Submitted:
29 November 2023
Posted:
29 November 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Antibodies
2.2. Plasmid Construction and Transfection
2.3. Flow Cytometry
3. Results
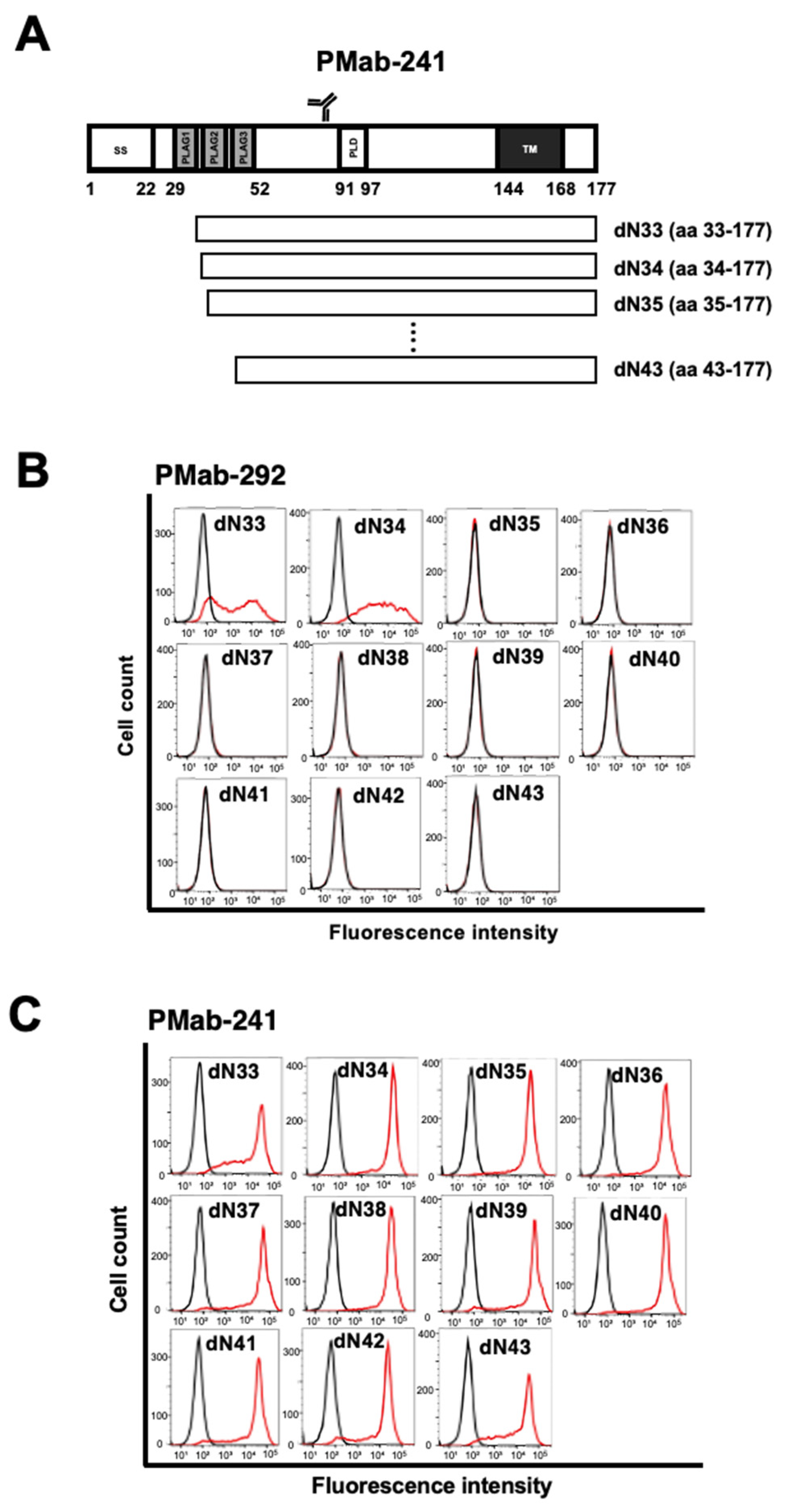
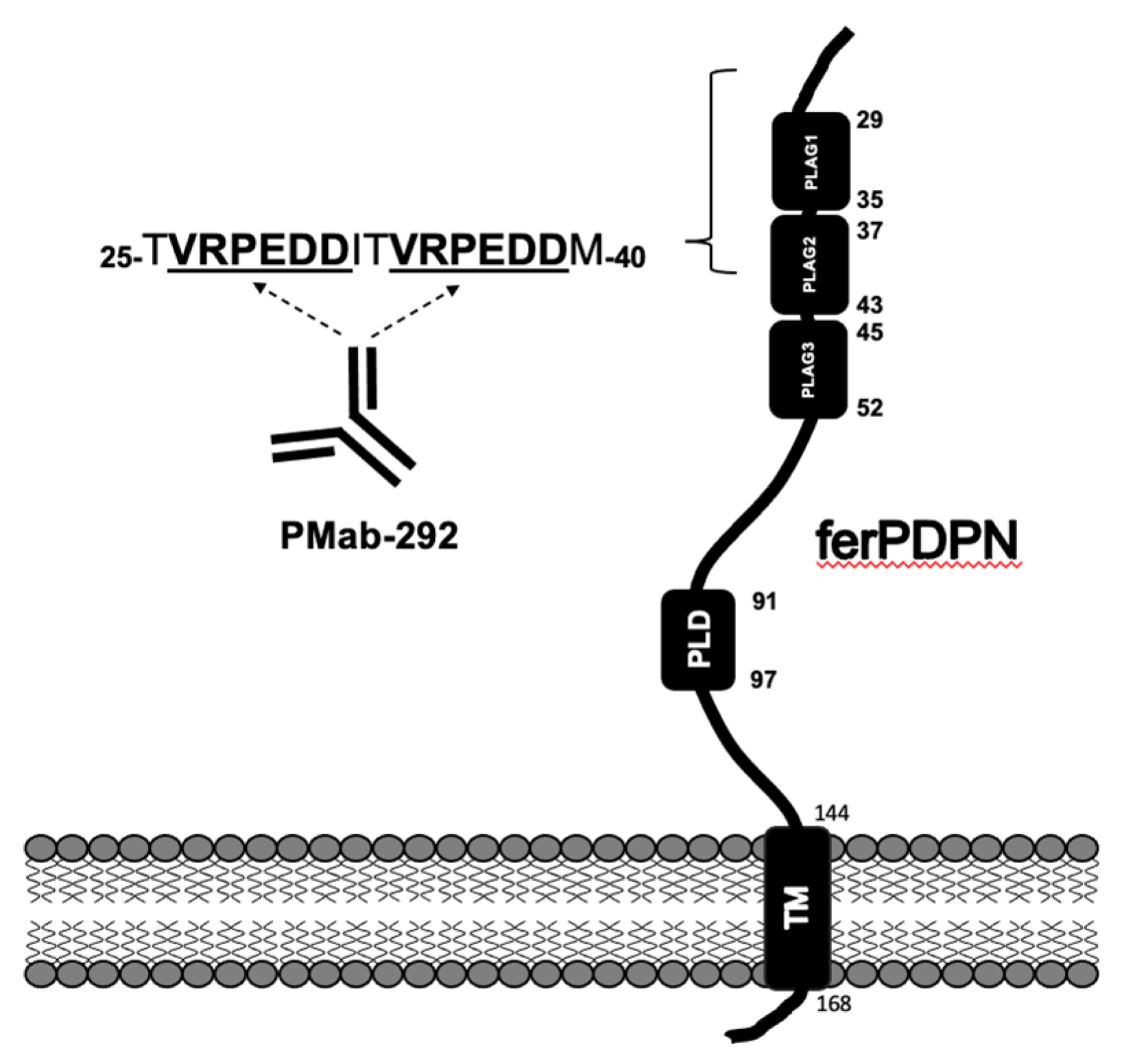
3.1. Epitope Mapping of PMab-292 Using the N-Terminal Deletion Mutants of ferPDPN
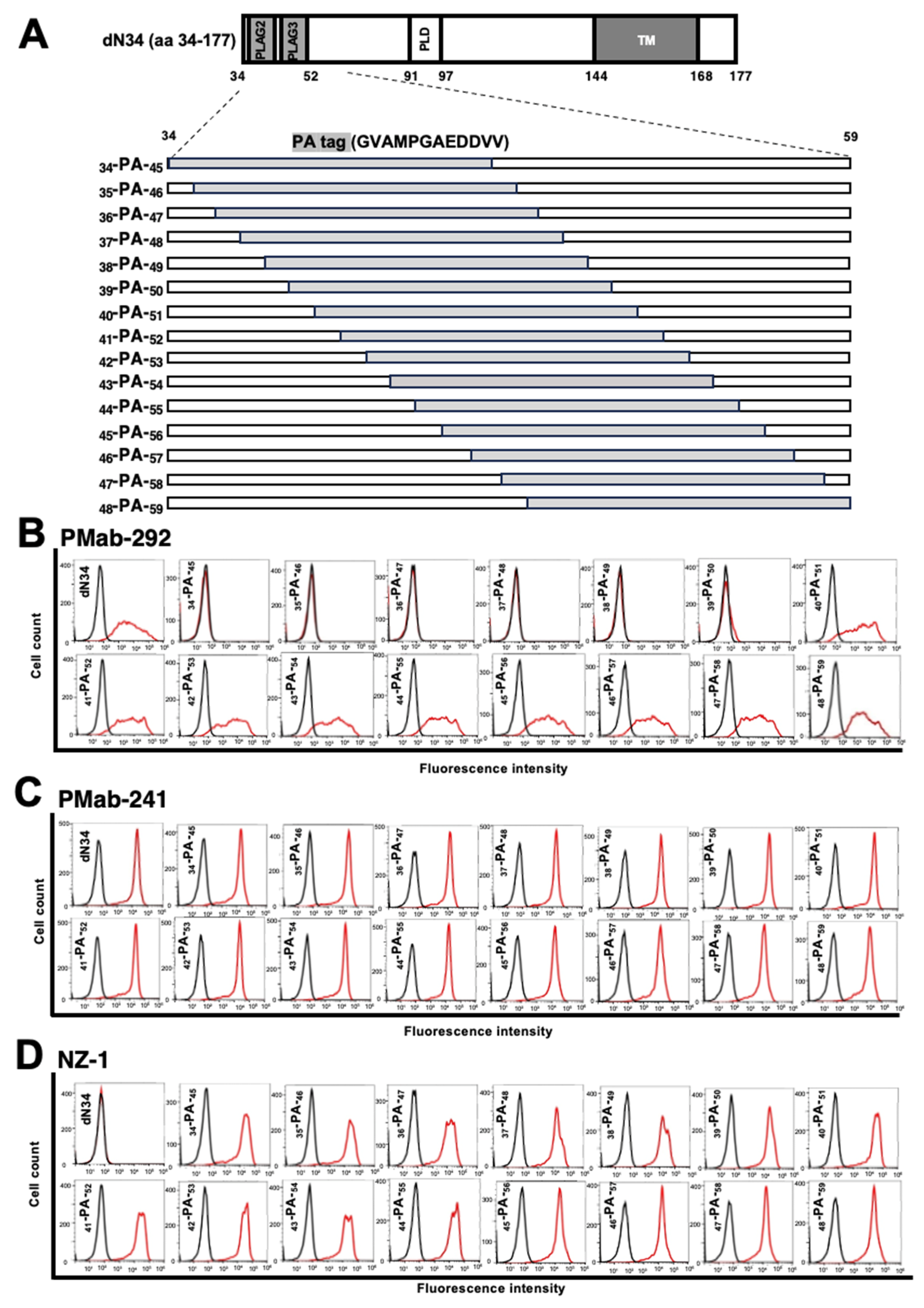
3.2. Flow Cytometry Using PMab-292 with PA Tag-Substituted ferPDPN
4. Discussion
References
- van den Brand, J.M.; Haagmans, B.L.; van Riel, D.; Osterhaus, A.D.; Kuiken, T. The pathology and pathogenesis of experimental severe acute respiratory syndrome and influenza in animal models. J. Comp. Pathol. 2014, 151, 83–112. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Fontela, C.; Dowling, W.E.; Funnell, S.G.P.; Gsell, P.S.; Riveros-Balta, A.X.; Albrecht, R.A.; Andersen, H.; Baric, R.S.; Carroll, M.W.; Cavaleri, M.; et al. Animal models for COVID-19. Nature 2020, 586, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Dobbs, L.G.; Williams, M.C.; Gonzalez, R. Monoclonal antibodies specific to apical surfaces of rat alveolar type I cells bind to surfaces of cultured, but not freshly isolated, type II cells. Biochim. Biophys. Acta 1988, 970, 146–156. [Google Scholar] [CrossRef] [PubMed]
- Rishi, A.K.; Joyce-Brady, M.; Fisher, J.; Dobbs, L.G.; Floros, J.; VanderSpek, J.; Brody, J.S.; Williams, M.C. Cloning, characterization, and development expression of a rat lung alveolar type I cell gene in embryonic endodermal and neural derivatives. Dev. Biol. 1995, 167, 294–306. [Google Scholar] [CrossRef] [PubMed]
