1. Introduction
Breast cancer (BC) is one of the greatest oncological challenges in developed countries. High morbidity, together with a high mortality rate, makes BC exceptionally important medically. Drug resistance (DR) is one of the major concerns, particularly in patients with advanced cancer. Different mechanisms of DR are responsible for tumor treatment failures, including decreased intracellular drug concentrations, disturbances in cell cycle, resistance to apoptosis, DNA repair deficiencies, activation of intracellular pathways related to cancer progression, epigenetic alterations, and changes in drug targets [
1]. One class of cellular compounds involved in the DR of cancer cells are glycosphingolipids (GSLs) [
2]. The involvement of GSLs in DR was first described by Lavie et al. [
3], who showed that DR cancer cells were characterized by the accumulation of glucosylceramide (GlcCer). Initially, it was proposed that increased synthesis of GlcCer highly decreases the intracellular content of pro-apoptotic ceramides, making cancer cells more resistant to drug-induced apoptosis [
4]. However, further studies showed that overexpression of GCS and accumulation of GlcCer correlated with high expression of P-glycoprotein (Pgp, MDR1), suggesting that GlcCer positively regulates the expression of
MDR1 gene [
5]. A subsequent study revealed that increased levels of globo-series GSLs (globotriaosylceramide and globotetraosylceramide), but not GlcCer by itself, are responsible for up-regulation of Pgp in cancer cells [
6,
7,
8]. In addition to globo-series GSLs, a high content of GM3 ganglioside was shown to protect murine lung carcinoma cells from cytotoxic effects of etoposide and doxorubicin [
9], and small-cell lung cancer cells from cytotoxic effects of DOX and cisplatin [
10]. However, in chronic myeloid leukemic K562 cells, increased GM3 content decreased resistance to etoposide and staurosporine [
11]. In the case of GM3, increased or decreased resistance to chemotherapy has been linked to differences in the resistance of cancer cells to apoptosis induced by anti-cancer drugs.
Galactosylceramide (GalCer) is a GSL synthesised by galactosylceramide synthase (UGT8, C.E. 2.4.2.62) [
12] encoded by the
UGT8 gene [
13], and is a major myelin-stabilizing component [
14]. However, it was recently shown that BC cells expressing high level of UGT8 and accumulating GalCer, form tumors and metastatic colonies in the lungs more efficiently than BC cells with suppressed UGT8 expression [
15]. In accordance with these results, tumor specimens with elevated GalCer content were characterized by higher Ki-67 proliferation index and a reduction in the number of apoptotic cells. Therefore, it was hypothesized that GalCer serves as an anti-apoptotic molecule, and its presence increases the ability of cancer cells to survive in the unfavourable tumor microenvironment [
15,
16]. In addition, GalCer was found to increase resistance of BC cells to doxorubicin, suggesting that GalCer can be an important factor in the BC cells resistance to chemotherapy DR. This hypothesis is endorsed by findings that GalCer content is increased in DR colon and ovarian cancer cells [
17,
18], and that Krabbe cells synthesizing large amounts of GalCer are more resistant to apoptosis induced by daunorubicin and cytosine arabinoside than Gaucher cells, which contain low levels of this GSL [
19]. Furthermore, inhibition of GlCer in U937 and HL60 cells with such inhibitors as D,L-threo-1-phenyl-2-palmitoylamino-3-morpholino-1-propanol (PDMP) or 1-phenyl-2-palmitoylamino-3-morpholino-1-propanol (PPMP), which was accompanied by increased levels of GalCer, protects them from daunorubicin-induced apoptosis [
19]. In addition, such cells with high content of exogenously added GalCer became more resistant to daunorubicin-induced apoptosis. Accumulation of GalCer was observed in Madin-Darby canine kidney cells exposed to hyperosmotic and heat stress [
20,
21]. Based on this data and generally accepted idea that ceramide is one of the important pro-apoptotic molecules [
22,
23], we proposed that the anti-apoptotic activity of GalCer can be liked to decreased levels of ceramide due to increased synthesis of GalCer [
15]. However, it was found that BC cells with various amounts of GalCer contain essentially the same amounts of ceramide [
16], suggesting that glycosylation of ceramide is not a major mechanism that increases resistance of tumor cells to drug-induced apoptosis. Consequently, a combination of molecular and pharmacological techniques were used in this research to examine the function of GalCer in BC cell resistance to anti-cancer agents and elucidate the mechanisms of its anti-apoptotic activity.
2. Materials and Methods
2.1. Cell lines and culture conditions
Human BC MDA-MB-231 and T47D cells were obtained from American Type Culture Collection (ATCC, VA, USA). The BC MCF7 and murine 4T1 mammary cancer cell lines were kindly provided by the Cell Line Collection of the Hirszfeld Institute of Immunology and Experimental Therapy (Wroclaw, Poland). MCF7 cell line were authenticated by the ATCC Cell Line Authentication Service. T47D.UGT8 cells overexpressing UGT8 and control T47D.C cells derived from T47D cells have previously been described by Suchanski et al. [
16]. MDA-MB-231 and MCF7 cells were cultured in α-MEM supplemented with 10% fetal bovine serum (FBS, Cytogen, Poland), 2 mM L-glutamine, and antibiotics. T47D and 4T1 cells were cultured in RPMI-1640 Medium with 10% FBS (Cytogen), 2 mM L-glutamine and antibiotics. For T47D cells, the culture medium was supplemented with bovine insulin (0.2 U/mL) (Merck, Germany).
2.2. Vectors construction, virus production and transductions
Using PCR, human UGT8 cDNA was generated from the MDA-MB-231 cDNA library as a template for primers: forXhoI-UGT8 and revBamHI-UGT8 (
Supplementary Table S1), and mouse galactosylceramide synthase (UGT8a) cDNA was amplified from the pCMV6-Entry vector containing UGT8a cDNA (MR208632, 1626 bp; OriGene, MD, USA) with forEcoRI-UGT8a and revMluI-UGT8a primers (
Supplementary Table S1). To generate UGT8 and UGT8a-expressing vectors, the resulting inserts were cloned into a pLVX-Puro Vector (Clontech, CA, USA) as previously described [
16] and named as pLVX-UGT8-Puro and pLVX-UGT8a-Puro, respectively. For knockout of the
UGT8 gene, a
UGT8 CRISPR/Cas9 vector encoding the Cas9 nuclease and UGT8-specific 20 nucleotide guide RNA (gRNA-TGTGATAGCTCATCTTTTAG) was purchased from Applied Biological Materials Inc. (Canada). For the generation of lentivirus, LentiX 293T cells (Clontech) were transfected at 50 – 60% confluence with Lenti-X Packaging Single Shots (Takara, Japan). The production of virus particles and cell transductions have been previously described [
15].
UGT8 knockout cells were selected using puromycin (1 μg/mL) (Thermo Fisher Scientific, UK). Antibiotic-resistant cells were subcloned using the limiting dilution technique. The mutation introduced by CRISPR-Cas9 editing was identified by sequencing genomic DNA with the primers: F1-exon2/UGT8 and R1-exon2/UGT8 (
Supplementary Table S1).
2.3. Quantitative PCR (qPCR) assay and apoptosis gene expression profiling with RT-qPCR array
RNA was purified from BC cells using an RNeasy Mini Kit (Qiagen, Germany). To synthesize cDNA, the SuperScipt RT Kit (Thermo Fisher Scientific) was used. The relative amounts of mRNAs were quantified by qPCR with iQ SYBR Green Supermix (Bio-Rad, CA, USA) using an iQ5 Optical System (Bio-Rad) and
ACTB as the reference gene. The primers for qPCR are shown in
Supplementary Table S1. The gene expression profiles of 46 apoptosis-related genes were determined by RT-qPCR (
Supplementary Table S2) using the EvaGreen dye-based detection system (Biotium, CA, USA). Two reference genes,
ACTB and
GAPDH, were used to normalise RNA levels. PCR amplification consisted of an initial denaturation for 3 min at 95ºC, followed by 35 cycles of 20 s denaturatio
n at 95ºC, annealing for 20 s at 5ºC and extension for 20 s at 72ºC. The relative changes in gene expression were calculated using the ΔΔCt (cycle threshold) method. Fold change values were calculated using the 2
-ΔΔCt formula.
2.4. Western blotting
Western blotting was performed as previously described [
16]. The following rabbit polyclonal antibodies were used to detect specific proteins: anti-UGT8 (Biorbyt, UK), anti-Bcl-2 (Abcam, UK), anti-TNFRSF1B (Merck), anti-TNFRSF9 (Biorbyt), anti-p53 (Cell Signaling, MA, USA), anti-CREB (Cell Signaling, MA, USA), anti NF-κB p65 (Abcam, UK) and mouse monoclonal anti-GAPDH antibody (Novus Biologicals, UK).
