; Andree, K.B.; Marshall, S.; Henriquez, V.; Hedrick, R.P. Genetic characterization and experimental pathogenesis of Piscirickettsia salmonis isolated from white sea bass Atractoscion nobilis. Dis. Aquat. Org. 2005, 63, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Zrnčić, S.; Vendramin, N.; Boutrup, T.S.; Boye, M.; Madsen, L.; Nonneman, B.; Brnić, D.; Oraić, D. First description and diagnostics of disease caused by Piscirickettsia salmonis in farmed European sea bass (Dicentrarchus labrax Linnaeus) from Croatia. J. Fish Dis. 2021, 44, 1033–1042. [Google Scholar] [CrossRef] [PubMed]
- Cvitanich, J.; Gárate, O.; Smith, C.E. Etiological agent in a Chilean coho disease isolated and confirmed by Koch’s postulates. Am. Fish Soc. Newsl. 1990, 18, 1–2. [Google Scholar]
- Cvitanich, J.; Gárate, O.; Smith, C.E. The isolation of a rickettsia-like organism causing disease and mortality in Chilean salmonids and its confirmation by Koch‘s postulate. J. Fish Dis. 1991, 14, 121–145. [Google Scholar] [CrossRef]
- Cvitanich, J.; Gárate, O.; Smith, C.E. Isolation of a new rickettsia-like organism from Atlantic salmon in Chile. Am. Fish Soc. Newsl. 1995, 23, 1–2. [Google Scholar]
- Karlsen, C.; Ottem, K.; Brevik, Ø.J.; Davey, M.; Sørum, H.; Winther-Larsen, H. The environmental and host-associated bacterial microbiota of Arctic seawater-farmed Atlantic salmon with ulcerative disorders. J. Fish. Dis. 2017, 40, 1645–1663. [Google Scholar] [CrossRef]
- Takle, H. Wounds and Skin Welfare in Atlantic Salmon and Rainbow Trout; Nofima: Tromsø, Norway, 2015. [Google Scholar]
- Salinas, I. The mucosal immune system of teleost fish. Biology 2015, 4, 525–539. [Google Scholar] [CrossRef]
- Salinas, I.; Fernández-Montero, A.; Ding, Y.; Sunyer, J.O. Mucosal immunoglobulins of teleost fish: A decade of advances. Dev. Comp. Immunol. 2021, 121, 104079. [Google Scholar] [CrossRef]
- Kelly, C.; Salinas, I. Under pressure: Interactions between commensal microbiota and the teleost immune system. Front. Immunol. 2017, 8, 132. [Google Scholar] [CrossRef] [PubMed]
- Hess, S.; Wenger, A.S.; Ainsworth, T.D.; Rummer, J.L. Exposure of clownfish larvae to suspended sediment levels found on the Great Barrier Reef: Impacts on gill structure and microbiome. Sci. Rep. 2015, 5, 10561. [Google Scholar] [CrossRef]
- Gomez, J.A.; Primm, T.P. A Slimy Business: The Future of Fish Skin Microbiome Studies. Microb. Ecol. 2021, 82, 275–287. [Google Scholar] [CrossRef] [PubMed]
- Koppang, E.O.; Kvellestad, A.; Fischer, U. 5—Fish mucosal immunity: Gill. In Mucosal Health in Aquaculture; Beck, B.H., Peatman, E., Eds.; Academic Press: San Diego, CA, USA, 2015; pp. 93–133. [Google Scholar]
- Streit, B. Bioaccumulation of contaminants in fish. In Fish Ecotoxicology; Braunbeck, T., Hinton, D.E., Streit, B., Eds.; Birkhauser Basel: Basel, Switzerland, 1998; pp. 353–387. [Google Scholar]
- Poleksic, V.; Mitrovic-Tutundzic, V. Fish Gills as a Monitor of Sublethal and Chronic Effects of Pollution. In Fishing News Books Oxford; Fishing News Books Ltd.: Farnham, UK, 1994. [Google Scholar]
- Legrand, T.P.; Catalano, S.R.; Wos-Oxley, M.L.; Stephens, F.; Landos, M.; Bansemer, M.S. The inner workings of the outer surface: Skin and gill microbiota as indicators of changing gut health in yellowtail kingfish. Front. Microbiol. 2018, 8, 26–64. [Google Scholar] [CrossRef] [PubMed]
