1. Introduction
Sustainable production of Persian walnuts (Juglans regia) is important for California’s agriculture and rural economy. In 2020, walnuts ranked 11th in California commodities, with an estimated value of $957.7 million1. Persian walnut trees in California are typically grown on interspecific hybrid rootstocks that promote vigor and increased productivity. Crown gall (caused by Agrobacterium tumefaciens), Phytophthora crown and root rot (Phytophthora spp.), and nematode root lesions (Pratylenchus vulnus) are important rootstock diseases of California walnuts. Agrobacterium tumefaciens is a rod-shaped, Gram-negative soil bacterium, while Phytophthora spp. are soilborne “water molds”. These microbes obstruct the vascular tissues with tumors (A. tumefaciens) or kill root tissues directly (Phytophthora spp.). Both pathogens cause disease by inhibiting the flow of nutrients and water to the grafted scion (upper portion of the plant), reducing crop productivity while imperiling plant health. Pratylenchus vulnus, an obligate migratory endoparasitic nematode, causes crop loss by damaging the root cortical tissue and consuming plant nutrients, which ultimately impedes the flow of nutrients and water to the scion.
Walnut growers spend substantial resources managing these phytopathogens. According to the 2017 UC Davis walnut production cost study, management costs for nematode suppression were $1,400 per acre before planting and $97 per acre per year for Phytophthora2. Costs to manage A. tumefaciens need to be more comprehensively estimated but include labor for surgical removal of tumors and follow-up topical treatments to prevent redevelopment.
Despite current cultural practices and treatments, these phytopathogens continue to cause severe yield losses. In one study, A. tumefaciens infection led to crop losses of 25% to 50% per infected tree or entire loss of the infected tree3. A second study found crown gall disease caused a 12% decrease in cumulative four-year yield per 25% of galled trunk diameter4. Aggressive species of Phytophthora, such as P. cinnamomi, can decimate most trees in walnut orchards infested with the pathogen unless a resistant rootstock is used5,6. To date, ‘RX1’ is the only commercially available rootstock that exhibits sufficient resistance to this pathogen7. Yield and/or survivability data for P. vulnus are less detailed but estimated at 15-20% and, in severe cases, total failure of an orchard. Nematode phytopathogens were estimated to be in 85% of walnut orchards8. It has been estimated that root system diseases cost the California walnut industry $241 million per year, an industry with an annual production value of approximately $1.24 billion9.
Another trait of interest is tree vigor. From the grower’s perspective, if trees become too large, they are hard to manage. For example, plant-protective sprays for pests and diseases become less effective as the tree gets larger due to the physical limitations of the equipment’s performance. Moreover, harvest can be more difficult. Walnuts are harvested by a machine that shakes the tree, usually near or at the trunk. As the trees get larger, shaking at the trunk is not sufficient and instead, individual scaffolds have to be shaken. Conversely, low vigor can be problematic in that the tree needs to reach a minimum size to bear fruit that is worth cultivating. Even in a dwarf orchard scenario, the trees must be planted closer together to maximize the efficiency of available sunlight usage compared to an orchard with higher vigor trees. It is also suspected that so-called ‘high-density’ plantings would be vulnerable to increased pest and disease pressure.
Persian walnut scions are grafted to rootstocks to physically combine benefits of the scion genotype, e.g., high nut quality, with beneficial traits of the rootstock genotypes. Early in California walnut production, selections of Northern California Black (Juglans hindsii) or English (J. regia) rootstocks highly susceptible to Phytophthora. spp and P. vulnus were used. In the early 1900s, Luther Burbank created the first hybrid walnut rootstock seedlings by crossing J. hindsii with J. regia10. Burbank termed the hybrid ‘paradox’ due to its unusual vigor and other ‘anomalies.’ Paradox became the most popular walnut rootstock due to its vigor and greater, though still limited, resistance to Phytophthora spp. and P. vulnus, compared to J. hindsii and J. regia10. Paradox is now understood to include any cross of a black walnut species (J. sect. Rhysocaryon) x J. regia. The currently most popular California scion cultivar, J. regia cv. ‘Chandler’ is highly susceptible to A. tumefaciens, Phytophthora. spp, and P. vulnus. Growers have looked to genetic resistance of rootstocks as a foundation for management of soilborne disease challenges. Interdisciplinary walnut rootstock breeding efforts led to the discovery of ‘RX1’ and ‘VX211’ rootstocks. Clonal Paradox ‘RX1’ (Juglans microcarpa x J. regia) is now the rootstock of choice when Phytophthora and A. tumefaciens are of concern due to its higher resistance to these pathogens7,11. Another clonal Paradox, ‘VX211’ (J. hindsii x J. regia), is the preferred rootstock when P. vulnus is a problem due to its greater tolerance than other commercially available rootstocks12. In current breeding work, selections from paradox-type crosses are made with the goal of creating elite hybrids resistant to A. tumefaciens, Phytophthora. spp, and P. vulnus infection. When analyzing breeding populations of Juglans microcarpa x J. regia, hybrids were discovered with improved resistance to A. tumefaciens and Phytophthora. spp.7,13.
In addition to traditional breeding, molecular approaches to walnut improvement are progressing. Expressing double-stranded RNA (RNAi) targeting key genes of A. tumefaciens and P. vulnus in walnuts resulted in resistance to these pathogens in vitro14,15,16. Years later, the publication of high-quality genomes of J. microcarpa and J. regia facilitated genomic and functional genomic analyses of the J. microcarpa x J. regia hybrids mentioned previously17,18. Juglans microcarpa is a short-statured black walnut native to riparian areas of North America, and its genome size and chromosome number are very similar to J. regia18. QTL analysis of these hybrids showed a significant region on chromosome 4D of J. microcarpa correlated with Phytophthora spp. and A. tumefaciens resistance19. These QTL results provided the first insights into the genetic basis of resistance to these key yield-limiting soilborne pathogens in walnut. These studies accelerated walnut rootstock development efforts for the industry.
The work that we report here complements the previous efforts through the use of a functional genomics approach. We hypothesized that genes associated with phenotypes of susceptibility, resistance, or vigor could be identified using RNA-seq. QTL analysis can provide valuable information on what genomic loci may be involved in phenotypes but gives little information on the genes involved or on their expression levels. Very little is understood about the molecular biology of basal host resistance or vigor in walnut rootstocks. The objectives of this research were to conduct an RNA-seq experiment (i) to determine loci with expression profiles related to basal resistance to A tumefaciens, Phytophthora spp., and P. vulnus; and (ii) to estimate the physiological function of these loci.
2. Results
Differential Expression Analysis
Roots of seven non-inoculated
J. microcarpa x
J. regia hybrids were selected to represent a range of phenotypes for vegetative vigor and resistance to
A. tumefaciens,
Phytophthora spp., and
P. vulnus. Phenotype units for “vigor” expressed lengths of shoot growth per unit of time; whereas units for response phenotype to
A. tumefaciens and
Phytophthora spp. expressed disease severity scores (with ranges of 1 to 4 and from 0 to 100, respectively) and units of response phenotype to
P. vulnus expressed counts of the phytopathogenic nematode per gram of root. To identify putative transcripts associated with these phenotypes, we performed RNA-seq analysis on seven unchallenged
J. microcarpa x
J. regia hybrid progeny. These progenies were selected from two breeding populations of seedling genotypes. One population of 368 individuals was derived from a cross between
J. microcarpa mother tree 31.01 and
J. regia father tree “Serr.” The other population of 42 individuals was derived from a cross between
J. microcarpa mother tree 29.11 and
J. regia father tree “Serr.” Hybrids MS1-36, MS1-41, MS1-56, and MS1-122 are progeny from the 31.01 x “Serr” cross, and 29JM-11, JMS-12, and STJM-4 are progeny from the 29.11 x “Serr” cross. The phenotypes of the seven hybrids are shown in
Figure S1. Note that phenotypic data for STJM-4 and JMS-12 are missing for
P. vulnus and vigor. Principal component analysis (PCA) of the counts per million (CPM) normalized reads revealed that PC2 strongly correlated with
A. tumefaciens phenotypic response (
