1. Introduction
Chronic Kidney Disease (CKD) is a term that evolves over time. Currently, the international guidelines define it as the condition on which the kidney function was decreased by Glomerular Filtration Rate (GFR) of less than 60 ml/min per 1.73 m
2 [
1]. As it can be observed,
Table 1 contains the five stages of the disease, which starts with a slight diminished function of the organ and goes to a total failure. Studies reveals that >10% of the global population suffers CKD at any stage; since the main two causes for a person to develop CKD are diabetes and hypertension, and both have an upward trend, the number is expected to grow in the following years [
2].
The traditional range of parameters for prioritization includes 6 components, whose levels will determine if the patient is considered low, medium, or high priority to receive a kidney transplantation.
Table 2 shows the ranges based on the analysis from 230 datasets from the Global Hospital of Chennai [
3].
According to Manns, et al. [
4], based on a cohort of 641 adults with CKD living in Canada, the 1-year median cumulative healthcare costs (including drugs, physician visits, emergency department visits, dialysis, and surgeries) was
$14,634.00 Canadian dollars, which is ∼
$11,500.00 US dollars. A patient with CKD will need to live with the condition until they get a kidney transplant, so this amount of money will be required year after year to increase the possibility of survival.
Due to the inequality in the global distribution of wealth, the amount of money in order to achieve basic needs is especially worrying.
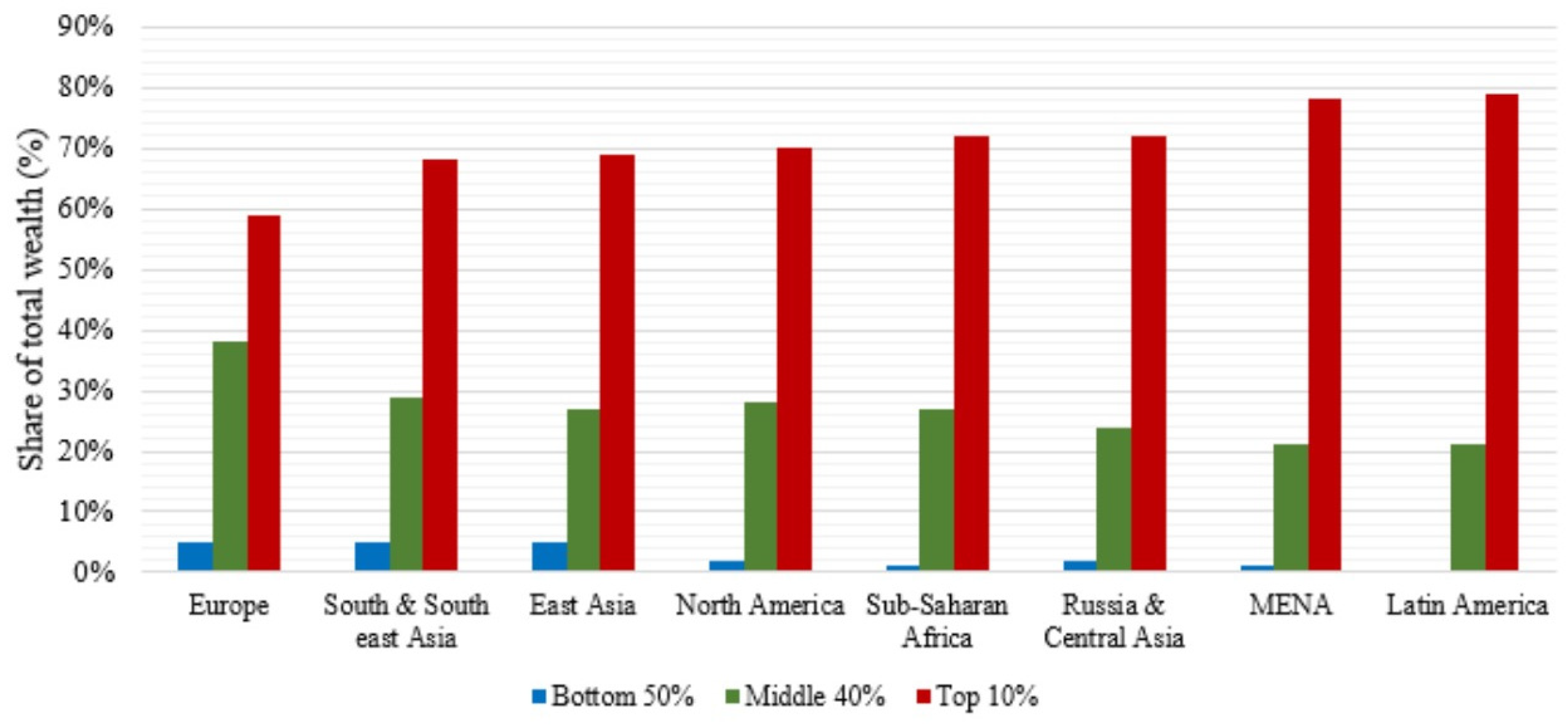
Figure 1 [
5] shows the distribution of the global wealth, showing that the richest 10% of the population owns between 60% to 80% of it. The gap between the top and the bottom is bigger on the undeveloped regions of the world (Latin America, Middle East & North Africa, Russia & Central Asia and Sub-Saharan Africa), but the inequality has the same trend on all the world.
CKD is an expensive disease that combined with an unequal distribution of wealth in the world and few people having access to medical care inhibits early detection which is vital for treating the disease before it progresses to advanced stages where medical costs are exponentially higher and chances of survival are lower.
With the purpose of positively impacting the most vulnerable sectors of society, it is necessary to develop cheaper diagnostic methods. In order to do this, it is important to use machine learning algorithms which offer medical staff alternative diagnostic methods to assist in decision making process.