- Pratte, Z.A.; Besson, M.; Hollman, R.D.; Stewart, F.J. The gills of reef fish support a distinct microbiome influenced by host-specific factors. Appl. Environ. Microbiol. 2018, 84, e00063-18. [Google Scholar] [CrossRef]
- Minich, J.J.; Poore, G.D.; Jantawongsri, K.; Johnston, C.; Bowie, K.; Bowman, J. Microbial ecology of Atlantic salmon (Salmo salar) hatcheries: Impacts of the built environment on fish mucosal microbiota. Appl. Environ. Microbiol. 2020, 86, e00411-20. [Google Scholar] [CrossRef] [PubMed]
- Sehnal, L.; Brammer-Robbins, E.; Wormington, A.M.; Blaha, L.; Bisesi, J.; Larkin, I. Microbiome composition and function in aquatic vertebrates: Small organisms making big impacts on aquatic animal health. Front. Microbiol. 2021, 12, 358. [Google Scholar] [CrossRef] [PubMed]
- Palaniappan, P.L.R.M.; Nishanth, T.; Renju, V.B. Bioconcentration of zinc and its effect on the biochemical constituents of the gill tissues of Labeo rohita: An ft-ir study. Infrared Phys. Technol. 2010, 53, 103–111. [Google Scholar] [CrossRef]
- Wu, S.; Wang, G.; Angert, E.R.; Wang, W.; Li, W.; Zou, H. Composition, diversity, and origin of the bacterial community in grass carp intestine. PLoS ONE 2012, 7, 30440. [Google Scholar] [CrossRef]
- Schommer, N.N.; Gallo, R.L. Structure and function of the human skin microbiome. Trends Microbiol. 2013, 21, 660–668. [Google Scholar] [CrossRef]
- Dorrestein, P.C.; Gallo, R.L.; Knight, R. Microbial skin inhabitants: Friends forever. Cell 2016, 165, 771–772. [Google Scholar] [CrossRef]
- Gallo, R.L. Human skin is the largest epithelial surface for interaction with microbes. J. Investig. Dermatol. 2017, 137, 1213–1214. [Google Scholar] [CrossRef] [PubMed]
- Lorgen-Ritchie, M.; Clarkson, M.; Chalmers, L.; Taylor, J.F.; Migaud, H.; Martin, S.A.M. Temporal changes in skin and gill microbiomes of Atlantic salmon in a recirculating aquaculture system—Why do they matter? Aquaculture 2022, 558, 738352. [Google Scholar] [CrossRef]
- Brown, R.M.; Wiens, G.D.; Salinas, I. Analysis of the gut and gill microbiome of resistant and susceptible lines of rainbow trout (Oncorhynchus mykiss). Fish Shellfish Immunol. 2019, 86, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Brown, R.; Moore, L.; Mani, A.; Patel, S.; Salinas, I. Effects of ploidy and salmonid alphavirus infection on the skin and gill microbiome of Atlantic Salmon (Salmo Salar). PLoS ONE 2021, 16, e0243684. [Google Scholar] [CrossRef]
- Casadei, E.; Mani, A.; Cisco, M.; Vågnes, Ø.; Salinas, I.; Patel, S. Sex-dependent effects of mechanical delousing on the skin microbiome of broodstock Atlantic salmon (Salmo salar L.). Sci. Rep. 2023, 13, 10824. [Google Scholar] [CrossRef]
- Valenzuela-Miranda, D.; Gonçalves, A.T.; Valenzuela-Muñoz, V.; Nuñez-Acuña, G.; Liachko, I.; Nelson, B.; Gallardo-Escarate, C. Proximity ligation strategy for the genomic reconstruction of microbial communities associated with the ectoparasite Caligus rogercresseyi. Sci. Rep. 2022, 12, 783. [Google Scholar] [CrossRef] [PubMed]
- Koch, R. Xth International Congress of Medicine, Berlin. 1890.