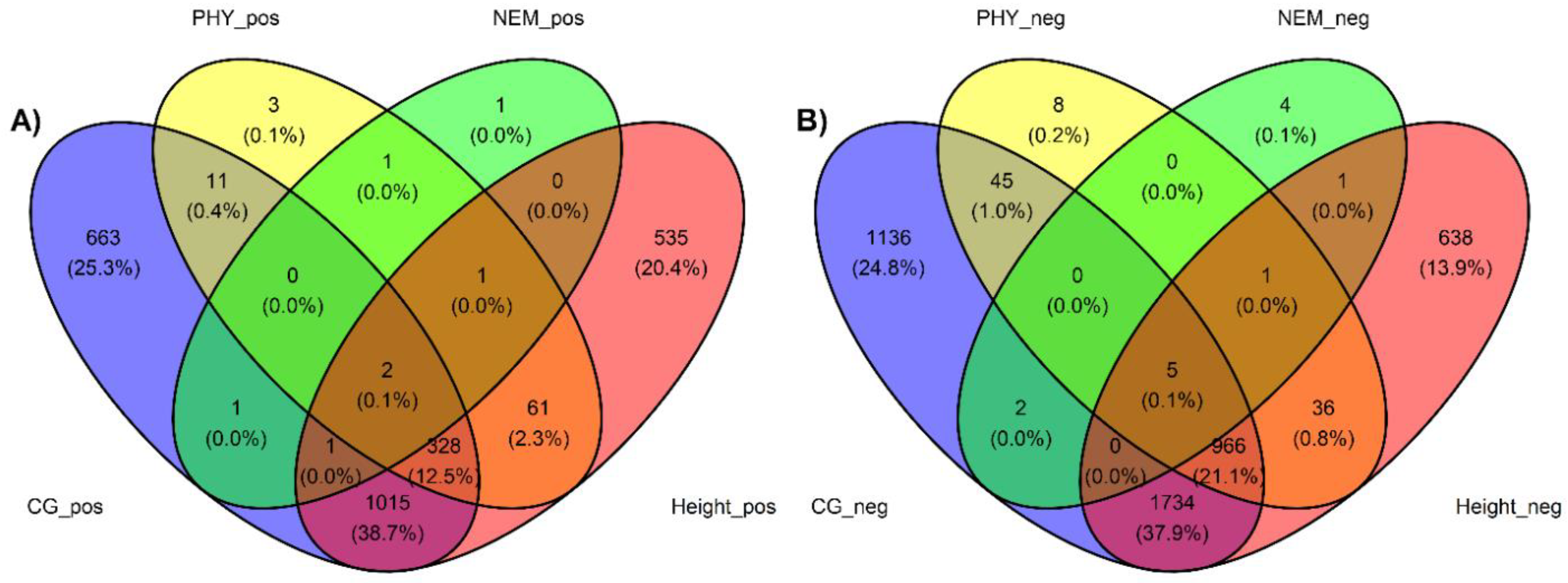
Table 1). Tree vigor showed a strong trending correlation with PC1 but was not significant (p-value 5.69E-02). Modeling of the expressed genes against each trait revealed hundreds of differentially expressed genes (DEGs), with many DEGs shared across traits (
Figure 1). We observed 3888 genes negatively associated with, 2021 genes positively associated with, and 13290 genes not associated with
A. tumefaciens phenotypic response, 1061 genes negatively associated with, 407 genes positively associated with, and 17731 genes not associated with
Phytophthora spp. phenotypic response, 13 genes negatively associated with, 7 genes positively associated with, and 17962 genes not associated with
P. vulnus phenotypic response, and 3381 genes negatively associated with, 1943 genes positively associated with, and 12658 genes not associated with tree height (
Table 2). Modeling of
P. vulnus phenotypic response yielded substantially fewer DEGs than the other traits (
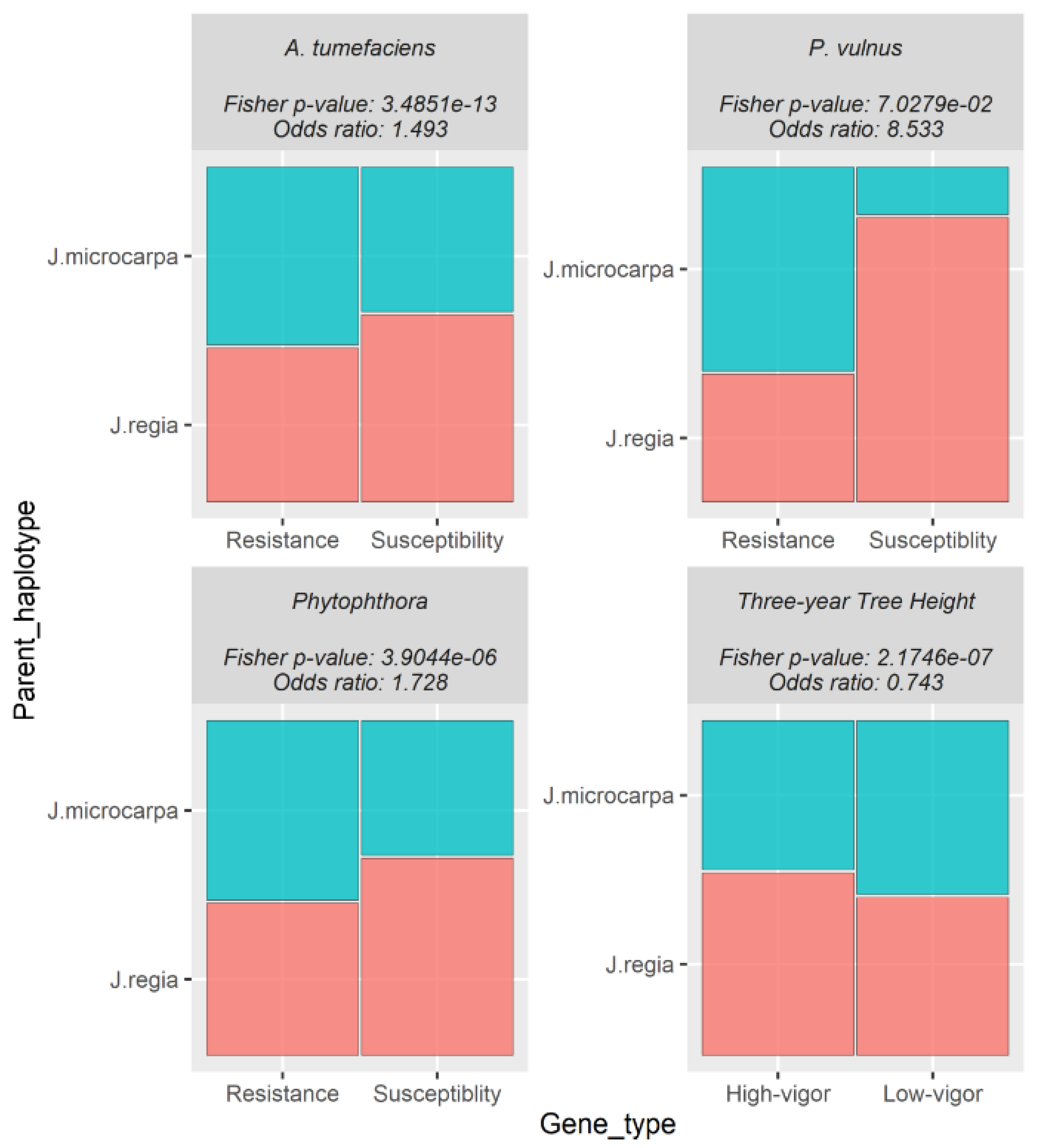
Table 2). In each trait analyzed, the odds of expressing genes positively associated with the trait were slightly higher in the
J. regia haplotype compared to the
J. microcarpa haplotype. Conversely, the odds of expressing genes negatively associated with the phenotype were slightly higher in the
J. microcarpa haplotype compared to that of the
J. regia haplotype. (
Figure 2). Regardless of haplotype, the number of differentially expressed genes negatively associated with each trait was always greater than differentially expressed genes positively associated with each trait (
Table 2). The full results of the differential expression analysis are available in file S3.
Biological Process Analysis
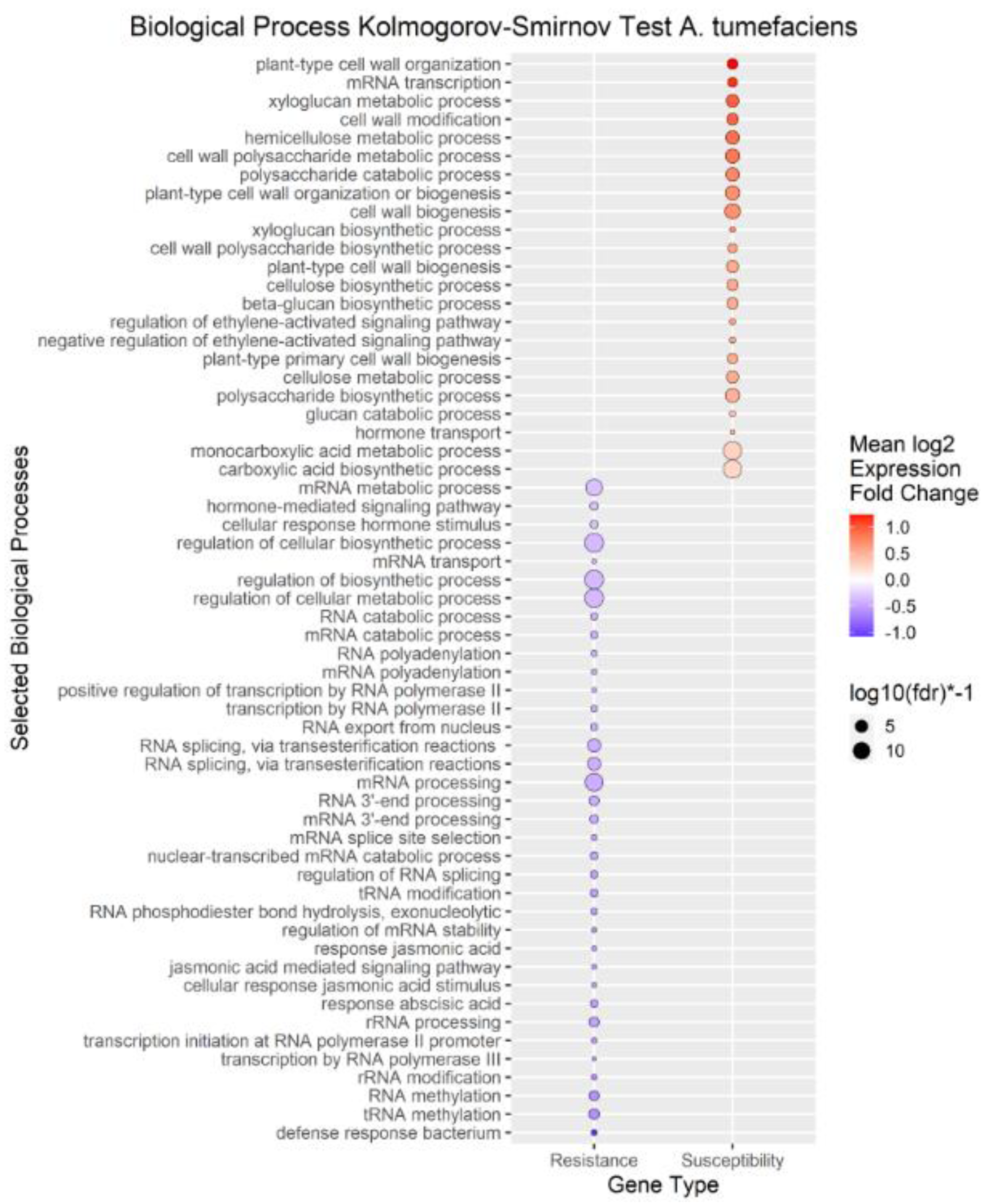
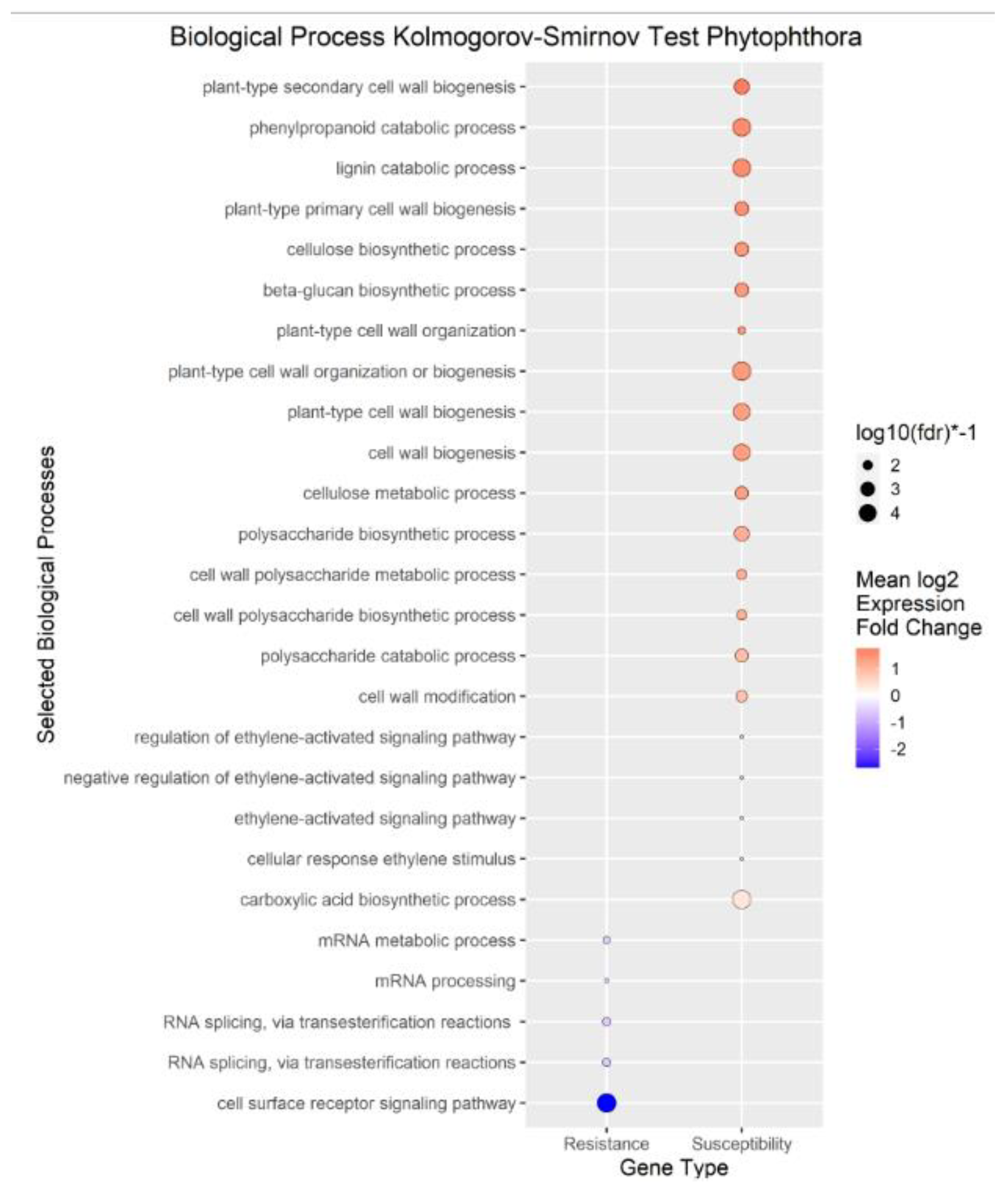
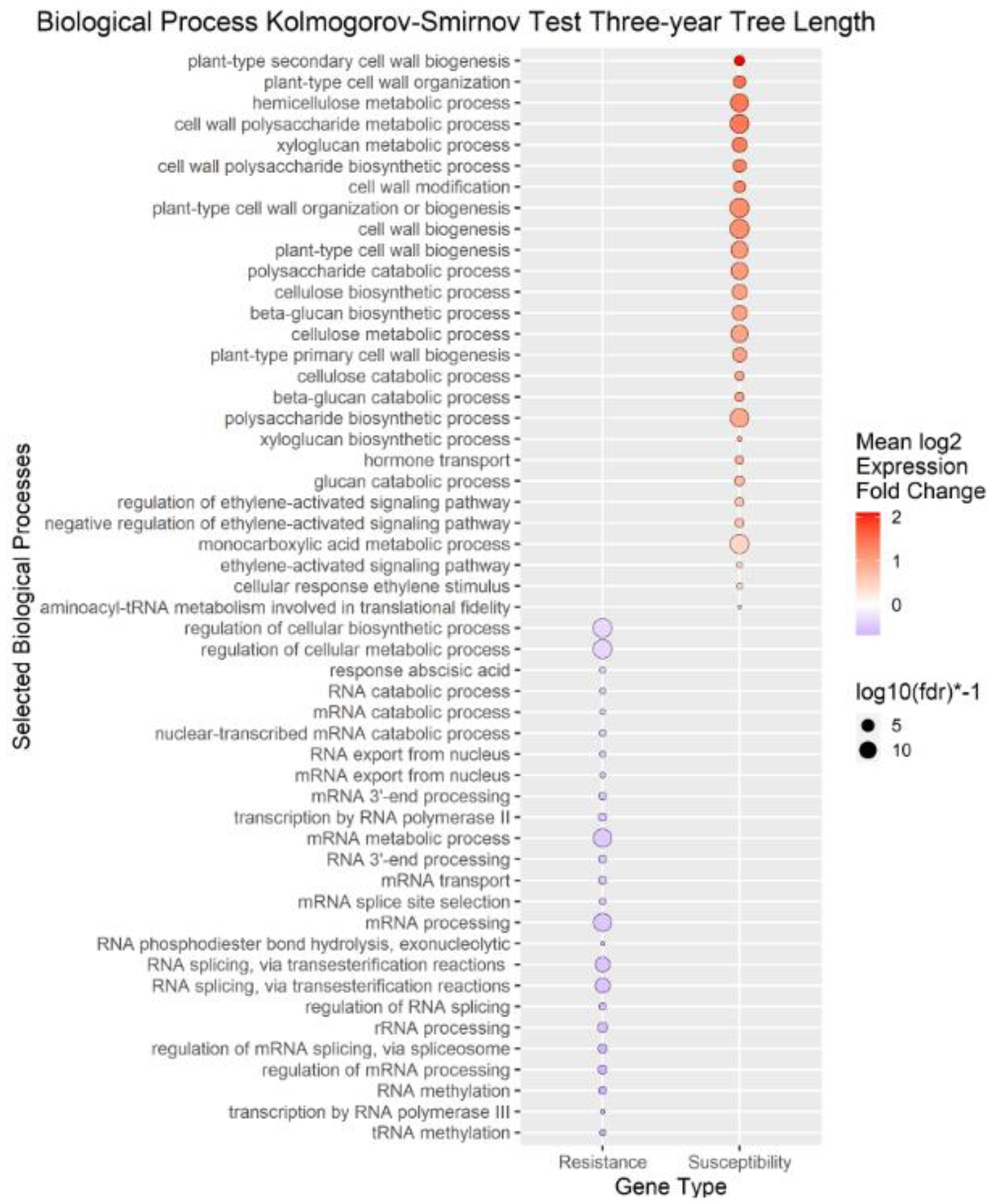
We performed enrichment analysis using Kolmogorov-Smirnov tests on the DEGs to reveal Gene Ontology biological processes (BPs) enriched in the DEGs of each trait. The enrichment analysis resulted in 2958 genes mapped to 322 biological processes for
A. tumefaciens phenotypic response, 464 genes mapped to 114 biological processes for
Phytophthora spp. phenotypic response, no significant results for
P. vulnus, and 2605 genes mapped to 296 biological processes for tree height (
Figure 3,
Figure 4 and
Figure 5). The differences in the number of enriched terms likely reflected the number of genes in the input of the analysis. Transcripts positively correlated with each trait were enriched in BPs involved in cell wall organization, polysaccharides, glucans, and cellulose. Genes that mapped to these terms included cellulose synthases, fasciclin-like arabinogalactan proteins, and xyloglucan endotransglucosylase/hydrolases (File S1). Conversely, transcripts negatively correlated with each trait were enriched in some form of RNA metabolic process. Regulation of RNA metabolic process is defined as “Any process that modulates the frequency, rate or extent of RNA biosynthetic process (
http://amigo.geneontology.org/amigo/term/GO:0051252).” “Defense response to bacterium” was significantly enriched with transcripts negatively correlated to
A. tumefaciens phenotypic response (
Figure 3). No terms mentioning defense response were significantly enriched in
Phytophthora spp. phenotypic response,
P. vulnus phenotypic response, or tree height. “Jasmonic acid-mediated signaling pathway,” “cellular response to jasmonic acid stimulus,” and “response to jasmonic acid” were enriched with transcripts negatively correlated to
A. tumefaciens phenotypic response (
Figure 3). These terms were not enriched in the other traits. Moreover, several terms mentioning ethylene were enriched with transcripts positively correlated to
A. tumefaciens phenotypic response,
Phytophthora spp. phenotypic response, and tree height (
Figure 3,
Figure 4 and
Figure 5). “Response to abscisic acid” was enriched with transcripts negatively correlated to
A. tumefaciens phenotypic response and tree height (
Figure 3 and
Figure 5). The expression of genes associated with the crucial plant hormone term salicylic acid was not enriched in any analysis (File S1). However, “hormone-mediated signaling pathway” and “cellular response to hormone stimulus were enriched with transcripts negatively correlated to
A. tumefaciens phenotypic response and “hormone transport” was enriched with transcripts positively correlated to tree height (
Figure 3 and
Figure 5).
