1. Introduction
Durong cell proliferation, organelles are inherited by the newly generated cells. The budding yeast
Saccharomyces cerevisiae proliferates by budding, a process characterized by the formation of a cytoplasmic protrusion called the ‘bud’, which grows as the cell cycle progresses and eventually separates to become an independent daughter cell. Proliferation by budding requires the bud-directed transport of DNA, RNA, proteins, organelles, and other materials essential for the daughter cell. The directed transport of organelles toward the growing bud is mainly mediated by class V myosins, Myo2 and Myo4, on actin filaments [
1,
2]; Cortical ER is carried by Myo4, while Myo2 transports most other organelles such as the Golgi apparatus, vacuoles, peroxisomes, and mitochondria [
3,
4,
5,
6,
7,
8,
9,
10,
11]. At the late stage of budding, namely, the cytokinesis stage, Myo2 translocates from the bud tip to the bud neck to deliver vesicles containing proteins and other materials required for the formation of the septum that physically separates the mother and daughter cells. Therefore, the transported organelles should be released from Myo2 once they arrive at the growing bud to avoid backward flow to the bud neck. At least for vacuoles, mitochondria, and presumably peroxisomes, unloading from the actin–myosin machinery after delivery to the bud involves the ubiquitin–proteasome system [
12,
13]. This review focuses on the mechanism and significance of the unloading of vacuoles, mitochondria, and peroxisomes from the actin–myosin machinery during their inheritance in yeast.
2. Vacuole Inheritance and E3 Ligases
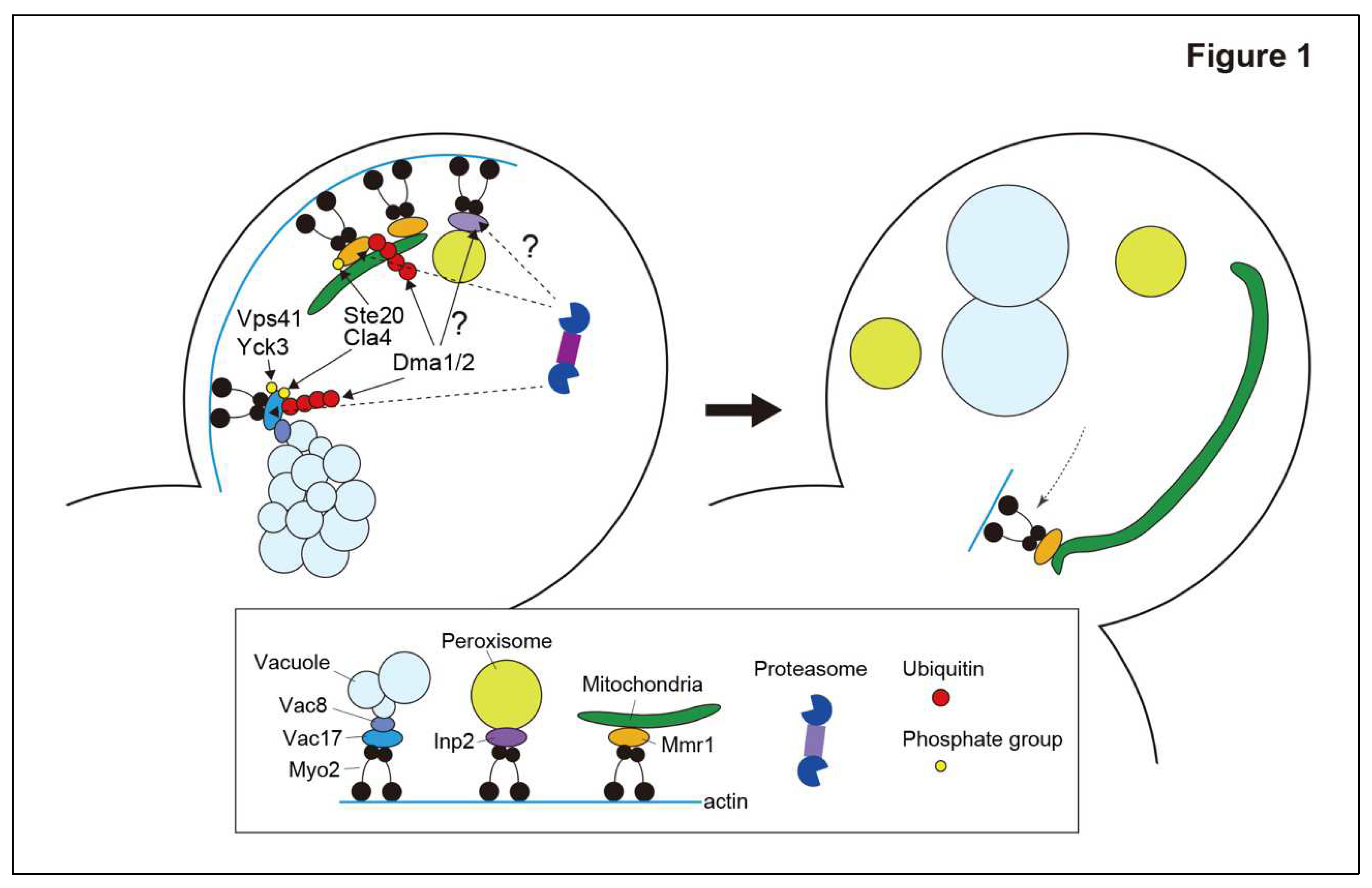
The involvement of ubiquitin ligases in organelle inheritance in yeast has been extensively studied for vacuoles. Prior to direct transport to the bud, vacuoles fragment and/or tubulate to form segregation structures that extend from the vacuole in the mother cell [
14]. The segregation structure is transported into the growing bud by Myo2 via actin filaments. Myo2 captures vacuoles through the vacuole-specific adaptor protein Vac17 [
5,
6]. The interaction between Myo2 and Vac17 is coordinated with cell cycle progression and is directly regulated by phosphorylation of Vac17 by th ecyclin-dependent kinase Cdk1/Cdc28 [
15]. Vac17 binds to the vacuole
via Vac8, which attaches to the vacuolar membrane via palmitoylation [
16,
17]. After vacuoles enter the growing bud, they are released from the actin–myosin machinery.
The release of vacuoles from the actin–myosin machinery in the bud is mediated by the proteolysis of Vac17 by the ubiquitin–proteasome system [
13] (
Figure 1). Vac17 is a short-lived protein containing a PEST motif, which often acts as a signal for protein degradation [