The rest of the paper is organized as follows: section two comprises a brief of literature review and machine-learning relevance researches on the healthcare industry. Next, in section three, we introduce the most important starting considerations to understand the scope and characteristics of the proposal. After that, section four presents the methodology by reviewing each of its stages. Algorithms and results are given in section five. Finally, in section six, some conclusions are drawn.
2. A Brief of Literature Review and Machine Learning in Healthcare
There are previous studies that use different machine learning algorithms, to detect different kidney related disorders. Vijarayani, et al. [
6] developed models using Support Vector Machine (SVM); they trained their model with the goal of classifying the patient’s kidney degradation stage (refer Table 1 for the five stages of CKD), and reached an accuracy of 70.96%.
Lakshmi, et al. [
7] used three different algorithms: decision trees, logical regression and neural networks, to classify kidney dialysis data. They concluded that the neural networks outperformed the other two algorithms in accuracy: 93.8% for neural network, 74.74% for logistic regression, and 78.44% for decision trees.
Subasi, et al. [
8] created a computational system using the following machine learning classifiers: SVM, NN, k-Nearest Neighbour (k-NN), and Decision Trees (DT), and concluded that Random Forest (RF) is better to predict similar diseases.
Kuo, et al. [
9] worked with ultrasound imaging, to predict CKD. They developed a deep learning approach that automatically determines the GFR. For their study, they utilized 4,505 ultrasound images derived from serum creatinine concentrations, and used a deep neural network to classify the patients. They obtained a 95% of accuracy.
Caocci [
10] made a comparison between a neural network and a linear regression, looking to measure sensitivity and specificity. Neural network showed a better performance with 85% of specificity vs. 68% for the linear regression, and sensitivity showed a 62% for neural network vs. 38% for the linear regression.
Al-Hyari [
11] used three algorithms to support the diagnosis of CKD: DT, Naive Bayes (NB) and Artificial Neural Networks (ANN), the results showed the best accuracy for DT 92.2%, compared to 88.2% for NB and 82.4% for ANN.
Krishnamurthy, et al. [
12] built a predictive model out of a total of 18,000 people with CKD and 72,000 people without CKD. For their model they worked with Convolutional Neural Networks (CNN), concluding that the study could be a useful tool for policymakers in predicting the trends of CKD in the population.
Kriplani, et al. [
13] worked creating a three layers deep NN, to predict the presence of CKD, and achieved an accuracy of 97.76%. For their study, they used the same dataset used on this paper, but they only utilized 18 of the 24 parameters.
Charleonnan, et al. [
14] worked with four machine-learning methods to predict CKD: k-NN, SVM, Logistic Regression (LR) and DT. They concluded that SVM has the highest accuracy with a result of 98%. They utilized the same dataset.
Elhoseny et al. [
15] worked with Density based Feature Selection (DFS) with an Ant Colony based Optimization (D-ACO) algorithm to be dominating a Olex-Geentic Algorithm (Olex-GA). They used the 24 attributes from the dataset, and obtained a model that gave 95% of accuracy.
Evidence proves that working with machine learning algorithms, lead to high accuracy levels for predicting CKD.
Machine learning is an evolving branch of computational algorithms, which are designed to emulate human intelligence by learning from the surrounding environments [
16]. Techniques based on machine learning have been applied successfully in multiple fields (pattern recognition, computer vision, spacecraft engineering, finances, entertainment, and especially important in biomedical and medical applications). Machine learning can be considered as the working horse of the new era. If applied properly, machine learning can help physicians make close to perfect diagnoses, choose the proper medications, determine the patients on risk, and improve patient’s health while reducing cost.
According to Bhardwaj [
17], around 90% of emergency room visits are preventable and 50% of the total costs of healthcare come from only 5% of the total patients. Machine learning can identify patients who may be more susceptible to recurring illnesses, and help diagnosing them. Healthcare is one of the fastest growing industries in the global economy; with the population growing and more people requiring care, this is quickly becoming expensive for governments. If we do not develop alternative methods for diagnosing and treating patients, the traditional methods will soon be insufficient, and make the healthcare industry to crash. The data is available, we just need to take a further step to process it and extract valuable information to support physicians’ decisions. For that purpose, we will work with machine learning algorithms.
3. Startup Considerations
3.1. Diagnosing CKD
To diagnose CKD, a patient will be subjected to clinical data, followed by multiple laboratory tests, expensive imaging studies, and finally a biopsy to confirm the condition. Biopsy is the last step, mainly because it is invasive, very expensive, time consuming and potentially risky due to the nature of the process. But it is not the only diagnosing step that symbolizes disadvantages; imaging studies (usually a magnetic resonance imaging of the kidney) is very expensive, means to be exposed to radiation effects and is insufficient to diagnose CKD. Instead of using imaging datasets, this study is focused on using the best possible combination of minimally invasive, inexpensive, non-risky, and accurate enough to provide physicians another tool for decision-making.
Clinical Data
Clinical data is the information that can be gathered from the historical records of the patient. It includes the electronic health records, which are usually available on the hospital systems, and collect the patient’s digital history. This should include rounds of diagnostics, and any medications that the patient is taking. Patient/Disease registries, which contains information, related specifically to the patient’s population, gathered for further research. Clinical trial data, which is data gathered as part of a clinical trial, it can contain information around new drug applications, treatment methods, and device testing.
Laboratory Tests
These are tests that use different samples like blood, urine, saliva, mucus, excrement, body tissues, or any other substance from the body of the patient [
18]. Depending on the desired type of test, there are different ways to collect the sample. Body tissues and blood are invasive, while saliva, mucus, urine, or excrement are obtained through natural body fluids, therefore without any invasive processes. The cost of the laboratory tests will also depend on the parameter to be analyzed.
