Submitted:
27 November 2023
Posted:
27 November 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
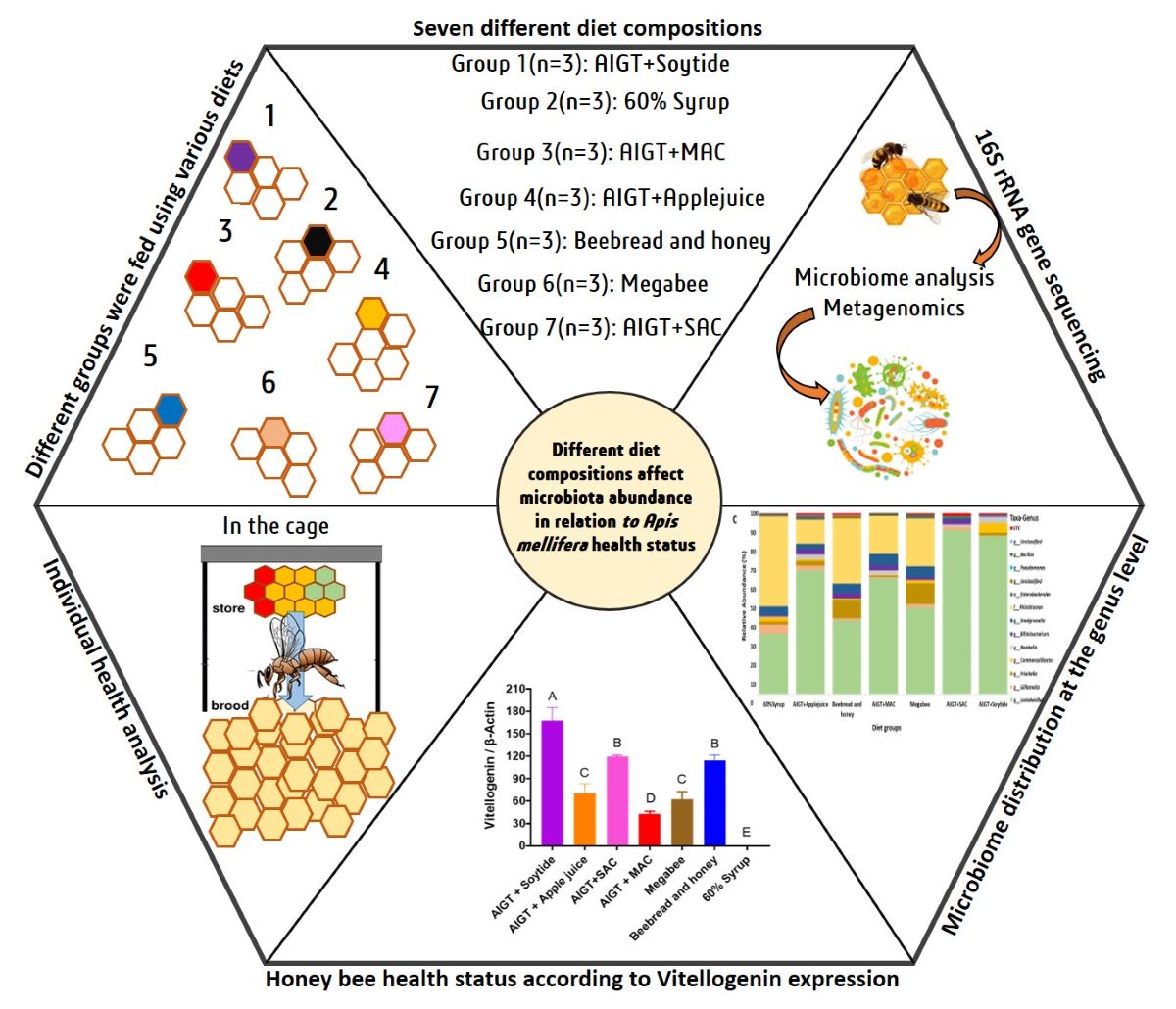
2. Material and Methods
2.1. Honey bee sampling
2.2. DNA extraction
2.3. RNA Extraction and Quantitative PCR (qPCR)
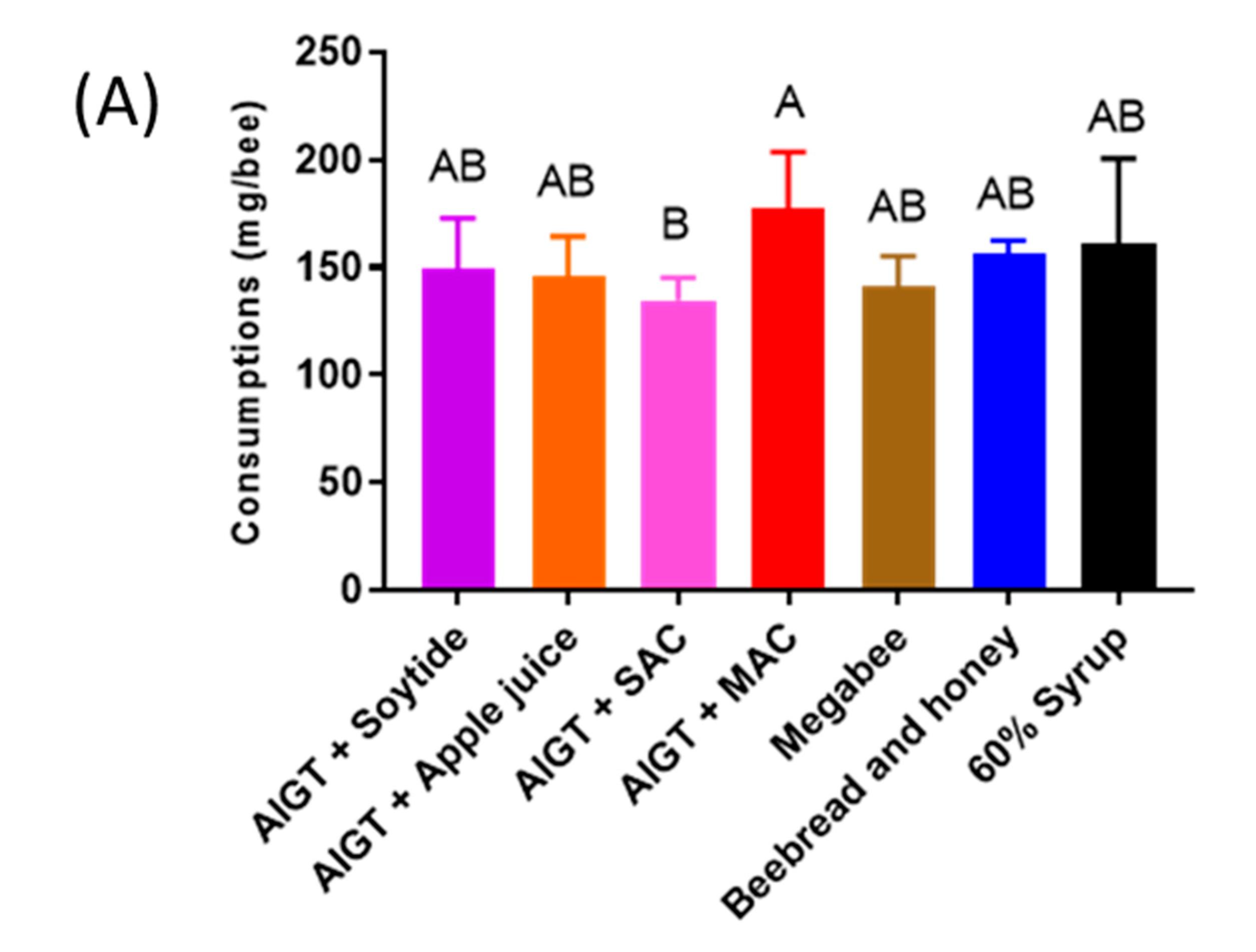
2.4. Consumption
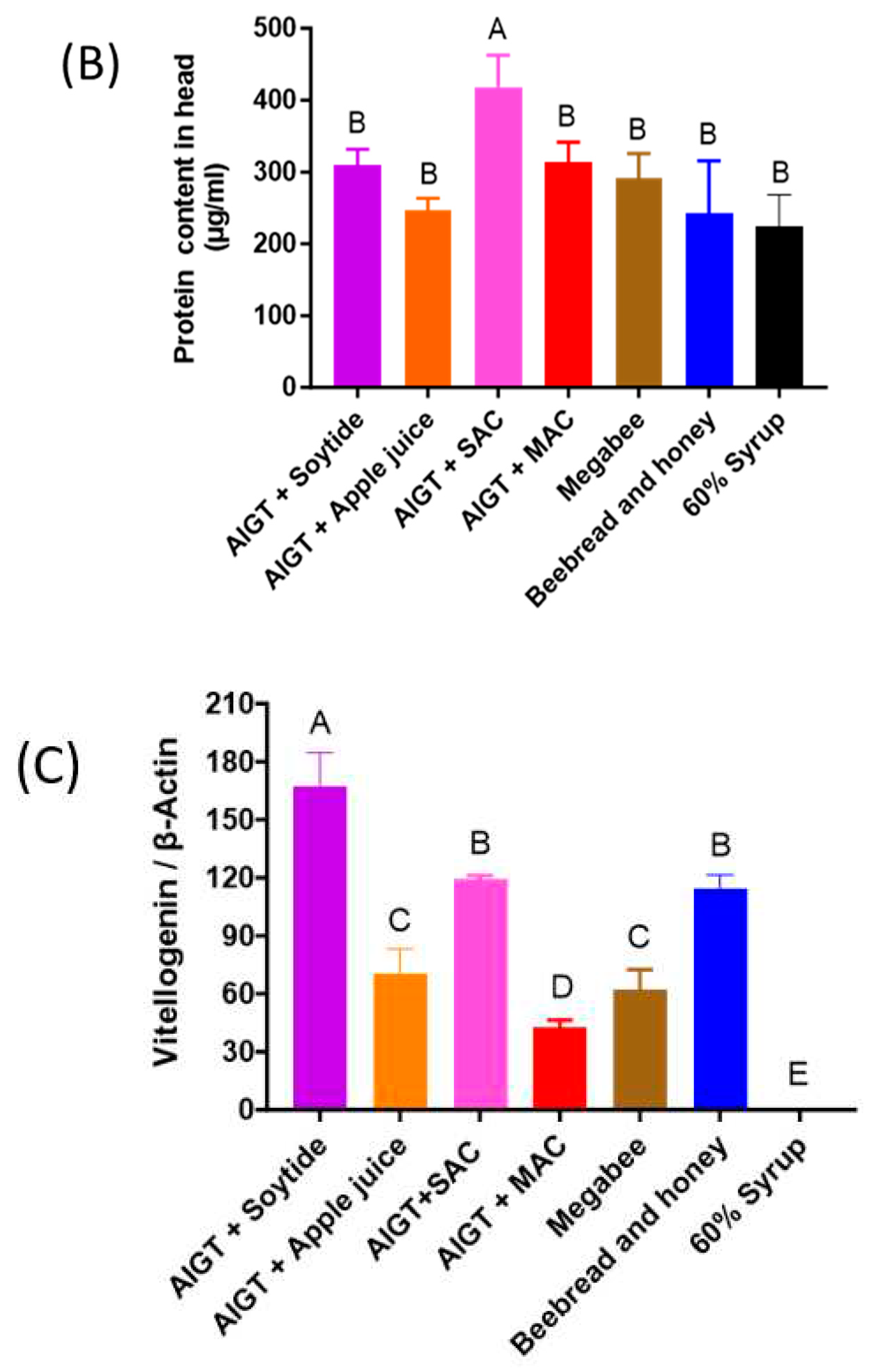
2.5. Soluble protein content
2.6. Sample preparation, 16s sequencing, and Taxonomic Analysis
2.7. Analysis in QIIME2
2.8. Bacterial profiles
2.9. Statistical Analysis
3. Results
3.1. Reads Profiling For Microbial Community
3.2. Gut Microbiota Diversity and Richness Between Different Diet Groups
3.2.1. Alpha diversity
3.2.2. Beta diversity
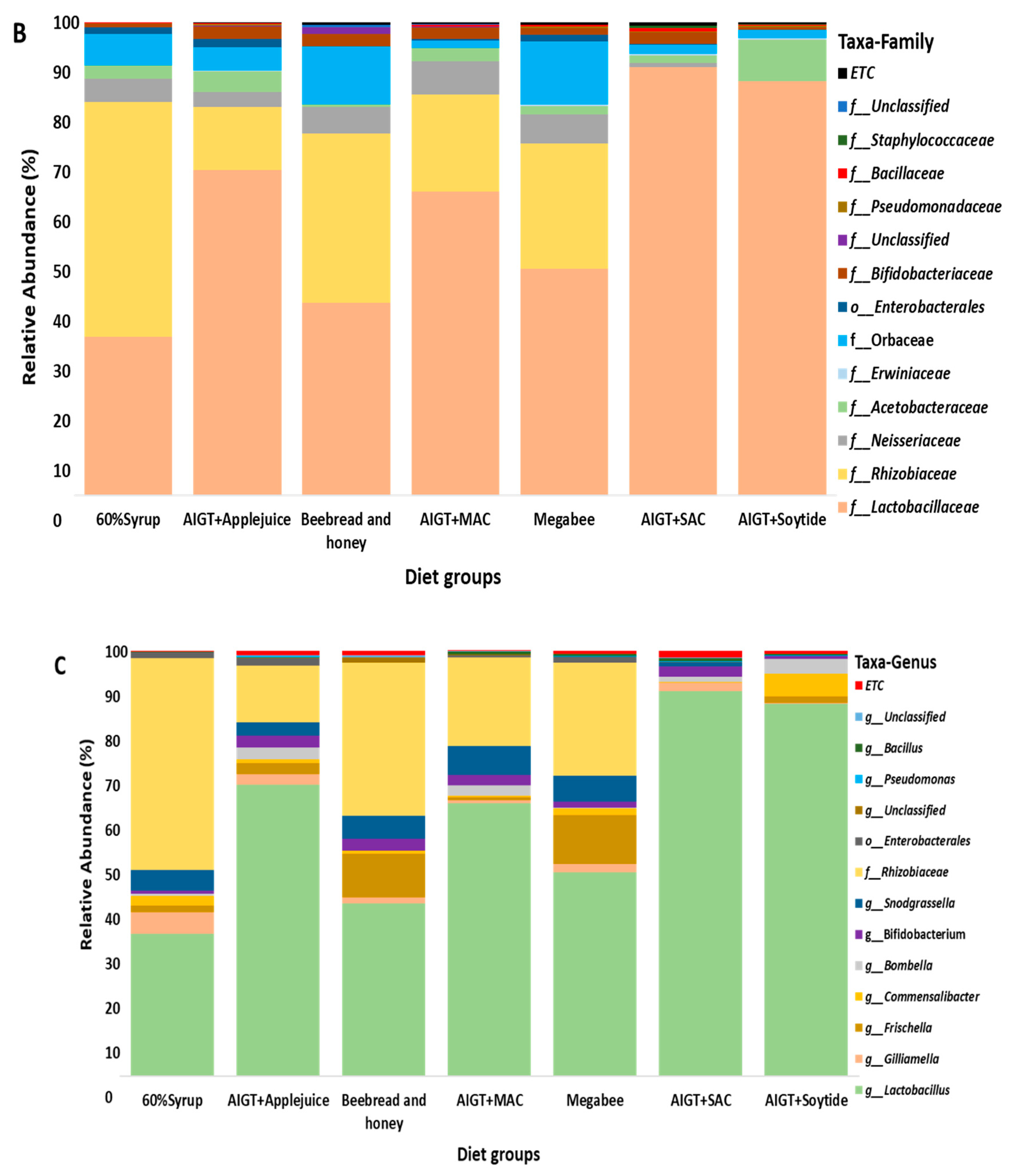
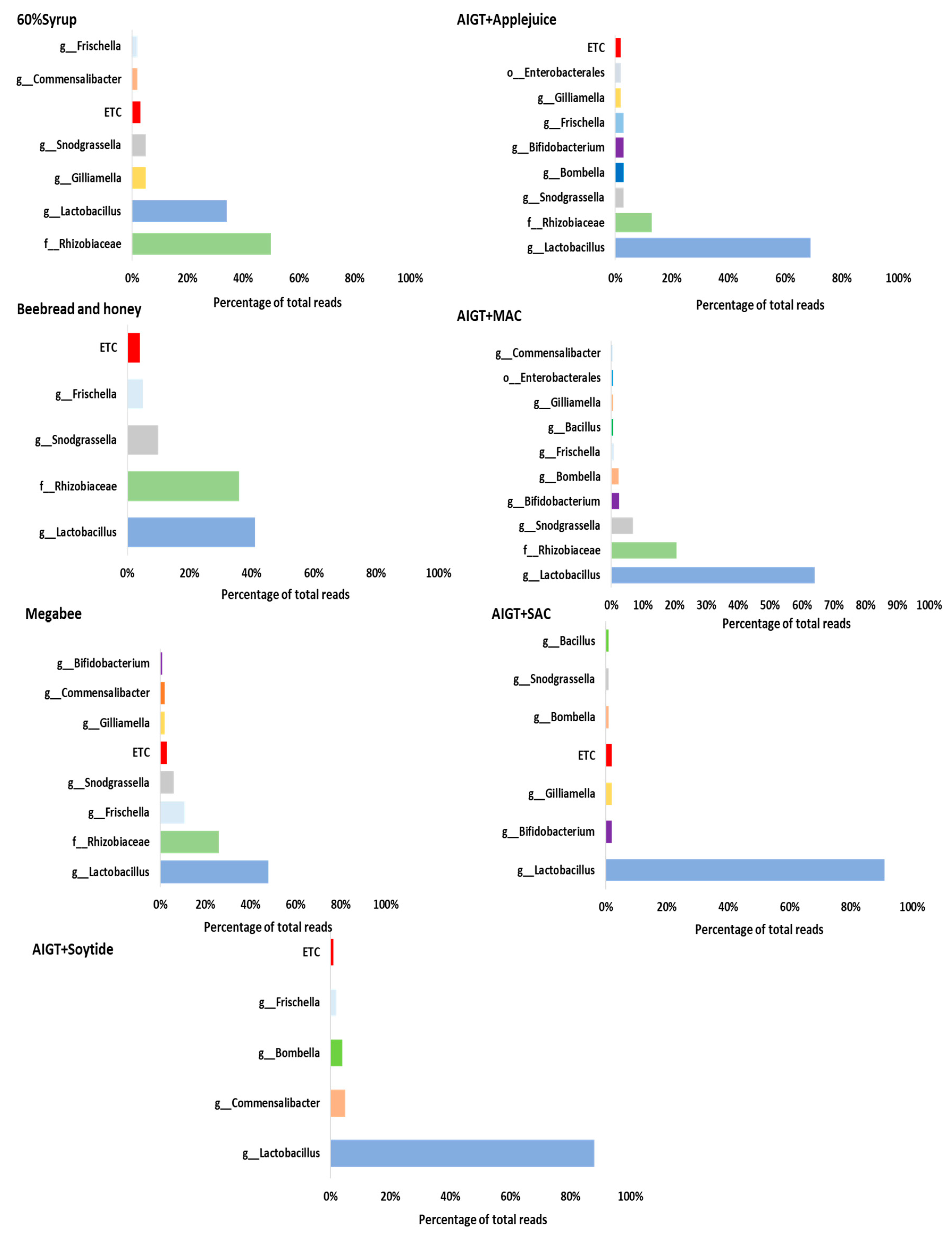
3.3. Gut Microbiota Taxonomy Diversity Based on Different Diets in Apis mellifera.
3.4. Honey bee Health Condition Depends on Different Diets
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Meixner, M.D.J.J.o.i.p. A historical review of managed honey bee populations in Europe and the United States and the factors that may affect them. 2010, 103, S80-S95.