- Brown, S.P.; Cornforth, D.M.; Mideo, N. Evolution of virulence in opportunistic pathogens: Generalism, plasticity, and control. Trends Microbiol. 2012, 20, 336–342. [Google Scholar] [CrossRef]
- Lhorente, J.P.; Gallardo, J.A.; Villanueva, B.; Carabaño, M.J.; Neira, R. Disease resistance in Atlantic salmon (Salmo salar): Coinfection of the intracellular bacterial pathogen Piscirickettsia salmonis and the sea louse Caligus rogercresseyi. PLoS ONE 2014, 9, e95397. [Google Scholar] [CrossRef]
- Arriagada, G.; Hamilton-West, C.; Nekouei, O.; Foerster, C.; Müller, A.; Lara, M.; Gallardo-Escárate, C. Caligus rogercresseyi infestation is associated with Piscirickettsia salmonis-attributed mortalities in farmed salmonids in Chile. Prev. Vet. Med. 2019, 171, 104771. [Google Scholar] [CrossRef]
- Bustos P, Figueroa C, Cádiz B, Santander T, Dixon B, Gallardo JA, Conejeros P. Immune response induced by coinfection of the sea louse Caligus rogercresseyi and the intracellular bacteria Piscirickettsia salmonis in vaccinated Atlantic salmon. J Fish Dis. 2023 Dec;46(12):1337-1342. [CrossRef]
- Rozas-Serri, M.; Peña, A.; Gardner, I.; Peñaloza, E.; Maldonado, L.; Muñoz, A.; Mardones, F.O.; Rodríguez, C.; Ildefonso, R.; Senn, C.; et al. Co-Infection by LF-89-Like and EM-90-Like Genogroups of Piscirickettsia Salmonis in Farmed Atlantic Salmon in Chile: Implications for Surveillance and Control of Piscirickettsiosis. Pathogens 2023, 12, 450. [Google Scholar] [CrossRef] [PubMed]
- Karatas, S.; Mikalsen, J.; Steinum, T.M.; Taksdal, T.; Bordevik, M.; Colquhoun, D.J. Real time PCR detection of Piscirickettsia salmonis from formalin-fixed paraffin-embedded tissues. J. Fish Dis. 2008, 31, 747–753. [Google Scholar] [CrossRef]
- Prophet, E.B.; Mills, B.; Arrington, J.B.; Sobin, L.H. Laboratory methods in histotechnology. Armed Forces Inst. Pathol. 1992, 56, 151–164. [Google Scholar]
- Noga, E.J. Fish Disease: Diagnosis and Treatment, 2nd ed.; Wiley-Blackwell: Oxford, UK, 2010; ISBN 978-0-8138-0697-6. [Google Scholar]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Dixon, P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- LEfSe. (Genome Biology, 2011 Jun 24;12(6):R60). Available online: http://huttenhower.sph.harvard.edu/lefse/.
- Griffith, D.M.; Veech, J.A.; Marsh, C.J. Cooccur: Probabilistic species co-occurrence analysis in R. J. Stat. Softw. 2016, 69, 1–17. [Google Scholar] [CrossRef]
- Kuntal, B.K.; Chandrakar, P.; Sadhu, S.; Mande, S.S. ‘NetShift’: A methodology for understanding ‘driver microbes’ from healthy and disease microbiome datasets. ISME J. 2019, 13, 442–454. [Google Scholar] [CrossRef]
- Friedman, J.; Alm, E.J. Inferring Correlation Networks from Genomic Survey Data. PLoS Comput. Biol. 2012, 8, e1002687. [Google Scholar] [CrossRef] [PubMed]
- Kashinskaya, E.N.; Simonov, E.P.; Andree, K.B.; Vlasenko, P.G.; Polenogova, O.V.; Kiriukhin, B.A.; Solovyev, M.M. Microbial community structure in a host-parasite system: The case of Prussian carp and its parasitic crustaceans. J. Appl. Microbiol. 2021, 131, 1722–1741. [Google Scholar] [CrossRef]