Imaging Studies
This is a type of test that creates detailed pictures of areas inside the body [
19]. To be able to capture a region that cannot be seen, the imaging tests require to use energy (like x-rays, ultrasound, radio waves, or radioactivity).
Biopsy
A biopsy is a test done by removing tissue from a living body, with the intention to discover the presence, cause or even the extent of a disease. There are multiple techniques to extract the sample, which depends on the location and the required tissue; there are needle biopsies, which just requires a needle, and open/close surgical biopsies, which by nature require a surgery [
20].
This study is seeking to create a neural network model, which can predict CKD with a high level of accuracy, and at the same time avoid the need of imaging studies and biopsies, to both reduce to a minimum level the invasive tests, and the high costs associated to detect the disease.
3.2. Neural Networks
Neural networks also known as artificial neural networks (ANNs) are structures inspired by the human brain, which mimic the way that biological neurons communicate with others. Each individual node has its own linear regression model, that is composed of input data, weights, a bias (or threshold) and an output.
The idea of neural networks as a model of how the neurons behave inside the human brain, started 80 years ago, with a simple electrical circuit created by the neurophysiologist McCulloch [
21]. Since then, neural networks have had multiple improvements, these is a summary of some important milestones:
- ■
1949: Hebb [
22] reinforced the concept of neurons. He pointed out that the neural pathways are strengthened each time they are used.
- ■
1974: Werbos [
23] contributed with the idea of backpropagation and its application when working with neural networks.
- ■
1989: LeCun, et al. [
24] illustrated how the use of constraints in backpropagation and its integration to the neural network architecture, can be used to get better results when training algorithms. This effort derived in a trained neural network capable to recognize handwritten zip code digits.
- ■
1998: LeCun, et al. [
24] used multilayer neural networks with a backpropagation algorithm to get handwritten character recognition, allowing automatic learning of segmentation and recognition.
- ■
2014: Schmidhuber [
25] makes a review of deep supervised, unsupervised, and reinforcement learning and indirect search for short programs encoding deep and large networks. He highlights the fact that deep artificial neural networks have won numerous contests in pattern recognition and machine learning.
- ■
2015: Agapitos et al. [
26] used a tree based Convolutional Neural Network (CNN), for handwritten digit recognition. They reported competitive results compared to state of art algorithms, creating a precedent in the evolution of CNN architectures.
In accordance with the literature, this work has concentrated the methodology towards the characteristics of the following neural networks: Back Propagation Neural Network (BPNN)[
27], Radial Basis Function Neural Network (RBFNN)[
28], General Regression Neural Network (GRNN)[
29], Probabilistic Neural Network (PNN)[
30] and Complementary Neural Network (CMNN)[
31].
Table 3 shows the evaluation of different characteristics of neural networks.
4. Methodology
As exposed on the literature review, there are multiple attempts to work with different algorithms, to try to improve the accuracy to detect CKD; however, there are no previous studies that consider creating an algorithm that will the best combination of attributes, with an optimal cost and reduction on the invasiveness and easiness to obtain them. To achieve this, we will include the following two enhancements on our methodology:
During the selection of the attributes, we will maintain the ones that are technically relevant (we obtained this by running a Random Forest algorithm) and create a reward for attributes that are medically relevant. This way we will keep only the attributes that are both medical and technically relevant.
Later in the process, we will run a genetic algorithm that will give as a result the combination of attributes that will provide the best accuracy, and the lowest cost and patient’s invasiveness.
As mentioned on the introduction, CKD is an expensive disease, which requires ongoing expenses. With the current global wealth distribution, the majority of the population does not have the possibility to cover the required amount of money to ensure survival. This paper, seeks to contribute with the society, by creating an algorithm to make the diagnostic of the disease less expensive, less invasive and easier to obtain, than making a dangerous biopsy.
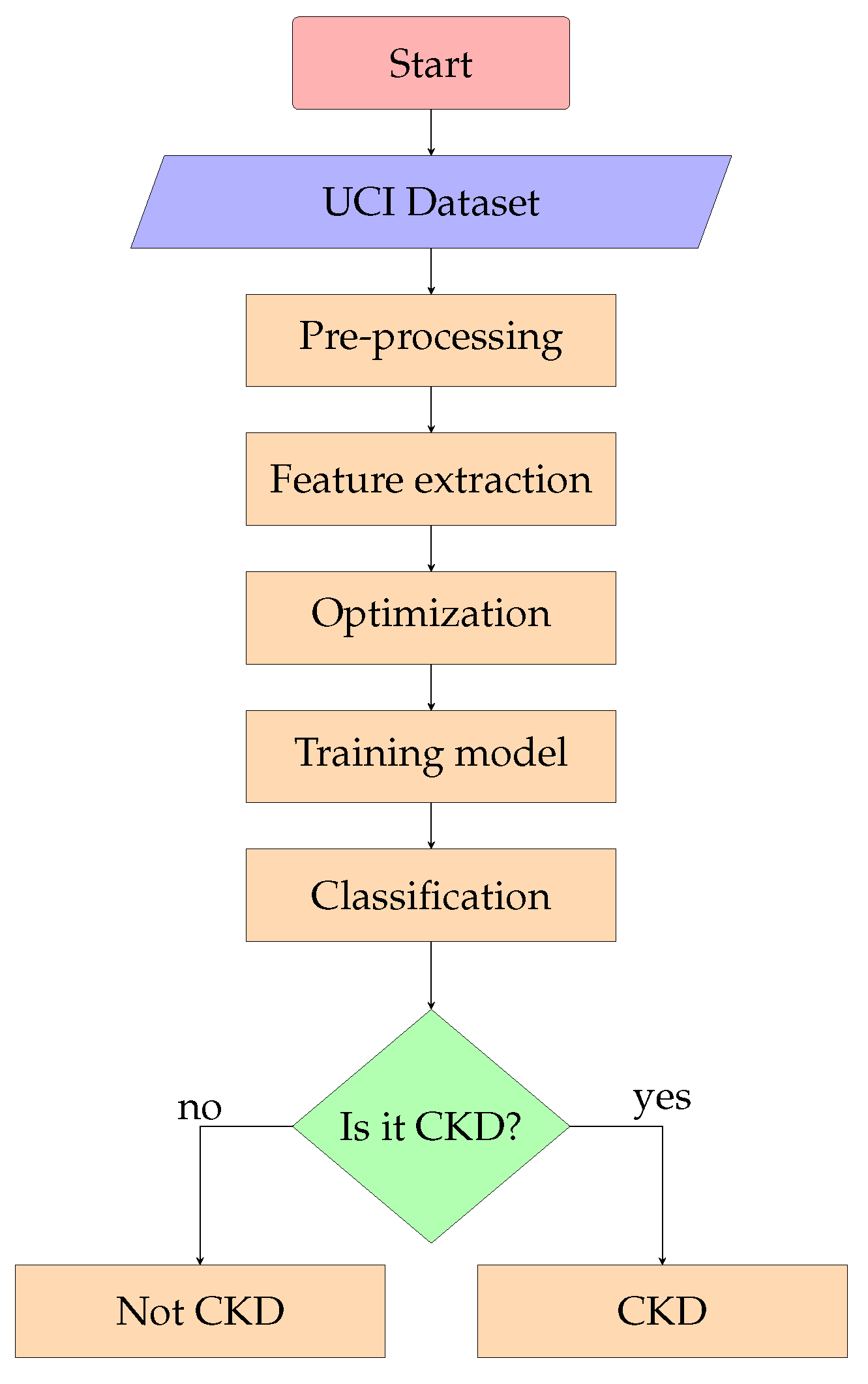
The sequence of steps and techniques used starting with the processing of the data, and finishing with an optimized algorithm, are represented by a flow diagram available on