- Koch, H.; Schmid-Hempel, P.J.P.o.t.N.A.o.S. Socially transmitted gut microbiota protect bumble bees against an intestinal parasite. 2011, 108, 19288-19292.
- Maigoro, A.Y.; Lee, S.J.N. Gut Microbiome-Based Analysis of Lipid A Biosynthesis in Individuals with Autism Spectrum Disorder: An In Silico Evaluation. 2021, 13, 688. [CrossRef]
- Lee, J.H.; Kim, H.-W.; Mustafa, B.; Lee, H.I.; Kwon, H.W.J.S.R. The relationships between microbiome diversity and epidemiology in domestic species of malaria-mediated mosquitoes of Korea. 2023, 13, 9081. [CrossRef]
- Maigoro, A.Y.; Muhammad, M.; Bello, B.; Useh, U.; Lee, S.J.J.o.M.F. Exploration of Gut Microbiome Research in Africa: A Scoping Review. 2023, 26, 616-623. [CrossRef]
- Evans, J.D.; Armstrong, T.-N.J.B.e. Antagonistic interactions between honey bee bacterial symbionts and implications for disease. 2006, 6, 1-9. [CrossRef]
- Wu, M.; Sugimura, Y.; Takaya, N.; Takamatsu, D.; Kobayashi, M.; Taylor, D.; Yoshiyama, M.J.J.o.i.p. Characterization of bifidobacteria in the digestive tract of the Japanese honeybee, Apis cerana japonica. 2013, 112, 88-93. [CrossRef]