Subcellular Localization Analysis
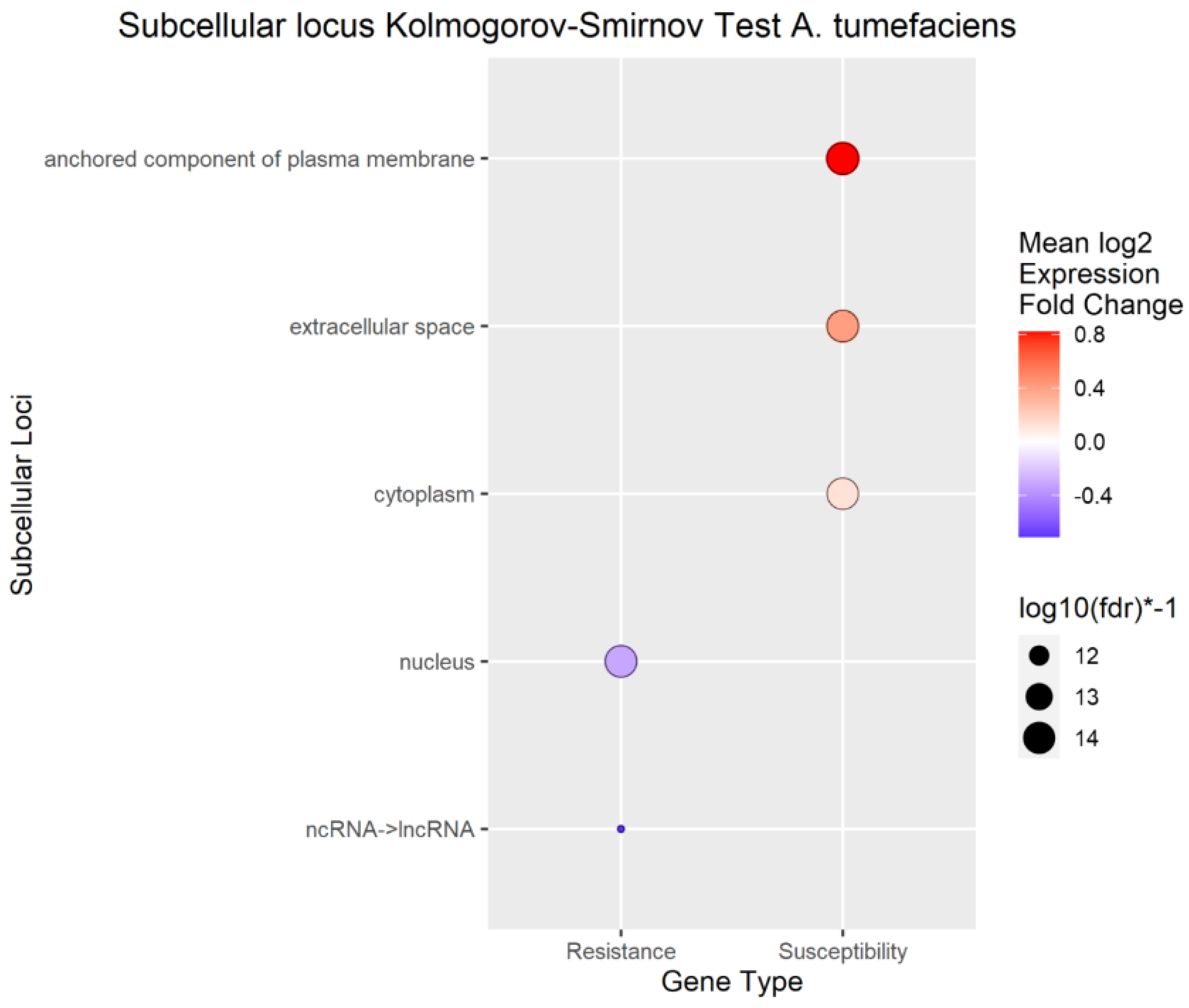
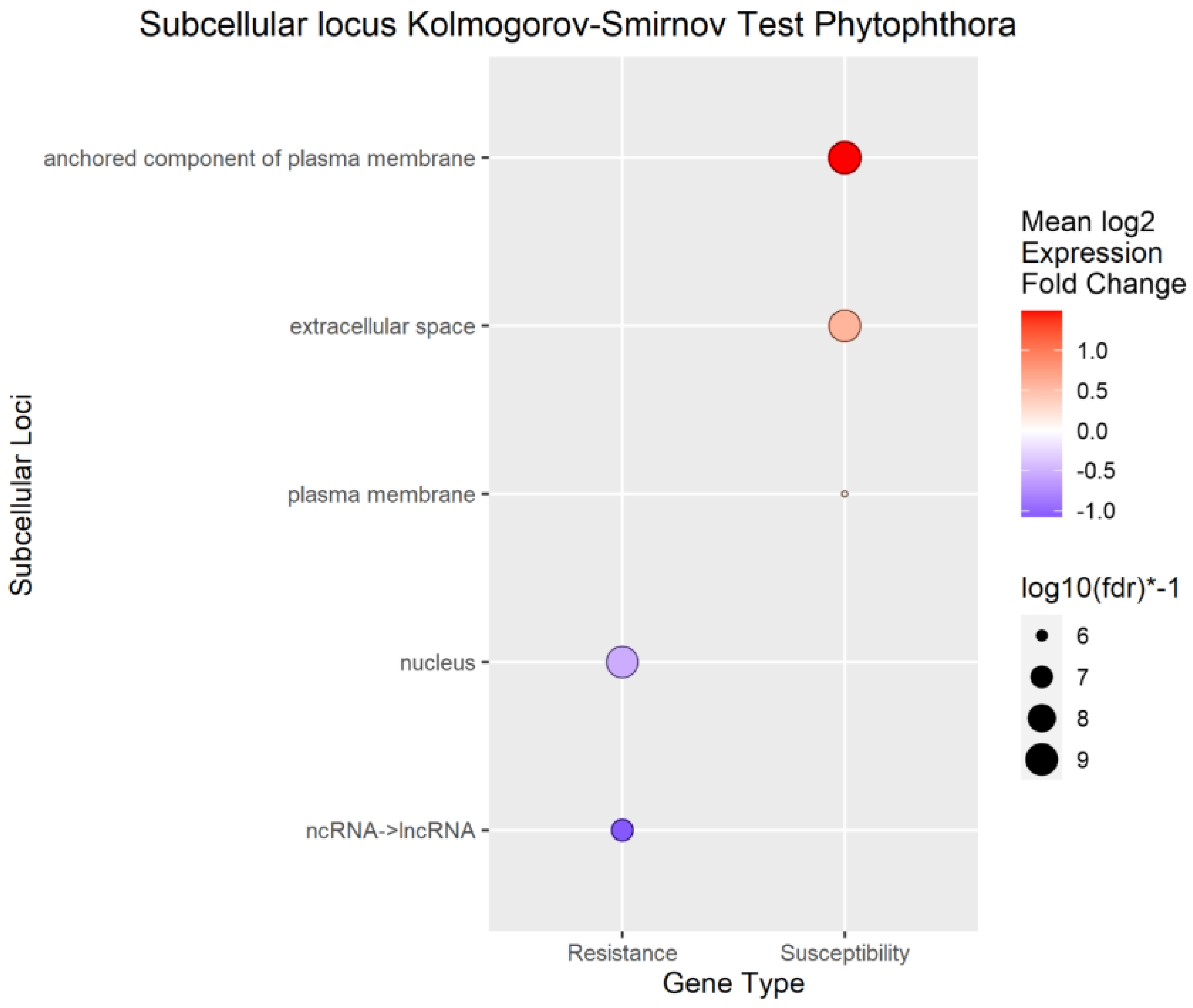
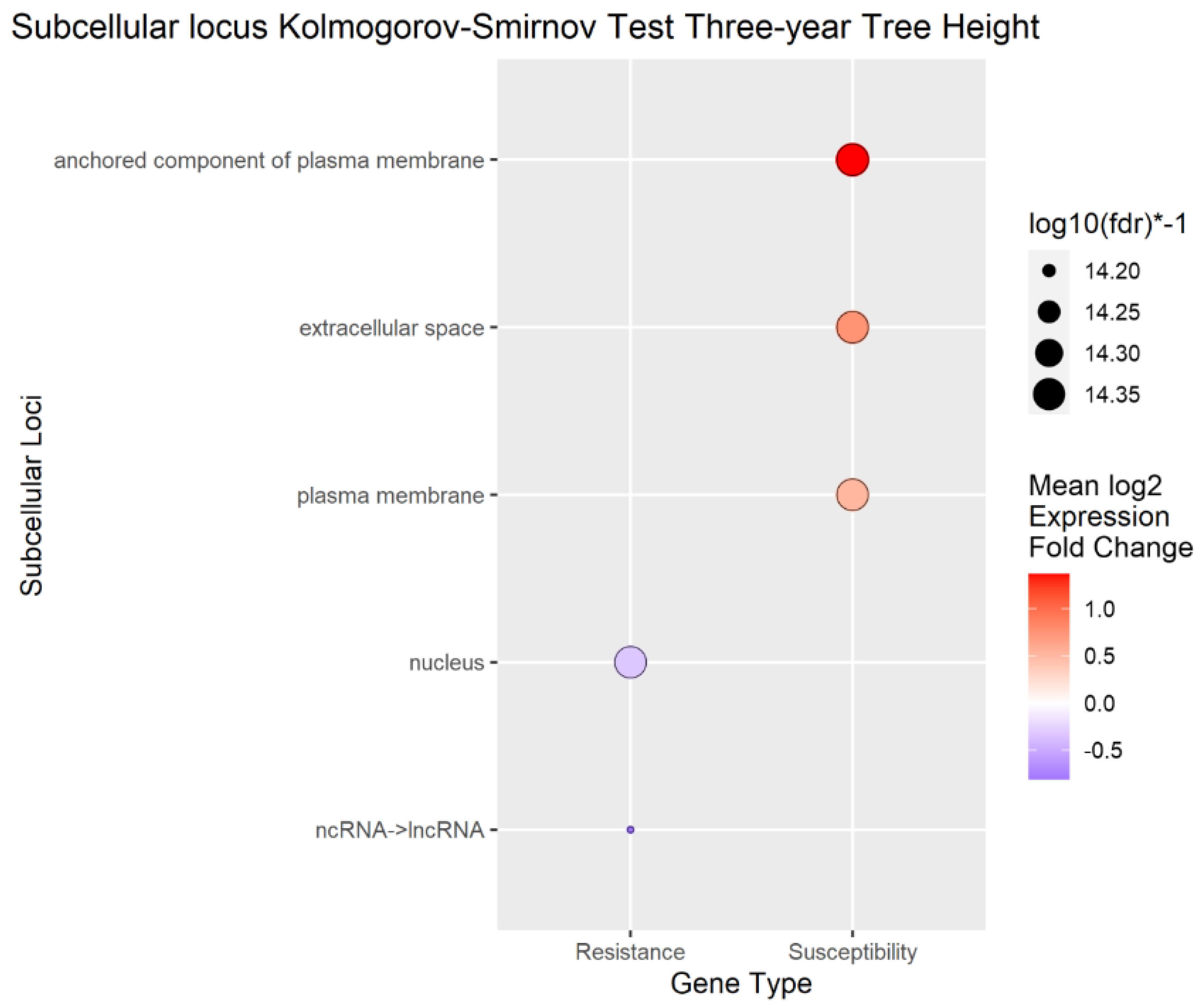
To better understand the overall localization of the DEGs within the cell, the computationally predicted subcellular localization of each set of DEGs was analyzed As with the biological process analysis, enrichment analysis using the Kolmogorov-Smirnov test was conducted on the subcellular localizations for the DEGs. Using a false discovery rate (FDR) threshold of 0.05 for all traits, the plasma membrane, anchored component of plasma membrane, and extracellular space were enriched in transcripts positively correlated to
A. tumefaciens,
Phytophthora spp., and vigor phenotypes (
Figure 6,
Figure 7 and
Figure 8). The cytoplasm was enriched only in the transcripts positively correlated to
A. tumefaciens, and the plasma membrane was enriched only in transcripts positively correlated to
Phytophthora. spp and vigor phenotypes (
Figure 6,
Figure 7 and
Figure 8). We observed no significant enrichment in subcellular loci for
P. vulnus phenotypic response. Alternatively, a theme of targeting the nucleus was observed for transcripts negatively correlated to each trait except for
P. vulnus phenotypic response (
Figure 6,
Figure 7 and
Figure 8). We included long non-coding RNAs in this analysis, and this class of genes was enriched with transcripts negatively correlated to each trait except for
P. vulnus phenotypic response. The full results of this analysis can be found in File S2.
QTL Region Analysis
QTLs for
A. tumefaciens and
Phytophthora spp. have been reported to peak between markers 31.01_Jm4D_26359154 and 31.01_Jm4D_26669075 for the breeding population our samples were derived from
19. We analyzed the DEGs within this region of the
J. microcarpa genome and found three transcripts negatively correlated to
A. tumefaciens phenotypic response and three transcripts negatively correlated to vigor. No DEGs from
Phytophthora spp. or
P. vulnus phenotypic responses were found in this region. Both small nuclear ribonucleoprotein SmD3b (NCBI ID 121260033) and pre-rRNA-processing protein TSR1 homolog (NCBI ID 121259960) were transcripts negatively correlated to
A. tumefaciens and vigor traits in this region of chromosome 4D (
Table 3). Probable acyl-activating enzyme 1, peroxisomal (NCBI ID 121259974), was a transcript negatively correlated uniquely to vigor, and dolichol-phosphate mannosyltransferase subunit 1 (NCBI ID 121260019) was a transcript negatively correlated uniquely to
A. tumefaciens (
Table 3).