- Morales-Rivera, M.F.; Valenzuela-Miranda, D.; Valenzuela-Muñoz, V.; Nuñez-Acuña, G.; Avendaño-Herrera, R.; Gallardo-Escárate, C. Nanopore sequencing evidenced the presence of fish bacterial pathogens in the sea louse (Caligus rogercresseyi) microbiota collected from distant salmon farms in Chile. Aquaculture 2022, 552, 738026. [Google Scholar] [CrossRef]
- Lunder, T.; Evensen, Ø.; Holstad, G.; Hastein, T. Winter ulcer in the Atlantic salmon Salmo Salar—Pathological and bacteriological investigations and transmission experiments. Dis. Aquat. Org. 1995, 23, 39–49. [Google Scholar] [CrossRef]
- Whitman, K.; Backman, S.; Benediktsdottir, E.; Coles, M.; Johnson, G.R. Isolation and characterization of a new Vibrio spp. (Vibrio wodanis) associated with winter ulcer disease in sea water raised Atlantic salmon (Salmo salar L.) in New Brunswick. Aquac. Can. 2001, 2000, 115. [Google Scholar]
- Karlsen, C.; Vanberg, C.; Mikkelsen, H.; Sørum, H. Co-infection of Atlantic salmon (Salmo salar), by Moritella viscosa and Aliivibrio wodanis, development of disease and host colonization. Vet. Microbiol. 2014, 171, 112–121. [Google Scholar] [CrossRef] [PubMed]
- Benediktsdóttir, E.; Helgason, S.; Sigurjónsdóttir, H. Vibrio spp. isolated from salmonids with shallow skin lesions and reared at low temperatures. J. Fish Dis. 1998, 21, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Bruno, D.W.; Griffiths, J.; Petrie, J.; Hastings, T.S. Vibrio viscosus in farmed Atlantic salmon Salmo salar in Scotland: Field and experimental observations. Dis. Aquat. Org. 1998, 34, 161–166. [Google Scholar] [CrossRef] [PubMed]
- Karlsen, C.; Sørum, H.; Willassen, N.P.; Asbakk, K. Moritella viscosa bypasses Atlantic salmon epidermal keratocyte clearing activity and might use skin surfaces as a port of infection. Vet. Microbiol. 2012, 154, 353–362. [Google Scholar] [CrossRef]
- Hjerde, E.; Karlsen, C.; Sørum, H.; Parkhill, J.; Willassen, N.P.; Thomson, N.R. Co-cultivation and transcriptome sequencing of two co-existing fish pathogens Moritella viscosa and Aliivibrio wodanis. BMC Genom. 2015, 10, 16–447. [Google Scholar] [CrossRef]
- Maharajan, A.D.; Hansen, H.; Khider, M.; Willassen, N.P. Quorum sensing in Aliivibrio wodanis 06/09/139 and its role in controlling various phenotypic traits. PeerJ 2021, 9, e11980. [Google Scholar] [CrossRef]
- Olsen, A.B.; Gulla, S.; Steinum, T.; Colquhoun, D.J.; Nilsen, H.K.; Duchaud, E. Multilocus sequence analysis reveals extensive genetic variety within Tenacibaculum spp. associated with ulcers in sea-farmed fish in Norway. Vet. Microbiol. 2017, 205, 39–45. [Google Scholar] [CrossRef]
- Wynne, J.W.; Thakur, K.K.; Slinger, J.; Samsing, F.; Milligan, B.; Powell, J.F.; Siah, A. Microbiome profiling reveals a microbial dysbiosis during a natural outbreak of tenacibaculosis (Yellow mouth) in Atlantic salmon. Front. Microb. 2020, 11, 586387. [Google Scholar] [CrossRef] [PubMed]
- Slinger, J.; Adams, M.B.; Wynne, J.W. Bacteriomic profiling of branchial lesions induced by Neoparamoeba perurans challenge reveals commensal dysbiosis and an association with Tenacibaculum dicentrarchi in AGD-affected Atlantic salmon (Salmo salar L.). Microorganisms 2020, 8, 1189. [Google Scholar] [CrossRef]