- Cox-Foster, D.L.; Conlan, S.; Holmes, E.C.; Palacios, G.; Evans, J.D.; Moran, N.A.; Quan, P.-L.; Briese, T.; Hornig, M.; Geiser, D.M.J.S. A metagenomic survey of microbes in honey bee colony collapse disorder. 2007, 318, 283-287. [CrossRef]
- Martinson, V.G.; Moy, J.; Moran, N.A.J.A.; microbiology, e. Establishment of characteristic gut bacteria during development of the honeybee worker. 2012, 78, 2830-2840. [CrossRef]
- Kwong, W.K.; Moran, N.A.J.N.r.m. Gut microbial communities of social bees. 2016, 14, 374-384.
- Jones, J.C.; Fruciano, C.; Hildebrand, F.; Al Toufalilia, H.; Balfour, N.J.; Bork, P.; Engel, P.; Ratnieks, F.L.; Hughes, W.O.J.E.; evolution. Gut microbiota composition is associated with environmental landscape in honey bees. 2018, 8, 441-451. [CrossRef]
- Raymann, K.; Moran, N.A.J.C.o.i.i.s. The role of the gut microbiome in health and disease of adult honey bee workers. 2018, 26, 97-104.
- Maes, P.W.; Rodrigues, P.A.; Oliver, R.; Mott, B.M.; Anderson, K.E.J.M.E. Diet-related gut bacterial dysbiosis correlates with impaired development, increased mortality and Nosema disease in the honeybee (Apis mellifera). 2016, 25, 5439-5450.
- Dillon, R.J.; Webster, G.; Weightman, A.J.; Keith Charnley, A.J.A.V.L. Diversity of gut microbiota increases with aging and starvation in the desert locust. 2010, 97, 69-77. [CrossRef]
- Ma, W.; Zheng, X.; Li, L.; Shen, J.; Li, W.; Gao, Y.J.E.; Safety, E. Changes in the gut microbiota of honey bees associated with jujube flower disease. 2020, 198, 110616. [CrossRef]
- Endo, A.; Salminen, S.J.S.; Microbiology, A. Honeybees and beehives are rich sources for fructophilic lactic acid bacteria. 2013, 36, 444-448. [CrossRef]
- Hubert, J.; Bicianova, M.; Ledvinka, O.; Kamler, M.; Lester, P.J.; Nesvorna, M.; Kopecky, J.; Erban, T.J.M.e. Changes in the bacteriome of honey bees associated with the parasite Varroa destructor, and pathogens Nosema and Lotmaria passim. 2017, 73, 685-698. [CrossRef]
- Engel, P.; Moran, N.A.J.F.m.r. The gut microbiota of insects–diversity in structure and function. 2013, 37, 699-735.