3. Discussion
This study follows up on a previous QTL analysis, which showed that resistance to
A. tumefaciens,
Phytophthora cinammomi, and
Phytophthora pini mapped to a ~300 kb region on chromosome 4D of the
J. microcarpa genome
19. With a small subset of the hybrids from theQTL study, we regressed the expressed genes against
A. tumefaciens,
Phytophthora spp.,
P. vulnus, and tree height (vigor) phenotypes. Increased disease severity scores with
A. tumefaciens and
Phytophthora spp. and increasing tree height was significantly enriched in Gene Ontology terms mentioning cell wall and polysaccharide biogenesis and ethylene signaling (
Figure 3,
Figure 4 and
Figure 5). Subcellular target predictions for these transcripts suggested that they trafficked to cellular barriers (
Figure 6,
Figure 7 and
Figure 8). Conversely, reduced disease severity scores with
A. tumefaciens,
Phytophthora spp. and decreasing tree height were significantly enriched in Gene Ontology terms mentioning RNA splicing and processing, hormone, and defense responses, dependening on the trait (
Figure 3,
Figure 4 and
Figure 5). Subcellular target predictions for transcripts associated with reduced disease severity scores for
A. tumefaciens,
Phytophthora spp. and decreasing tree height suggested they trafficked to the nucleus. This result supported the biological process results, further suggesting their involvement in RNA splicing and processing (
Figure 6,
Figure 7 and
Figure 8). We found four unique DEGs associated with resistance to
A. tumefaciens and decreasing tree height within the QTL region reported in
19. Two of these genes, small nuclear ribonucleoprotein SmD3b and pre-rRNA-processing protein TSR1 homolog, were shared with resistance to
A. tumefaciens and decreasing tree height (
Table 3). Given that these genes are within the QTL region and are differentially expressed, they are likely candidates for the gene(s) causal for resistance to
A. tumefaciens and modulators of tree vigor. Moreover, if these are genes with SNPs associated with resistance or are being affected by said SNP, perhaps they are causing the global change in RNA splicing and processing that is associated with resistance as they are genes involved in such processes. We further hypothesize that RNA splicing may affect cell wall biogenesis, as we observed that the two processes are inversely related.
Uninfected Transcriptional Repertoires May Predict Susceptibility
One component from PCA of the gene counts correlated strongly with
A. tumefaciens gall size in this study (
Table 1), suggesting the transcriptional repertoire was critical for the host response. The correlation between tree height and PC1 was strong but with two fewer replicates than
A. tumefaciens or
Phytophthora spp. it did not meet the significance threshold (
Table 1,
Figure S1). Given this result, RNA-seq may be able to predict disease outcomes. The use of RNA-seq for the genomic prediction of traits in plants has been successful for carotenoid and seed oil biosynthesis in sweet corn and rapeseed, respectively
20,21. This result suggested that there is potential for deciphering the mechanisms underlying phenotypic phenomena and using the expression of candidate genes as biomarkers for genomic selection.
Low Differential Expression Signal in P. vulnus
While all other traits analyzed in this study showed a strong signal in the expression data, modeling against P. vulnus only resulted in a handful of DEGs. The top resistance gene was an E3 ubiquitin-protein ligase UPL1-like, and the top susceptibility gene was an endoplasmic reticulum-Golgi intermediate compartment protein 3-like. These genes and others should be further analyzed for fitness as candidate genes to be altered in genetic experiments to confirm their association with P. vulnus susceptibility. There could be many reasons for the low signal. Perhaps the P. vulnus phenotype is mediated only by a few genes. Also, P. vulnus susceptibility was phenotyped by counting the number of nematodes per gram of root and, thus, was not a direct measure of the host response. That is, the root lesions caused by P. vulnus were not measured, but the nematode population was. Also, nematode numbers are notoriously variable and thus offer relatively low precision. The phenotyping for all other traits in this study were direct measures of the host, such as percent crown and root rot due to Phytophthora spp., size of gall caused by A. tumefaciens, or tree height. Given that we analyzed biochemicals (mRNAs) of the host, it is sensible how direct measures of the host phenotypes would garner greater signal to mRNAs than nematode counts, which may not correlate well with the root lesions caused by P. vulnus.
J. regia May be a Source of Vigor and Susceptibility
The use of a combined genome reference in our differential expression analysis revealed the expression of significantly more transcripts positively correlated with each susceptibility and vigor trait from the
J. regia haplotype than from the
J. microcarpa haplotype (
Figure 2). The
P. vulnus phenotypic response was not significant in this analysis. Conversely, the
J. microcarpa haplotype expressed significantly more transcripts negatively correlated with each trait. This seems consistent with the fact that, initially, own-rooted plantings of English walnuts (
J. regia) were soon found to be susceptible to
A. tumefaciens,
Phytophthora spp., and
P. vulnus. To remedy this problem, rootstocks sourced from
J. hindsii were used
10. Eventually, hybrids of
J. hindsii x
J. regia and
J. microcarpa x
J. regia were introduced for improved disease tolerance and, in the former, at least, vigor.. Therefore,
J. regia seems to be a source of disease susceptibility in rootstocks, which is consistent with our RNA-seq analysis. On a different note, significantly more genes associated with high vigor were expressed from the
J. regia haplotype than from the
J. microcarpa haplotype (
Figure 2), indicating that it may be contributing more to the height of the hybrids than
J. microcarpa. High-vigor trees seem to be preferred in the California walnut industry, or at least a minimum amount of vigor is required for realistic productivity. Perhaps there is a “middle ground” between the contribution of each haplotype to gene expression as it relates to agronomic desirability.
Defense Response and Hormones Involved
One potential pathogen resistance mechanism could be the upregulation of genes associated with defense responses, such as the family of Pathogenesis-Related (PR), Nonexpresser of PR genes (NPR) genes, or any plant hormone responses. Genoontology.org defines “defense response” as “Reactions, triggered in response to the presence of a foreign body or the occurrence of an injury, which result in restriction of damage to the organism attacked or prevention/recovery from the infection caused by the attack.” A complete list of genes associated with this term when filtered for “
Juglans regia” can be found at
http://amigo.geneontology.org/amigo/term/GO:0006952 The walnut expression data suggested that the resistance mechanism may involve defense-related genes or plant hormones. We observed that “Defense response to bacterium” was significantly associated with resistance and not susceptibility in
A. tumefaciens (
Figure 3). This result is unsurprising given that
A. tumefaciens is a bacterium. Oddly, no terms mentioning defense response were significantly enriched in
Phytophthora spp. or
P. vulnus. Interestingly, jasmonate signaling was associated exclusively with resistance to
A. tumefaciens (
Figure 3). A recent review on jasmonic acid in plants reports protective effects of both endogenous and exogenous jasmonic acid against necrotrophic pathogens
22. Therefore, it is possible that jasmonic acid signaling is contributing to resistance to
A. tumefaciens in these hybrids. Moreover, several terms mentioning ethylene were enriched with transcripts positively associated with
A. tumefaciens and
Phytophthora spp. phenotypic responses, and vigor, suggesting roles for ethylene in susceptibility to these pathogens and increased vigor (
Figure 3,
Figure 4 and
Figure 5). The literature of ethylene’s effect on plant growth seems mixed as plants can respond with increased or decreased growth depending on the species and concentrations analyzed
23. Similar trends have been noted for ethylene’s role in defense, where treatment with ethylene or its inhibitor, or mutants with impaired ethylene signaling elicited conflicting results depending on the plant and pathogen combinations
24,25. Our results also suggested abscisic acid is involved in resistance to
A. tumefaciens and decreased vigor (
Figure 3). Like ethylene, abscisic acid’s effects on plant-pathogen interactions are mixed
26. Of course, abscisic acid’s role in plant growth is clear; for example, it regulates stomatal closure and thus limits growth at higher concentrations by limiting carbon acquisition
27. This matches well with the observations that abscisic acid-related genes were highly expressed in low-vigor hybrids, potentially resulting in greater stomatal closure and less growth (
Figure 5).
Potential Link Between Cell Wall Biogenesis, Pathogenesis, and Vigor
Cell wall biogenesis transcripts were linked to increased disease susceptibility and plant vigor in linear modeling and enrichment analysis of the transcriptional repertoires. Cell wall biogenesis is defined as “A cellular process that results in the biosynthesis of constituent macromolecules, assembly, and arrangement of constituent parts of a cell wall. It includes biosynthesis of constituent macromolecules, such as proteins and polysaccharides, and those macromolecular modifications that are involved in synthesis or assembly of the cellular component. A cell wall is the rigid or semi-rigid envelope lying outside the cell membrane of plant, fungal and most prokaryotic cells, maintaining their shape and protecting them from osmotic lysis (
http://amigo.geneontology.org/amigo/term/GO:0042546).”
A recent review on plant cell walls stated, “Cell wall composition is one of the most important issues affecting cell wall structure and function.”28. Plant cell walls are perhaps the most critical component of plant immunity and often the first obstacle a pathogen must overcome. Host manipulation, carbon acquisition, effector delivery, and defense response suppression often require breaching the cell wall29. Cellulose, hemicelluloses like xyloglucan and xylan, pectins, and cell wall proteins, in order of abundance, represent the predominant constituents of the cell wall30,31. Highlighting the importance of these polysaccharides are the myriad of cell wall degrading enzymes that many pathogens use to breach this barrier. These enzymes are key virulence factors and include such enzymes as; cellulase, xylanase, pectin methylesterase, polygalacturonase, and pectate lyase32.
In terms of vigor, every plant cell in a growing tissue must expand in volume to achieve this growth. For example, meristematic xylem cells can expand >30,000 times their original size, and superficial cotton hair cells elongate 1,000 times their original size before maturity33. The cell wall must undergo biogenesis in order to cope with such expansion. Plant cell walls determine the functional properties, shape, communication, and overall expansion and growth of plant cells34.
Our subcellular localization results support the hypothesis that cell wall biogenesis is involved in the pathogenesis of
A. tumefaciens and
Phytophthora spp. and vigor in walnuts. The transcripts positively associated with
A. tumefaciens and
Phytophthora spp. phenotypic responses, and vigor, targeted significantly more cellular barriers, such as the plasma membrane, its anchored components, and the extracellular space, and likely the cell wall, than the with transcripts negatively associated with
A. tumefaciens and
Phytophthora spp. phenotypic responses, and vigor (
Figure 6,
Figure 7 and
Figure 8). In other words, significant increases in the expression of genes targeting the cellular barriers occurred as the plants became taller and more susceptible to these pathogens. In the biological processes and subcellular loci associated with the up genes, increased expression of genes associated with cell wall biogenesis (e.g., cellulose synthase, xyloglucan endotransglucosylase, arabinogalactan proteins) resulted in increased vigor and susceptibility to
A. tumefaciens and
Phytophthora spp. (
Figure 3,
Figure 4 and
Figure 5, File S1 & S2). The predicted functions, biological processes, and subcellular localizations of these up genes strongly suggest that increased activity of cell wall biosynthetic enzymes contributes to vigor and pathogen susceptibility in these hybrids.
Cell Wall Biosynthetic Genes are Known to Affect Pathogenesis and Plant Growth
Tree height,
A. tumefaciens phenotypic response, and
Phytophthora spp. phenotypic response are strongly correlated in our phenotypic data (
Table 4). The correlation between these traits is likely the reason each trait’s set of differentially expressed genes includes some form of cell wall biogenesis. In other words, tree height,
A. tumefaciens phenotypic response, and
Phytophthora spp. phenotypic response may be linked through a common mechanism relating to the expression of cell wall biogenetic genes. Alterations in cell wall biogenesis can affect a plant’s susceptibility to pathogens. For example, in
Arabidopsis thaliana, impairment in five genes necessary for cellulose synthesis in primary cell walls resulted in enhanced resistance to
Fusarium oxysporum, a root-infecting hemibiotrophic fungal pathogen
35. These genes included
cesa3-3,
cobra-6,
ctl1-2, kor
1-4, and
prc1-1.
