1. Introduction
Breast cancer is a highly heterogenous disease and classified in distinct subgroups based on transcriptomic profiles that are associated with outcome [
1]. Expression experiments initially identified four breast cancer ‘intrinsic’ subtypes (basal-like, HER2-enriched, luminal and normal breast-like) [
2], and subsequent studies have led to the sub-stratification of luminal breast cancers into luminal A and luminal B, and shown that this classification system is of prognostic significance [
3].
The overlap ratio can be as high as 60–90% between basal-like breast cancer (BLBC) and triple negative breast cancer (without expression of hormonal receptors) (TNBC), compared to only 11.5% between non-TNBC and BLBC [
2,
4]. High risk early triple-negative breast cancer is frequently associated with early recurrence and high mortality [
5]. In this subgroup of aggressive breast cancer, neoadjuvant chemotherapy is the preferred treatment approach [
6]. Immune checkpoint inhibition may enhance endogenous anticancer immunity after increased release of tumor-specific antigens with chemotherapy [
7]. Pembrolizumab is an anti–programmed death 1 (PD-1) monoclonal antibody. Among patients with early triple-negative breast cancer, the percentage with a pathological complete response was significantly higher among those who received pembrolizumab plus neoadjuvant chemotherapy than among those who received placebo plus neoadjuvant chemotherapy [
8].
Ferroptosis is a non-apoptosis regulating cell death. Unlike autophagy and apoptosis, ferroptosis is an iron-dependent and reactive oxygen species (ROS)-reliant cell death with characteristics mainly of cytological changes, including decreased or vanished mitochondria cristae, a ruptured outer mitochondrial membrane, and a condensed mitochondrial membrane [
9]. Accumulating evidence supports that it regulates tumor progression by releasing multiple signal molecules in the tumor microenvironment, and plays a key role in cancer biology and drug resistance [
10]. Expression correlation between PD-L1 and ferroptosis-related genes have already observed in transcriptome of TNBC especially a strong positive correlation with ACSL4 expression [
11].
In this present work, we identify that erastin and RSL3 ferroptosis inducers up-regulated CD274 in TNBC cells (MDA-MB-231 and HCC38). In breast tumors from TCGA cohort, expression of CD274 is associated to overall survival and tumors presenting high expression level of CD274 up-regulated some ferroptosis driver genes also associated to prognosis, like: IDO1, IFNG and TNFAIP3. CD274-ferroptosis driver score computed on breast tumor transcriptome stratified patients on their prognosis: low score was observed in basal subgroup presenting higher level of recurrent risk scores such as: oncotypeDx risk, genomic grade index (ggi) and gene70 score. In cohort METABRIC, CD274, IDO1, IFNG and TNFAIP3 were confirmed to be overexpressed in TNBC subgroup. CD274-ferroptosis driver score was confirmed as an independent parameter of overall survival of the patients according inclusion of age at diagnosis and Tumor, Nodes, Metastasis stage classification in the multivariate model. Tumoral expression of CD274, TNFAIP3, IFNG and IDO1 in biopsy of breast ductal carcinoma was confirmed at protein level by immunohistochemistry imaging.
2. Materials and Methods
2.1. Transcriptome Datasets of Cell Lines Stimulated with Ferroptosis Inducers
2.1.1. MDA-MB-231 RNA-Sequencing Transcriptome
RNA-sequencing FPKM quantification performed on MDA-MB-231 triple negative breast cancer cell lines from dataset GSE173905 [
12] was downloaded on Gene Expression Omnibus website [
13] at the following web address:
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE173905, (accessed on 2023, November 22th). During these triplicate experiments, MDA-MB-231 cell line was treated during 72 hours with two distinct ferroptosis inducers: erastin and RSL3.
2.1.2. HCC38 Microarray Transcriptome
Normalized matrix of microarray from dataset GSE154425 [
14] was downloaded at the following web addressed:
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE154425, (accessed on 2023, November 22th) and annotated with the corresponding technology platform: GPL17692, Affymetrix Human Gene 2.1 ST Array, available at the web address:
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL17692, (accessed on 2023, November 22th). These triplicated experiments were performed on HCC38 triple negative breast cancer cell line. Experimental conditions of this transcriptome dataset comprised vehicle control and in vitro stimulations during 18 hours with erastin (ferroptosis inducer), with tubacin (HDAC6 inhibitor) and with combination of the two drugs.
2.2. Cohorts of Breast Cancer Tumor Samples
2.2.1. Training Cohort: TCGA Firehose Invasive Breast Cancer Cohort
TCGA invasive breast cancer [
15] on Cbioportal [
16]. RNA-sequencing transcriptome data were normalized by voom transformation in edgeR R-package version 3.38.4 [
17,
18]. Genefu R-package version 2.28.0 [
19] was computed to perform three molecular classification: pam50 subtypes [
3,
20], scmod2 subtypes [
21], claudin-low characterized by low expression of tight junction proteins claudins 3, 4 and 7 and E-cadherin [
22], prediction and three distinct predictive scores: oncotype.Dx, gene70 prediction [
23], genomic grade index (ggi) [
24] prediction. These scoring metadata were added to phenotype data of the samples in an experiment set (eset) also containing normalized RNA-sequencing data. A R-package entitled “tcga.breast” was compilated under version 1.0.0 with inclusion of this eset data [
25] .
2.2.2. Validation Cohort: METABRIC Breast Cancer Cohort
METABRIC breast cancer transcriptome cohort [
26,
27,
28] was investigated as validation cohort through the website Cbioportal [
16]. This interactive data portal allowed to perform oncoprint expression representation for CD274 and retained ferroptosis driver genes. Relapse free survival analysis was performed according stratification transcriptome alterations observed for CD274 and retained ferroptosis driver genes.
2.3. Transcriptome Immune Modulation Scoring
Bioinformatics development and analyses were performed in R software environment 4.2.1. Based on gene lists implicated in immune landscape of cancer [
29], a R-package was built to perform single sample expression enrichment on classes and subclasses of molecules identified as immune modulators [
30] in transcriptome experiments. This package is named “immunemod” (version 1.0.0) and is available at the following web address:
https://github.com/cdesterke/immunemod, (accessed on 2023, November 22th).
2.4. Ferroptosis Visualization in Transcriptome Enrichment
On TCGA breast cancer cohort, CD274 (PD-L1) expression was used to stratify the cohort in two groups (patients CD274-low and patients CD274-high) according overall survival of the patients. This stratification was obtained with survminer R-package version 0.49 and survival was fitted with survival R-package version 3.5.7. Differential expression gene analysis between CD274-low and CD274-high patient samples was done with LIMMA R-package version 3.52.4 [
31]. Based on ferrDb V2 database [
32], a R-package was built to improve visualization of ferroptosis related genes among differential expressed genes.
2.5. CD274-Ferroptosis Driver Gene Score Quantification
Loop of univariate overall survival analyses against expression of immune marker (CD274) and ferroptosis drivers were computed with “loopcolcox” R-package (version 1.0.1) available at the following web address:
https://github.com/cdesterke/loopcolcox, accessed on 2023, November 22th). For each significant marker (univariate Cox p-value <= 0.1), Cox beta coefficients were extracted to computed an expression score comprising CD274 and 3 ferroptosis drivers: TNFAIP3, IFNG and IDO1. On TCGA breast cancer transcriptome cohort, the exact equation of the score was: (Expression(IFNG)*-0.127036521465874) + (Expression(IDO1)*-0.0711938750267523) + (Expression(TNFAIP3)*-0.152997397587969) + (Expression(CD274) *-0.102709239674522). Optimal score threshold stratification according overall survival of the patients was obtained with survminer R-package version 0.49 and survival model was fitted with survival R-package version 3.5.7. Transcriptome expression heatmap and associated unsupervised clustering (Euclidean distances) were drawn with pheatmap R-package version 1.0.12. Multivariate overall survival model was done with survival R-package version 3.5.7 by including CD274-ferroptosis score categories with age at diagnosis and Tumor, Nodes, Metastasis (TNM) classification of the tumors. The multivariate overall survival model was assessed by performing individual and global Schoenfeld test. Univariate binomial analyses on clinical data were done with Publish R-package version 2023.01.17. Graphical representations (boxplots and scatterplots) were drawn with ggplot2 R-package version 3.4.4 [
33].
2.6. Immunohistochemistry
For markers employed to compute the CD274-ferroptosis driver score (CD274, IDO1, IFNG and TNFAIP3), their respective protein expression in human breast tumor was verified by immunohistochemistry on samples processed by protein-atlas consortium [
34].