- Apablaza, P.; Frisch, K.; Brevik, Ø.J.; Småge, S.B.; Vallestad, C.; Duesund, H.; Mendoza, J.; Nylund, A. Primary Isolation and Characterization of Tenacibaculum maritimum from Chilean Atlantic Salmon Mortalities Associated with a Pseudochattonella spp. Algal Bloom. J. Aquat. Anim. Health 2017, 29, 143–149. [Google Scholar] [CrossRef]
- Avendaño-Herrera, R.; Toranzo, A.; Magariños, B. Tenacibaculosis infection in marine fish caused by Tenacibaculum maritimum: A review. Dis. Aquat. Org. 2006, 71, 255–266. [Google Scholar] [CrossRef]
- Småge, S.B.; Brevik, Ø.J.; Duesund, H.; Ottem, K.F.; Watanabe, K.; Nylund, A. Tenacibaculum finnmarkense sp. nov., a fish pathogenic bacterium of the family Flavobacteriaceae isolated from Atlantic salmon. Antonie Van Leeuwenhoek 2016, 109, 273–285. [Google Scholar] [CrossRef] [PubMed]
- Benediktsdóttir, E.; Verdonck, L.; Sproer, C.; Helgason, S.; Swings, J. Characterization of Vibrio viscosus and Vibrio wodanis isolated at different geographical locations: A proposal for reclassification of Vibrio viscosus as Moritella viscosa comb. nov. Int. J. Syst. Evol. Microbiol. 2000, 50, 479–488. [Google Scholar] [CrossRef] [PubMed]
- Lunder, T.; Sørum, H.; Holstad, G.; Steigerwalt, A.G.; Mowinc-kel, P.; Brenner, D.J. Phenotypic and genotypic characterization of Vibrio viscosus sp. nov. and Vibrio wodanis sp. nov. isolated from Atlantic salmon (Salmo salar) with ‘winter ulcer’. Int. J. Syst. Evol. Microbiol. 2000, 50, 427–450. [Google Scholar] [CrossRef]
- Hovda, M.B.; Fontanillas, R.; McGurk, C.; Obach, A.; Rosnes, J.T. Seasonal variations in the intestinal microbiota of farmed Atlantic salmon (Salmo salar L.). Aquac. Res. 2012, 43, 154–159. [Google Scholar] [CrossRef]
- Hamdan, K.; Littman, D.R. The microbiome in infectious disease and inflammation. Annu. Rev. Immunol. 2013, 30, 759–795. [Google Scholar] [CrossRef]
- Rudi, K.; Angell, I.L.; Pope, P.B.; Vik, J.O.; Sandve, S.R.; Snipen, L.G. Stable core gut microbiota across the freshwater-to-saltwater transition for farmed Atlantic salmon. Appl. Environ. Microbiol. 2018, 84, e01974-17. [Google Scholar] [CrossRef] [PubMed]
- Llewellyn, M.S.; McGinnity, P.; Dionne, M.; Letourneau, J.; Thonier, F.; Carvalho, G.R.; Derome, N. The biogeography of the Atlantic salmon (Salmo salar) gut microbiome. Multidiscip. J. Microb. Ecol. 2015, 10, 1280–1284. [Google Scholar] [CrossRef] [PubMed]
- Austin, B.; Austin, D.A. Bacterial Fish Pathogens. Disease of Farmed and Wild Fish; Springer International Publishing: Cham, Switzerland, 2016; pp. 475–498. [Google Scholar] [CrossRef]
- Attia, M.M.; Abdelsalam, M.; Elgendy, M.Y.; Sherif, A.H. Dactylogyrus extensus and Pseudomonas fluorescens dual infection in farmed common carp (Cyprinus carpio). Microb. Pathog. 2022, 173, 105867. [Google Scholar] [CrossRef] [PubMed]
- Oh, W.T.; Kim, J.H.; Jun, J.W.; Giri, S.S.; Yun, S.; Kim, H.J.; Kim, S.G.; Kim, S.W.; Han, S.J.; Kwon, J.; et al. Genetic characterization and pathological analysis of a novel bacterial pathogen, Pseudomonas tructae, in rainbow trout (Oncorhynchus mykiss). Microorganisms 2019, 7, 432. [Google Scholar] [CrossRef] [PubMed]
- Rao, Q.; Liu, Y.; Chen, C.; Lin, Q.; Ren, L.; Huang, M.; Tu, J.; Luo, T. Pseudomonas ovata sp. nov., isolated from the skin of the tail of Farmed Murray cod (Maccullochella peelii peelii) with a profound ulceration. Curr. Microbiol. 2019, 76, 1168–1174. [Google Scholar] [CrossRef] [PubMed]