Cesa3-3 and prc1-1 are cellulose synthases
36,37.
Ctl1-2 is an apoplastic chitinase-like protein regulating cellulose biosynthesis
38.
Cobra-6 affects cellulose biosynthesis; its mechanism is nebulous but may interact with the orientation and synthesis of cellulose microfibrils
39.
Kor1-4 is a plasma membrane-bound endo-1,4, -β-glucanase
40. A significant phenotype of these mutants is cellulose deficiency, which is likely involved in enhanced resistance to
F. oxysporum. While we did not measure cellulose accumulation in these hybrids, the transcriptomic data indicated positive relationships between the expression of cellulose biosynthetic enzymes and susceptibility to all diseases (
Figure 3,
Figure 4 and
Figure 5, File S1). Moreover, the transcriptional changes in the
Fusarium-resistant
A. thaliana mutants mirrored that of our resistant hybrids. Specifically, we observed reduced expression of many
cesas (cellulose synthases) and
cobras (protein COBRA) in the resistant walnut hybrids (Files S1). Downregulation of cell wall biosynthetic transcripts was observed in RNAs of wild-type
A. thaliana challenged with
F. oxysporum vs. unchallenged
35. Interestingly, the impairment of cellulose synthases in
A. thaliana also impairs growth
35,41. In fact, mutations in
cobra-6,
ctl1-2, and
kor1-4 all resulted in reduced plant growth
38,39,40. Our observations of transcripts associated with vigor agree very well with this, suggesting increasing expression of cell wall biosynthetic genes results in increasing vigor in walnuts (File S1). Moreover, we could also hypothesize a potential link between walnut vigor and disease resistance. This could be tested by knocking out a cell wall biosynthetic gene or inhibiting its protein product; resulting in reduced growth and increased disease resistance, would contribute to the integrity of this theory.
While it is functional to understand how reduced cellulose biosynthesis affects plant growth, the mechanism of its role in disease resistance is less clear. Perhaps the recomposition of cell wall polysaccharides creates a matrix that is more resistant to pathogenic cell wall degrading enzymes. For example, the proportion of pectin or hemicellulose could increase relative to that of cellulose, changing the characteristics of the cell wall considerably and possibly making a pathogenic cellulase less effective.
The arabinogalactan proteins are another gene family significant for cell wall biogenesis, growth, and potentially pathogen defense31. In Arabidopsis, downregulation of the arabinogalactan protein gene AtAGP17 resulted in decreased efficiency of Agrobacterium transformation42. In microscopic investigation of the Arabidopsis AtAGP17 mutant, A. tumefaciens could not bind to its root surface compared to wild-type A. thaliana roots. Moreover, nonspecific inhibition of arabinogalactan proteins with the Yariv reagent also reduced A. tumefaciens transformation of Arabidopsis roots42. Like the other cell wall biogenesis-related genes mentioned above, arabinogalactan protein knockouts tend to have a reduced growth or growth defect phenotype, and the growth of plants on Yariv reagent-containing media is also reduced43. Similarly, in our analysis, two arabinogalactan proteins were lowly expressed in resistant and low-vigor hybrids (File S1). Perhaps the expression of these genes is contributing to our observed phenotypes.
4. Methods
Sample Collection, RNA Isolation, and Sequencing
Hybrids of J. microcarpa x J. regia varying in resistance to A. tumefaciens, Phytophthora spp., P. vulnus and tree height at three years of age were used in this experiment. Plants of the specific genotypes MS1-36, MS1-41, MS1-56, MS1-122, STJM-4, 29JM-11, and JMS-12 were grown in pots in a greenhouse for six months. Healthy white actively growing root samples were collected from six plants of each genotype. A total of 42 root samples were chosen for RNA extraction. All samples were collected in the lab and flash-frozen in liquid nitrogen immediately after harvesting. Collected samples were stored at -80 ℃ before the analysis. For transcriptome analysis by deep sequencing, tissue samples were ground to a fine powder in liquid nitrogen. Total RNA was extracted using the MagMAX™ mirVana™ total RNA isolation kit (Thermo Fisher Scientific, USA). Library preparation and transcriptome analysis were carried out by Seqmatic LLC (Fremont, CA). At Seqmatic, RNA library preparation was done using Illumina Stranded mRNA library preparation kit, and transcriptome analysis was carried out using NextSeq High Output Run (Paired-End Read 2x150bp). The raw .fastq files were deposited in SRA under BioProject PRJNA726720.
Phenotypic Analysis
The phenotyping of these hybrids for resistance to A. tumefaciens and Phytophthora spp. has been described in detail19. The pathogen resistance phenotype used for A. tumefaciens was a score on a scale of one (no gall) to four (complete girdling of the plant from gall). The pathogen resistance phenotype used for Phytophthora spp. was a visually assessed percentage of crown or root rotted after pathogen inoculation of P. cinnamomi and P. pini. The average percentages of crown and root rot for P. cinnamomi and P. pini was entered into the analysis and were then referred to as “Phytophthora spp.”
To phenotype for P. vulnus, at least eight clonal saplings of experimental Juglans genotypes and commercial comparatives RX1 and VX21111,45 were planted in field plots in March 2014. The experimental plants were propagated from tissue cultures and developed into saplings in greenhouse culture. At the Kearney Agricultural Research and Extension Center, saplings were planted in their genotype group in rows of 3.35 m distance at 1.65 m spacing within the row. About one month after planting, every tree was concomitantly inoculated with ~1,000 vermiform P. vulnus and second-stage juveniles (J2) of Meloidogyne incognita by dispensing infested field soil from underneath infected perennial crops at the base of the tree. Selected trees of the genotype groups were chosen for root collections for nematode evaluations. A 20-25-cm deep trench was dug next to the tree to collect young roots of the respective tree genotype avoiding suberized roots. Kept cool in plastic bags until processing within 48 hours of collection, the roots were chopped into 1.2-cm pieces, and 20-g portions placed on top of Baermann funnels. In a mist chamber apparatus, the roots were intermittently sprayed with water at 27 °C for five days. After this, the extracts were collected, and P. vulnus were identified and counted12. Nematode numbers were recorded on a per 1-gram basis. In each dormant season, the height of each of the trees in these nematode field screens was measured from the ground level perpendicularly to the maximum height of the trees with an extended ruler. Thus, the tree height was measured under nematode infested conditions.
Differential Expression Analysis
The gene counts were analyzed using the R programming language for statistical computing and graphics version 4.1.2 in R studio
47,48. For PCA, all gene counts were normalized by the counts per million mapped reads (CPM) method, and any gene with maximum expression under 30 CPM was excluded from the downstream analysis. Principal Component Analysis (PCA) was conducted on the filtered expression matrix and the Principal Component (PC) scores were averaged for each hybrid and used for regression against each trait (
Figure 1). A linear model was fit to each gene in the filtered expression matrix for
A. tumefaciens (crown gall score),
P. cinnamomi and
P. pini (average percent root and crown rot),
P. vulnus (root-lesion nematode count per gram of root), and tree height at three years of age. To help reduce type I error, limma-voom and empirical Bayes smoothing of standard errors were employed. Moreover, to account for pseudoreplication, the biological replicates within each hybrid were treated as random effects within the model. After modeling, genes were considered differentially expressed if their false discovery rate (FDR) was less than or equal to 0.05.
Expression by Haplotype
We used Fisher’s exact test in R to analyze the differences in the number of DEGs between the
J. microcarpa and
J. regia haplotypes. Unique lists of DEGs positively and negatively correlated with each trait were extracted from the DEA and used to make a two-by-two contingency matrix. This matrix was used to test the alternative hypothesis that the odds ratio of up genes to down genes between
J. microcarpa and
J. regia was not equal to one. An example of the contingency matrix is in
Figure S2.
Gene Ontology Analysis
Unique lists of DEGs with their fold changes were extracted from the DEA and used as inputs for the Kolmogorov-Smirnov test to test for coordinated shifts in gene expression by biological process as is used in the PANTHER enrichment Test49,50,51,52. Juglans regia was used as the species from which to derive the annotation set for this analysis. Juglans regia genes were inferred from Juglans microcarpa genes using BLAST (Basic Local Alignment Search Tool). GO biological process complete was used as the annotation data set, and only terms with FDR less than 0.05 were considered enriched. In figures three, four, and five, the term labels are biased and limited to those terms mentioning “RNA,” “cell wall,” “polysaccharide,” “cellulose,” “glucan,” “defense,” “jasmonic acid,” “abscisic acid,” “salicylic acid,” or “ethylene” and FDR less than or equal to 0.05. This was done for two reasons (i) the results are so large that visualizing and reporting all of them is unreasonable. Therefore, we looked for the biggest patterns in the results and pursued those. (ii) Make priori hypotheses about certain biological processes of interest and report on those results.
Subcellular Localization Analysis
R Packages and Code Availability
The R packages Limma
54, edgeR
55, base
47, data.table
56, tidytable
57, dplyr
58, stats
47, ggpubr
59, ggplot2
60, graphics
47, sjPlot
61, tibble
62, utils
47, tidyr
63, gridExtra
64, OmicsAnalyst
65, tidytext
66, stringr
67, knitr
68, and rmarkdown
69 were used to facilitate all analyses done in R
, . All code used for the differential expression analysis, gene ontology analysis, expression by haplotype analysis, and subcellular localization analysis can be found at
https://github.com/hsaxe/SCRI_ROOT_R.
5. Conclusion
This study provided a starting point for functional genomic insights on potential host mechanisms of resistance and susceptibility to A. tumefaciens, Phytophthora. spp, and vigor in J. microcarpa x J. regia hybrids. Principal component analysis identified some transcriptional repertoires associated with each trait, highlighting the ability of RNA-seq to capture genetic variation related to the traits of interest. This study focused on cell wall biogenesis and cellular barriers. However, other biological processes and subcellular loci were significant in pathogenesis or vigor, such as jasmonic acid, ethylene, abscisic acid, the nucleus, and long non-coding RNAs. Moreover, we point out quality candidate genes that are likely to be causal in resistance to A. tumefaciens as they are differentially expressed as resistance genes and within the QTL region for resistance reported in a previous QTL study19. Current breeding efforts seem well suited to focus on black walnut species because J. regia may be a source of susceptibility to A. tumefaciens and Phytophthora spp. However, J. microcarpa may be a source of low vigor. Therefore, careful consideration of the potential tradeoff for desired agronomic traits is suggested when determining the usefulness of these hybrids in future breeding. Aside from P. vulnus, the transcripts positively correlated with all traits were enriched in the GO terms cell wall organization/biogenesis. Moreover, these same genes targeted the cellular barriers significantly more than the transcripts negatively correlated with these traits. These results suggested that increased activity in gene expression related to cell wall biogenesis may be a susceptibility factor in these diseases and a factor promoting vigor in these hybrids. Therefore, modulating cell wall biogenesis may modulate pathogenesis and vigor in walnuts. This new hypothesis must be validated by knocking out genes involved in cell wall biogenesis, such as the cellulose synthases or arabinogalactan proteins. Alternatively, these genes could be inhibited via isoxaben for the cellulose synthases or the yariv reagent for the arabinogalactan proteins42,44.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org.