- Shu, Q.; Wang, Y.; Gu, H.; Zhu, Q.; Liu, W.; Dai, Y.; Li, F.; Li, B.J.A.o.I.B.; Physiology. Effects of artificial diet breeding on intestinal microbial populations at the young stage of silkworm (Bombyx mori). 2023, e22019. [CrossRef]
- Wang, Y.; Li, Z.; Ma, L.; Li, G.; Han, K.; Liu, Z.; Wang, H.; Xu, B.J.F.i.M. The native dietary habits of the two sympatric bee species and their effects on shaping midgut microorganisms. 2021, 12, 738226. [CrossRef]
- Su, Q.; Tang, M.; Hu, J.; Tang, J.; Zhang, X.; Li, X.; Niu, Q.; Zhou, X.; Luo, S.; Zhou, X.J.b. Diet outweighs genetics in shaping gut microbiomes in Asian honeybee. 2022, 2022.2001. 2023.477436.
- Kim, H.J.; Hwang, J.; Ullah, Z.; Mustafa, B.; Kwon, H.W.J.J.o.A.-P.e. Comparison of physicochemical properties of pollen substitute diet for honey bee (Apis mellifera). 2022, 25, 101967. [CrossRef]
- Lim, S.; Jung, J.; Yunusbaev, U.; Ilyasov, R.; Kwon, H.W.J.S.r. Characterization and its implication of a novel taste receptor detecting nutrients in the honey bee, Apis mellifera. 2019, 9, 11620.
- Noordyke, E.R.; van Santen, E.; Ellis, J.D. Tracing the Fate of Pollen Substitute Patties in Western Honey Bee (Hymenoptera: Apidae) Colonies. J Econ Entomol 2021, 114, 1421–1430. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.; Jung, J.; Yunusbaev, U.; Ilyasov, R.; Kwon, H.W. Characterization and its implication of a novel taste receptor detecting nutrients in the honey bee, Apis mellifera. Scientific Reports 2019, 9, 11620. [Google Scholar] [CrossRef] [PubMed]
- Omar, E.M.; Darwish, H.Y.A.; Othman, A.A.; El-Seedi, H.R.; Al Naggar, Y. Crushing corn pollen grains increased diet digestibility and hemolymph protein content while decreasing honey bee consumption. Apidologie 2022, 53, 52. [Google Scholar] [CrossRef]
- Ketnawa, S.; Ogawa, Y. Evaluation of protein digestibility of fermented soybeans and changes in biochemical characteristics of digested fractions. Journal of Functional Foods 2019, 52, 640–647. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nature biotechnology 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nature methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Team, R.C. R: A language and environment for statistical computing. 2013.
- Gkantiragas, A.G.; Gabrielli, J.J.b. A Meta-Analysis of the 16S-rRNA Gut Microbiome Data in Honeybees (Apis Mellifera). 2021, 2021.2012. 2018.473299.
- Berg, G.; Rybakova, D.; Fischer, D.; Cernava, T.; Vergès, M.-C.C.; Charles, T.; Chen, X.; Cocolin, L.; Eversole, K.; Corral, G.H.J.M. Microbiome definition re-visited: old concepts and new challenges. 2020, 8, 1-22. [CrossRef]
- Maigoro, A.; Lee, S. Gut microbiome-based analysis of lipid a biosynthesis in individuals with autism spectrum disorder: an in silico evaluation. Nutrients. 2021; 13: 688. 2021. [CrossRef]
- Yun, J.-H.; Roh, S.W.; Whon, T.W.; Jung, M.-J.; Kim, M.-S.; Park, D.-S.; Yoon, C.; Nam, Y.-D.; Kim, Y.-J.; Choi, J.-H.J.A.; et al. Insect gut bacterial diversity determined by environmental habitat, diet, developmental stage, and phylogeny of host. 2014, 80, 5254-5264. [CrossRef]
- Krams, I.A.; Kecko, S.; Jõers, P.; Trakimas, G.; Elferts, D.; Krams, R.; Luoto, S.; Rantala, M.J.; Inashkina, I.; Gudrā, D.J.J.o.E.B. Microbiome symbionts and diet diversity incur costs on the immune system of insect larvae. 2017, 220, 4204-4212. [CrossRef]
- Montagna, M.; Mereghetti, V.; Gargari, G.; Guglielmetti, S.; Faoro, F.; Lozzia, G.; Locatelli, D.; Limonta, L.J.E.M. Evidence of a bacterial core in the stored products pest Plodia interpunctella: the influence of different diets. 2016, 18, 4961-4973. [CrossRef]
- Chandler, J.A.; Morgan Lang, J.; Bhatnagar, S.; Eisen, J.A.; Kopp, A.J.P.g. Bacterial communities of diverse Drosophila species: ecological context of a host–microbe model system. 2011, 7, e1002272. [CrossRef]
- Pérez-Cobas, A.E.; Maiques, E.; Angelova, A.; Carrasco, P.; Moya, A.; Latorre, A.J.F.m.e. Diet shapes the gut microbiota of the omnivorous cockroach Blattella germanica. 2015, 91, fiv022. [CrossRef]
- Jones, A.G.; Mason, C.J.; Felton, G.W.; Hoover, K.J.S.R. Host plant and population source drive diversity of microbial gut communities in two polyphagous insects. 2019, 9, 2792. [CrossRef]
- Lazzaro, B.P.; Rolff, J.J.S. Danger, microbes, and homeostasis. 2011, 332, 43-44.
- Kwong, W.K.; Moran, N.A.J.I.j.o.s.; microbiology, e. Cultivation and characterization of the gut symbionts of honey bees and bumble bees: description of Snodgrassella alvi gen. nov., sp. nov., a member of the family Neisseriaceae of the Betaproteobacteria, and Gilliamella apicola gen. nov., sp. nov., a member of Orbaceae fam. nov., Orbales ord. nov., a sister taxon to the order ‘Enterobacteriales’ of the Gammaproteobacteria. 2013, 63, 2008-2018.
- Ellegaard, K.M.; Engel, P.J.N.c. Genomic diversity landscape of the honey bee gut microbiota. 2019, 10, 446.
- Jia, H.-R.; Geng, L.-L.; Li, Y.-H.; Wang, Q.; Diao, Q.-Y.; Zhou, T.; Dai, P.-L.J.S.R. The effects of Bt Cry1Ie toxin on bacterial diversity in the midgut of Apis mellifera ligustica (Hymenoptera: Apidae). 2016, 6, 24664. [CrossRef]
- Wang, H.; Liu, C.; Liu, Z.; Wang, Y.; Ma, L.; Xu, B.J.B.m. The different dietary sugars modulate the composition of the gut microbiota in honeybee during overwintering. 2020, 20, 1-14. [CrossRef]
- Kane, M.D.; Breznak, J.A.J.A.; Microbiology, E. Effect of host diet on production of organic acids and methane by cockroach gut bacteria. 1991, 57, 2628-2634.