6,
18]. The PEST motif of Vac17 is required for rapid Vac17 degradation and the release of vacuoles from the actin–myosin machinery. E3 ubiquitin ligase Dma1 is required for Vac17 degradation and vacuole detachment from Myo2 [
13]. Dma1 is a RING-type E3 ubiquitin ligase. Dma1 and its paralogous protein Dma2 have been implicated in the regulation of septin dynamics and the spindle position checkpoint [
19]. This finding sheds light on a novel and unexpected role of Dma1/2 in organelle inheritance. In
dma1D cells, Vac17 levels are elevated, and vacuoles are mistargeted to the bud neck, to which Myo2 translocates at the cytokinesis stage [
13]. Dma1 and Dma2 are involved in Vac17 ubiquitination
in vivo, and Dma1 appears to play a major role in vacuole release compared to Dma2. Thus, vacuole release from the actin–myosin machinery requires ubiquitination of Vac17 by Dma1 and Dma2 and subsequent proteolysis.
The degradation of Vac17 must be regulated spatially and temporally to avoid unloading of the vacuole before it reaches the growing bud. Vac17 phosphorylation during vacuole inheritance plays a critical role in spatiotemporal regulation of Vac17 ubiquitination and release. During the initial step of vacuole inheritance, Cdk1/Cdc28 phosphorylates Vac17 to mediate Vac17-Myo2 attachment [
15]. The next regulatory step is phosphorylation of Vac17 at Thr240 in the PEST motif by a yet-to-be-identified kinase, which is required for recruiting Dma1 to Vac17. Interestingly, the recruitment of Dma1 alone is not sufficient for Vac17 ubiquitination. Recruited Dma1 is activated by additional phosphorylation of Vac17 at Ser222 by Cla4, which is a member of the p21-activated kinase family [
20]. Because Cla4 localizes to the bud cortex and is nearly absent from the mother cell, Cla4-dependent phosphorylation of Vac17 Ser222 is thought to be the spatial cue that ensures Vac17 ubiquitination only after the vacuole has entered the growing bud [
20]. The mechanism by which the phosphorylation of Vac17 Ser222 activates Dma1 remains unknown. Ubiquitination of Vac17 by Dma1 and Dma2 is insufficient for Vac17 degradation and vacuole release from the actin–myosin machinery. Vac17 degradation requires additional phosphorylation of Vac17 involving Yck3 and Vps41 [
21]. In
yck3D and
vps41D cells, ubiquitinated Vac17 accumulates and vacuoles are mistargeted to the bud neck at the cytokinesis stage. Yck3- and Vps41-dependent Vac17 phosphorylation is proposed to facilitate the dissociation of Vac17 from Myo2 [
22]. Although Vps41 is a subunit of the homotypic fusion and vacuole protein sorting (HOPS) tethering complex involved in vacuole fusion [
23,
24], the role of Vps41 in vacuole release from the actin–myosin machinery is independent of the HOPS complex [
21].