2.5. Purification of neutral GSLs
GSLs were extracted from 108–109 cells by the chloroform–methanol extraction method and neutral GSLs were purified as previously described .
2.6. Thin-layer chromatogram (TLC)-binding assay
High-performance-thin layer chromatography (HP-TLC) was performed on silica gel 60 HP-TLC plates (Merck). Neutral GSLs were separated using a 2-isopropanol:methyl acetate:15 M ammonium hydroxide:water solvent system (75:10:5:15, v/v/v/v). GSLs were visualised by spraying the plate with primuline reagent (0.05% primuline in acetone/water, 4:1 v/v) and heated for 1 min at 120°C. GalCer was detected by a TLC-binding assay as described previously [
15], using rabbit polyclonal anti-GalCer antibodies (Merck).
2.7. Cell survival assay
The viability of the cells was assessed by MTT assay. Briefly, 5 × 103 cells/well were seeded in a 96-well plate (Nunc, NY USA) and incubated with increasing concentrations of doxorubicin hydrochloride (DOX, Pfizer, NY, USA), paclitaxel (PAX, Ebewe Pharma, Austria) or cisplatin cis-diamminedichloroplatinum (II) (CDDP, Merck) for 48 h. Then, the medium was discarded, and the cells were incubated with MTT solution (0.5 mg/mL, Merck) for 4 h at 37°C. After removing the MTT solution, 200 μL of DMSO (Chempur, Poland) was added for 15 min at 22°C and the optical density was measured at 570 nm.
2.8. Apoptotic assay
The cells (5 × 105) were seeded in multi-well plates (Nunc). The following day, the cells were treated with DOX with or without the selective inhibitor ABT-199 of Bcl-2 protein (Cayman Chemical, MI, USA) or TNFα (Thermo Fisher Scientific) for 48 h. The percentages of apoptotic cells were determined using the Dead Cell Apoptosis Kit (Thermo Fisher Scientific). Fluorescence was measured on a BD FACS Lyric (Becton-Dickinson, NJ, USA) and data were processed using the BD FACSuite™ Software (Becton-Dickinson). The cells in early apoptosis were stained with Annexin V APC , whereas cells in late apoptosis were stained with Annexin V APC and SYTOX Green.
2.9. In-vivo tumor growth assay
The animal experiments were approved by the Second Local Ethics Committee for Animal Experimentation (033/2019/P1,Wroclaw, Poland). Six-week old female BALB/c mice were obtained from the Mossakowski Medical Research Institute, Polish Academy of Sciences (Warsaw, Poland) and kept under specific pathogen-free conditions at room temperature. Suspensions of 4T1 cells (2 x 104 cells/100 µL PBS) were mixed with the equal volume of ice-cold BD Matrigel Matrix High Concentration (Becton-Dickinson) and injected subcutaneously (s.c.). Growth of tumors was monitored once a week by measuring tumour diameter using a calliper. The tumor volume (TV) was calculated as TV (mm3) = (d2x D)/2, where d is the shortest diameter and D is the longest diameter. For the therapeutic experiments, mice were injected intravenously (i.v.) twice a week with DOX (1.5 mg/kg body weight). Mice were euthanised by cervical dislocation following light anaesthesia by isoflurane (Forane, Abott Laboratories, IL, USA) inhalation. Tissue samples were collected in 10% buffered formalin and subjected to histological analyses.
2.10. Cloning of human TNFRSF1B, TNFRSF9 and BCL2 gene promoters and determination of their activity using luciferase reporter vectors
The Genomatix MatInspector software (
http://www. genomatix.de) was applied to identify the nucleotide sequences of TNFRSF1B, TNFRSF9 and BCL2 promoters, which were generated by PCR using genomic DNA purified from MDA-MB-231 cells as a template and primers listed in
Supplementary Table S1. They were amplified as follows: 3 min of initial denaturation at 95°C, followed by 30 cycles of 20 s denaturation at 95ºC, annealing at 55°C for 20 s, and 1 min of final extension at 72ºC. The resulting PCR products containing additional sequences corresponding to MluI and HindIII restriction sites were cloned into pGL3-Basic luciferase vector (Promega, USA). The obtained constructs were named pGL3-TNFRSF1B/LUC, pGL3-TNFRSF9/LUC, and pGL3-BCL2//LUC for the TNFRSF1B, TNFRSF9, and BCL2 promoters, respectively.
To determine the activity of the promoters, pGL3-TNFRSF1B/LUC, pGL3-TNFRSF9/LUC, and pGL3-BCL2/LUC constructs (2 μg) were co-transfected with control Renilla luciferase-expressing pRL-TK vector (2 μg) using polyethylenimine (Merck) into BC cells seeded into multi-well plates and grown to 70% confluence. After 48 h, to measure promoter activity, cells were analyzed using the Dual-Luciferase Reporter Assay System (Promega) following recommendations provided by the manufacturer.
2.10. mRNA stability assay
Cells (5 x 105) were treated with 2 μg/mL of actinomycin D (Thermo Fisher Scientific) in DMSO for 2, 4 and 6 h at 37°C. At the indicated time points, RNA was extracted using Fenozol reagent (A&A Biotechnology, Poland) and the level of Bcl-2 transcripts was analysed using RT-qPCR with iQ SYBR Green Supermix (Bio-Rad) as described above in qPCR assay section.
2.11. siRNA transfections
Transfection with mixture of p53 siRNAs (Santa Cruz Biotechnology, USA) was performed using Lipofectamine RNAiMAX transfection reagent (Thermo Fisher Scientific). Briefly, cells (2.5x105) were seeded in a 6-well plates in complete α-MEM and at 60% confluence incubated with siRNAs for 72 h. The transfection reagent was prepared according to RNAiMAX Transfection Procedure (Thermo Fisher Scientific). Cells were harvested by trypsinization and subjected to further experiments.
2.12. Statistical analysis
All statistical analyses were performed using Prism 5.0 (GraphPad, CA, USA). The two-way Anova Dunnetts post-test was used for statistical analysis of in vivo tumor growth experiments. The Mann–Whitney test was used to compare the groups of data that did not meet the assumptions of the parametric test. In all analyses, the results were considered statistically significant at p < 0.05.
4. Discussion
We previously showed that BC MDA-MB-231 cells, which synthesize large amounts of GalCer become more resistant to DOX [
15]. In the present study, using three human BC cellular models with
loss-of-function (MDA-MB-231 cells) or
gain-of-function (MCF7 and T47D cells) phenotypes, we extended this finding and proved that BC cells are also more resistant to other anti-cancer drugs such as PTX and CDDP. These
in vitro results are strongly supported by an
in vivo study using mouse model. Interestingly, not only human, but also murine mammary carcinoma 4T1 cells overexpressing murine UGT8a and accumulating GalCer become more resistant to DOX. When such cells were injected into BALB/c mice, they showed increased resistance to DOX therapy than the control 4T1 cells. Taken together, these data strongly suggest that GalCer is a marker of DR in BC cells and a new target for breast cancer treatment. In agreement with the general view that the majority of anticancer drugs induce apoptosis in cancer cells [
34], we also showed that GalCer serves as an anti-apoptotic molecule, increasing the resistance of BC cells to apoptosis induced by DOX [
15]. Therefore, the present study aimed to further define the mechanisms by which GalCer increases the resistance of BC cells to drug-induced apoptosis. We found that GalCer specifically downregulated the expression of
TNFRSF1B and
TNFRSF9 genes and increased
BCL-2 gene expression at the mRNA and protein levels, proving that changes in their expression directly affect the apoptotic properties of BC cells. Therefore, this results showed for the first time that the same species of GSL could produce simultaneous but opposite effects on the extrinsic and intrinsic (mitochondrial) apoptotic pathways. In contrast to GalCer, other GSLs involved in the regulation of apoptotic gene expression belong to the ganglio-series of GSLs and act mainly as pro-apoptotic molecules. One of these gangliosides, GD3, is involved in suppression of the
BCL-2 gene expression at the transcriptional level
via dephosphorylation of AKT and CREB [
35]. The forced expression of Bcl-2 protected from GD3-induced apoptosis in T cell lymphoma CEM cells and mouse oligodendrocytes, suggesting that relevant GD3 targets are under Bcl-2 control [
36], which we observed in case of GalCer, but with an opposite effect. However, GD3, in contrast to GalCer, activates the mitochondrial pathway at the level of the permeability transition pore complex causing cytochrome c release and caspase activation [
37]. In addition, GD3 acts as an pro-apoptotic agents by preventing (through a currently unknown mechanism) the nuclear localisation of active NF-κB that suppresses the NF-κB-dependent survival pathway [
38]. Sequestration of NF-κB in the cytoplasm protects the activation of genes dependent on NF-κB, sensitises hepatocytes against apoptotic stimuli such as TNFα, and sensitises human hepatoma cells to daunorubicin and/or ionising radiation [
39]. In contrast, overexpression of ganglioside sialidase (Neu3) in colon carcinoma cells was associated with increased Bcl-2 and decreased caspase expression, which is another example of apoptosis suppression associated with GSLs metabolism [
40]. Increased resistance to drug-induced apoptosis associated with Bcl-2 expression was observed in mouse 3LL Lewis lung cancer cells overexpressing GM3 ganglioside compared with GM3-negative parental cells [
9]. In this case, upregulation of Bcl-2 was linked to the post-translational protein modifications.