3. Results
3.1. CD274 (PD-L1) Is Up-Regulated by Ferroptosis Inducers in Triple Negative Breast Cancer Cells
Triple negative breast cancer MDA-MB-231 cell line was stimulated with ferroptosis inducers (erastin, RSL3) during 72 hours and RNA-sequencing was processed on these experimental conditions [
12]. Immune modulation signature extracted from cancer immune landscape [
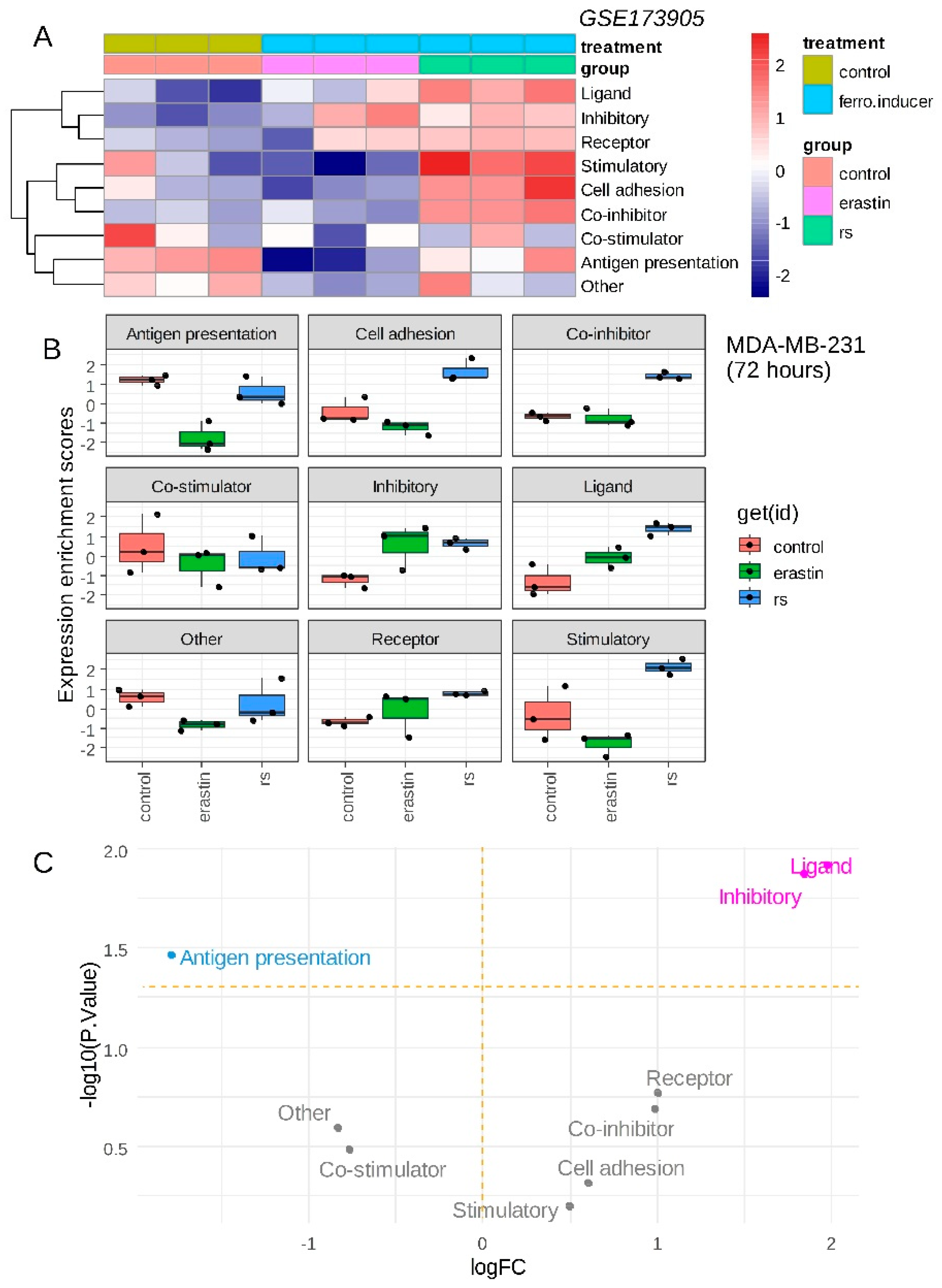
29] was investigated in these experiments. Single sample immune scoring showed potential immune regulation in presence of ferroptosis inducers: erastin and RSL3 as compared to the vehicle control (
Figure 1A). RSL3 ferroptosis inducer showed a more important immune regulation than erastin (
Figure 1A). A common increase of “inhibitory” immune checkpoints was observed for erastin and RSL3 stimulation as compared to control (
Figure 1B) and it appeared to increase significantly with “ligand subcategory” for both ferroptosis inducers (
Figure 1C). Increase of immune ligand score appeared more pronounced for RSL3 than for erastin stimulation (
Figure 1B). These results suggest that ferroptosis inducers could increase inhibitory immune checkpoint score in transcriptome of triple negative breast cancer cells.
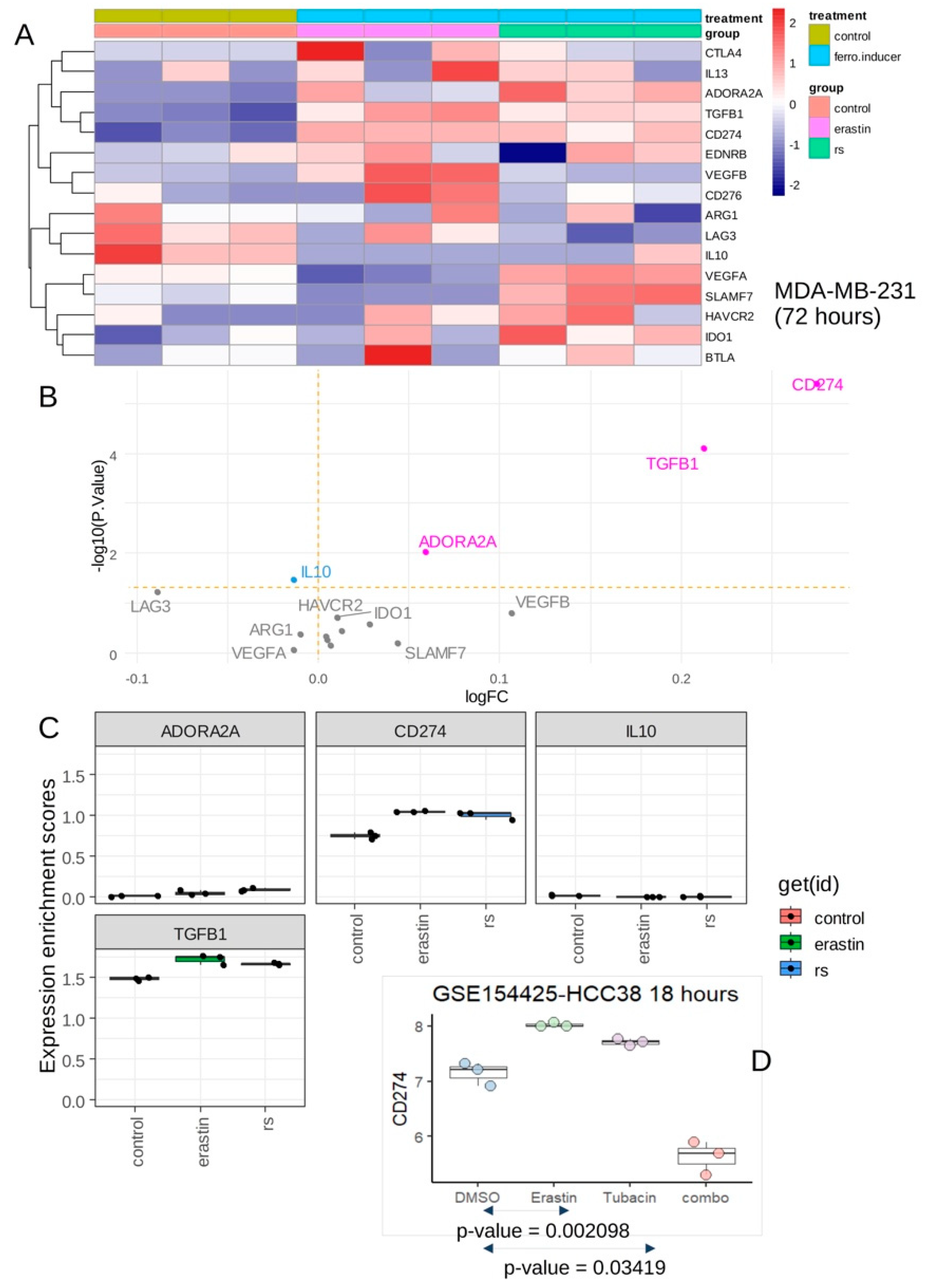
Expression of inhibitory immune checkpoints was investigated in same transcriptome dataset and both ferroptosis inducers showed to induce significant overexpression of CD274, TGFB1 and ADORA2A (
Figure 2A,B) with especially a marked overexpression of CD274 (
Figure 2B,C). CD274 expression was investigated in an independent dataset of transcriptome GSE154425 (Alothaim, Charbonneau and Tang, 2021). These experiments were done on HCC38 triple negative breast cancer cell line with stimulation during eighteen hours by erastin (ferroptosis inducer), tubacin (HDAC6 inhibitor) and combination of the two molecules (combo) (
Figure 2D). These independent experiments confirmed that erastin could induce overexpression of CD274 in HCC38 TNBC cell line. These results suggest that ferroptosis inducers (erastin, RSL3) stimulation on TNBC cells increased expression of CD274, a known target of cancer immunotherapy.