- Breiteneder-Geleff, S.; Matsui, K.; Soleiman, A.; Meraner, P.; Poczewski, H.; Kalt, R.; Schaffner, G.; Kerjaschki, D. Podoplanin, novel 43-kd membrane protein of glomerular epithelial cells, is down-regulated in puromycin nephrosis. Am. J. Pathol. 1997, 151, 1141–1152. [Google Scholar] [PubMed]
- Hirakawa, S.; Hong, Y.K.; Harvey, N.; Schacht, V.; Matsuda, K.; Libermann, T.; Detmar, M. Identification of vascular lineage-specific genes by transcriptional profiling of isolated blood vascular and lymphatic endothelial cells. Am. J. Pathol. 2003, 162, 575–586. [Google Scholar] [CrossRef] [PubMed]
- Petrova, T.V.; Mäkinen, T.; Mäkelä, T.P.; Saarela, J.; Virtanen, I.; Ferrell, R.E.; Finegold, D.N.; Kerjaschki, D.; Ylä-Herttuala, S.; Alitalo, K. Lymphatic endothelial reprogramming of vascular endothelial cells by the Prox-1 homeobox transcription factor. Embo J. 2002, 21, 4593–4599. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, H.; Rayes, J.; Miyashita, T.; Ishii, G.; Retzbach, E.P.; Sheehan, S.A.; Takemoto, A.; Chang, Y.W.; Yoneda, K.; Asai, J.; et al. Podoplanin: An emerging cancer biomarker and therapeutic target. Cancer Sci. 2018, 109, 1292–1299. [Google Scholar] [CrossRef] [PubMed]
- Quintanilla, M.; Montero-Montero, L.; Renart, J.; Martín-Villar, E. Podoplanin in Inflammation and Cancer. Int. J. Mol. Sci. 2019, 20, 707. [Google Scholar] [CrossRef] [PubMed]