To further prove that GalCer affects the expression of specific apoptotic genes at the transcriptional level, the promoters of
TNFRSF1B, TNFRSF9, and
BCL2 genes were cloned and their activities were determined using MDA-MB-231 and MCF7 cellular models. Consistent with our hypothesis, MDA.Δ.C and MCF7.UGT8 cells, both with high GalCer content, showed lower activity of
TNFRSF1B and
TNFRSF9 promoters than MDA.Δ.UGT8.4 and MCF7.C cells lacking GalCer. In MCF7.UGT8 cells, activity of the
BCL2 promoter was higher than in MCF7.C cells.
In silico analysis performed to identify potential transcription factors that bind to promoter sequences and affect the activities of
TNFRSF1B, TNFRSF9 and
BCL2 promoters revealed that the binding sites for CREB1, NF-ĸB1, and p53 were present in all three promoters, indicating that they can be simultaneously involved in the regulation of
TNFRSF1B, TNFRSF9 and
BCL2 gene expression. No data about transcription factors involved in regulation of
TNFRSF1B and
TNFRSF9 gene are available, however, CREB1, NF-ĸB1 and p53 play significant regulatory roles in case of other TNFR family members, upregulating their expression [
28,
29]. In contrast, much more is known about the regulation of
BCL2 by these three transcription factors [
30,
31,
32]. CREB1 and NF-ĸB1 increase [
30,
41], and p53 downregulates [
33] the expression of
BCL2 gene. With regards to MCF7.UGT8 and T47D.UGT8 cells, accumulation of GalCer leads to downregulation of
TNFRSF1B and
TNFRSF9 genes and upregulation of the
BCL2 gene in comparison to MCF7.C and T47D.C cells that do not produce GalCer, therefore the involvement of p53 in their simultaneous but opposite regulation seems to be probable. This assumption is further supported by data obtained from MDA-MB-231 cells. In this cellular model, no difference in promoter activity of the
BCL2 gene was observed between MDA.Δ.C and MDA.Δ.UGT8.4 cells, despite significant differences in mRNA and protein levels. To explain these apparent contradictions, we assumed, based on the available information that p53 can be engaged in post-transcriptional regulation of Bcl-2 expression by increasing the Bcl-2 transcript stability [
42]. Therefore, an actinomycin D assay was performed on MDA.Δ.UGT8.4 and control MDA.C cells, which revealed that the stability of Bcl-2 mRNA was considerably lower in BC cells that did not express GalCer. These data also revealed for the first time that one species of GSL can regulate the expression of specific genes through different mechanisms. The involvement of p53 in the regulation of
TNFRSF1B, TNFRSF9, and
BCL2 genes was supported by the Western blot analysis, which revealed that the accumulation of GalCer in BC cells is associated with decreased level of p53. These correlational observations are strongly supported by the siRNA-based RNA interference study, in which p53 expression was silenced. Direct evidence that p53 is involved in the regulation of
TNFRSF1B,
TNFRSF9 and
BCL2 expression came from the experiments with siRNA-mediated suppression of p53 in MCF7.C and MDA.Δ.UGT8.4 cells. First, it was found that inhibition of p53 strongly reduced the expression of the
TNFRSF1B and
TNFRSF9 genes. Importantly, the activities of the
TNFRSF1B and
TNFRSF9 promoters were also greatly reduced in these cells, confirming our proposal that the expression of these genes is regulated at the transcriptional level. Second, inhibition of p53 increased the expression of the
BCL2 gene. In MCF7.C cells, this was due to increased promoter activity. However, p53 inhibition had no effect on
BCL2 gene expression at the mRNA level and its promoter activity in MDA.Δ.UGT8.4 cells. Therefore, in light of our results showing that p53 can regulate Bcl-2 expression by increasing Bcl-2 transcript stability, siRNA-treated MDA.Δ.UGT8.4 cells were subjected to an actinomycin D assay, which showed that p53 is indeed involved in the post-transcriptional regulation of Bcl-2.
Since p53 functions primarily as a transcription factor [
43], it is not surprising that in MCF7 cells carrying wild-type p53 [
44], regulation of all three target genes takes place at the level of promoter activity. However, in the case of MDA-MB-231 cells carrying a mutant form of p53 [
45], only
TNFRSF1B and
TNFRSF9 genes were regulated at the level of transcription, which was not observed in the case of
BCl2 gene. These differences can be explained by the studies showing that different p53 variants have an altered promoter activity spectrum [
46]. In our case, the R280K mutation did not affect p53 binding to the
TNFRSF1B and
TNFRSF9 promoters, but prevented it from binding to the
BCL2 promoter. Instead, such mutant p53 acquires new properties and binds to the 5'UTR of Bcl2 mRNA as it was shown in case of genetic variant of p53 with the substitution of proline by arginine within the proline-rich domain expressed by mucous cells [
42].
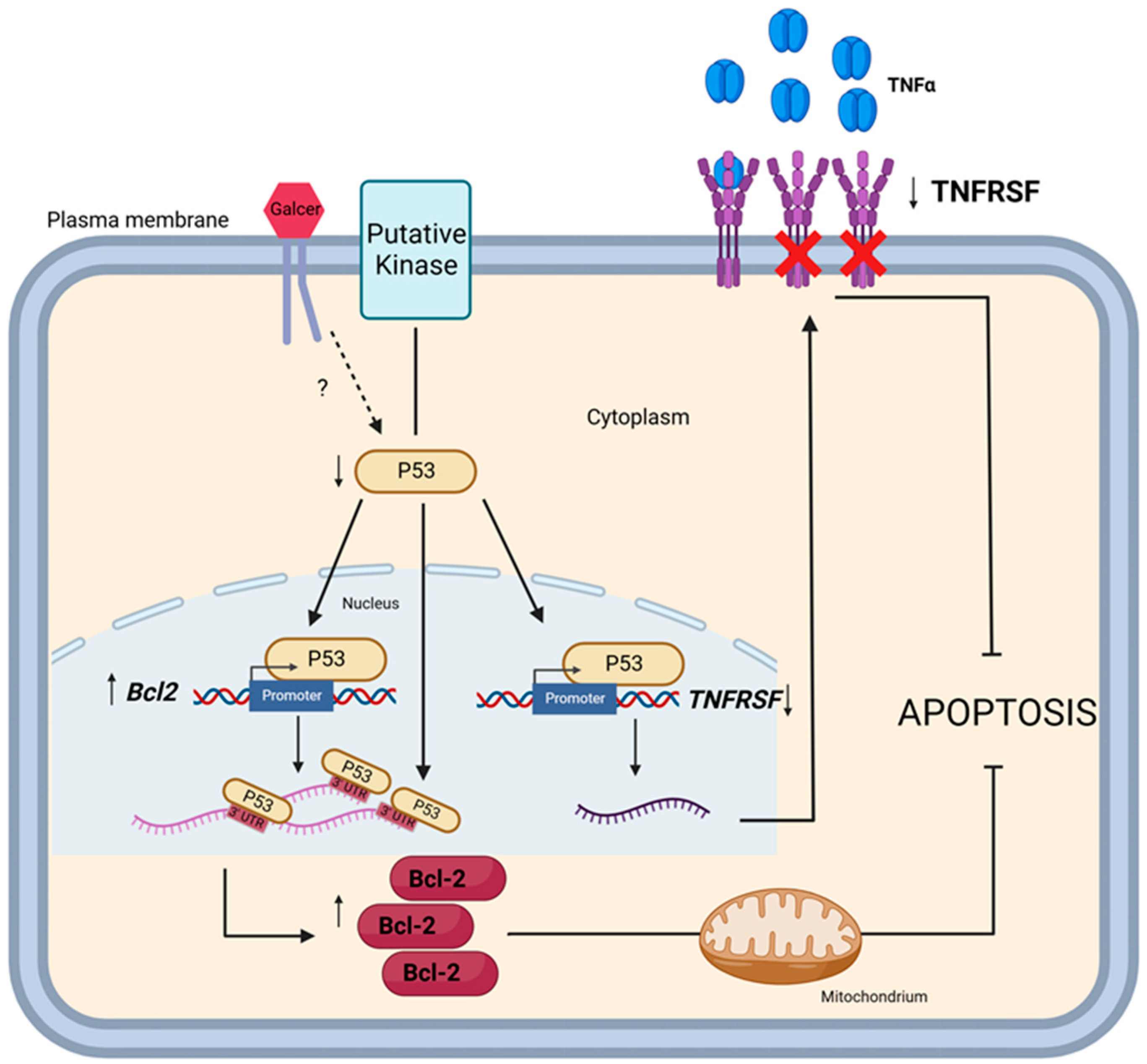
Studies are in progress to identify the specific mechanisms by which GalCer decreases the level of p53 and in this way subsequently affects
TNFRSF1B, TNFRSF9, and
BCL2 expression. At this stage of our research, we propose the following hypothesis: GalCer localised in lipid rafts interacts with a specific protein, most likely a protein kinase, to trigger signalling pathway leading to reduce expression of p53, which in turn causes decreased expression of
TNFRSF1B and
TNFRSF9 and increased expression of
BCL2 genes (
Figure 7).