3.2. Breast Tumors with High Expression of CD274 Harbored Upregulation of Ferroptosis Drivers
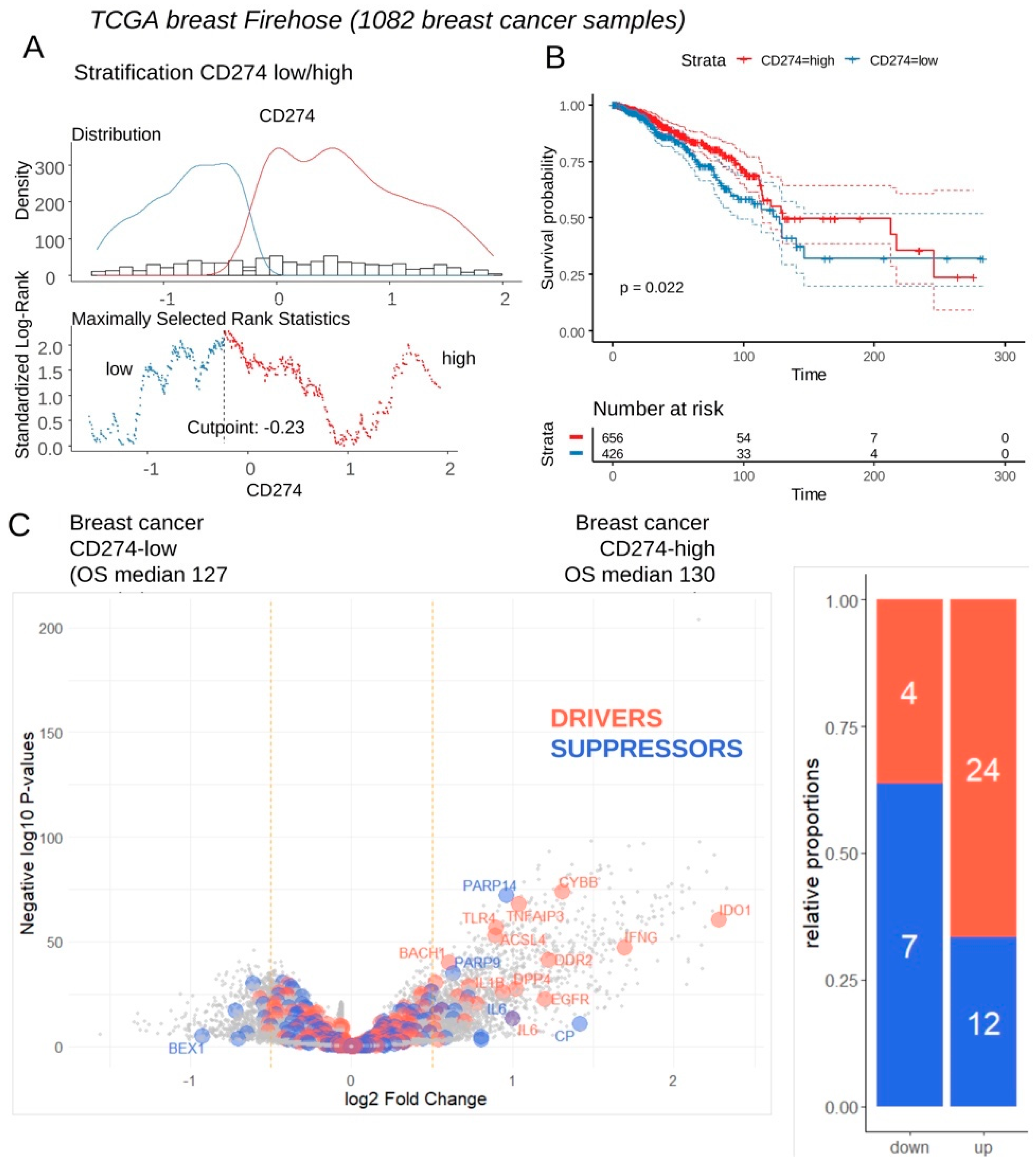
Expression of CD274 was investigated in transcriptome of breast tumors according clinical associated data of TCGA invasive breast cohort. This transcriptome cohort composed of 1082 cases was stratified in two subgroups of samples according CD274 expression and overall survival of the patients (
Figure 3A). Patients with CD274 low expression (n=426) in their tumor harbored a significant shorter overall survival than patients with CD274 high expression (n=656) in their tumor (log rank test p-value=0.022,
Figure 3B). These two groups of patients did not show significant difference in term of age at diagnosis (p-value=0.1871019,
Table 1), also in term of TNM classification (
Table 1). In term of molecular classification, a significant difference was found between CD274-low and CD274-high patients (p-value=0.0002416,
Table 1) with higher proportion of basal and Her2 subgroups in CD274-high patients. Among predictive score, a significant difference was found for G70 score (p-value=0.0005523,
Table 1) but nothing significant for OncotypeDx and ggi scores (
Table 1). Differential expression gene analysis highlighted a significant increase of expression for ferroptosis drivers in samples CD274-high as compared to CD274-low samples (
Figure 3C). These results confirm a high connection between ferroptosis induction and CD274 expression in breast cancer.
3.3. CD274-Ferroptosis Driver Score Predicts Recurrence of Breast Cancer
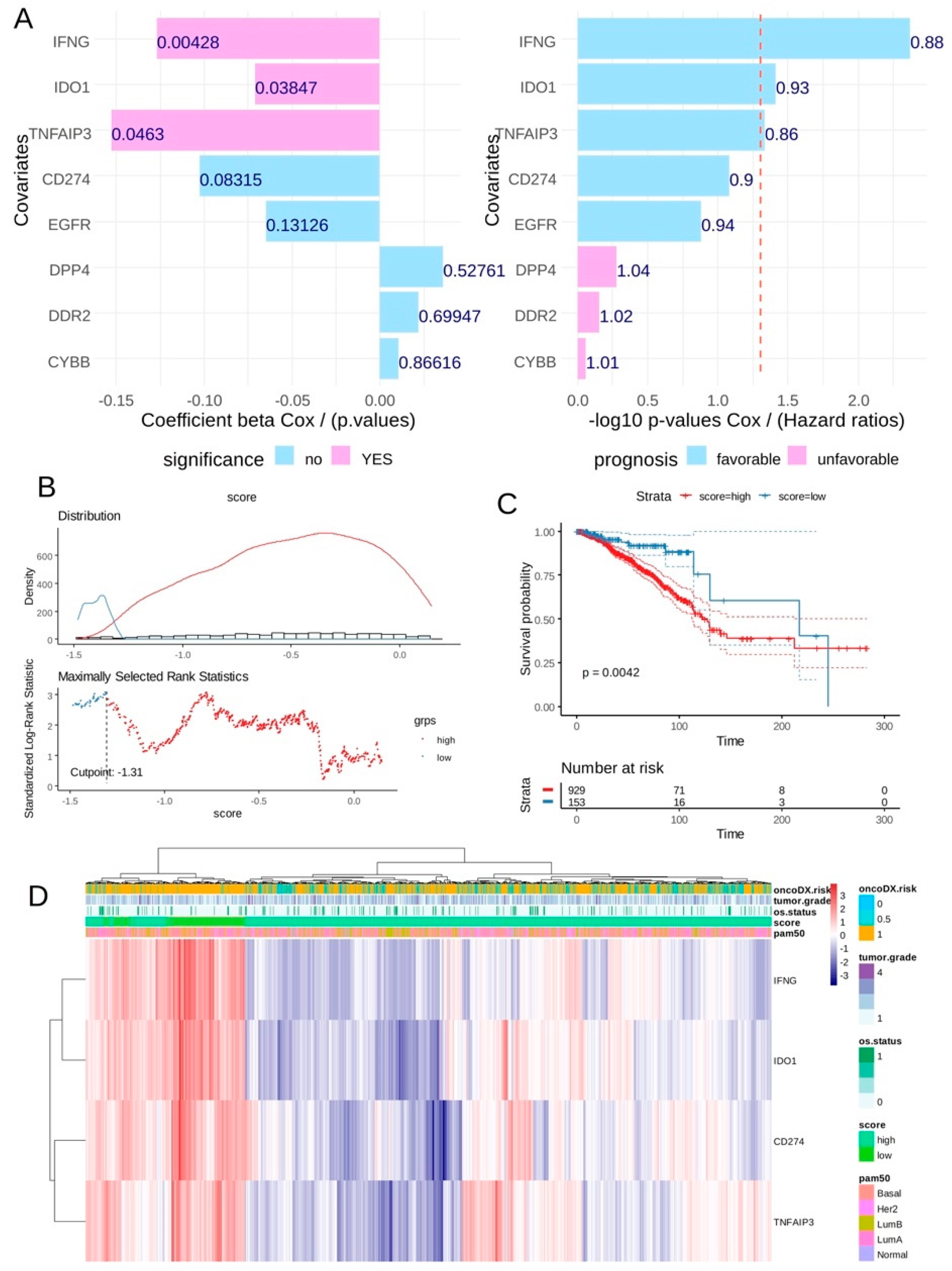
In breast tumor transcriptome from TCGA cohort, univariate overall survival analyses according expression of ferroptosis drivers highlighted a significant favorable expression for three of them: IFNG, IDO1 and TNFAIP3 (
Figure 4A) conjointly with CD274 expression (univariate Cox p-value=0.08). A CD274-ferroptosis driver score combining expression and Cox beta-coefficients of CD274, IFNG, TNFAIP3 and IDO1 was computed. According overall survival of the patients an optimal threshold cutpoint was determined in order to stratify the cohort in two groups in relation with this score (
Figure 4B): patients with low score (n=153), patients with high score (n=929). Log rank test confirmed significant difference in term of overall survival between these two groups of patients with longer survival time for patients in low-score category (log rank test p-value=0.0042,
Figure 4C). In term of TNM classification, no significant difference was observed in tumor stage (p-value = 0.06,
Table 2) and metastasis stage (p-value=0.29,
Table 2), but a significant higher proportion of node stage 2 and 3 was observed in score-high group (p-value=0.006,
Table 2). Unsupervised clustering performed on expression of the four markers confirmed score stratification of patients with a left cluster enriched in patients with low score and high expression of the four molecules (