Figure 2.
4.1. Dataset
The dataset [
32] was obtained from the UCI repository, created from data of the Apollo Hospital in Tamil India. It contains 400 instances, and has 24 medical related attributes, and a class attribute:
| Age |
Sugar |
Pus cell |
Blood urea |
Sodium |
White blood cell |
| Blood pressure |
Red bood cells |
Pus cell clumps |
Serum creatinine |
Potassium |
Red blood cell |
| Specific gravity |
Hypertension |
Bacteria |
Coronary artery disease |
Hemoglobin |
Pedal edema |
| Albumin |
Diabetes mellitus |
Blood glucose |
Appetite |
Packed cell volume |
Anemia |
4.2. Pre-Processing
Cleaning
The dataset has close to 5% of missing values. These values were filled using the median of the attribute value. Then we normalized the dataset using a range of values between zero and one.
Penalizing Variables
To be able to decide the variables that will be included in the model, they were given a weighted average grade that considers the following metrics:
Invasiveness level: this metric depends on how meddling is the process to obtain the desired attribute. One is the least invasive, and three the most invasive.
Cost level: this metric is telling us how much will a patient need to spend. The number was obtained from an average cost of tests in laboratories, with the following ranges:
| Level |
Range |
| 0 |
No cost-10.00 USD |
| 1 |
$10.01 USD-$20.00 USD |
| 2 |
$20.01 USD-$50.00 USD |
| 3 |
$50.01 USD-More |
Accessibility level: this metric measures how hard is to obtain the attribute. One is the most accessible and three is hard to obtain.
The weighted grade is composed of 40% of invasiveness level, 40% of cost level and 20% of the accessibility level. The detailed values are available on
Table 4.
The penalization value will be used by the genetic algorithm, to punish the variables with higher grades. This way, the model will include the variables that are more relevant, and also the ones that will accomplish the main objectives from this research; to develop a model to predict with high reliability whether a patient is likely to develop CKD while reducing the amount of invasiveness, cost, and least accessible tests.
4.3. Feature Extraction
Relevance
Not all of the attributes are relevant for the model. To decide which attributes to use, we ran a random forest: this machine-learning algorithm combines the output of a set of multiple decision trees, with the goal of reaching one single result. According to [
33], it is the result of a combination of decision trees. Each of those trees, has a comprised of data sample from a training set with replacement (this is called the bootstrap sample).
On the proposed model, the random forest graded the variables of our data set, using the random forest classifier method.
Table 5 shows the results in order of relevance.
Reward Variables
It is clear which are the variables contributing the most on the algorithm model. However, their medical relevance is important and therefore considered in the model. We made weighted average to decide the value that will feed the genetic algorithm.
Table 6 shows the reward points, which is a weighted average of the algorithm and medical relevance.
The reward points are a combination of the algorithm’s relevance of the variable (previously obtained through the random forest) and the medical relevance of the variable (this value was given by a certified medic with a specialty in kidney studies). The combination of both is providing a number that can be used to reward the relevant variables on the genetic algorithm.
4.4. Optimization
We used a genetic algorithm to optimize our solution, and seek for the biggest impact in the society by the creation of an algorithm capable to predict CKD, using the best combination of low cost and invasiveness with easy accessibility, and at the same time have the variables that are medically relevant to increase the accuracy of the algorithm.
A genetic algorithm uses operators that are inspired on biology (mutation, crossover, and selection) seeking to optimize the best possible solution for a problem.
There are two weighted values for each of the variables:
Penalization: calculated as a function of undesired features (level of invasiveness, cost of getting the data, inaccessibility to the data).
Reward points: calculated as a function of desired features (how much of the variance is accounted by the variable on the model according to the random forest algorithm and the medical relevance according to a medical expert).
A genetic algorithm helps us to select among all the possible combination of variables, the one that will have the best positive values, without reaching a defined threshold of negative values. This is achieved using a "fitness function" that uses the operators described above.