Figure 1.
Characteristics of BC cells with knock-out or overexpression of UGT8 gene. A(I) Level of UGT8 protein expression in parental MDA-MB-231, MDA-MB-231 cells transduced with “empty” vector (MDA.Δ.C), and MDA-MB-231 cell clones with knockout of UGT8 gene (MDA.Δ.UGT8.1 and MDA.Δ.UGT8.4). A(II) Immunostaining of neutral GSLs from MDA-MB-231, MDA.Δ.C, MDA.Δ.UGT8.1 and MDA.Δ.UGT8.4 cells. B(I) Level of UGT8 protein expression in parental MCF7 cells, MCF7 cells transduced with “empty” vector (MCF7.C), and MCF7 cells overexpressing UGT8 (MCF7.UGT8). B(II) Immunostaining of neutral GSLs from MCF7, MCF7.C, and MCF7.UGT8. The expression of UGT8 was analysed by Western blotting using rabbit polyclonal antibodies directed against UGT8. Cellular proteins (40 µg) separated by SDS-PAGE on 12% gel were transferred onto nitrocellulose membrane. GAPDH was used as the internal control. For GalCer immunostaining, samples of neutral GSLs that had been purified from 1×107 cells were separated on HP-TLC plate. (C) Viability of BC MDA.Δ.UGT8.1 and MDA.Δ.UGT8.4 cells with knockout of the UGT8 gene and MCF7.UGT8 and T47D.UGT8 cells overexpressing UGT8 and GalCer exposed to increased concentrations of anti-cancer drugs: doxorubicin (DOX) (I), paclitaxel (PTX) (II), and cisplatin cis-diamminedichloroplatinum (III) (CDDP) (III) for 48 h. Percentage of viable cells was determined by MTT assay. Data represent the mean ± standard deviation (SD) of 6 replicates from 2 independent measurements. Differences that are statistically significant (*p < 0.1, **p < 0.01).
Figure 1.
Characteristics of BC cells with knock-out or overexpression of UGT8 gene. A(I) Level of UGT8 protein expression in parental MDA-MB-231, MDA-MB-231 cells transduced with “empty” vector (MDA.Δ.C), and MDA-MB-231 cell clones with knockout of UGT8 gene (MDA.Δ.UGT8.1 and MDA.Δ.UGT8.4). A(II) Immunostaining of neutral GSLs from MDA-MB-231, MDA.Δ.C, MDA.Δ.UGT8.1 and MDA.Δ.UGT8.4 cells. B(I) Level of UGT8 protein expression in parental MCF7 cells, MCF7 cells transduced with “empty” vector (MCF7.C), and MCF7 cells overexpressing UGT8 (MCF7.UGT8). B(II) Immunostaining of neutral GSLs from MCF7, MCF7.C, and MCF7.UGT8. The expression of UGT8 was analysed by Western blotting using rabbit polyclonal antibodies directed against UGT8. Cellular proteins (40 µg) separated by SDS-PAGE on 12% gel were transferred onto nitrocellulose membrane. GAPDH was used as the internal control. For GalCer immunostaining, samples of neutral GSLs that had been purified from 1×107 cells were separated on HP-TLC plate. (C) Viability of BC MDA.Δ.UGT8.1 and MDA.Δ.UGT8.4 cells with knockout of the UGT8 gene and MCF7.UGT8 and T47D.UGT8 cells overexpressing UGT8 and GalCer exposed to increased concentrations of anti-cancer drugs: doxorubicin (DOX) (I), paclitaxel (PTX) (II), and cisplatin cis-diamminedichloroplatinum (III) (CDDP) (III) for 48 h. Percentage of viable cells was determined by MTT assay. Data represent the mean ± standard deviation (SD) of 6 replicates from 2 independent measurements. Differences that are statistically significant (*p < 0.1, **p < 0.01).
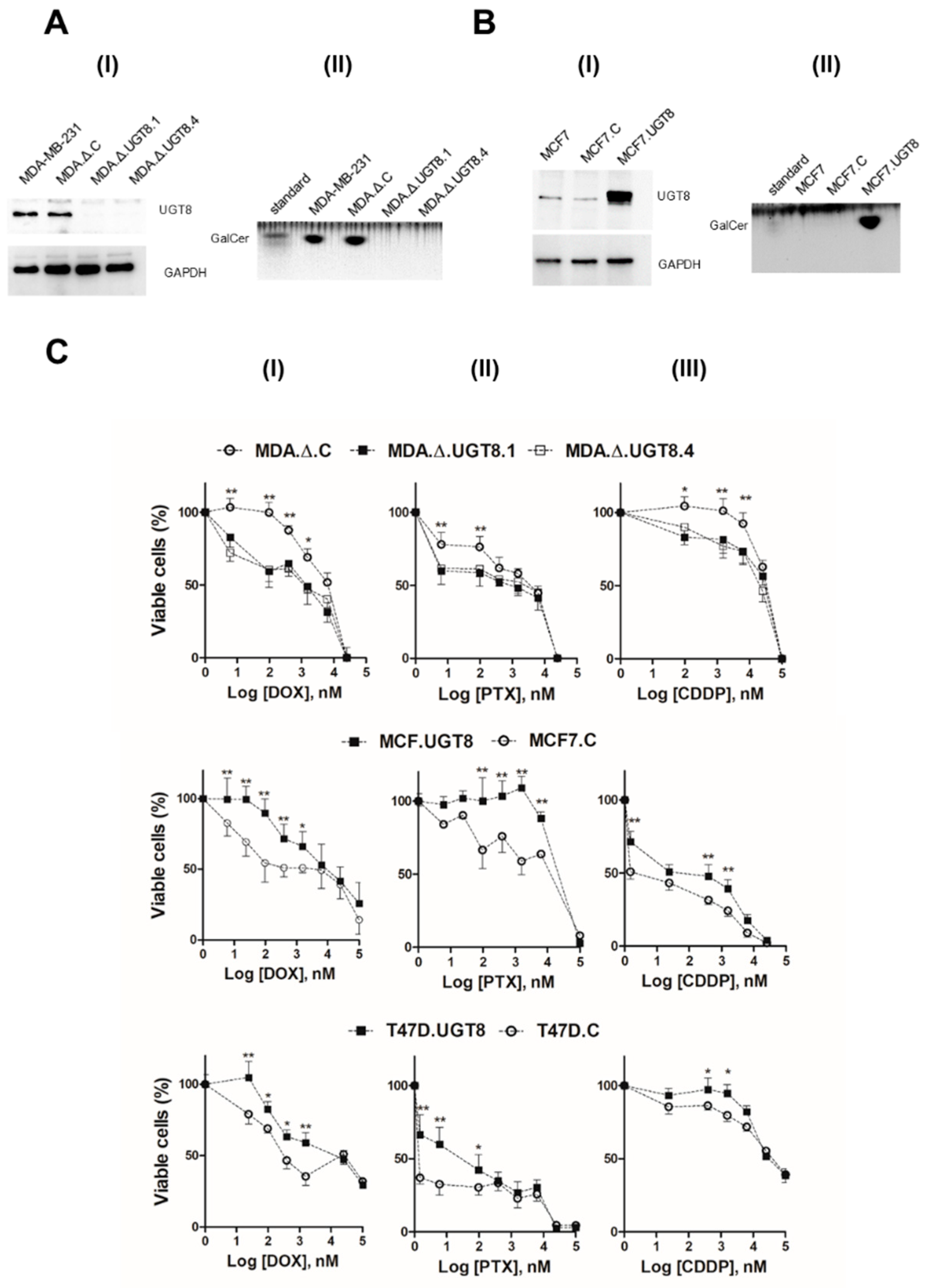
Figure 2.