- Kato, Y.; Fujita, N.; Kunita, A.; Sato, S.; Kaneko, M.; Osawa, M.; Tsuruo, T. Molecular identification of Aggrus/T1alpha as a platelet aggregation-inducing factor expressed in colorectal tumors. J. Biol. Chem. 2003, 278, 51599–51605. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Kaneko, M.K.; Kato, Y. Roles of Podoplanin in Malignant Progression of Tumor. Cells 2022, 11, 575. [Google Scholar] [CrossRef] [PubMed]
- Goto, N.; Suzuki, H.; Tanaka, T.; Asano, T.; Kaneko, M.K.; Kato, Y. Development of a Monoclonal Antibody PMab-292 Against Ferret Podoplanin. Monoclon. Antib. Immunodiagn. Immunother. 2022, 41, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Takei, J.; Yamada, S.; Konnai, S.; Ishinazaka, T.; Shimozuru, M.; Kaneko, M.K.; Kato, Y. PMab-241 Specifically Detects Bear Podoplanin of Lymphatic Endothelial Cells in the Lung of Brown Bear. Monoclon. Antib. Immunodiagn. Immunother. 2019, 38, 282–284. [Google Scholar] [CrossRef] [PubMed]
- Kaji, C.; Tsujimoto, Y.; Kato Kaneko, M.; Kato, Y.; Sawa, Y. Immunohistochemical Examination of Novel Rat Monoclonal Antibodies against Mouse and Human Podoplanin. Acta Histochem. Cytochem. 2012, 45, 227–237. [Google Scholar] [CrossRef] [PubMed]
- Kato, Y.; Kaneko, M.K.; Kuno, A.; Uchiyama, N.; Amano, K.; Chiba, Y.; Hasegawa, Y.; Hirabayashi, J.; Narimatsu, H.; Mishima, K.; et al. Inhibition of tumor cell-induced platelet aggregation using a novel anti-podoplanin antibody reacting with its platelet-aggregation-stimulating domain. Biochem. Biophys. Res. Commun. 2006, 349, 1301–1307. [Google Scholar] [CrossRef] [PubMed]