Figure 4D). In this left cluster of score-low patients tends to be of basal type with less events of death and presenting an ontypeDX risk (
Figure 4D). Concerning pam50 molecular classification, significant higher proportion of basal and Her2+ was observed in score-low group (p-value<0.0001,
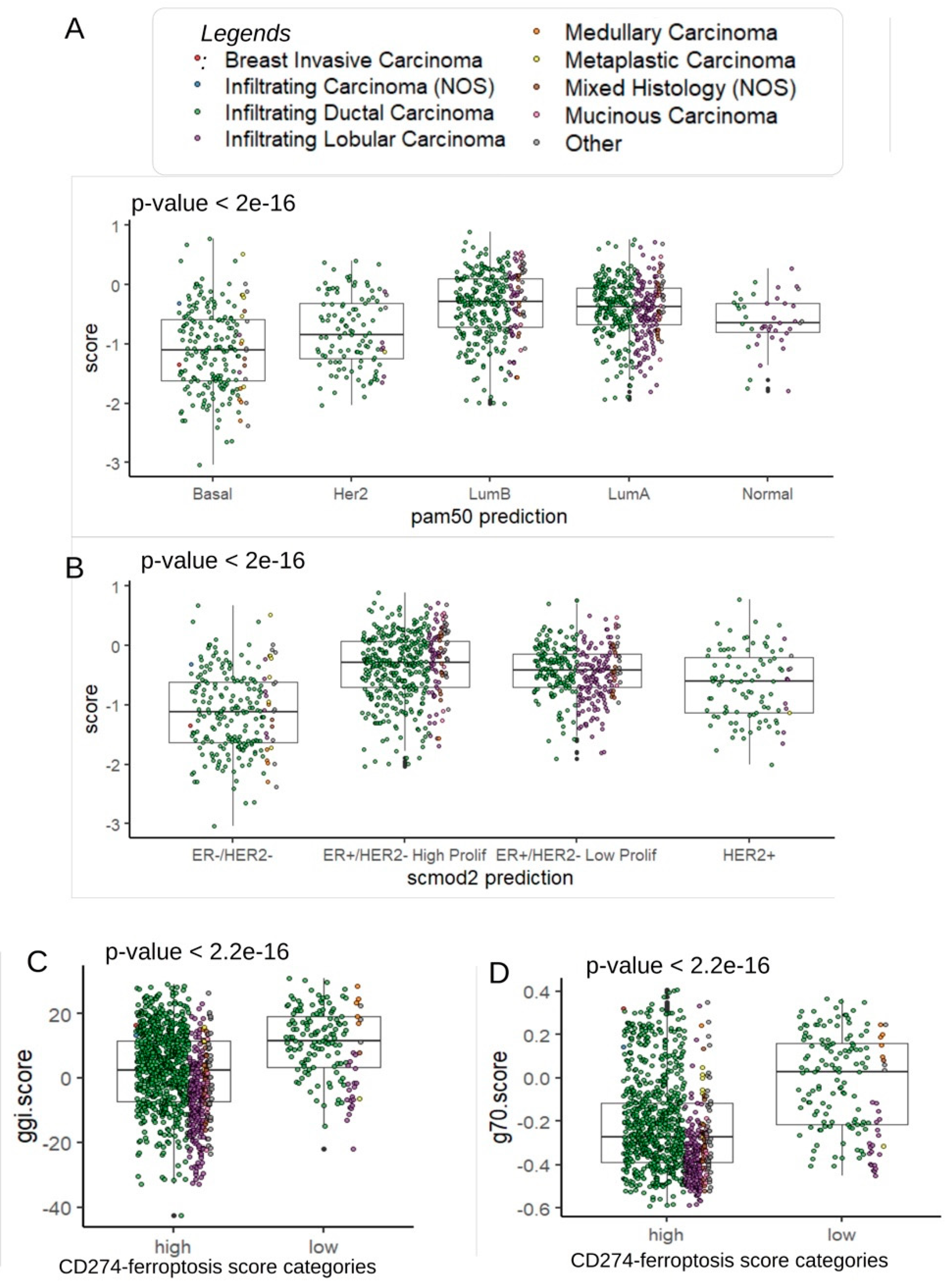
Table 2). Effectively, patients with lower score were confirmed to be in basal and Her2 subtypes (
Figure 5A). In basal and Her2 subgroups, a high expression was observed for CD274, IFNG and IDO1 (
Supplemental Figure S1). A high expression TNFAIP3 was observed only in basal and normal-like subgroups (
Supplemental Figure S2). Concerning SCMOD2 classifier, low level of CD274-ferroptosis driver was observed in ER-/Her2- group (
Figure 5B). Interestingly, this CD274-ferroptosis driver score was found significantly associated to distinct prognostic scores. Patients with low score harbored higher values of Genomic Grade Index (ggi) (p-value< 2.2.e-16,
Figure 5C and
Table 2), higher values of OncotypeDx score (p-value< 2.2.e-16,
Figure 5D,
Supplemental Figure S2A and
Table 2) and higher values of gene70 score (p-value<0.0001,
Table 2).
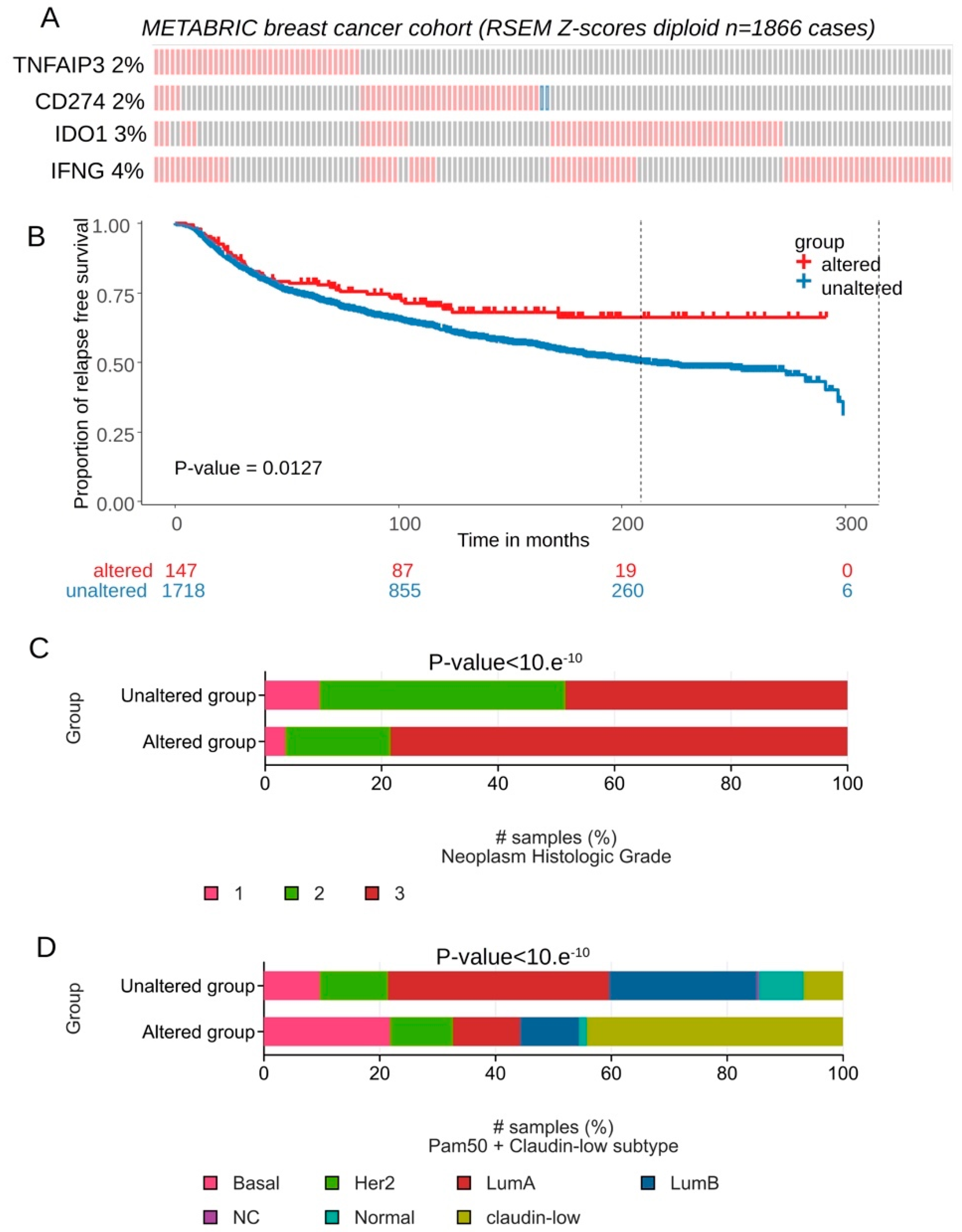
Expression of CD274, IFNG, TNFAIP3 and IDO1 was investigated in an independent cohort of transcriptome: METABRIC cohort comprising 1866 breast tumors. Majority of transcriptional alterations observed for these markers concerned upregulation with a frequency superior or equal to 2 percent of the tumors (
Figure 6A). In term of relapse free survival patients presenting combined transcriptional alteration for these four markers presented significant longer survival time during the follow-up (log rank test p=0.0127,
Figure 6B). In term of tumor grade for this validation cohort, a significant increase of tumor stage 3 was found associated in group of patients affected by transcriptional alterations of these four genes (p-value<10.e-10,
Figure 6C). Patients affected by transcriptional alterations of these four genes presented also a significant increase of basal and claudin-low subgroups (p-value<10.e-10,
Figure 6D). Higher expression of CD274 was found TNBC subgroup of METABRIC cohort as compared to other breast cancer samples (p-value=1.25e-10,
Supplemental Figure S3A). Higher expression of IDO1 was found TNBC subgroup of METABRIC cohort as compared to other breast cancer samples (p-value<2.2e-16,
Supplemental Figure S3B). Higher expression of IFNG was found TNBC subgroup of METABRIC cohort as compared to other breast cancer samples (p-value<2.2e-16,
Supplemental Figure S3C). Higher expression of TNFAIP3 was found TNBC subgroup of METABRIC cohort as compared to other breast cancer samples (p-value<2.2e-16,
Supplemental Figure S3D).