Characteristics of murine mammary carcinoma 4T1 cells overexpressing murine UGT8a and GalCer. A(I) Expression of UGT8a protein in parental 4T1 cells, control 4T1 cells transduced with vector alone (4T1.C), and 4T1 cells overexpressing UGT8a (4T1.UGT8a); A(II) immunostaining of neutral GSLs from 4T1, 4T1.C and 4T1.UGTa cells. Western blotting with anti-human UGT8 antibodies was used to analyze the expression of UGT8a. Cellular proteins (40 µg) separated by SDS-PAGE on 12% gel were transferred onto nitrocellulose membrane. GAPDH was used as the internal control For GalCer detection, samples of neutral GSLs purified from 1×107 cells was applied to an HP-TLC plate. (B) Viability of 4T1.C and 4T1.UGT8a cells overexpressing UGT8a and GalCer incubated with increased concentrations of DOX for 48 h. Percentage of viable cells was determined using MTT assay. Data represent the mean ±SD of 6 replicates from 2 independent measurements. Statistically significant differences (*p < 0.1). (C) Impact of i. v. administration of DOX on the growth of 4T1.C and 4T1.UGT8a tumors in BALB/c mice. 4T1.UGT8a – mice with 4T1.UGT8a tumors overexpressing UGT8a and GalCer treated with placebo (PBS); 4T1.UGT8 + DOX – mice with 4T1.UGT8a tumors treated with DOX; 4T1.C - mice with control 4T1.C tumors treated with placebo (PBS); 4T1.C + DOX - mice with 4T1.C tumours treated with DOX. Data was presented as mean tumor volume (±SD) for group of mice (n = 5) of the specified time intervals. Data was analysed using Prism 5.0 Two-way Anova Dunnetts post-test (p* < 0.05).
Figure 2.
Characteristics of murine mammary carcinoma 4T1 cells overexpressing murine UGT8a and GalCer. A(I) Expression of UGT8a protein in parental 4T1 cells, control 4T1 cells transduced with vector alone (4T1.C), and 4T1 cells overexpressing UGT8a (4T1.UGT8a); A(II) immunostaining of neutral GSLs from 4T1, 4T1.C and 4T1.UGTa cells. Western blotting with anti-human UGT8 antibodies was used to analyze the expression of UGT8a. Cellular proteins (40 µg) separated by SDS-PAGE on 12% gel were transferred onto nitrocellulose membrane. GAPDH was used as the internal control For GalCer detection, samples of neutral GSLs purified from 1×107 cells was applied to an HP-TLC plate. (B) Viability of 4T1.C and 4T1.UGT8a cells overexpressing UGT8a and GalCer incubated with increased concentrations of DOX for 48 h. Percentage of viable cells was determined using MTT assay. Data represent the mean ±SD of 6 replicates from 2 independent measurements. Statistically significant differences (*p < 0.1). (C) Impact of i. v. administration of DOX on the growth of 4T1.C and 4T1.UGT8a tumors in BALB/c mice. 4T1.UGT8a – mice with 4T1.UGT8a tumors overexpressing UGT8a and GalCer treated with placebo (PBS); 4T1.UGT8 + DOX – mice with 4T1.UGT8a tumors treated with DOX; 4T1.C - mice with control 4T1.C tumors treated with placebo (PBS); 4T1.C + DOX - mice with 4T1.C tumours treated with DOX. Data was presented as mean tumor volume (±SD) for group of mice (n = 5) of the specified time intervals. Data was analysed using Prism 5.0 Two-way Anova Dunnetts post-test (p* < 0.05).
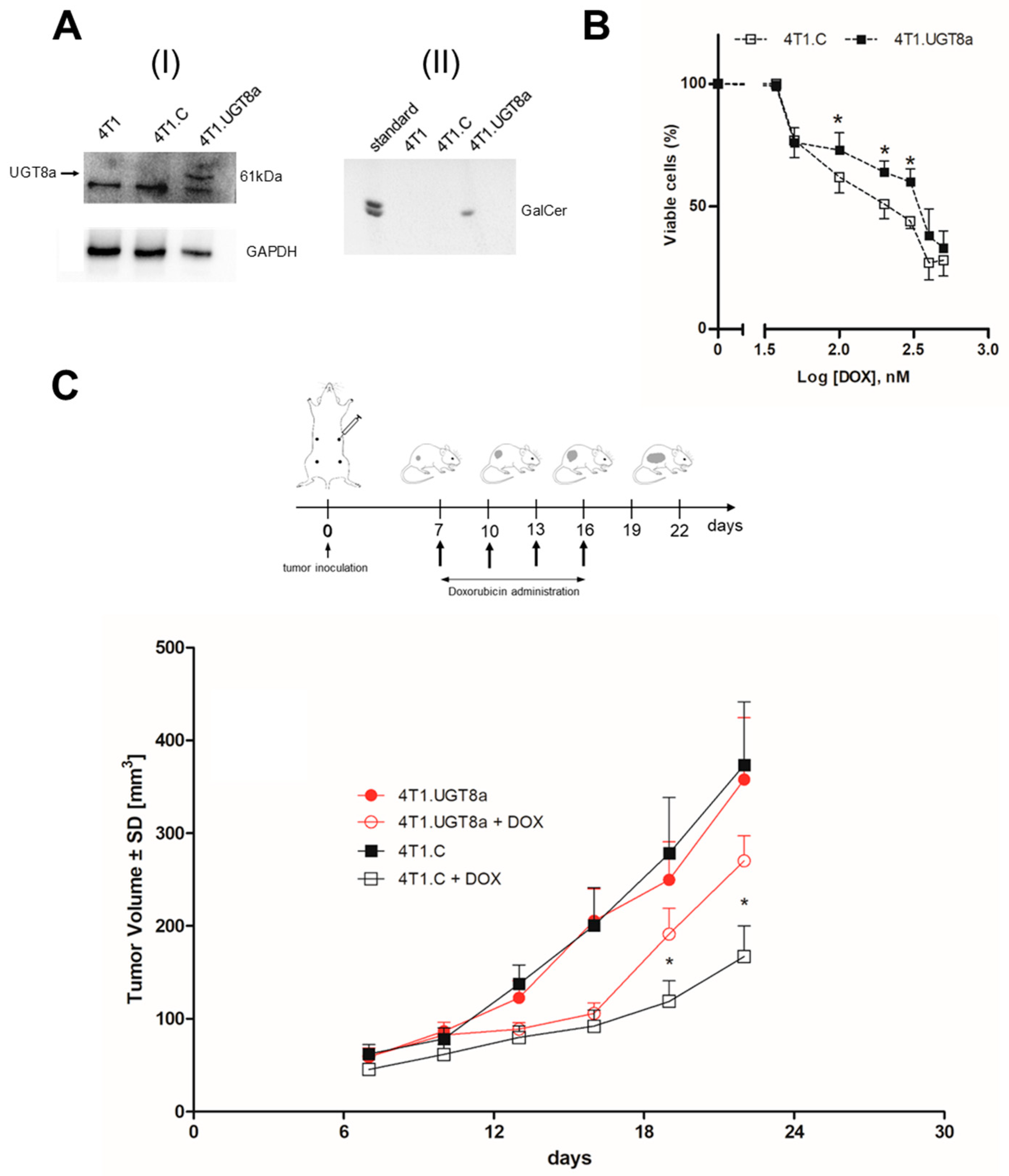
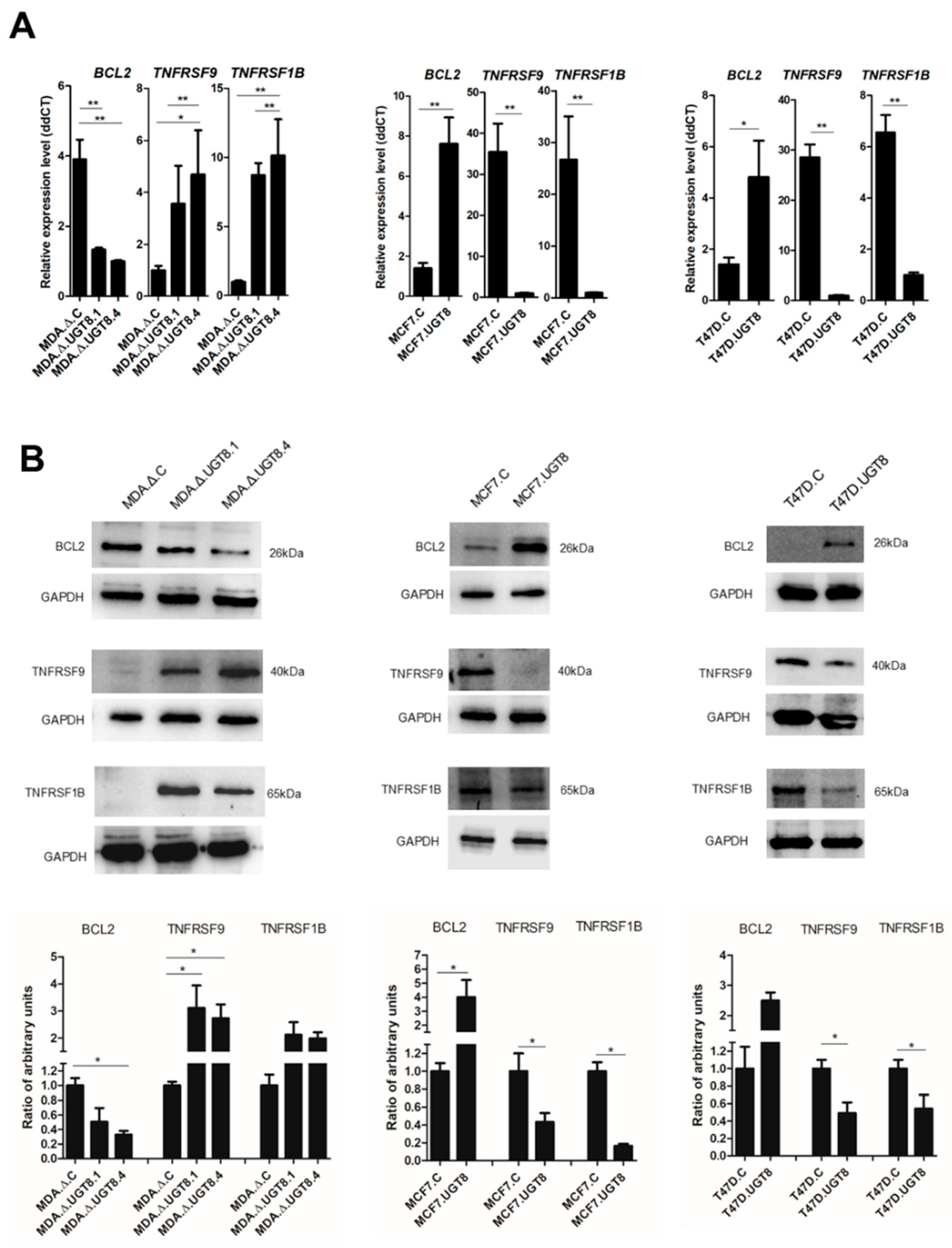
Figure 3.