After running the genetic algorithm with the following settings:
- ■
Type: binary chromosome
- ■
Population size: 200
- ■
Number of generations: 5000
- ■
Elitism: 40
- ■
Mutation chance: 0.04
The result of the best combination of variables (with a limit of negative threshold of 20 points) is:
| Age |
Blood urea |
Packed cell volumen |
| Blood pressure |
Serum creatinine |
Hypertension |
| Albumin |
Sodium |
Diabetes |
| Red blood cells |
Potasium |
Appetite |
| Blood glucose |
Hemoglobin |
Pedal edema |
5. Algorithms and Results
5.1. Training a PNN
Due to the positive characteristics of the probabilistic neural networks (good handling scarce data, and handling noise) the first iteration used this type of neural network.
5.1.1. Considerations for PNN
Some additional factors to consider about the probabilistic neural network:
It is sensitive for cases when one input feature has higher values than another one.
The data must be normalized before doing the training of the algorithm.
For training, it uses lazy learning, which means that it does not require multiple iterations to be trained, the algorithm will store the parameters, and will use them to predict.
It is not only good with small datasets, but actually, the prediction is not good with large datasets.
5.1.2. Architecture PNN
The neural network was developed using NeuPy [
34], which is a Python-based library that helps creating neural networks. As a backend, it uses TensorFlow [
35]. After multiple iterations, the model with the best accuracy was created with the following architecture:
Number of vertices (neurons) = 320 in the first hidden layer.
Number of classes = 2 in the second hidden layer.
Threshold = NA (PNN is not trained by a backpropagation method).
Size of the training data = 80% of the dataset.
Size of the testing data = 20% of the dataset.
5.1.3. Results
After the iterations, the test data set was used to try the algorithm, getting 89% of accuracy. The algorithm had 3% of false positives, and 8% of false negatives.
5.2. Second Iteration BPNN
5.2.1. Architecture BPNN
Seeking for better accuracy, a second model was done using a back propagation model. The model was trained in R using the package "Neuralnet" [
36] which allows the use of flexible settings according to custom choices of parameters for training the neural network.
After multiple iterations, the model with the best accuracy had the following architecture:
Number of vertices (neurons) = 10
Threshold = 0.01
Iterations = 1,000,000
Size of the training data = 80% of the dataset.
Size of the testing data = 20% of the dataset.
5.2.2. Results
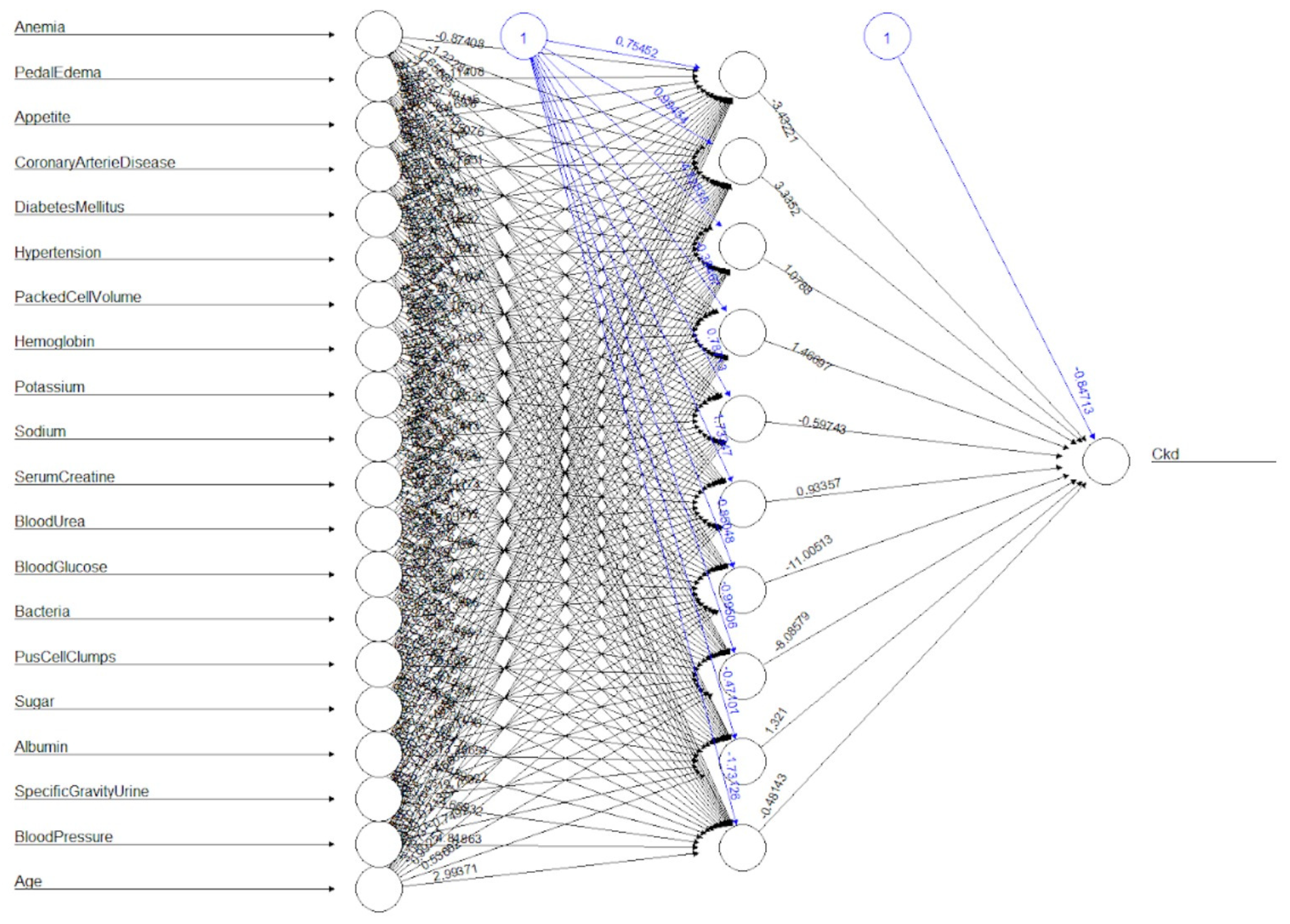
After training, the test data set was used to try the algorithm, giving 97% of accuracy. The algorithm had 1% of false positives, and 2% of false negatives. The graphical representation of the neural network is shown in
Figure 3.