Contributions
A.W. led efforts to acquire extramural funding. C.A.L, P.J.B., provided the plant material. D.A.K., G.T.B and A.W., provided the phenotypic data. A.M.D and C.A.L developed the study design. S.W., generated the RNAseq data. H.S., B.B., analyzed the RNAseq data and performed functional analysis. H.S. and A.M.D. generated the first draft of the paper. All authors read, edited and approved the final draft.
Acknowledgments
This work was supported in part by USDA NIFA-SCRI grant no. 2018-51181-28437 and grants from the California Walnut Board. We also thank: T. Buzo, Z.T.Z Maung and M. McKenry for their invaluable help in the testing and phenotyping of the nematode data; N.J. Ott and H. Forbes for help in phenotyping resistance to Phytophthora spp.; and A. McClean for help in phenotyping resistance to A. tumefaciens.
Conflicts of Interests
The authors state no conflict of interest.
References
- CDFA. California Agricultural Statistics Review 2020-2021. https://www.nass.usda.gov/Statistics_by_State/California/index.php (2021). last accessed July 24, 2023.
- Grant, J.A., Caprile, J.L., Doll, D.A., and Murdock, J. Sample costs to establish an orchard and produce walnuts, San Joaquin Valley - North (Cost Studies UC Davis). https://coststudyfiles.ucdavis.edu/uploads/cs_public/1d/05/1d05d59d-706e-4937-8e05-5563b7468e23/2017-walnutssjvn-final_draft.pdf (2017). last accessed: July 24, 2023.
- Olson, B., Walton, J., Laminen, B., and Sam, M. The Effect of Crown Gall on Tree Growth and Productivity (Walnut Research UC Davis) (2002).
- Epstein, L. et al. Crown Gall Can Spread Between Walnut Trees in Nurseries and Reduce Future Yields. Hilgardia 62, 111–115 (2008). [CrossRef]
- Mircetich, S.M. and M.E. Matheron. Phytophthora root and crown rot of walnut trees. Phytopathology 73:1481–1488 (1983).
- Matheron, M.E. and S.M. Mircetich. Pathogenicity and relative virulence of Phytophthora spp. from walnut and other plants to rootstocks of English walnut trees. Phytopathology 75:977–981 (1985). [CrossRef]
- Browne, G.T. et al. Resistance to species of Phytophthora identified among clones of Juglans microcarpa × J. regia. HortScience 50, 1136–1142 (2015). [CrossRef]
- McKenry, M.V. MEthyl bromide altenatives and improvements - year five. California Walnut Board - Annual Walnut Research Reports, 399-403 (1997).
- Westphal, A. et al. Putting Phenotypic and Genotypic Tools to Work for Improving Walnut Rootstocks (2020).
- Preece, J.E., and McGranahan, G. Luther burbank’s contributions to walnuts. HortScience 50, 201–204 (2015). [CrossRef]
- McGranahan, G., Browne, G., Leslie, C., Hackett, W., and McKenna, J. WALNUT ROOTSTOCK “RX1” (2010a).
- Buzo, T., Mckenna, J., Kaku, S., Anwar, S.A., and Mckenry, M. V. VX211, A vigorous new walnut hybrid clone with nematode tolerance and a useful resistance mechanism. J. Nematol. 41, 211–216 (2009).
- Kluepfel, D. A. et al. Evaluation of wild walnut Juglans spp. for resistance to crown gall disease. Phytopathology 101, S92–S92 (2011).
- Escobar, M.A., Civerolo, E.L., Summerfelt, K.R., and Dandekar, A.M. RNAi-mediated oncogene silencing confers resistance to crown gall tumorigenesis. Proc. Nat. Acad. Sci. U.S.A. 98, 13437-13442 (2001). [CrossRef]
- Escobar, M.A., Leslie, C.A., McGranahan, G.H., and Dandekar, A.M. Silencing crown gall disease in walnut (Juglans regia L.). Plant Sci. 163(3), 591-597 (2002). [CrossRef]
- Walawage, S.L. et al. Stacking resistance to crown gall and nematodes in walnut rootstocks. BMC Genomics 14, 1–13 (2013). [CrossRef]
- Marrano, A. et al. High-quality chromosome-scale assembly of the walnut (Juglans regia L.) reference genome. Gigascience 9, 1–16 (2020). [CrossRef]
- Zhu, T. et al. Sequencing a Juglans regia × J. microcarpa hybrid yields high-quality genome assemblies of parental species. Hortic. Res. 6, 55 (2019). [CrossRef]
- Ramasamy, R.K. et al. Co-located quantitative trait loci mediate resistance to Agrobacterium tumefaciens, Phytophthora cinnamomi, and P. pini in Juglans microcarpa × J. regia hybrids. Hortic. Res. 8 (2021). [CrossRef]
- Hershberger, J. et al. The Plant Genome - 2022 - Hershberger - Transcriptome-wide association and prediction for carotenoids and tocochromanols in.pdf. Plant Genome 15, 2 (2022).
- Tang, S. et al. Genome- and transcriptome-wide association studies provide insights into the genetic basis of natural variation of seed oil content in Brassica napus. Mol. Plant 14, 470–487 (2021). [CrossRef]
- Wang, Y., Mostafa, S., Zeng, W., and Jin, B. Function and Mechanism of Jasmonic Acid in Plant Responses to Abiotic and Biotic Stresses. International Journal of Molecular Sciences 22, 8568 (2021). [CrossRef]
- Iqbal, N. et al. Ethylene Role in Plant Growth, Development and Senescence: Interaction with Other Phytohormones. Frontiers in plant science, 8, 475. https://doi.org/10.3389/fpls.2017.00475 (2017). [CrossRef]
- Thomma, B. P., Eggermont, K., Tierens, K. F., & Broekaert, W. F. Requirement of functional ethylene-insensitive 2 gene for efficient resistance of Arabidopsis to infection by Botrytis cinerea. Plant physiology, 121(4), 1093–1102. https://doi.org/10.1104/pp.121.4.1093 (1999). [CrossRef]
- van Loon, L.C., Geraats, B.P.J., Linthorst, H.J.M. Ethylene as a modulator of disease resistance in plants. Trends in Plant Science, Volume 11, Issue 4, April 2006, Pages 184-191 (2006). [CrossRef]
- Lievens, L., Pollier, J., Goossens, A., Beyaert, R., & Staal, J. (2017). Abscisic Acid as Pathogen Effector and Immune Regulator. Frontiers in plant science, 8, 587. [CrossRef]
- Kavi Kishor, P. B., Tiozon, R. N., Jr, Fernie, A. R., & Sreenivasulu, N. Abscisic acid and its role in the modulation of plant growth, development, and yield stability. Trends in plant science, 27(12), 1283–1295. [CrossRef]
- Zhang, B., Gao, Y., Zhang, L., and Zhou, Y. The plant cell wall: Biosynthesis, construction, and functions. J. Integr. Plant Biol. 63, 251–272 (2021). [CrossRef]
- Nühse, T.S. Cell wall integrity signaling and innate immunity in plants. Front. Plant Sci. 3, 1–5 (2012). [CrossRef]
- Lampugnani, E.R., Khan, G.A., Somssich, M., and Persson, S. Building a plant cell wall at a glance. J. Cell Sci. 131 (2018). [CrossRef]
- Silva, J., Ferraz, R., Dupree, P., Showalter, A.M., and Coimbra, S. Three Decades of Advances in Arabinogalactan-Protein Biosynthesis. Front. Plant Sci. 11 (2020). [CrossRef]
- Malinovsky, F.G., Fangel, J.U., and Willats, W.G.T. The role of the cell wall in plant immunity. Front. Plant Sci. 5, 1–12 (2014). [CrossRef]
- Cosgrove, D. Growth of the plant cell wall. Nat Rev Mol Cell Biol 6, 850–861 https://doi.org/10.1038/nrm1746 (2005). [CrossRef]
- Maleki, S.S., Mohammadi, K., Ji, K. Characterization of Cellulose Synthesis in Plant Cells, The Scientific World Journal, vol. 2016, Article ID 8641373, 8 pages.
- Menna, A. et al. A primary cell wall cellulose-dependent defense mechanism against vascular pathogens revealed by time-resolved dual transcriptomics. 1–20 (2021). [CrossRef]
- Fagard, M. et al. Procuste1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of arabidopsis. Plant Cell 12, 2409–2423 (2000). [CrossRef]
- Hernández-Blanco, C. et al. Impairment of cellulose synthases required for Arabidopsis secondary cell wall formation enhances disease resistance. Plant Cell 19, 890–903 (2007). [CrossRef]
- Sánchez-Rodríguez, C. et al. CHITINASE-LIKE1/POM-POM1 and its homolog CTL2 are glucan-interacting proteins important for cellulose biosynthesis in Arabidopsis. Plant Cell 24, 589–607 (2012). [CrossRef]
- Roudier, F. et al. COBRA, an Arabidopsis extracellular glycosyl-phosphatidyl inositol-anchored protein, specifically controls highly anisotropic expansion through its involvement in cellulose microfibril orientation. Plant Cell 17, 1749–1763 (2005). [CrossRef]
- Nicol, F.et al. A plasma membrane-bound putative endo-1,4-β-D-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO J. 17, 5563–5576 (1998). [CrossRef]
- Hu, H. et al. Cellulose Synthase Mutants Distinctively Affect Cell Growth and Cell Wall Integrity for Plant Biomass Production in Arabidopsis, Plant and Cell Physiology, Volume 59, Issue 6, June, Pages 1144–1157. [CrossRef]
- Gaspar, Y.M. et al. Characterization of the arabidopsis lysine-rich arabinogalactan-protein AtAGP17 mutant (rat1) that results in a decreased efficiency of agrobacterium transformation. Plant Physiol. 135, 2162–2171 (2004). [CrossRef]
- Hromadová, D., Soukup, A., & Tylová, E. Arabinogalactan Proteins in Plant Roots - An Update on Possible Functions. Frontiers in plant science, 12, 674010. https://doi.org/10.3389/fpls.2021.674010 (2021). [CrossRef]
- Desprez, T. et al. Resistance against herbicide isoxaben and cellulose deficiency caused by distinct mutations in same cellulose synthase isoform CESA6. Plant physiology, 128(2), 482–490. [CrossRef]
- McGranahan, G. et al. WALNUT ROOTSTOCK “VX211”. (2010b).
- Dobin, A. et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. https://www.r-project.org/ last accessed: July 24, 2023 (2022).
- RStudio Team. RStudio: Integrated Development for R. https://posit.co/download/rstudio-desktop/ last accessed: July 24, 2023. (2022).