Levels of Bcl-2, TNFRS9, and TNFRS1B expression in BC MDA-MB-231, MCF7, and T47D cells with various contents of GalCer. (A) qPCR was used to determine mRNA levels, which were normalized against β-actin and cells with low expression levels of UGT8 mRNA served as calibrator sample. Data is expressed as mean ±SD. (B) Western blotting combined with densitometric analysis were used to quantify protein expression levels in BC cell lines. Specific cellular proteins (40 µg) separated by SDS-PAGE on 12% gel were transferred onto nitrocellulose membrane. GAPDH was used as the internal control. Band intensities were analyzed with Image Lab software (Bio-Rad Laboratories) and GraphPad Prism version 5. (*p < 0.05). All values are means ±SD.
Figure 3.
Levels of Bcl-2, TNFRS9, and TNFRS1B expression in BC MDA-MB-231, MCF7, and T47D cells with various contents of GalCer. (A) qPCR was used to determine mRNA levels, which were normalized against β-actin and cells with low expression levels of UGT8 mRNA served as calibrator sample. Data is expressed as mean ±SD. (B) Western blotting combined with densitometric analysis were used to quantify protein expression levels in BC cell lines. Specific cellular proteins (40 µg) separated by SDS-PAGE on 12% gel were transferred onto nitrocellulose membrane. GAPDH was used as the internal control. Band intensities were analyzed with Image Lab software (Bio-Rad Laboratories) and GraphPad Prism version 5. (*p < 0.05). All values are means ±SD.
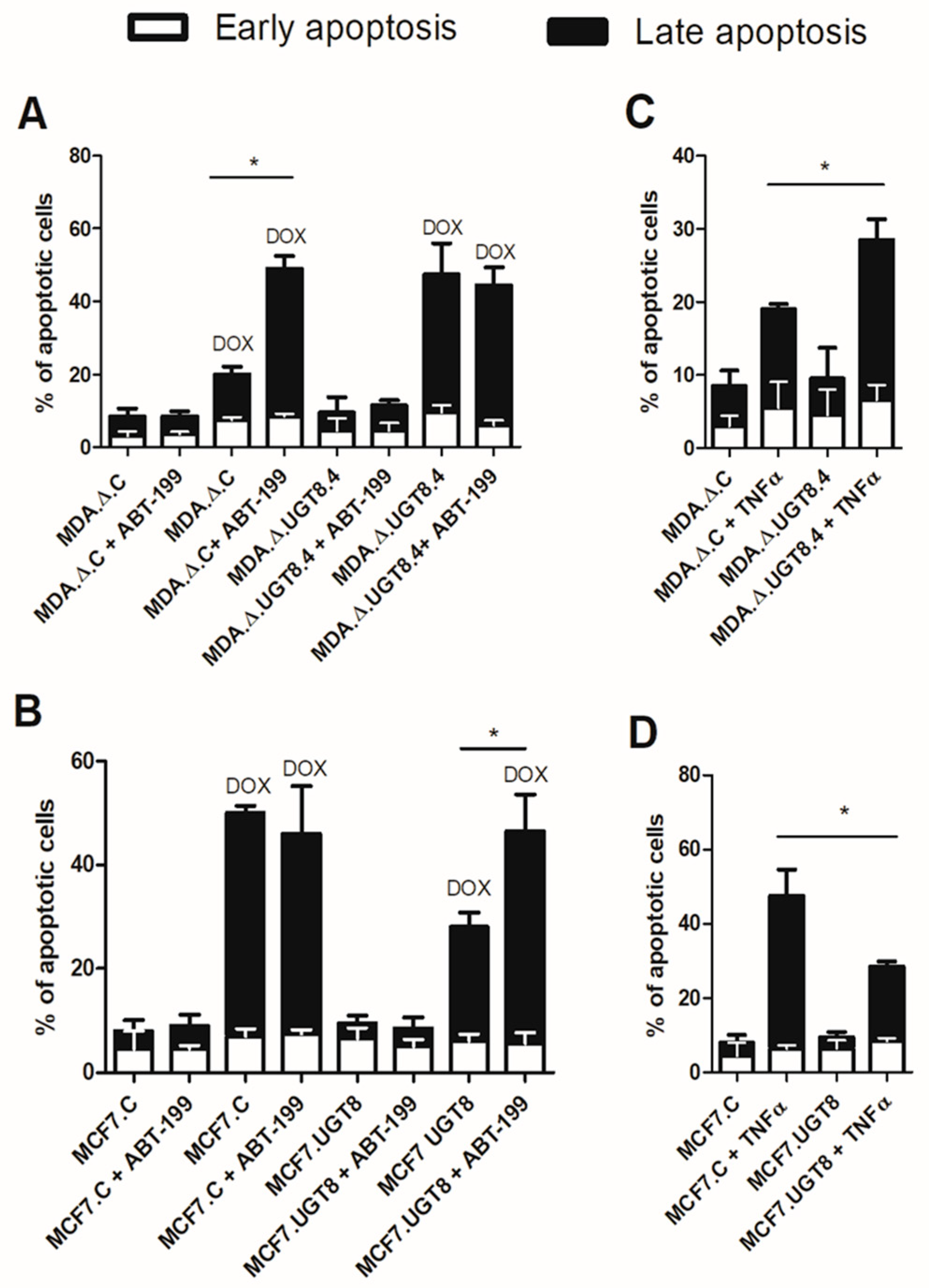
Figure 4.
Sensitivity to DOX-induced apoptosis of BC (A) MDA-MB-231 and (B) MCF7 cells with various GalCer levels and incubated with ABT-199 (specific inhibitor of Bcl-2). Cells were cultured with ABT-199 at a concentration of 0.125 µM or ABT-199 and DOX at concentrations of 0.125 µM and 1 µM, respectively, for 48 h. Sensitivity to TNFα-induced apoptosis of BC (C) MDA-MB-231 and (D) MCF7 cells with different GalCer content. Cells were cultured in the presence of TNFα at a concentration of 25 µM for 48 h. Apoptotic cells were determined by flow cytometry using Annexin V and SYTOX Green stain. MDA.Δ.C - MDA-MB-231 cells transduced with vector alone; MDA.Δ.UGT8.4 - MDA-MB-231 cell clones with knockout of the UGT8 gene; MCF7.C - MCF7 cells transduced with vector alone; MCF7.UGT8 – MCF7 cells overexpressing UGT8 and GalCer. Data represent the mean ±SD of 6 replicates from 2 independent experiments. Statistically significant differences (*p < 0.1).