For this dataset, the backpropagation neural network has proven to give better accuracy than the probabilistic neural network.
5.3. Third Iteration BPNN with Optimal Variables
The genetic algorithm provided the set of variables that are both relevant on the algorithm model, and medically relevant to predict CKD. A new iteration of a back propagation neural network was trained, with the goal of obtaining the results that anyone could expect, by only considering the variables that the genetic algorithm provided as the optimized ones.
5.3.1. Architecture Optimized BPNN
The same model that was previously done before running the genetic algorithm was selected; the neural network was trained in R using the package "Neuralnet" [
36] using the following parameters:
Number of vertices (neurons) = 10
Threshold = 0.01
Size of the training data = 80% of the dataset.
Size of the testing data = 20% of the dataset.
5.3.2. Results
After this iteration, the neural network had the same 1% of false positives (FP), and 3% of false negatives (FN), which represent an increase of 1% of false negatives.
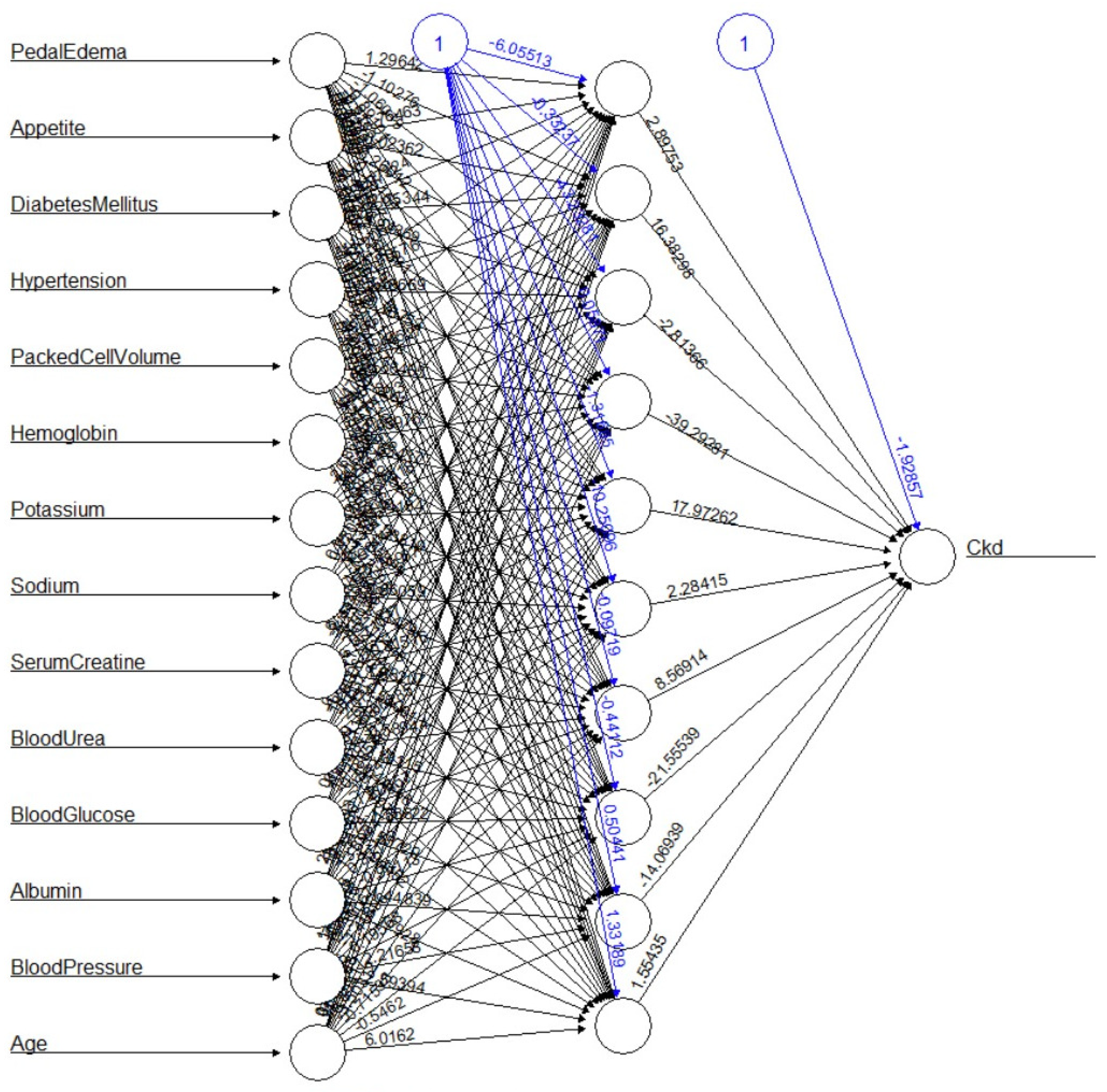
The graphical representation of the neural network is shown in
Figure 4. After removing 6 variables (suggested by the genetic algorithm), the results are promising.
Accuracy is the ratio of the correct predicted observations True Negatives (TN), and True Positives (TP), compared to the total amount of observations:
The proposed method gave 97% of accuracy.
Precision is the proportion of correct predicted positives, compared to the total amount of true positives plus false positives.