- Moradali, M.F.; Ghods, S.; Rehm, B.H.A. Pseudomonas aeruginosa lifestyle: A paradigm for adaptation, survival, and persistence. Front. Cell Infect. Microbiol. 2017, 7, 39. [Google Scholar] [CrossRef] [PubMed]
- Levipan, H.A.; Irgang, R.; Tapia-Cammas, D.; Avendaño-Herrera, R. A high-throughput analysis of biofilm formation by the fish pathogen Tenacibaculum dicentrarchi. J. Fish Dis. 2019, 42, 617–621. [Google Scholar] [CrossRef]
- Cornelis, P.; Bodilis, J. A survey of TonB-dependent receptors in fluorescent pseudomonads. Environ. Microbiol. Rep. 2009, 1, 256–262. [Google Scholar] [CrossRef]
- Avendaño-Herrera, R.; Toranzo, A.E.; Romalde, J.L.; Lemos, M.L.; Magariños, B. Iron uptake mechanisms in the fish pathogen Tenacibaculum maritimum. Appl. Environ. Microbiol. 2005, 71, 6947–6953. [Google Scholar] [CrossRef]
- Calquín, P.; Ruiz, P.; Oliver, C.; Sánchez, P.; Haro, R.; Oliva, H. Physiological evidence that Piscirickettsia salmonis produces siderophores and uses iron from different sources. J. Fish Dis. 2017, 41, 553–558. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.A.; Pizarro, P.; Ojeda, P.; Contreras, J.; Oyanedel, S.; Larenas, J. Routes of entry of Piscirickettsia salmonis in rainbow trout Oncorhynchus mykiss. Dis. Aquat. Org. 1999, 37, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.A.; Rojas, M.E.; Guajardo, A.; Contreras, J.; Morales, M.A.; Larenas, J. Experimental infection of coho salmon Oncorhynchus kisutch by exposure of skin, gills, and intestine with Piscirickettsia salmonis. Dis. Aquat. Org. 2004, 61, 53–57. [Google Scholar] [CrossRef] [PubMed]
- Turnbull, J.F.; Richards, R.H.; Robertson, D.A. Gross, histological and scanning electron microscopic appearance of dorsal fin rot in farmed Atlantic salmon, Salmo salar L., parr. J. Fish Dis. 1996, 19, 415–427. [Google Scholar] [CrossRef]
- Yañez, J.M.; Lhorente, J.P.; Bassini, L.N.; Oyarzún, M.; Neira, R.; Newman, S. Genetic co-variation between resistance against both Caligus rogercresseyi and Piscirickettsia salmonis, and body weight in Atlantic salmon (Salmo salar). Aquaculture 2014, 433, 295–298. [Google Scholar] [CrossRef]
- González, L.; Robles, C.; San Martín, M.C. Management issues regarding caligidosis treatment on salmon farms in Chile affected by infection salmon anaemia virus (ISAV), Piscirickettsia salmonis and Neoparamoeba perurans. Ocean Coast. Manag. 2016, 123, 74–83. [Google Scholar] [CrossRef]



| Farmed Sampled | Health Condition | Sampled Type | Region |
| C1 | Infected with P. salmonis | Skin, gill, and ulcer | Los Lagos |
| C2 | Infected with P. salmonis | Skin, gill, and ulcer | Los Lagos |
| C3 | Infected with P. salmonis | Skin, gill, and ulcer | Aysén |
| C4 | Infected with P. salmonis | Skin, gill, and ulcer | Los Lagos |
| C5 | Infected with P. salmonis | Skin, gill, and ulcer | Los Lagos |
| C6 | Infected with P. salmonis | Skin, gill, and ulcer | Los Lagos |
| C7 | Infected with P. salmonis | Skin, gill, and ulcer | Aysén |
| C8 | Control Farm | Skin and gill | Aysén |
| C9 | Control Farm | Skin and gill | Los Lagos |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