3. Mitochondria Inheritance and E3 Ligases
Mitochondria are transported to the growing bud by Myo2 along actin filaments [
9,
10]. Myo2 captures mitochondria via the adaptor protein Mmr1 [
25]. Mmr1 binds to mitochondria by interacting with lipid molecules on the outer membrane via its basic amino acid residues [
26]. After being transported to the growing bud, mitochondria are released from the actin–myosin machinery and move dynamically in the daughter cells. Mitochondria are mainly inherited by daughter cells via this pathway. Ypt11 is a small G protein involved in a parallel pathway for passing on mitochondria to the daughter cell [
9,
10]. Cells lacking both
MMR1 and
YPT11 are lethal, whereas each of the single-deletion mutants is viable.
In budding yeast, daughter cells preferentially receive healthy mitochondria, in other words, mitochondria that have a greater reducing power and that generate lower levels of reactive oxygen species [
27]. Yeast cells are also equipped with a system to preserve a portion of the healthy mitochondria in the mother cell to avoid complete loss of high-quality mitochondria. The retention of healthy mitochondria is mediated by their anchoring to the mother cell via Mfb1 [
28,
29].
The ubiquitin–proteasome system has recently been reported to be involved in mitochondrial inheritance [
12]. The adaptor protein Mmr1 is a short-lived protein [
30] that is degraded rapidly by the proteasome (
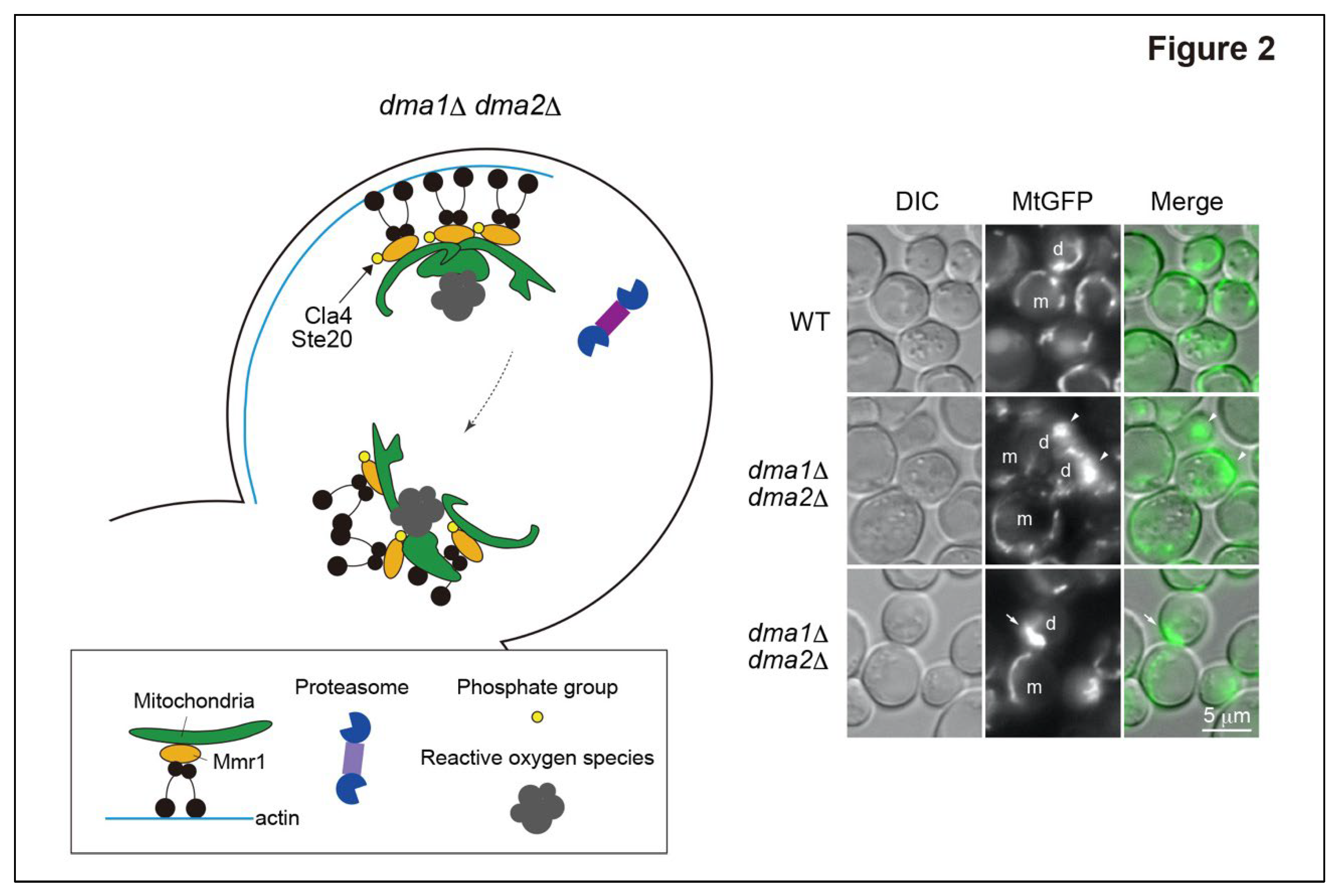
Figure 1). The redundant E3 ligases Dma1 and Dma2 were detected in the fraction of Mmr1-interacting proteins and shown to ubiquitinate Mmr1 both in vivo and in vitro. Defects in Mmr1 ubiquitination in
dma1D
dma2D cells lead to a failure to unload mitochondria from the actin–myosin machinery in the bud. Accordingly, mitochondria in
dma1D
dma2D cells are first stacked at the bud tip, then exhibit backward movement from the bud tip to the bud neck together with Myo2, and they become stacked again at the stage of cytokinesis (
Figure 2). Mitochondria stacked at the bud tip and bud neck in
dma1D
dma2D cells are intricately entwined and expanded or deformed into an abnormal shape (see Discussion and future perspectives). The respitory activity is abnormally high in the deformed mitochondria in
dma1D
dma2D cells, and higher levels of reactive oxygen species are generated in deformed mitochondria compared to in normal mitochondria of WT cells. As a result,
dma1D
dma2D cells are hypersensitive to oxidative stress caused by the reactive oxygen species-generating reagent paraquat or by the deletion of genes encoding superoxide dismutases
SOD1 and
SOD2. In summary, ubiquitination and degradation of Mmr1 are essential for the normal dynamics, morphology, and function of mitochondria.