- Ashburner, M. et al. Gene Ontology: tool for the unification of biology. Nat. Genet. 25, 25–29 (2000). [CrossRef]
- Carbon, S. et al. The Gene Ontology resource: Enriching a GOld mine. Nucleic Acids Res. 49, D325–D334 (2021). [CrossRef]
- Mi, H., Muruganujan, A., Ebert, D., Huang, X., and Thomas, P.D. PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 47, D419–D426. (2019) (a). [CrossRef]
- Mi, H., et al. Protocol Update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0) Nat Protoc 14, 703–721. (2019) (b). [CrossRef]
- Savojardo, Castrense et al. BUSCA: an integrative web server to predict subcellular localization of proteins. Nucleic acids research vol. 46,W1: W459-W466. [CrossRef]
- Ritchie, M.E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015). [CrossRef]
- Robinson MD, McCarthy DJ, Smyth GK. “edgeR: a Bioconductor package for differential expression analysis of digital gene expression data.” Bioinformatics, 26(1), 139-140. (2010). [CrossRef]
- Dowle, M., and Srinivasan, A. data.table: Extension of `data.frame`. https://cran.r-project.org/package=data.table last accessed: July 24, 2023 (2022).
- Fairbanks, M. tidytable: Tidy Interface to data.table. https://github.com/markfairbanks/tidytable last accessed: July 24, 2023 (2022).
- Wickham, H., François, R., Henry, L., and Müller, K. dplyr: A Grammar of Data Manipulation. https://cran.r-project.org/package=dplyr last accessed: July 24, 2023 (2022a).
- Kassambara, A. ggpubr: ggplot2 Based Publication Ready Plots. https://rpkgs.datanovia.com/ggpubr/ last accessed: July 24, 2023 (2022).
- Wickham, H. et al. ggplot2: Create Elegant Data Visualisations Using the Grammar of Graphics. https://cran.r-project.org/package=ggplot2 last accessed: July 24, 2023 (2022b).
- Lüdecke, D. sjPlot: Data Visualization for Statistics in Social Science. https://strengejacke.github.io/sjPlot/ last accessed: July 24, 2023 (2022).
- Müller, K., and Wickham, H. tibble: Simple Data Frames. https://cran.r-project.org/package=tibble last accessed: July 24, 2023 (2022).
- Wickham, H., and Girlich, M. tidyr: Tidy Messy Data. https://cran.r-project.org/package=tidyr last accessed: July 24, 2023 (2022).
- Auguie, B. gridExtra: Miscellaneous Functions for “Grid” Graphics. https://cran.r-project.org/package=gridExtra last accessed: July 24, 2023 (2017).
- Saxe, H. OmicsAnalyst: Functions to facilitate the statistical analysis of omics data. https://github.com/hsaxe/Omics-Analyst last accessed: July 24, 2023 (2023).
- Robinson, D., and Silge, J. tidytext: Text Mining using dplyr, ggplot2, and Other Tidy Tools. https://github.com/juliasilge/tidytext last accessed: July 24, 2023 (2022).
- Wickham, H. stringr: Simple, Consistent Wrappers for Common String Operations. https://cran.r-project.org/package=stringr last accessed: July 24, 2023 (2022).
- Xie, Y. knitr: A General-Purpose Package for Dynamic Report Generation in R. https://yihui.org/knitr/ last accessed: July 24, 2023 (2022).
- Allaire, J.J. et al. rmarkdown: Dynamic Documents for R. https://www.r-project.org/ last accessed: July 24, 2023 (2022).
Figure 1.
A) Venn diagram of differentially expressed genes positively correlated with A. tumefaciens (CG), Phytophthora. spp (PHY), P. vulnus (NEM), and tree height at three years (height) phenotype scores. B) Venn diagram of differentially expressed genes negatively correlated with A. tumefaciens (CG), Phytophthora. spp (PHY), P. vulnus (NEM), and tree height at three years (height) phenotype scores.
Figure 1.
A) Venn diagram of differentially expressed genes positively correlated with A. tumefaciens (CG), Phytophthora. spp (PHY), P. vulnus (NEM), and tree height at three years (height) phenotype scores. B) Venn diagram of differentially expressed genes negatively correlated with A. tumefaciens (CG), Phytophthora. spp (PHY), P. vulnus (NEM), and tree height at three years (height) phenotype scores.
Figure 2.
Mosaic plot representing proportions of differentially expressed genes (DEGs) from each trait colored by the haplotype the genes mapped to. Each plot is labeled with the pathogen, Fisher’s exact p-value, and Fisher’s exact odds ratio. The odds ratio represents the ratio of the odds of the J. regia haplotype expressing a gene positively correlated to the trait compared to the odds of the J. microcarpa haplotype expressing a gene negatively correlated to the trait.
Figure 2.
Mosaic plot representing proportions of differentially expressed genes (DEGs) from each trait colored by the haplotype the genes mapped to. Each plot is labeled with the pathogen, Fisher’s exact p-value, and Fisher’s exact odds ratio. The odds ratio represents the ratio of the odds of the J. regia haplotype expressing a gene positively correlated to the trait compared to the odds of the J. microcarpa haplotype expressing a gene negatively correlated to the trait.
Figure 3.
Enrichment Analysis of Gene Ontology terms mapped to differentially expressed genes (DEGs) for A. tumefaciens phenotypic response. Each point represents a biological process term. See methods for term labeling.
Figure 3.
Enrichment Analysis of Gene Ontology terms mapped to differentially expressed genes (DEGs) for A. tumefaciens phenotypic response. Each point represents a biological process term. See methods for term labeling.
Figure 4.
Enrichment Analysis of Gene Ontology terms mapped to differentially expressed genes (DEGs) for Phytophthora. spp phenotypic response. Each point represents a biological process term. See methods for term labeling.
Figure 4.
Enrichment Analysis of Gene Ontology terms mapped to differentially expressed genes (DEGs) for Phytophthora. spp phenotypic response. Each point represents a biological process term. See methods for term labeling.
Figure 5.
Enrichment Analysis of Gene Ontology terms mapped to differentially expressed genes (DEGs) for tree height. Each point represents a biological process term. See methods for term labeling.
Figure 5.
Enrichment Analysis of Gene Ontology terms mapped to differentially expressed genes (DEGs) for tree height. Each point represents a biological process term. See methods for term labeling.
Figure 6.
Enrichment Analysis of BUSCA subcellular loci mapped to differentially expressed genes (DEGs) for A. tumefaciens phenotypic response.
Figure 6.
Enrichment Analysis of BUSCA subcellular loci mapped to differentially expressed genes (DEGs) for A. tumefaciens phenotypic response.
Figure 7.
Enrichment Analysis of BUSCA subcellular loci mapped to differentially expressed genes (DEGs) Phytophthora .spp phenotypic response.
Figure 7.
Enrichment Analysis of BUSCA subcellular loci mapped to differentially expressed genes (DEGs) Phytophthora .spp phenotypic response.
Figure 8.
Enrichment Analysis of BUSCA subcellular loci mapped to differentially expressed genes (DEGs) for tree height.
Figure 8.
Enrichment Analysis of BUSCA subcellular loci mapped to differentially expressed genes (DEGs) for tree height.
Table 1.
Correlation of disease traits against principal components of variation from PCA of RNA data. Correlation coefficient and p-value are displayed in the plot. CG_Avg. = A. tumefaciens phenotypic response, Height_3Y = tree height at three years of age, PHY_Avg = Phytophthora spp. phenotypic response, RLN_3Y = P. vulnus phenotypic response.
Table 1.
Correlation of disease traits against principal components of variation from PCA of RNA data. Correlation coefficient and p-value are displayed in the plot. CG_Avg. = A. tumefaciens phenotypic response, Height_3Y = tree height at three years of age, PHY_Avg = Phytophthora spp. phenotypic response, RLN_3Y = P. vulnus phenotypic response.
| Cor |
PC |
P.val |
Trait |
| -0.804 |
PC2 |
2.94E-02 |
CG |
| -0.639 |
PC5 |
1.22E-01 |
PHY |
| -0.698 |
PC4 |
1.90E-01 |
RLN_3Y |
| -0.867 |
PC1 |
5.69E-02 |
length_3Y |
Table 2.
Summary of the results of the differential expression analysis. CG_Avg. = A. tumefaciens phenotypic response, Height_3Y = tree height at three years of age, PHY_Avg = Phytophthora spp. phenotypic response, RLN_3Y = P. vulnus phenotypic response.
Table 2.
Summary of the results of the differential expression analysis. CG_Avg. = A. tumefaciens phenotypic response, Height_3Y = tree height at three years of age, PHY_Avg = Phytophthora spp. phenotypic response, RLN_3Y = P. vulnus phenotypic response.
| Statistic |
CG_Avg |
PHY_Avg |
RLN_3Y |
Height _3Y |
| Down |
3888 |
1061 |
13 |
3381 |
| NotSig |
13290 |
17731 |
17962 |
12658 |
| Up |
2021 |
407 |
7 |
1943 |
Table 3.
DEGs observed within QTL region of chromosome 4D from 19.
Table 3.
DEGs observed within QTL region of chromosome 4D from 19.
| GeneID |
name |
Trait |
logFC |
start |
end |
| 121260033 |
small nuclear ribonucleoprotein SmD3b |
CG |
-0.647 |
26403410 |
26405628 |
| 121260019 |
dolichol-phosphate mannosyltransferase subunit 1 isoform X1 |
CG |
-0.346 |
26421312 |
26423874 |
| 121259960 |
pre-rRNA-processing protein TSR1 homolog |
CG |
-0.295 |
26449293 |
26457407 |
| 121260033 |
small nuclear ribonucleoprotein SmD3b |
Height_3Y |
-0.530 |
26403410 |
26405628 |
| 121259974 |
probable acyl-activating enzyme 1, peroxisomal |
Height_3Y |
-0.512 |
26477730 |
26481055 |
| 121259960 |
pre-rRNA-processing protein TSR1 homolog |
Height_3Y |
-0.256 |
26449293 |
26457407 |
Table 4.
Correlation analysis of traits analyzed in this study. CG_Avg. = A. tumefaciens phenotypic response, Height_3Y = tree height at three years of age, PHY_Avg = Phytophthora spp. phenotypic response, RLN_3Y = P. vulnus phenotypic response.
Table 4.
Correlation analysis of traits analyzed in this study. CG_Avg. = A. tumefaciens phenotypic response, Height_3Y = tree height at three years of age, PHY_Avg = Phytophthora spp. phenotypic response, RLN_3Y = P. vulnus phenotypic response.
| Relationship |
Cor |
P.val |
| Height _3Y |
PHY_Avg |
0.983 |
0.003 |
| Height _3Y |
CG_Avg |
0.936 |
0.019 |
| PHY_Avg |
CG_Avg |
0.705 |
0.077 |
| RLN_3Y |
PHY_Avg |
0.663 |
0.222 |
| RLN_3Y |
Height _3Y |
0.574 |
0.311 |
| RLN_3Y |
CG_Avg |
0.547 |
0.341 |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).