3.4. CD274-Ferroptosis Driver Score Is an Independent Prognosis Marker in Breast Cancer Overall Survival
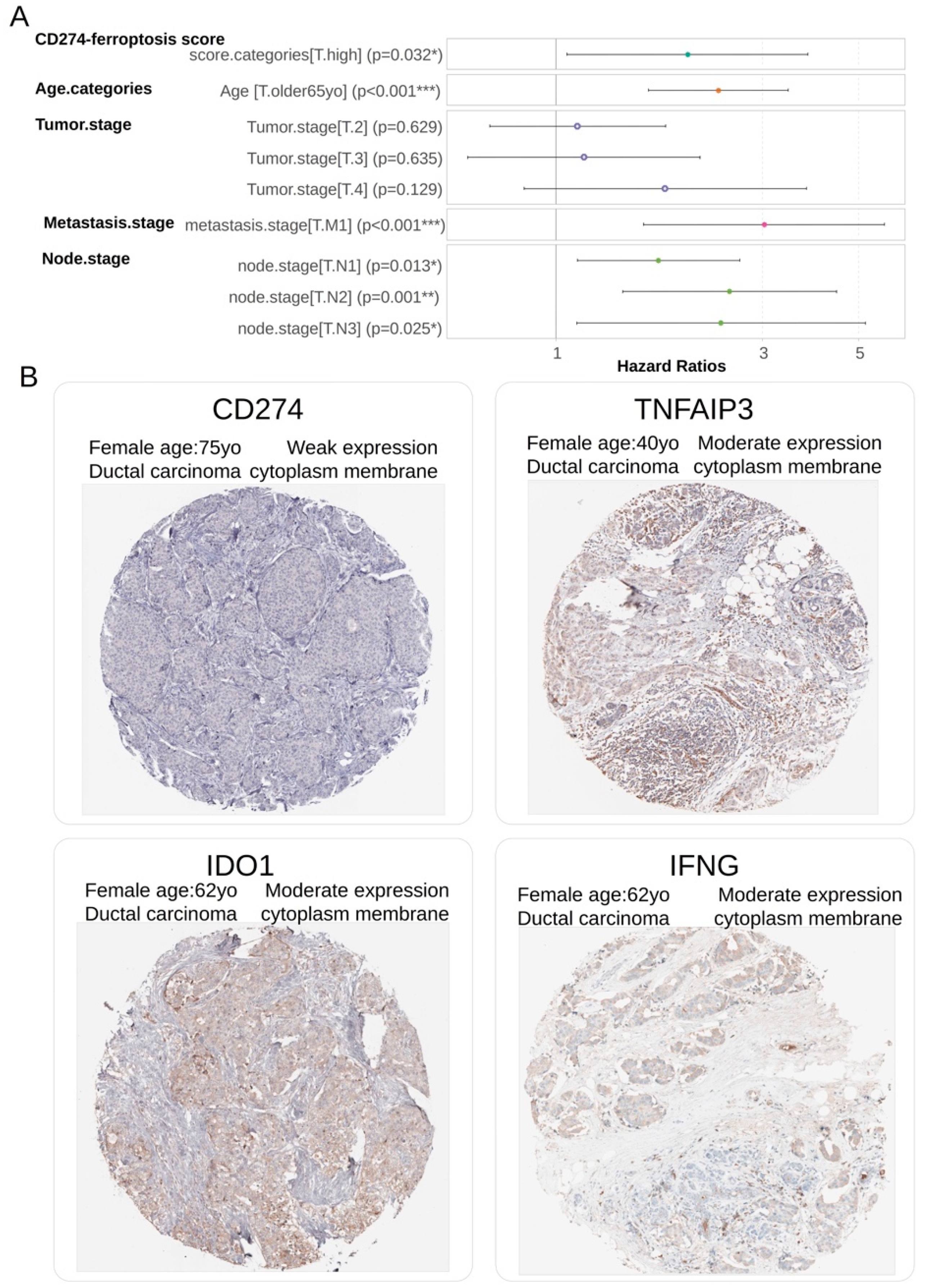
The independence of CD274-ferroptosis driver score in breast cancer prognosis was investigated through building a multivariable overall survival model on TCGA transcriptome cohort. This Cox multivariable model was performed with inclusion of score stratification (
Table 2), age at diagnosis stratification (threshold 65 years old) and TNM stage classification. Global Schoenfeld test assessed linearity of the residuals in the model at multivariable level (Global Schoenfeld test, p-value=0.07,
Supplemental Figure S2B). Concordance index of overall multivariate model reached a Ci equal to 0.722 (with standard error = 0.025). In the multivariate overall survival model, patients quantified with high value of CD274-ferroptosis driver score harbored a significant positive hazard ratio (2.012+/-1.063, multivariable p-value=3.18.e-2,
Table 3,
Figure 7A). These results suggest/support that CD274-ferroptosis driver score is an adverse independent prognosis marker of the overall survival in breast cancer. At protein level by immunohistochemistry, a weak expression of CD274 and moderate expression of ferroptosis drivers (IFNG, TNFAIP3 and IDO1) were observed in biopsy of ductal breast carcinoma (
Figure 7B).
4. Discussion
In the present work, we observed that ferroptosis inducers such as erastin and RSL3 increased expression of PD-L1 in distinct triple negative breast cancer cells. Anti-PD-L1 immunotherapy have been shown to improved pathological complete response of high risk early triple negative breast cancer patient which received neo-adjuvant chemotherapy [
8].
Some synergy between induction of ferroptosis and immunogenic cell death (ICD) have been already described to potentiate cancer anti-PD-L1 immunotherapy. For example, a tannic acid (TA)-Fe3+-coated doxorubicin (DOX)-encapsulated 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(poly(ethylene glycol))-2000] (ammonium salt) (DSPE-PEG) micelle (TFDD) for apoptosis/ferroptosis-mediated immunogenic cell death (ICD) is reported. By coating TA-Fe3+ on the surface of DOX-loaded micelles, an apoptotic agent and a ferroptotic agent are simultaneously delivered into the cancer cells and induce cell death. Furthermore, the intracellular oxidative environment generated by the apoptosis/ferroptosis hybrid pathway stimulates the endoplasmic reticulum (ER) and leads to ICD induction. The associated in vivo results show that the combination treatment of TFDD and anti-programmed death-ligand 1 antibodies (anti-PD-L1) considerably inhibits tumor growth and improves antitumor immunity by activating CD4+ and CD8+ T cells and decreasing the ratio of regulatory T cells (Treg) to CD4+ T cells [
35]. Expression of PD-L1 have been also described to be positively correlated to the expression of ferroptosis-related gene such ACSL4 [
11]. In breast tumor, we observed that breast tumors which harbored high level of CD274 up-regulated expression of ferroptosis driver genes. Among these up-regulated ferroptosis driver genes, some of them were also found associated in their expression to prognosis of the patients: IFNG, TNFAIP3 and IDO1. In breast tumor transcriptome, CD274-ferroptosis driver score was found associated to the prognosis of breast cancer patients and also to the risk of recurrence (g70, ggi and oncotypeDx scores) [
36]. Interferon gamma (IFNγ) released from CD8+ T cells downregulates the expression of SLC3A2 and SLC7A11, two subunits of the glutamate-cystine antiporter system xc-, impairs the uptake of cystine by tumour cells, and as a consequence, promotes tumour cell lipid peroxidation and ferroptosis [
37]. A20/TNFAIP3 is significantly up-regulated in TNBC and its expression level is highly correlated with low/poor survival of metastasis-free patients, and promotes cancer metastasis via multi-monoubiquitylation, which activates the functions of Snail [
38]. In BV-2 microglial cell line, knock-out of TNFAIP3 (A20) attenuates cell susceptibility to OGD/R-induced ferroptosis and upregulates inflammatory responses [
39]. TNFAIP3, also known as A20, is an ubiquitin-editing enzyme and functions as an endogenous suppressor of NF-κB, which activates inflammation (Priem, van Loo and Bertrand, 2020). A20 restricts NF-κB signals through its deubiquitinase activity. A20 is regulated by microRNA (miRNA) and acts as a regulator of endothelial cell ferroptosis [
40] and the A20/TNFAIP3-CDC20-CASP1 axis promotes inflammation-mediated metastatic disease in triple-negative breast cancer [
41]. Indoleamine 2,3-dioxygenase 1 (IDO1) is an immunosuppressive enzyme involved in tumor immune escape. Blockade of the IDO1 pathway is an emerging modality of cancer immunotherapy. IDO1 expression in basal-like TNBCs is considered as an immune inhibitory signal that counterbalances active immunity and may reflect the high mutational load of these tumors [
42]. IDO1 oxidizes tryptophan (TRP) to generate kynurenine (KYN) and KYN serves as the source for molecules that inhibit ferroptotic cell death [
43].
5. Conclusions
Ferroptosis inducers up-regulated PD-L1 in TNBC cells, known as effective target of immunotherapy in high risk early TNBC which received neo-adjuvant therapy. Basal and TNBC tumors highly expressed CD274 and ferroptosis drivers: IFNG, TNFAIP3 and IDO1. CD274-ferroptosis driver score is associated to prognosis and to the risk of recurrence in breast cancer. A potential synergy of ferroptosis inducers with anti-PD-L1 immunotherapy is suggested in recurrent TNBC.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org, Figure S1. Breast tumor expression of markers included in CD274-ferrptosis score (TCGA cohort); Figure S2. OncotypeDx quantification in breast tumor sample and Schoenfeld test for multivariable survival model (TCGA cohort); Figure S3. Overexpression of CD274, IDO1, IFNG and TNFAIP3 in TNBC subgroup of METABRIC cohort:
Author Contributions
Conceptualization, CD, YC, AH; methodology, CD, YC, AH; software, CD; validation, CD, YC, AH; formal analysis, CD; investigation, CD, YC, AH; resources, CD, AH; data curation, CD; writing—original draft preparation, CD, YC, AH; writing—review and editing, CD, YC, AH; visualization, CD, YC, AH; supervision, AH; project administration, AH; funding acquisition, YC, AH. YX, RE, CDu provided a critical review of the data and manuscript.
Funding
This work was supported by core funding from University, CNRS and INSERM. A.H. was supported by grants from the Comité de Paris de la ligue contre le cancer (LCC RS23/75-75).
Institutional Review Board Statement
Not applicable for this work.
Informed Consent Statement
Not applicable for this work.
Data Availability Statement
Acknowledgments
Many thanks to TCGA and METABRIC cohort consortium for availability of breast cancer data.