- Fujii, Y.; Kaneko, M.K.; Kato, Y. MAP Tag: A Novel Tagging System for Protein Purification and Detection. Monoclon. Antib. Immunodiagn. Immunother. 2016, 35, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Wakasa, A.; Kaneko, M.K.; Kato, Y.; Takagi, J.; Arimori, T. Site-specific epitope insertion into recombinant proteins using the MAP tag system. J. Biochem. 2020, 168, 375–384. [Google Scholar] [CrossRef] [PubMed]
- Sayama, Y.; Sano, M.; Asano, T.; Furusawa, Y.; Takei, J.; Nakamura, T.; Yanaka, M.; Okamoto, S.; Handa, S.; Komatsu, Y.; et al. Epitope Mapping of PMab-241, a Lymphatic Endothelial Cell-Specific Anti-Bear Podoplanin Monoclonal Antibody. Monoclon. Antib. Immunodiagn. Immunother. 2020, 39, 77–81. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, M.K.; Kato, Y.; Kameyama, A.; Ito, H.; Kuno, A.; Hirabayashi, J.; Kubota, T.; Amano, K.; Chiba, Y.; Hasegawa, Y.; et al. Functional glycosylation of human podoplanin: Glycan structure of platelet aggregation-inducing factor. FEBS Lett. 2007, 581, 331–336. [Google Scholar] [CrossRef] [PubMed]
- Sekiguchi, T.; Takemoto, A.; Takagi, S.; Takatori, K.; Sato, S.; Takami, M.; Fujita, N. Targeting a novel domain in podoplanin for inhibiting platelet-mediated tumor metastasis. Oncotarget 2016, 7, 3934–3946. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