Similar to vacuole inheritance, the release of mitochondria from the actin–myosin machinery is spatially and temporally regulated by phosphorylation [
12,
20]. The bud-localized kinases Cla4 and Ste20 phosphorylate Ser414 of Mmr1. Ser414 phosphorylation is a prerequisite for Mmr1 ubiquitination by Dma1 and Dma2. The Mmr1-S414A mutant protein has a prolonged lifetime compared to that of the WT Mmr1 protein, and cells expressing this mutant Mmr1 exhit mitochondrial stacking at the bud tip and bud neck, similar to
dma1D
dma2D cells. Restricted localization of Cla4 and Ste20 to the bud cortex may guarantee the unloading of mitochondria from the actin–myosin machinery only after the mitochondria enter the growing bud. As mentioned above, degradation of the vacuolar-specific adaptor Vac17 requires its Yck3- and Vps41-dependent phosphorylation. In contrast, Yck3 and Vps41 are dispensable for rapid degradation of Mmr1. Whether additional phosphorylation of Mmr1 by a pathway other than the Yck3–Vps41 axis is required for the release of mitochondria from the actin–myosin machinery remains unknown and is an important issue for future consideration.
Interestingly, a minor fraction of Mmr1 on the mitochondria, particularly around one of the two ends of tubular mitochondria, appears to escape degradation and maintain association with Myo2 during cytokinesis [
12]. Residual Mmr1 is observed at the bud neck, probably because of the translocation of Myo2 to the bud neck. Consequently, one end of the tubular mitochondria is anchored to the bud neck and acts as a fulcrum that supports the dynamic movement of the other part of the mitochondria in the daughter cell (
Figure 1). The mechanism and biological function of the selective protection of Mmr1 from degradation are unknown and will make for an intriguing future study.
4. Peroxisome Inheritance and E3 Ligases
Bud-directed transport of peroxisomes is also mediated by Myo2 along actin filments. Similar to vacuoles and mitochondria, an organelle-specific adaptor protein, Inp2, bridges Myo2 with the peroxisome [
8,
31]. Inp2 is a peroxisome membrane protein containing a PEST motif and several substrate recognition sites for Cdk1/Cdc28 [
8,
32]. A fraction of peroxisomes are anchored to the mother cell to maintain the balance of the peroxisome population between the mother and daughter cells. The retention of peroxisomes in the mother cells is considered to be mediated by Inp1, which bridges Pex3 on both the peroxisome membrane and the mother cortical ER membrane [
32,
33]. Overexpression of Inp1 results in the loss of peroxisomes in growing buds [
34]. Inp1 is also implicated in the anchoring of peroxisomes to the bud after transport; loss of Inp1 causes backward translocation of peroxisomes to mother cells [
34].