- Bauer, S.; Tholen, A.; Overmann, J.; Brune, A.J.A.o.M. Characterization of abundance and diversity of lactic acid bacteria in the hindgut of wood-and soil-feeding termites by molecular and culture-dependent techniques. 2000, 173, 126-137. [CrossRef]
- Vásquez, A.; Olofsson, T.C.; Sammataro, D.J.A. A scientific note on the lactic acid bacterial flora in honeybees in the USA–A comparison with bees from Sweden. 2009, 40, 26-28. [CrossRef]
- Vásquez, A.; Forsgren, E.; Fries, I.; Paxton, R.J.; Flaberg, E.; Szekely, L.; Olofsson, T.C.J.P.o. Symbionts as major modulators of insect health: lactic acid bacteria and honeybees. 2012, 7, e33188.
- Coenye, T.; Vandamme, P.J.M. Extracting phylogenetic information from whole-genome sequencing projects: the lactic acid bacteria as a test case. 2003, 149, 3507-3517. [CrossRef]
- Anderson, K.E.; Johansson, A.; Sheehan, T.H.; Mott, B.M.; Corby-Harris, V.; Johnstone, L.; Sprissler, R.; Fitz, W.J.G.P. Draft genome sequences of two Bifidobacterium sp. from the honey bee (Apis mellifera). 2013, 5, 1–3. [Google Scholar] [CrossRef]
- Crotti, E.; Rizzi, A.; Chouaia, B.; Ricci, I.; Favia, G.; Alma, A.; Sacchi, L.; Bourtzis, K.; Mandrioli, M.; Cherif, A.J.A.; et al. Acetic acid bacteria, newly emerging symbionts of insects. 2010, 76, 6963-6970. [CrossRef]
- Butler, È.; Alsterfjord, M.; Olofsson, T.C.; Karlsson, C.; Malmström, J.; Vásquez, A.J.B.m. Proteins of novel lactic acid bacteria from Apis mellifera mellifera: an insight into the production of known extra-cellular proteins during microbial stress. 2013, 13, 1-12. [CrossRef]
- Hamdi, C.; Balloi, A.; Essanaa, J.; Crotti, E.; Gonella, E.; Raddadi, N.; Ricci, I.; Boudabous, A.; Borin, S.; Manino, A.J.J.o.a.e. Gut microbiome dysbiosis and honeybee health. 2011, 135, 524-533. [CrossRef]
- Olofsson, T.C.; Vásquez, A.J.C.m. Detection and identification of a novel lactic acid bacterial flora within the honey stomach of the honeybee Apis mellifera. 2008, 57, 356-363.
- Kešnerová, L.; Emery, O.; Troilo, M.; Liberti, J.; Erkosar, B.; Engel, P.J.T.I.j. Gut microbiota structure differs between honeybees in winter and summer. 2020, 14, 801-814. [CrossRef]
- Liu, P.; Zhu, Y.; Ye, L.; Shi, T.; Li, L.; Cao, H.; Yu, L.J.S.R. Overwintering honeybees maintained dynamic and stable intestinal bacteria. 2021, 11, 22233. [CrossRef]
- Ribière, C.; Hegarty, C.; Stephenson, H.; Whelan, P.; O’Toole, P.W.J.M.e. Gut and whole-body microbiota of the honey bee separate thriving and non-thriving hives. 2019, 78, 195-205. [CrossRef]
- Zanni, V.; Galbraith, D.A.; Annoscia, D.; Grozinger, C.M.; Nazzi, F. Transcriptional signatures of parasitization and markers of colony decline in Varroa-infested honey bees (Apis mellifera). Insect Biochemistry and Molecular Biology 2017, 87, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Aurori, C.M.; Buttstedt, A.; Dezmirean, D.S.; Mărghitaş, L.A.; Moritz, R.F.; Erler, S.J.J.o.G.S.A.B.S.; Sciences, M. What is the main driver of ageing in long-lived winter honeybees: antioxidant enzymes, innate immunity, or vitellogenin? 2014, 69, 633-639. [CrossRef]
- Wang, H.; Zhang, S.-W.; Zeng, Z.-J.; Yan, W.-Y.J.A. Nutrition affects longevity and gene expression in honey bee (Apis mellifera) workers. 2014, 45, 618-625. [CrossRef]
- Bitondi, M.; Simoes, Z.P.J.J.o.A.R. The relationship between level of pollen in the diet, vitellogenin and juvenile hormone titres in Africanized Apis mellifera workers. 1996, 35, 27-36.
- D’alvise, P.; Böhme, F.; Codrea, M.C.; Seitz, A.; Nahnsen, S.; Binzer, M.; Rosenkranz, P.; Hasselmann, M.J.A. The impact of winter feed type on intestinal microbiota and parasites in honey bees. 2018, 49, 252-264. [CrossRef]
- Bleau, N.; Bouslama, S.; Giovenazzo, P.; Derome, N.J.M. Dynamics of the honeybee (Apis mellifera) gut microbiota throughout the overwintering period in Canada. 2020, 8, 1146. [CrossRef]
- Cornet, L.; Cleenwerck, I.; Praet, J.; Leonard, R.R.; Vereecken, N.J.; Michez, D.; Smagghe, G.; Baurain, D.; Vandamme, P.J.M. Phylogenomic analyses of Snodgrassella isolates from honeybees and bumblebees reveal taxonomic and functional diversity. 2022, 7, e01500-01521. [CrossRef]
- Taylor, M. Microbiota in the honey bee gut and their association with bee health: a thesis presented in partial fulfilment of the requirements for the degree of Doctor of Philosophy (PhD) in Ecology at Massey University School of Agriculture and Environment, Palmerston North, New Zealand. Massey University, 2020.