The proposed method model gave 98% of precision.
Recall is the proportion of correct positives, compared to the number of true positives plus false negatives.
The proposed method model gave 94.34% of recall.
6. Conclusions
Derived from the fact that 10% of the worldwide population owns between 60 and 80% of the world wealth, health and particularly CKD becomes a real problem, because most of the population does not have access to a traditional diagnosing method. In the case of CKD, the early detection of the disease is key to increasing chances of survival, and improve quality of life.
The use of machine learning algorithms is a powerful tool that can allow medical doctors with alternative methods for properly diagnosing and treating patients, while minimizing the cost and invasiveness level.
Although there are guidelines that helps to understand which are the best algorithms to use depending on the characteristics of the dataset, the results can vary, so it is important to test them before getting to anticipated conclusions. On this particular case, due to the nature of the dataset a PNN was theoretically the best type of neural network; however, our model got better results using a back propagation one.
It is always important to analyze if the dataset can be simplified, there is a high chance that a big dataset has correlated variables that can be excluded from the model. This has a bigger impact on the healthcare industry, as it means that a good prediction model can be trained without the need of certain laboratory tests, reducing unnecessary invasiveness and cost to the patients.
Technology will continue to become an ally of the diagnosis for patients; engineers require to properly train the right algorithms, while using the right input. In the near future, the medical industry will experience a positive change that will incorporate machine learning algorithms, as its core of diagnosis. This is a blue ocean opportunity, yet to become exploited.
Author Contributions
Conceptualization, Omar García-González and Iván Villalón-Turrubiates; Data curation, Luis E. Chávez-Camarena; Methodology, Omar García-González, Iván Villalón-Turrubiates, Luis E. Chávez-Camarena and Carlos Hernández-Mejía; Formal analysis, Omar García-González, Iván Villalón-Turrubiates, Luis E. Chávez-Camarena and Carlos Hernández-Mejía; Investigation, Omar García-González, Iván Villalón-Turrubiates, Luis E. Chávez-Camarena and Carlos Hernández-Mejía; Resources, Luis E. Chávez-Camarena and Carlos Hernández-Mejía; Visualization, Omar García-González, Luis E. Chávez-Camarena and Carlos Hernández-Mejía; Writing-original draft preparation, Omar García-González and Iván Villalón-Turrubiates; Writing-review and editing, Omar García-González, Iván Villalón-Turrubiates, Luis E. Chávez-Camarena and Carlos Hernández-Mejía. All authors have read and agreed to the published version of the manuscript.
Conflicts of Interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
References
- Webster, A.C.; Nagler, E.V.; Morton, R.L.; Masson, P. Chronic Kidney Disease. Lancet 2017, 389, 1238–1252. [Google Scholar] [CrossRef] [PubMed]
- Kovesdy, C.P. Epidemiology of chronic kidney disease: an update 2022. Kidney international supplements 2022, 12, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Ravindra, B.V.; Sriraam, N.; Geetha, M. Discovery of significant parameters in kidney dialysis data sets by K-means algorithm. In Proceedings of the International Conference on Circuits, Communication, Control and Computing; 2014; pp. 452–454. [Google Scholar]
- Manns, B.; Hemmelgarn, B.; Tonelli, M.; Au, F.; So, H.; Weaver, R.; Quinn, A.E.; Klarenbach, S. The Cost of Care for People With Chronic Kidney Disease. Canadian journal of kidney health and disease 2019, 6. [Google Scholar] [CrossRef]
- Feenstra, R.C.; Inklaar, R.; Timmer, M. The Next Generation of the Penn World Table. American Economic Review 2015, 105. [Google Scholar] [CrossRef]
- Vijayarani, S.; Dhayanand, S. Data mining classification algorithms for kidney disease prediction. International Journal on Cyber-rithms and Informatics (IJCI) 2015, 4, 13–25. [Google Scholar] [CrossRef]
- Lakshmi, K.R.; Nagesh, Y.; VeeraKrishna, M. Performance comparison of three data mining techniques for predicting kidney dialysis survivability. International Journal of Advances in Engineering and Technology 2014, 7, 242–254. [Google Scholar]
- Subasi, A.; Alickovic, E.; Kevric, J. Diagnosis of Chronic Kidney Disease by Using Random Forest. In Proceedings of the CMBEBIH 2017; Badnjevic, A., Ed.; Springer Singapore: Singapore, 2017; pp. 589–594. [Google Scholar]
- Chin-Chi, K.; Chun-Min, C.; Kuan-Ting, L.; Wei-Kai, L.; Hsiu-Yin, C.; Chih-Wei, C.; Meng-Ru, H.; Pei-Ran, S.; Rong-Lin, Y.; Kuan-Ta, C. Automation of the kidney function prediction and classification through ultrasound-based kidney imaging using deep learning. npj Digital Medicine 2019, 2. [Google Scholar] [CrossRef]
- Caocci, G.; Baccoli, R.; Littera, R.; Orru, S.; Carcassi, C.; La Nasa, G. Comparison Between an Artificial Neural Network and Logistic Regression in Predicting Long Term Kidney Transplantation Outcome. In Artificial Neural Networks; Suzuki, K., Ed.; IntechOpen: Rijeka, 2013; chapter 5. [Google Scholar]