Conflicts of Interest
Not applicable for this work
References
- Sotiriou, C.; Neo, S.-Y.; McShane, L.M.; Korn, E.L.; Long, P.M.; Jazaeri, A.; Martiat, P.; Fox, S.B.; Harris, A.L.; Liu, E.T. Breast Cancer Classification and Prognosis Based on Gene Expression Profiles from a Population-Based Study. Proc. Natl. Acad. Sci. USA 2003, 100, 10393–10398. [Google Scholar] [CrossRef] [PubMed]
- Perou, C.M.; Sørlie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular Portraits of Human Breast Tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Sorlie, T.; Tibshirani, R.; Parker, J.; Hastie, T.; Marron, J.S.; Nobel, A.; Deng, S.; Johnsen, H.; Pesich, R.; Geisler, S.; et al. Repeated Observation of Breast Tumor Subtypes in Independent Gene Expression Data Sets. Proc. Natl. Acad. Sci. USA 2003, 100, 8418–8423. [Google Scholar] [CrossRef]
- Prat, A.; Pineda, E.; Adamo, B.; Galván, P.; Fernández, A.; Gaba, L.; Díez, M.; Viladot, M.; Arance, A.; Muñoz, M. Clinical Implications of the Intrinsic Molecular Subtypes of Breast Cancer. Breast Edinb. Scotl. 2015, 24 Suppl. 2, S26–35. [Google Scholar] [CrossRef]
- Hudis, C.A.; Gianni, L. Triple-Negative Breast Cancer: An Unmet Medical Need. The Oncologist 2011, 16, 1–11. [Google Scholar] [CrossRef]
- Cardoso, F.; Kyriakides, S.; Ohno, S.; Penault-Llorca, F.; Poortmans, P.; Rubio, I.T.; Zackrisson, S.; Senkus, E.; ESMO Guidelines Committee. Electronic address: clinicalguidelines@esmo.org Early Breast Cancer: ESMO Clinical Practice Guidelines for Diagnosis, Treatment and Follow-Up†. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2019, 30, 1194–1220. [Google Scholar] [CrossRef]
- Farkona, S.; Diamandis, E.P.; Blasutig, I.M. Cancer Immunotherapy: The Beginning of the End of Cancer? BMC Med. 2016, 14, 73. [Google Scholar] [CrossRef]
- Schmid, P.; Cortes, J.; Pusztai, L.; McArthur, H.; Kümmel, S.; Bergh, J.; Denkert, C.; Park, Y.H.; Hui, R.; Harbeck, N.; et al. Pembrolizumab for Early Triple-Negative Breast Cancer. N. Engl. J. Med. 2020, 382, 810–821. [Google Scholar] [CrossRef]
- Yu, H.; Guo, P.; Xie, X.; Wang, Y.; Chen, G. Ferroptosis, a New Form of Cell Death, and Its Relationships with Tumourous Diseases. J. Cell. Mol. Med. 2017, 21, 648–657. [Google Scholar] [CrossRef] [PubMed]
- Mou, Y.; Wang, J.; Wu, J.; He, D.; Zhang, C.; Duan, C.; Li, B. Ferroptosis, a New Form of Cell Death: Opportunities and Challenges in Cancer. J. Hematol. Oncol.J Hematol Oncol 2019, 12, 34. [Google Scholar] [CrossRef] [PubMed]
- Wuguo, T.; Jianjie, Z.; Donglin, L. Correlation between Ferroptosis-Related Genes and PDL-1 Immunotherapy in Triple-Negative Breast Cancer. Asian J. Surg. 2023, 46, 4595–4597. [Google Scholar] [CrossRef]
- Li, P.; Lin, Q.; Sun, S.; Yang, N.; Xia, Y.; Cao, S.; Zhang, W.; Li, Q.; Guo, H.; Zhu, M.; et al. Inhibition of Cannabinoid Receptor Type 1 Sensitizes Triple-Negative Breast Cancer Cells to Ferroptosis via Regulating Fatty Acid Metabolism. Cell Death Dis. 2022, 13, 808. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for Functional Genomics Data Sets--Update. Nucleic Acids Res. 2013, 41, D991–995. [Google Scholar] [CrossRef]
- Alothaim, T.; Charbonneau, M.; Tang, X. HDAC6 Inhibitors Sensitize Non-Mesenchymal Triple-Negative Breast Cancer Cells to Cysteine Deprivation. Sci. Rep. 2021, 11, 10956. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Lichtenberg, T.; Hoadley, K.A.; Poisson, L.M.; Lazar, A.J.; Cherniack, A.D.; Kovatich, A.J.; Benz, C.C.; Levine, D.A.; Lee, A.V.; et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 2018, 173, 400–416.e11. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinforma. Oxf. Engl. 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Law, C.W.; Chen, Y.; Shi, W.; Smyth, G.K. Voom: Precision Weights Unlock Linear Model Analysis Tools for RNA-Seq Read Counts. Genome Biol. 2014, 15, R29. [Google Scholar] [CrossRef]
- Gendoo, D.M.A.; Ratanasirigulchai, N.; Schröder, M.S.; Paré, L.; Parker, J.S.; Prat, A.; Haibe-Kains, B. Genefu: An R/Bioconductor Package for Computation of Gene Expression-Based Signatures in Breast Cancer. Bioinforma. Oxf. Engl. 2016, 32, 1097–1099. [Google Scholar] [CrossRef]
- Parker, J.S.; Mullins, M.; Cheang, M.C.U.; Leung, S.; Voduc, D.; Vickery, T.; Davies, S.; Fauron, C.; He, X.; Hu, Z.; et al. Supervised Risk Predictor of Breast Cancer Based on Intrinsic Subtypes. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2009, 27, 1160–1167. [Google Scholar] [CrossRef]
- Wirapati, P.; Sotiriou, C.; Kunkel, S.; Farmer, P.; Pradervand, S.; Haibe-Kains, B.; Desmedt, C.; Ignatiadis, M.; Sengstag, T.; Schütz, F.; et al. Meta-Analysis of Gene Expression Profiles in Breast Cancer: Toward a Unified Understanding of Breast Cancer Subtyping and Prognosis Signatures. Breast Cancer Res. 2008, 10, R65. [Google Scholar] [CrossRef]
- Prat, A.; Parker, J.S.; Karginova, O.; Fan, C.; Livasy, C.; Herschkowitz, J.I.; He, X.; Perou, C.M. Phenotypic and Molecular Characterization of the Claudin-Low Intrinsic Subtype of Breast Cancer. Breast Cancer Res. BCR 2010, 12, R68. [Google Scholar] [CrossRef]
- van ’t Veer, L.J.; Dai, H.; van de Vijver, M.J.; He, Y.D.; Hart, A.A.M.; Mao, M.; Peterse, H.L.; van der Kooy, K.; Marton, M.J.; Witteveen, A.T.; et al. Gene Expression Profiling Predicts Clinical Outcome of Breast Cancer. Nature 2002, 415, 530–536. [Google Scholar] [CrossRef]
- Sotiriou, C.; Wirapati, P.; Loi, S.; Harris, A.; Fox, S.; Smeds, J.; Nordgren, H.; Farmer, P.; Praz, V.; Haibe-Kains, B.; et al. Gene Expression Profiling in Breast Cancer: Understanding the Molecular Basis of Histologic Grade To Improve Prognosis. JNCI J. Natl. Cancer Inst. 2006, 98, 262–272. [Google Scholar] [CrossRef] [PubMed]
- Desterke, C. Tcga.Breast a R Package Composed of Eset (RNAseq) - TCGA Breast Cancer with Clinical Scores Computed. 2023, 102630601 Bytes.