- Horton, M.A.; Oliver, R.; Newton, I.L.J.P. No apparent correlation between honey bee forager gut microbiota and honey production. 2015, 3, e1329. [CrossRef]
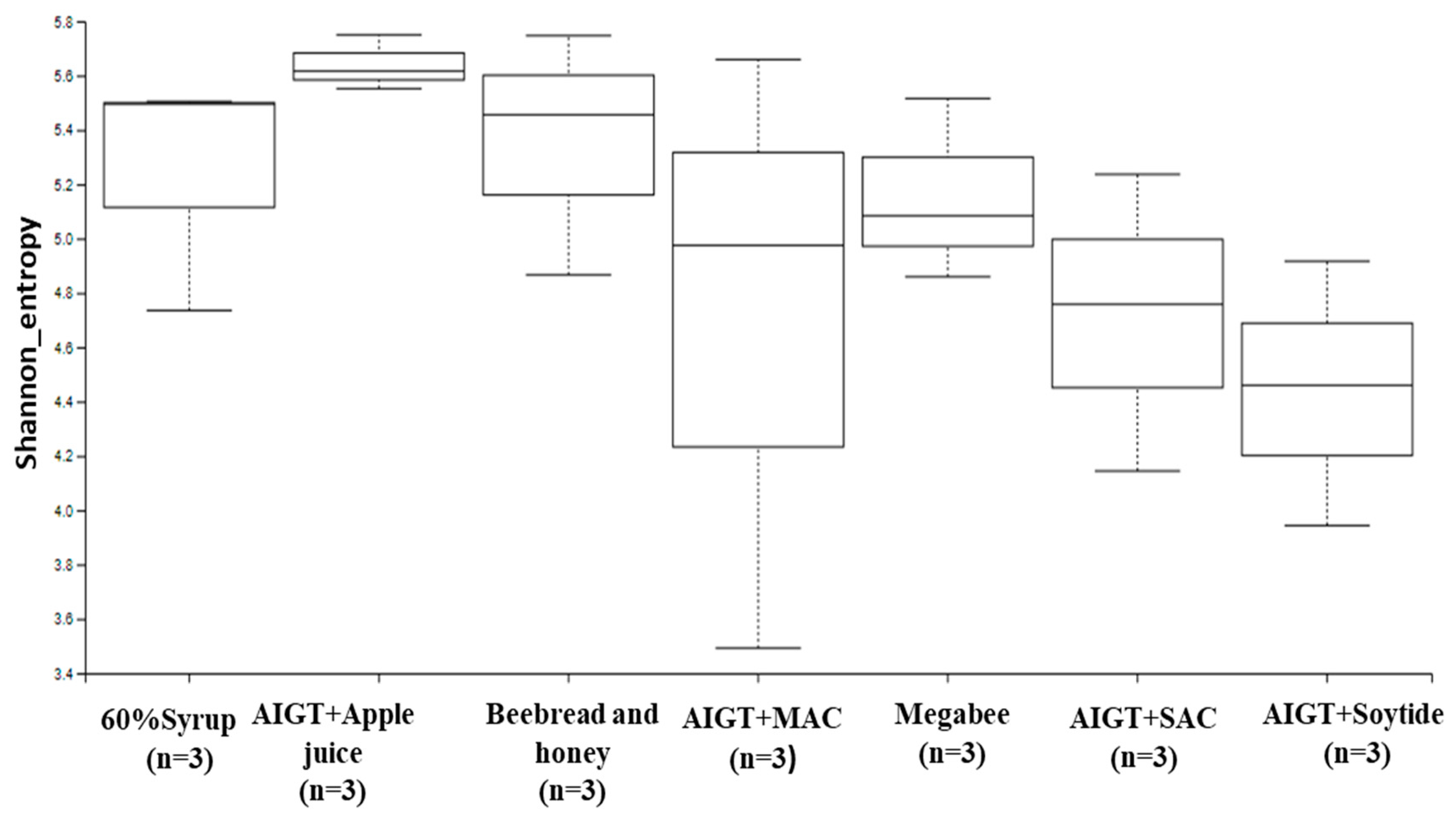

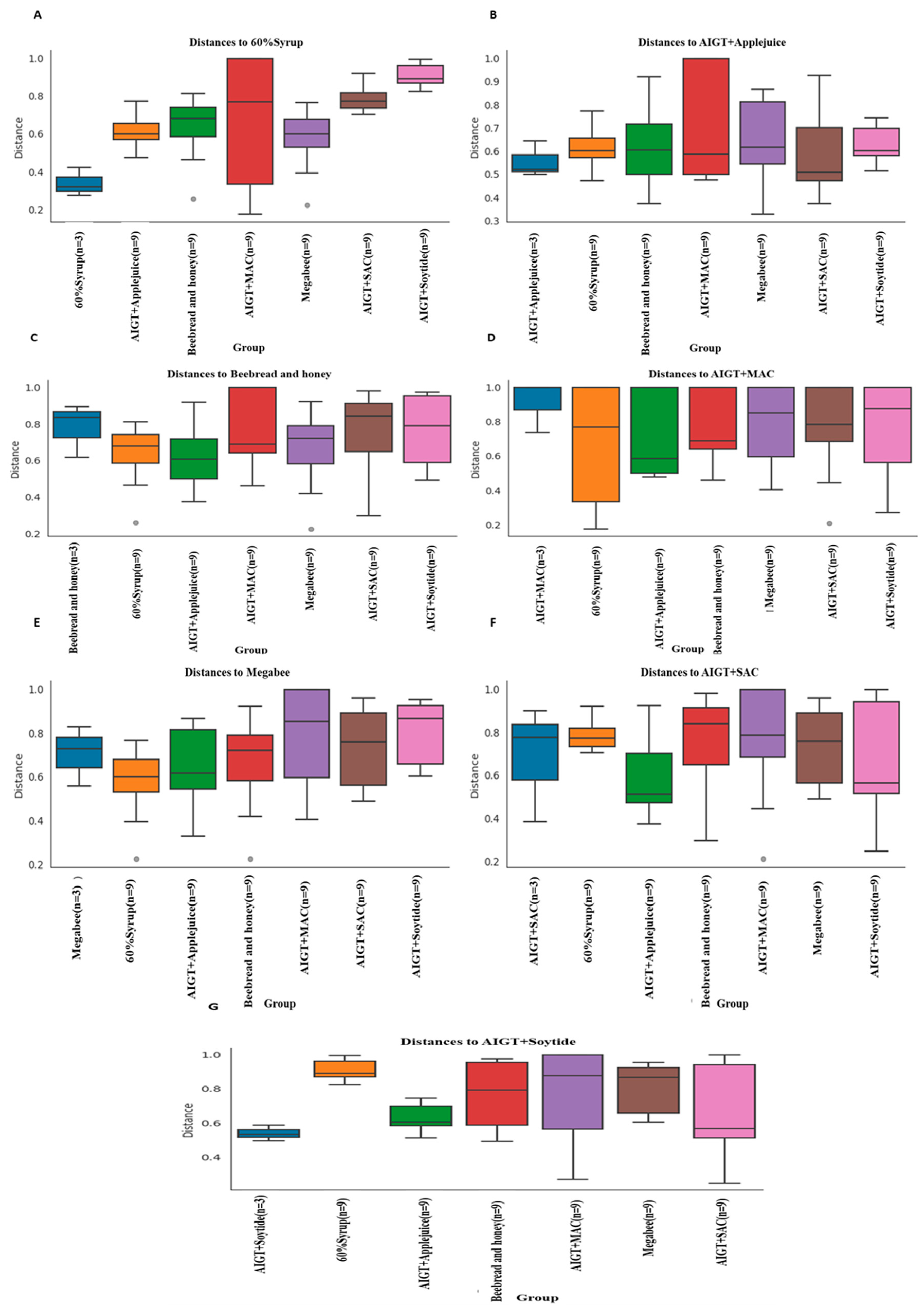
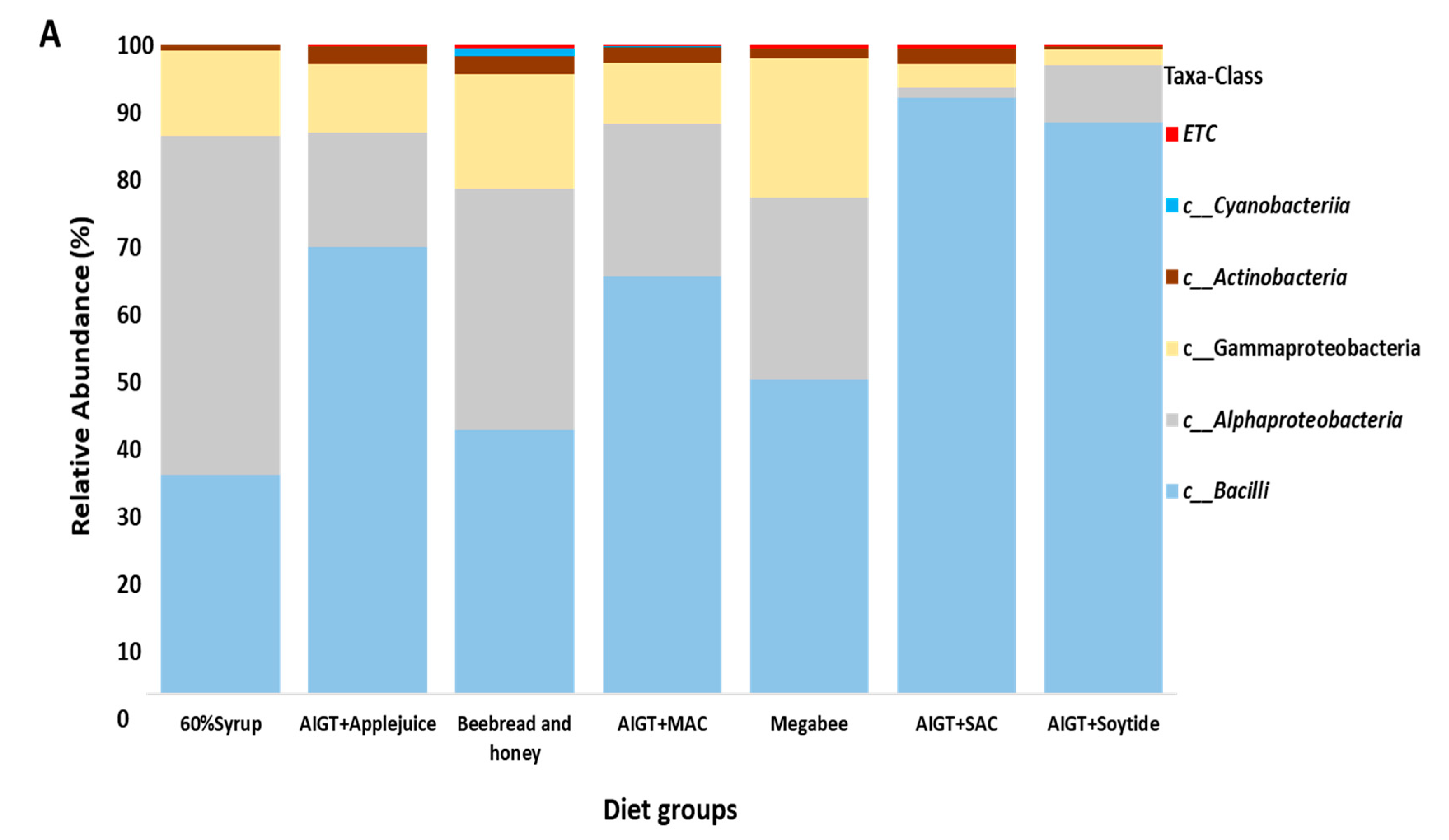
| Group 1 | Group 2 | P-value | q-value |
|---|---|---|---|
| 60% Syrup (n=3) | AIGT+Apple juice (n=3) | 0.049 | 0.260 |
| AIGT+Apple juice (n=3) | Megabee (n=3) | 0.049 | 0.260 |
| AIGT+Apple juice (n=3) | AIGT+SAC (n=3) | 0.049 | 0.260 |
| AIGT+Apple juice (n=3) | AIGT+Soytide (n=3) | 0.049 | 0.260 |
| Beebread and honey (n=3) | Megabee (n=3) | 0.512 | 0.672 |
| Megabee (n=3) | AIGT+SAC (n=3) | 0.275 | 0.481 |
| AIGT+SAC (n=3) | AIGT+Soytide (n=3) | 0.512 | 0.672 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