Figure 4.
Sensitivity to DOX-induced apoptosis of BC (A) MDA-MB-231 and (B) MCF7 cells with various GalCer levels and incubated with ABT-199 (specific inhibitor of Bcl-2). Cells were cultured with ABT-199 at a concentration of 0.125 µM or ABT-199 and DOX at concentrations of 0.125 µM and 1 µM, respectively, for 48 h. Sensitivity to TNFα-induced apoptosis of BC (C) MDA-MB-231 and (D) MCF7 cells with different GalCer content. Cells were cultured in the presence of TNFα at a concentration of 25 µM for 48 h. Apoptotic cells were determined by flow cytometry using Annexin V and SYTOX Green stain. MDA.Δ.C - MDA-MB-231 cells transduced with vector alone; MDA.Δ.UGT8.4 - MDA-MB-231 cell clones with knockout of the UGT8 gene; MCF7.C - MCF7 cells transduced with vector alone; MCF7.UGT8 – MCF7 cells overexpressing UGT8 and GalCer. Data represent the mean ±SD of 6 replicates from 2 independent experiments. Statistically significant differences (*p < 0.1).
Figure 5.
Promoter activities of (A) TNFRS1B, (B) TNFRS9, and (C) BCL2 genes cloned into pGL3 basic vector after transfection into BC cells with various contents of GalCer. MDA.Δ.C - MDA-MB-231 cells transduced with “empty” vector; MDA.Δ.UGT8.4 - MDA-MB-231 cell clones with knock-out of the UGT8 gene; MCF7.C – MCF7 cells transduced with “empty” vector; MCF7.UGT8 – MCF7 cells overexpressing UGT8 and GalCer. The promoter activities were measured using the dual-luciferase reporter assay system. The bars represent average luciferase activities compared with the control pGL3 vector. Data represent the mean ±SD of 3 replicates from 2 independent experiments. Statistically significant differences (*p < 0.1). (D) GalCer increases the stability of Bcl-2 mRNA in BC cells. MDA.Δ.C and MDA.Δ.UGT8.4 cells were treated with actinomycin D for 2 - 6 h and after indicated time points, the Bcl-2 mRNA expression levels, which were normalised against β-actin were evaluated by qPCR. MDA MB 231.C and MDA MB 231.Δ4 cells not treated with actinomycin D were used as calibrator sample. The results are shown as the percentage of the control (0 h). Data represent the mean ±SD of 6 replicates from 2 independent experiments. Statistically significant differences (*p < 0.1).
Figure 5.
Promoter activities of (A) TNFRS1B, (B) TNFRS9, and (C) BCL2 genes cloned into pGL3 basic vector after transfection into BC cells with various contents of GalCer. MDA.Δ.C - MDA-MB-231 cells transduced with “empty” vector; MDA.Δ.UGT8.4 - MDA-MB-231 cell clones with knock-out of the UGT8 gene; MCF7.C – MCF7 cells transduced with “empty” vector; MCF7.UGT8 – MCF7 cells overexpressing UGT8 and GalCer. The promoter activities were measured using the dual-luciferase reporter assay system. The bars represent average luciferase activities compared with the control pGL3 vector. Data represent the mean ±SD of 3 replicates from 2 independent experiments. Statistically significant differences (*p < 0.1). (D) GalCer increases the stability of Bcl-2 mRNA in BC cells. MDA.Δ.C and MDA.Δ.UGT8.4 cells were treated with actinomycin D for 2 - 6 h and after indicated time points, the Bcl-2 mRNA expression levels, which were normalised against β-actin were evaluated by qPCR. MDA MB 231.C and MDA MB 231.Δ4 cells not treated with actinomycin D were used as calibrator sample. The results are shown as the percentage of the control (0 h). Data represent the mean ±SD of 6 replicates from 2 independent experiments. Statistically significant differences (*p < 0.1).
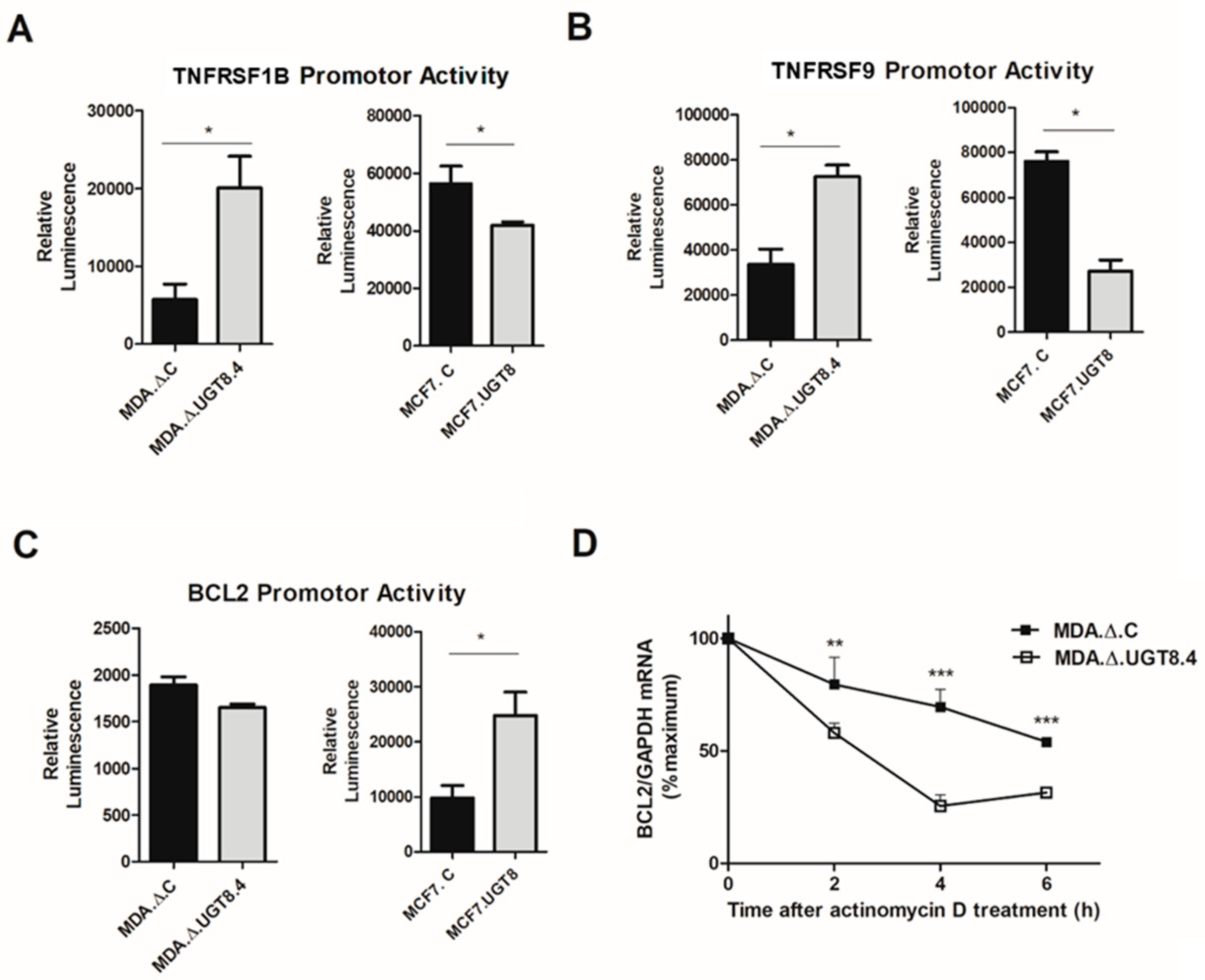
Figure 6.