- Al-Hyari, A.Y.; Al-Taee, A.M.; Al-Taee, M.A. Clinical decision support system for diagnosis and management of Chronic Renal Failure. In Proceedings of the 2013 IEEE Jordan Conference on Applied Electrical Engineering and Computing Technologies (AEECT); 2013; pp. 1–6. [Google Scholar]
- Krishnamurthy, S.; KS, K.; Dovgan, E.; Luštrek, M.; Gradišek Piletič, B.; Srinivasan, K.; Li, Y.C.J.; Gradišek, A.; Syed-Abdul, S. Machine Learning Prediction Models for Chronic Kidney Disease Using National Health Insurance Claim Data in Taiwan. Healthcare 2021, 9. [Google Scholar] [CrossRef] [PubMed]
- Kriplani, H.; Patel, B.; Roy, S. Prediction of Chronic Kidney Diseases Using Deep Artificial Neural Network Technique. In Proceedings of the Computer Aided Intervention and Diagnostics in Clinical and Medical Images; Peter, J.D.; Fernandes, S.L.; Eduardo Thomaz, C.; Viriri, S., Eds.; Springer International Publishing: Cham, 2019; pp. 179–187. [Google Scholar]
- Charleonnan, A.; Fufaung, T.; Niyomwong, T.; Chokchueypattanakit, W.; Suwannawach, S.; Ninchawee, N. Predictive analytics for chronic kidney disease using machine learning techniques. In Proceedings of the 2016 Management and Innovation Technology International Conference (MITicon), pp. MIT–80–MIT–83. 2016. [Google Scholar]
- Elhoseny, M.; Shankar, K.; Uthayakumar, J. Intelligent Diagnostic Prediction and Classification System for Chronic Kidney Disease. Sci Rep 2019, 9. [Google Scholar] [CrossRef] [PubMed]
- El Naqa, I.; Murphy, M.J. What Is Machine Learning? In Machine Learning in Radiation Oncology: Theory and Applications; El Naqa, I., Li, R., Murphy, M.J., Eds.; Springer International Publishing: Cham, 2015; pp. 3–11. [Google Scholar]
- Bhardwaj, R.; Nambiar, A.R.; Dutta, D. A Study of Machine Learning in Healthcare. In Proceedings of the 2017 IEEE 41st Annual Computer Software and Applications Conference (COMPSAC), Vol. 2; 2017; pp. 236–241. [Google Scholar]
- Levey, A.S.; Coresh, J. Chronic kidney disease. Lancet 2012, 349. [Google Scholar] [CrossRef] [PubMed]
- Pimentel, A.; Bover, J.; Elder, G.; Cohen-Solal, M.; Ureña-Torres, P.A. The Use of Imaging Techniques in Chronic Kidney Disease-Mineral and Bone Disorders (CKD-MBD)—A Systematic Review. Diagnostics 2021, 11. [Google Scholar] [CrossRef] [PubMed]
- Zaza, G.; Bernich, P.; Lupo, A.; of Renal Biopsies (TVRRB), T.R. Renal biopsy in chronic kidney disease: lessons from a large Italian registry. American journal of nephrology 2013, 37. [Google Scholar] [CrossRef] [PubMed]
- McCulloch, W.; Pitts, W. A logical calculus of the ideas immanent in nervous activity. Bulletin of Mathematical Biophysics 1943, 5. [Google Scholar] [CrossRef]
- Hebb, D. Summation and learning in perception. In The Organization of Behavior: A Neuropsychological Theory; Psychology Press, 2002; chapter 2. [Google Scholar]
- Werbos, P. Beyond Regression: New Tools for Prediction and Analysis in the Behavioral Sciences; Harvard University, 1975. [Google Scholar]
- LeCun, Y.; Boser, B.; Denker, J.S.; Henderson, D.; Howard, R.E.; Hubbard, W.; Jackel, L.D. Backpropagation Applied to Handwritten Zip Code Recognition. Neural Computation 1989, 1, 541–551. [Google Scholar] [CrossRef]
- Schmidhuber, J. Deep learning in neural networks: An overview. Neural Networks 2015, 61, 85–117. [Google Scholar] [CrossRef] [PubMed]
- Agapitos, A.; O’Neill, M.; Nicolau, M.; Fagan, D.; Kattan, A.; Brabazon, A.; Curran, K. Deep evolution of image representations for handwritten digit recognition. In Proceedings of the 2015 IEEE Congress on Evolutionary Computation (CEC); 2015; pp. 2452–2459. [Google Scholar]
- Jeatrakul, P.; Wong, K. Comparing the performance of different neural networks for binary classification problems. In Proceedings of the 2009 Eighth International Symposium on Natural Language Processing; 2009; pp. 111–115. [Google Scholar]
- Bors, A.; Pitas, I. Median radial basis function neural network. IEEE Transactions on Neural Networks 1996, 7, 1351–1364. [Google Scholar] [CrossRef] [PubMed]
- Xudong, S.; Junbin, L.; Ke, Z.; Jun, H.; Xiaogang, J.; Yande, L. Generalized regression neural network association with terahertz spectroscopy for quantitative analysis of benzoic acid additive in wheat flour. Royal Society Open Science 2019, 6. [Google Scholar] [CrossRef]
- Kokkinos, Y.; Margaritis, K.G. Parallel and Local Learning for Fast Probabilistic Neural Networks in Scalable Data Mining. In Proceedings of the Proceedings of the 6th Balkan Conference in Informatics; Association for Computing Machinery: New York, NY, USA, 2013; pp. 47–52. [Google Scholar]
- Kraipeerapun, P.; Amornsamankul, S. Applying Multiple Complementary Neural Networks to Solve Multiclass Classification Problem. International Journal of Applied Mathematics and Informatics 2012, 6, 134–141. [Google Scholar]
- Rubini, L.; Soundarapandian, P.; Eswaran, P. Chronic Kidney Disease. UCI Machine Learning Repository. 2015. [Google Scholar]
- What is random forest? Accessed on September 12, 2023.
- Sharp, M. Neural Network Programming with Python: Create Your Own Neural Network! CreateSpace Independent Publishing Platform. 2016.
- Singh, P.; Manure, A. Learn TensorFlow 2.0; Apress Berkeley, 2020.
- Grolemund, G.; Wickham, H. R for Data Science; O’Reilly Media, 2017.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).