- Curtis, C.; Shah, S.P.; Chin, S.-F.; Turashvili, G.; Rueda, O.M.; Dunning, M.J.; Speed, D.; Lynch, A.G.; Samarajiwa, S.; Yuan, Y.; et al. The Genomic and Transcriptomic Architecture of 2,000 Breast Tumours Reveals Novel Subgroups. Nature 2012, 486, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Pereira, B.; Chin, S.-F.; Rueda, O.M.; Vollan, H.-K.M.; Provenzano, E.; Bardwell, H.A.; Pugh, M.; Jones, L.; Russell, R.; Sammut, S.-J.; et al. The Somatic Mutation Profiles of 2,433 Breast Cancers Refines Their Genomic and Transcriptomic Landscapes. Nat. Commun. 2016, 7, 11479. [Google Scholar] [CrossRef]
- Rueda, O.M.; Sammut, S.-J.; Seoane, J.A.; Chin, S.-F.; Caswell-Jin, J.L.; Callari, M.; Batra, R.; Pereira, B.; Bruna, A.; Ali, H.R.; et al. Dynamics of Breast-Cancer Relapse Reveal Late-Recurring ER-Positive Genomic Subgroups. Nature 2019, 567, 399–404. [Google Scholar] [CrossRef]
- Thorsson, V.; Gibbs, D.L.; Brown, S.D.; Wolf, D.; Bortone, D.S.; Ou Yang, T.-H.; Porta-Pardo, E.; Gao, G.F.; Plaisier, C.L.; Eddy, J.A.; et al. The Immune Landscape of Cancer. Immunity 2018, 48, 812–830.e14. [Google Scholar] [CrossRef] [PubMed]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene Set Variation Analysis for Microarray and RNA-Seq Data. BMC Bioinformatics 2013, 14, 7. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma Powers Differential Expression Analyses for RNA-Sequencing and Microarray Studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Zhou, N.; Yuan, X.; Du, Q.; Zhang, Z.; Shi, X.; Bao, J.; Ning, Y.; Peng, L. FerrDb V2: Update of the Manually Curated Database of Ferroptosis Regulators and Ferroptosis-Disease Associations. Nucleic Acids Res. 2023, 51, D571–D582. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2009. [Google Scholar]
- Uhlén, M.; Björling, E.; Agaton, C.; Szigyarto, C.A.-K.; Amini, B.; Andersen, E.; Andersson, A.-C.; Angelidou, P.; Asplund, A.; Asplund, C.; et al. A Human Protein Atlas for Normal and Cancer Tissues Based on Antibody Proteomics. Mol. Cell. Proteomics MCP 2005, 4, 1920–1932. [Google Scholar] [CrossRef]
- Jeong, S.D.; Jung, B.-K.; Lee, D.; Ha, J.; Chang, H.-G.; Lee, J.; Lee, S.; Yun, C.-O.; Kim, Y.-C. Enhanced Immunogenic Cell Death by Apoptosis/Ferroptosis Hybrid Pathway Potentiates PD-L1 Blockade Cancer Immunotherapy. ACS Biomater. Sci. Eng. 2022, 8, 5188–5198. [Google Scholar] [CrossRef]
- Reis-Filho, J.S.; Pusztai, L. Gene Expression Profiling in Breast Cancer: Classification, Prognostication, and Prediction. Lancet Lond. Engl. 2011, 378, 1812–1823. [Google Scholar] [CrossRef]
- Wang, W.; Green, M.; Choi, J.E.; Gijón, M.; Kennedy, P.D.; Johnson, J.K.; Liao, P.; Lang, X.; Kryczek, I.; Sell, A.; et al. CD8+ T Cells Regulate Tumour Ferroptosis during Cancer Immunotherapy. Nature 2019, 569, 270–274. [Google Scholar] [CrossRef]
- Lee, J.-H.; Jung, S.M.; Yang, K.-M.; Bae, E.; Ahn, S.G.; Park, J.S.; Seo, D.; Kim, M.; Ha, J.; Lee, J.; et al. A20 Promotes Metastasis of Aggressive Basal-like Breast Cancers through Multi-Monoubiquitylation of Snail1. Nat. Cell Biol. 2017, 19, 1260–1273. [Google Scholar] [CrossRef]
- Liu, X.; Jin, X.; Wang, X.; Yan, X.; Wang, C.; Wang, K.; He, X.; Zhai, W. Knockdown of A20 Attenuates Microglial Susceptibility to OGD/R-Induced Ferroptosis and Upregulates Inflammatory Responses. Immunopharmacol. Immunotoxicol. 2023, 45, 539–548. [Google Scholar] [CrossRef]
- Xiao, F.-J.; Zhang, D.; Wu, Y.; Jia, Q.-H.; Zhang, L.; Li, Y.-X.; Yang, Y.-F.; Wang, H.; Wu, C.-T.; Wang, L.-S. miRNA-17-92 Protects Endothelial Cells from Erastin-Induced Ferroptosis through Targeting the A20-ACSL4 Axis. Biochem. Biophys. Res. Commun. 2019, 515, 448–454. [Google Scholar] [CrossRef]
- Song, C.; Kendi, A.T.; Lowe, V.J.; Lee, S. The A20/TNFAIP3-CDC20-CASP1 Axis Promotes Inflammation-Mediated Metastatic Disease in Triple-Negative Breast Cancer. Anticancer Res. 2022, 42, 681–695. [Google Scholar] [CrossRef]
- Kim, S.; Park, S.; Cho, M.S.; Lim, W.; Moon, B.-I.; Sung, S.H. Strong Correlation of Indoleamine 2,3-Dioxygenase 1 Expression with Basal-Like Phenotype and Increased Lymphocytic Infiltration in Triple-Negative Breast Cancer. J. Cancer 2017, 8, 124–130. [Google Scholar] [CrossRef]
- Fiore, A.; Zeitler, L.; Russier, M.; Groß, A.; Hiller, M.-K.; Parker, J.L.; Stier, L.; Köcher, T.; Newstead, S.; Murray, P.J. Kynurenine Importation by SLC7A11 Propagates Anti-Ferroptotic Signaling. Mol. Cell 2022, 82, 920–932.e7. [Google Scholar] [CrossRef] [PubMed]
Figure 1.
Ferroptosis inducers up-regulated inhibitory immune checkpoints in triple negative breast cancer cells: A/ Heatmap of immune modulation scores computed on transcriptome of MDA-MB-231 cells stimulated during 72 hours with ferroptosis inducers: erastin and RSL3 (rs); B/ Boxplot of immune modulation scores computed on transcriptome of MDA-MB-231 cells stimulated during 72 hours with ferroptosis inducers: erastin and RSL3 (rs); C/ Volcanoplot testing regulation of immune modulation scores computed on transcriptome of MDA-MB-231 cells stimulated or not during 72 hours with ferroptosis inducers.
Figure 1.
Ferroptosis inducers up-regulated inhibitory immune checkpoints in triple negative breast cancer cells: A/ Heatmap of immune modulation scores computed on transcriptome of MDA-MB-231 cells stimulated during 72 hours with ferroptosis inducers: erastin and RSL3 (rs); B/ Boxplot of immune modulation scores computed on transcriptome of MDA-MB-231 cells stimulated during 72 hours with ferroptosis inducers: erastin and RSL3 (rs); C/ Volcanoplot testing regulation of immune modulation scores computed on transcriptome of MDA-MB-231 cells stimulated or not during 72 hours with ferroptosis inducers.
Figure 2.
Ferroptosis inducers up-regulated PD-L1 (CD274) inhibitory immune checkpoint in triple negative breast cancer cells: A/ Expression heatmap of inhibitory immune checkpoints in transcriptome of MDA-MB-231 stimulated or not with ferroptosis inducers: erastin and RSL3 (rs); B/ Volcanoplot testing regulation of inhibitory immune checkpoints in MDA-MB-231 stimulated or not with ferroptosis inducers; C/ Boxplot of expression for inhibitory immune checkpoints in transcriptome of MDA-MB-231 stimulated or not with ferroptosis inducers: erastin and RSL3 (rs); D/ Expression boxplot CD274 in HCC38 triple negative breast cancer cells according stimulation with erastin ferroptosis inducer, tubacin (HDAC6 inhibitor) and combo (combination of erastin and tubacin).
Figure 2.
Ferroptosis inducers up-regulated PD-L1 (CD274) inhibitory immune checkpoint in triple negative breast cancer cells: A/ Expression heatmap of inhibitory immune checkpoints in transcriptome of MDA-MB-231 stimulated or not with ferroptosis inducers: erastin and RSL3 (rs); B/ Volcanoplot testing regulation of inhibitory immune checkpoints in MDA-MB-231 stimulated or not with ferroptosis inducers; C/ Boxplot of expression for inhibitory immune checkpoints in transcriptome of MDA-MB-231 stimulated or not with ferroptosis inducers: erastin and RSL3 (rs); D/ Expression boxplot CD274 in HCC38 triple negative breast cancer cells according stimulation with erastin ferroptosis inducer, tubacin (HDAC6 inhibitor) and combo (combination of erastin and tubacin).
Figure 3.
Breast tumors of patients with longer survival conjointly overexpressed CD274 and ferroptosis drivers: A/ Optimal threshold for CD274 expression according overall survival of the patient from Firehose TCGA invasive breast cancer transcriptome cohort (n=1082 tumors); B/ Kaplan Meier and log rank test for breast cancer TCGA cohort according overall survival outcome and stratification on CD274 expression groups; C/ Volcanoplot and barplot of ferroptosis actors (FerrDbV2: drivers in orange and suppressor in blue) regulated between breast tumors according their low or high level of CD274 expression.
Figure 3.
Breast tumors of patients with longer survival conjointly overexpressed CD274 and ferroptosis drivers: A/ Optimal threshold for CD274 expression according overall survival of the patient from Firehose TCGA invasive breast cancer transcriptome cohort (n=1082 tumors); B/ Kaplan Meier and log rank test for breast cancer TCGA cohort according overall survival outcome and stratification on CD274 expression groups; C/ Volcanoplot and barplot of ferroptosis actors (FerrDbV2: drivers in orange and suppressor in blue) regulated between breast tumors according their low or high level of CD274 expression.
Figure 4.
CD274-ferrptosis score allowed to stratified breast tumors according their prognostic in TCGA training cohort: A/ Barplot of univariate Cox (beta coefficients and Negative log10 p-values) analysis for CD274 and ferroptosis drivers expression according overall survival of patients in TCGA breast cancer cohort; B/ Optimal threshold determination for CD274-ferroptosis score according overall survival of the patient (TCGA cohort); C/ Kaplan Meier and log rank test on overall survival (TCGA cohort) according CD274-ferroptosis score group stratification; D/ Unsupervised clustering (Euclidean distances) and expression heatmap of CD274 and ferroptosis drivers used to compute expression score.
Figure 4.
CD274-ferrptosis score allowed to stratified breast tumors according their prognostic in TCGA training cohort: A/ Barplot of univariate Cox (beta coefficients and Negative log10 p-values) analysis for CD274 and ferroptosis drivers expression according overall survival of patients in TCGA breast cancer cohort; B/ Optimal threshold determination for CD274-ferroptosis score according overall survival of the patient (TCGA cohort); C/ Kaplan Meier and log rank test on overall survival (TCGA cohort) according CD274-ferroptosis score group stratification; D/ Unsupervised clustering (Euclidean distances) and expression heatmap of CD274 and ferroptosis drivers used to compute expression score.
Figure 5.
Clinical associations of CD274-ferroptosis driver score in TCGA breast cancer cohort: A/ Boxplot of CD274-ferroptosis driver score stratified on pam50 molecular classification (Fisher ANOVA p-value); B/ Boxplot of CD274-ferroptosis driver score stratified on scmod2 molecular classification (Fisher ANOVA p-value); C/ Boxplot of ggi molecular score stratified on categories of CD274-ferroptosis driver score (Student t test p-value); D/ Boxplot of ggi molecular score stratified on categories of CD274-ferroptosis driver score (Student t test p-value).
Figure 5.