(A) P53 expression in MDA-MB-231 cells transduced with “empty” vector (MDA.Δ.C), MDA-MB-231 cells with knockout of the UGT8 gene and lack of GalCer (MDA.Δ.UGT8.4), MCF7 cells transduced with vector alone (MCF7.C), and MCF7 cells overexpressing UGT8 and GalCer (MCF7.UGT8). Western blotting was used to analyse p53 expression in BC cell lines. Rabbit anti-p53 polyclonal antibodies were used to detect p53. Proteins (40 µg) separated by SDS-PAGE on 12% gel were transferred onto a nitrocellulose membrane. GAPDH was used as an internal control. (B) TNFRS1B, TNFRS9, and BCL2-2 gene expression levels in MDA.Δ.UGT8.4 and MCF.7 cells transfected with siRNA directed against p53 mRNA. qPCR was used to determine mRNA levels, normalized against β-actin. Cells with low expression levels of UGT8 mRNA were used as calibrators. Data are expressed as mean ±SD. (C) TNFRS1B, TNFRS9 and BCL2 promoter activities in MDA.Δ.UGT8.4 and MCF.7 cells transfected with siRNA targeting p53 mRNA. The activities of the promoters cloned into the pGL3 basic vector were measured using the dual-luciferase reporter assay system. The bars represent the average luciferase activities compared to the control pGL3 vector. (D) The stability of Bcl-2 mRNA in MDA.Δ.UGT8.4 cells transfected with siRNA targeting p53 mRNA. Cells were treated with actinomycin D for 2 - 6 h and after indicated time points, the Bcl-2 mRNA expression levels, which were normalized against β-actin were evaluated by qPCR. MDA.Δ.UGT8.4 cells not treated with actinomycin D were used as calibrator samples. The results are shown as the percentage of the control (0 h). MDA.Δ.UGT8.4/siTP53 and MCF7.C/siTP53 – BC cells transfected with siRNA directed against p53 mRNA; MDA.Δ.UGT8.4 and MCF7.C – control BC cells transfected with scrambled siRNA. Data represent the mean ±SD of 3 replicates from 2 independent experiments. Statistically significant differences (*p < 0.1). Data represent the mean ±SD of 3 replicates from 2 independent experiments. Statistically significant differences (*p < 0.1).
Figure 6.
(A) P53 expression in MDA-MB-231 cells transduced with “empty” vector (MDA.Δ.C), MDA-MB-231 cells with knockout of the UGT8 gene and lack of GalCer (MDA.Δ.UGT8.4), MCF7 cells transduced with vector alone (MCF7.C), and MCF7 cells overexpressing UGT8 and GalCer (MCF7.UGT8). Western blotting was used to analyse p53 expression in BC cell lines. Rabbit anti-p53 polyclonal antibodies were used to detect p53. Proteins (40 µg) separated by SDS-PAGE on 12% gel were transferred onto a nitrocellulose membrane. GAPDH was used as an internal control. (B) TNFRS1B, TNFRS9, and BCL2-2 gene expression levels in MDA.Δ.UGT8.4 and MCF.7 cells transfected with siRNA directed against p53 mRNA. qPCR was used to determine mRNA levels, normalized against β-actin. Cells with low expression levels of UGT8 mRNA were used as calibrators. Data are expressed as mean ±SD. (C) TNFRS1B, TNFRS9 and BCL2 promoter activities in MDA.Δ.UGT8.4 and MCF.7 cells transfected with siRNA targeting p53 mRNA. The activities of the promoters cloned into the pGL3 basic vector were measured using the dual-luciferase reporter assay system. The bars represent the average luciferase activities compared to the control pGL3 vector. (D) The stability of Bcl-2 mRNA in MDA.Δ.UGT8.4 cells transfected with siRNA targeting p53 mRNA. Cells were treated with actinomycin D for 2 - 6 h and after indicated time points, the Bcl-2 mRNA expression levels, which were normalized against β-actin were evaluated by qPCR. MDA.Δ.UGT8.4 cells not treated with actinomycin D were used as calibrator samples. The results are shown as the percentage of the control (0 h). MDA.Δ.UGT8.4/siTP53 and MCF7.C/siTP53 – BC cells transfected with siRNA directed against p53 mRNA; MDA.Δ.UGT8.4 and MCF7.C – control BC cells transfected with scrambled siRNA. Data represent the mean ±SD of 3 replicates from 2 independent experiments. Statistically significant differences (*p < 0.1). Data represent the mean ±SD of 3 replicates from 2 independent experiments. Statistically significant differences (*p < 0.1).
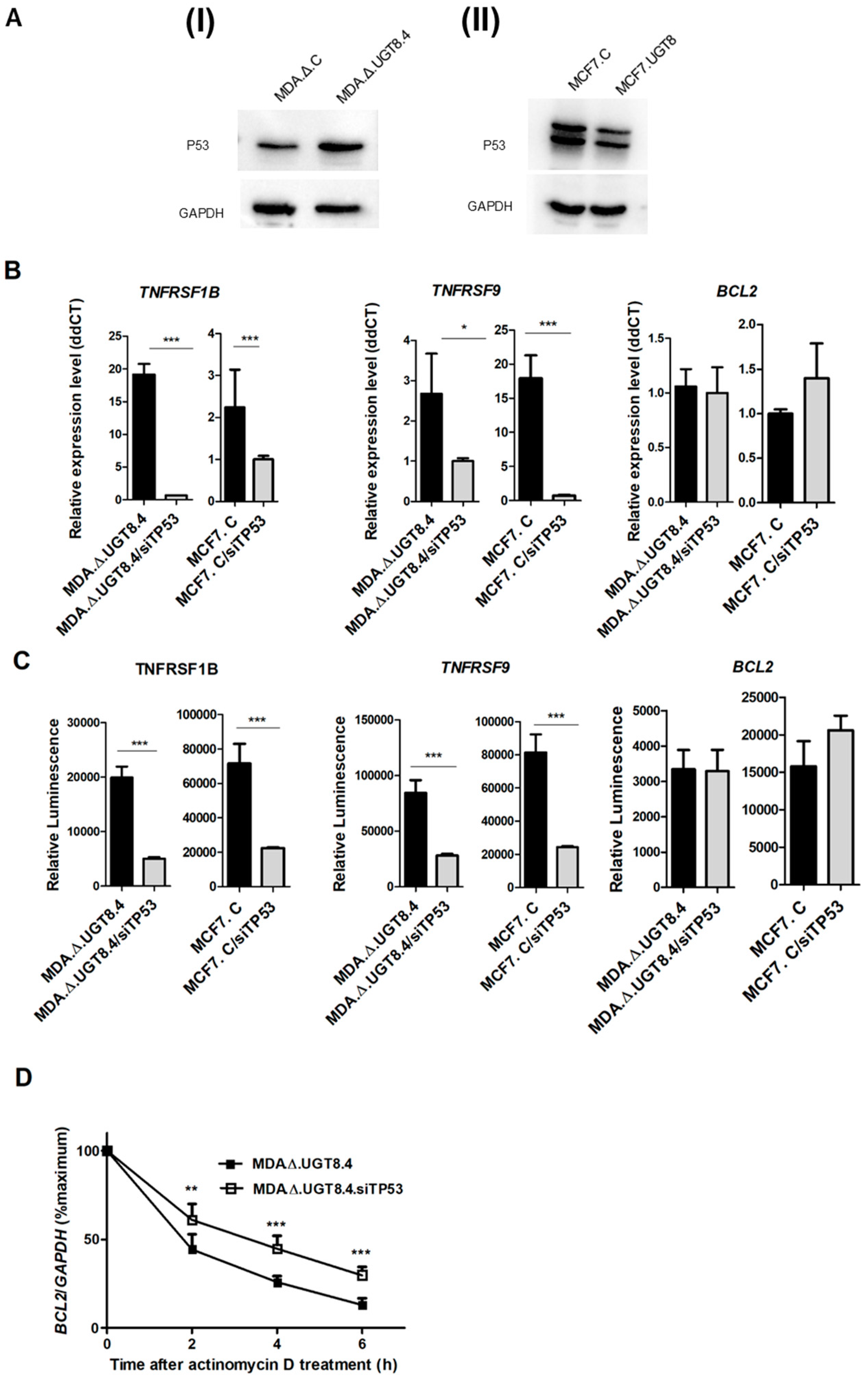
Figure 7.
Diagram showing the putative regulatory role of GalCer in regulation of TNFRSF1B, TNFRSF9 and BCL2 gene expression that results in resistance of BC cells to apoptosis (Created with BioRender.com). As hypothesized, GalCer modulates the activities of signaling pathway/pathways which leads to decreased expression of p53. Depending on the cellular context, p53, acting as a transcription factor, upregulates expression of the BCL2 gene and downregulates the expression of TNFRSF1B and TNFRSF9 genes at the transcriptional level, or p53 acts as mRNA-binding protein (RBP) and upregulates expression of Bcl-2 protein at the post-translational level. ↑ - upregulation, ↓ - downregulation.
Figure 7.
Diagram showing the putative regulatory role of GalCer in regulation of TNFRSF1B, TNFRSF9 and BCL2 gene expression that results in resistance of BC cells to apoptosis (Created with BioRender.com). As hypothesized, GalCer modulates the activities of signaling pathway/pathways which leads to decreased expression of p53. Depending on the cellular context, p53, acting as a transcription factor, upregulates expression of the BCL2 gene and downregulates the expression of TNFRSF1B and TNFRSF9 genes at the transcriptional level, or p53 acts as mRNA-binding protein (RBP) and upregulates expression of Bcl-2 protein at the post-translational level. ↑ - upregulation, ↓ - downregulation.