Clinical associations of CD274-ferroptosis driver score in TCGA breast cancer cohort: A/ Boxplot of CD274-ferroptosis driver score stratified on pam50 molecular classification (Fisher ANOVA p-value); B/ Boxplot of CD274-ferroptosis driver score stratified on scmod2 molecular classification (Fisher ANOVA p-value); C/ Boxplot of ggi molecular score stratified on categories of CD274-ferroptosis driver score (Student t test p-value); D/ Boxplot of ggi molecular score stratified on categories of CD274-ferroptosis driver score (Student t test p-value).
Figure 6.
CD274 and ferroptosis drivers over expression in breast tumors was confirmed in patients with longer relapse free survival (METABRIC cohort): A/ Oncoprint of transcriptional alterations observed for CD274 and ferroptosis drivers (red: up expression, blue: down regulation, grey: unaltered); B/ Kaplan Meier and log rank test with relapse free survival as outcome and stratified on patient groups presenting or not alterations of CD274 and ferroptosis drivers in their transcriptome; C/ Barplot of Neoplasm histologic grade group proportions stratified on presence or absence of CD274-ferroptosis alterations; D/ Barplot of Neoplasm histologic grade group proportions stratified on presence or absence of CD274-ferroptosis alterations.
Figure 6.
CD274 and ferroptosis drivers over expression in breast tumors was confirmed in patients with longer relapse free survival (METABRIC cohort): A/ Oncoprint of transcriptional alterations observed for CD274 and ferroptosis drivers (red: up expression, blue: down regulation, grey: unaltered); B/ Kaplan Meier and log rank test with relapse free survival as outcome and stratified on patient groups presenting or not alterations of CD274 and ferroptosis drivers in their transcriptome; C/ Barplot of Neoplasm histologic grade group proportions stratified on presence or absence of CD274-ferroptosis alterations; D/ Barplot of Neoplasm histologic grade group proportions stratified on presence or absence of CD274-ferroptosis alterations.
Figure 7.
CD274-ferroptosis driver score is independent prognostic marker in breast cancer: A/ Forest plot of multivariable Cox model with overall survival as outcome (TCGA cohort) and included CD274-ferroptosis score with other covariates such as age categories (threshold 65 yo), and TNM (Tumor, Nodes, Metastasis) classification; B/ Representative detection at protein level (immunohistochemistry) in breast tumors for molecules included in computation of CD274-ferroptosis driver score.
Figure 7.
CD274-ferroptosis driver score is independent prognostic marker in breast cancer: A/ Forest plot of multivariable Cox model with overall survival as outcome (TCGA cohort) and included CD274-ferroptosis score with other covariates such as age categories (threshold 65 yo), and TNM (Tumor, Nodes, Metastasis) classification; B/ Representative detection at protein level (immunohistochemistry) in breast tumors for molecules included in computation of CD274-ferroptosis driver score.
Table 1.
Univariate analyses on CD274 expression groups according clinical parameters of patients in TCGA invasive breast cancer cohort.
Table 1.
Univariate analyses on CD274 expression groups according clinical parameters of patients in TCGA invasive breast cancer cohort.
| Variable |
Level |
low (n=426) |
high (n=656) |
Total (n=1,082) |
p-value |
| age.diagnosis |
younger65yo |
283 (66.4) |
462 (70.4) |
745 (68.9) |
|
| |
older65yo |
143 (33.6) |
194 (29.6) |
337 (31.1) |
0.1871019 |
| tumor.stage |
T1 |
102 (24.1) |
174 (26.5) |
276 (25.6) |
|
| |
T2 |
239 (56.5) |
388 (59.1) |
627 (58.1) |
|
| |
T3 |
66 (15.6) |
71 (10.8) |
137 (12.7) |
|
| |
T4 |
16 (3.8) |
23 (3.5) |
39 (3.6) |
0.1341858 |
| |
missing |
3 |
0 |
3 |
|
| node.stage |
N1 |
141 (33.9) |
214 (33.1) |
355 (33.4) |
|
| |
N0 |
196 (47.1) |
316 (48.9) |
512 (48.2) |
|
| |
N2 |
46 (11.1) |
73 (11.3) |
119 (11.2) |
|
| |
N3 |
33 (7.9) |
43 (6.7) |
76 (7.2) |
0.8484744 |
| |
missing |
10 |
10 |
20 |
|
| metastasis.stage |
M0 |
336 (97.4) |
558 (97.9) |
894 (97.7) |
|
| |
M1 |
9 (2.6) |
12 (2.1) |
21 (2.3) |
0.7909348 |
| |
missing |
81 |
86 |
167 |
|
| pam50.robust |
LumB |
154 (36.2) |
169 (25.8) |
323 (29.9) |
|
| |
Her2 |
31 (7.3) |
80 (12.2) |
111 (10.3) |
|
| |
LumA |
166 (39.0) |
245 (37.3) |
411 (38.0) |
|
| |
Normal |
14 (3.3) |
28 (4.3) |
42 (3.9) |
|
| |
Basal |
61 (14.3) |
134 (20.4) |
195 (18.0) |
0.0002416 |
| ggi.score |
mean (sd) |
2.1 (12.6) |
3.4 (13) |
2.9 (12.9) |
0.1112944 |
| g70.score |
mean (sd) |
-0.23 (0.2) |
-0.18 (0.2) |
-0.2 (0.2) |
0.0005523 |
| oncotypedx.score |
mean (sd) |
61 (35.9) |
65 (36.4) |
63.5 (36.2) |
0.0772518 |
| OS_STATUS |
Alive |
356 (83.6) |
575 (87.7) |
931 (86.0) |
|
| |
Dead |
70 (16.4) |
81 (12.3) |
151 (14.0) |
0.0711620 |
Table 2.
Univariate analyses of CD274-ferroptosis score categories according clinical parameters of patients in TCGA invasive breast cancer cohort.
Table 2.
Univariate analyses of CD274-ferroptosis score categories according clinical parameters of patients in TCGA invasive breast cancer cohort.
| Variable |
Level |
low (n=153) |
high (n=929) |
Total (n=1,082) |
p-value |
| age.at diagnosis |
younger65yo |
114 (74.5) |
631 (67.9) |
745 (68.9) |
|
| |
older65yo |
39 (25.5) |
298 (32.1) |
337 (31.1) |
0.124501 |
| tumor.stage |
T1 |
45 (29.4) |
231 (24.9) |
276 (25.6) |
|
| |
T2 |
93 (60.8) |
534 (57.7) |
627 (58.1) |
|
| |
T3 |
14 (9.2) |
123 (13.3) |
137 (12.7) |
|
| |
T4 |
1 (0.7) |
38 (4.1) |
39 (3.6) |
0.061994 |
| |
missing |
0 |
3 |
3 |
|
| node.stage |
N1 |
46 (30.1) |
309 (34.0) |
355 (33.4) |
|
| |
N0 |
91 (59.5) |
421 (46.3) |
512 (48.2) |
|
| |
N2 |
12 (7.8) |
107 (11.8) |
119 (11.2) |
|
| |
N3 |
4 (2.6) |
72 (7.9) |
76 (7.2) |
0.006523 |
| |
missing |
0 |
20 |
20 |
|
| metastasis.stage |
M0 |
140 (99.3) |
754 (97.4) |
894 (97.7) |
|
| |
M1 |
1 (0.7) |
20 (2.6) |
21 (2.3) |
0.288441 |
| |
missing |
12 |
155 |
167 |
|
| pam50.robust |
LumB |
26 (17.0) |
297 (32.0) |
323 (29.9) |
|
| |
Her2 |
26 (17.0) |
85 (9.1) |
111 (10.3) |
|
| |
LumA |
26 (17.0) |
385 (41.4) |
411 (38.0) |
|
| |
Normal |
4 (2.6) |
38 (4.1) |
42 (3.9) |
|
| |
Basal |
71 (46.4) |
124 (13.3) |
195 (18.0) |
< 0.0001 |
| ggi.score |
mean (sd) |
10.5 (10.9) |
1.6 (12.7) |
2.9 (12.9) |
< 0.0001 |
| g70.score |
mean (sd) |
0 (0.2) |
-0.2 (0.2) |
-0.2 (0.2) |
< 0.0001 |
| oncotypedx.score |
mean (sd) |
84.8 (25.8) |
59.9 (36.5) |
63.5 (36.2) |
< 0.0001 |
| OS_STATUS |
Alive |
141 (92.2) |
790 (85.0) |
931 (86.0) |
|
| |
Dead |
12 (7.8) |
139 (15.0) |
151 (14.0) |
0.025827 |
Table 3.
Cox overall survival multivariate on TCGA invasive breast cancer cohort including CD274-ferroptosis score categories and clinical parameters.
Table 3.
Cox overall survival multivariate on TCGA invasive breast cancer cohort including CD274-ferroptosis score categories and clinical parameters.
| Covariates |
Hazard ratios |
CI95 Low |
CI95 High |
P-value |
| CD274-ferroptosis score[T.high] |
2.012 |
1.063 |
3.810 |
3.18E-02 |
| age.categories [T.older65yo] |
2.368 |
1.635 |
3.430 |
5.13E-06 |
| tumor_grade[T.2] |
1.122 |
0.703 |
1.790 |
6.29E-01 |
| tumor_grade[T.3] |
1.161 |
0.627 |
2.149 |
6.35E-01 |
| tumor_grade[T.4] |
1.787 |
0.845 |
3.782 |
1.29E-01 |
| metastasis.stage[T.M1] |
3.021 |
1.594 |
5.726 |
6.98E-04 |
| node.stage[T.N1] |
1.726 |
1.123 |
2.653 |
1.28E-02 |
| node.stage[T.N2] |
2.517 |
1.426 |
4.445 |
1.46E-03 |
| node.stage[T.N3] |
2.405 |
1.118 |
5.174 |
2.48E-02 |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).