E3 ubiquitin ligases may be involved in unloading peroxisomes from the actin–myosin machinery after they have traveled to the growing bud. In
dma1D
dma2D cells, peroxisomes are stacked at the bud neck during cytokinesis, similar to vacuoles and mitochondria [
13]. How Dma1 and Dma2 are involved in the regulation of peroxisome dynamics in the bud remains unknown.
5. Discussion and Future Perspectives
Unloading of vacuoles and mitochondria largely share the same mechanism (
Figure 1). First, degradation of the adaptor proteins Vac17 and Mmr1 in vacuoles and mitochondria, respectively, is required for organelle release from the actin–myosin machinery. Secondly, the same E3 ubiquitin ligases, Dma1 and Dma2, are involved in the ubiquitination of adaptor proteins. Thirdly, phosphorylation of adaptor proteins serves as a spatial cue to trigger their ubiquitination in order for adaptor proteins to only be degraded once the cargo has reached the growing bud. Fourth, the kinases that lead to the ubiquitination of adaptors, such as the p21-activated kinases Cla4 and Ste20, are shared during the unloading of vacuoles and mitochondria. Although the mechanism of peroxisome release from Myo2 is still unknown, it is noteworthy that peroxisomes are stacked at the bud neck in
dma1D
dma2D cells, like vacuols and mitochondria. Overall, it is likely that a common strategy for yeast cells is to degrade adaptor proteins using the ubiquitin–proteasome system to unload inherited organelles.
We discuss whether this mechanism can be applied more broadly,
e.g., to organelle dynamics in mammalian cells. In mammalian cells, mitochondrial location is controlled by microtubules and actin filaments [
35,
36]. Mammalian cells proliferate by cell division, which is characterized by the partitioning of a preexisting space and, therefore, does not require the formation of a novel space. Symmetrical segregation of mitochondria during mammalian cell division requires the mitochondrial myosin Myo19 [
37]. However, it is also reported that mitochondrial segregation during mammalian cell division is passive and does not require mitochondrial transport by the cytoskeleton [
38]. In this passive mode of mitochondrial segregation, the mitochondria dissociate from the cytoskeleton before the onset of cell division. Interestingly, prior to the passive mitochondrial segregation during mammalian cell division, mitochondrial motor proteins dissociate from the mitochondria, leading to the release of the mitochondria from microtubules [
38]. This motor shedding requires the action of the protein kinases CDK1 and Aurora A. It would be particularly interesting to determine whether targeted degradation of mitochondrial motor proteins and/or adaptor proteins is involved in the release of mitochondria from the cytoskeleton for passive segregation during mammalian cell division.
Intracellular transport of melanosomes is another interesting model for studying directed organelle transport. The transport and correct positioning of melanosomes in mammalian cells are dependent on the cytoskeleton. Melanosomes are first directionally transported by the motor protein kinesin on microtubules, followed by switching over to actin filaments and then being carried by myosin toward the cell surface for secretion [
39]. It would be natural to consider that a series of such elaborate transport processes should be spatially and temporally highly coordinated. Therefore, it would be interesting for future research to investigate whether ubiquitin-mediated degradation of adaptor proteins is involved in directed melanosome movement.
In
dma1D and
dma2D cells, mitochondria are stacked at the bud tip and bud neck (
Figure 2). Electron microscopy has revealed that such stacked mitochondria are expanded or abnormally deformed [
12]. Mmr1, in addition to connecting mitochondria and Myo2, has been reported to have a role in anchoring transported mitochondria to the bud cortex and in facilitating mitochondrial fusion [
40]. Failure of Mmr1 degradation in
dma1D
dma2D cells may lead to abnormally accelerated fusion of mitochondria, which in turn results in mitochondrial deformation. It is generally thought that mitochondrial fusion contributes to elevated respiratory activity [
41], which is in good agreement with the mitochondria in
dma1D
dma2D cells. It would also be intriguing to determine whether the E3 ubiquitin ligases Dma1 and Dma2 indirectly regulate mitochondrial fusion through ubiquitination and subsequent degradation of Mmr1.
Author Contributions
K.O. and T.K. conceived the idea and framework of the manuscript. K.O. prepared the initial draft. All authors contributed to the manuscript editing.
Funding
No external funding was obtained for preparing this manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data availability Statement
Not applicable.
Acknowledgments
We would like to thank Editage (
www.editage.com) for English language editing.
Conflicts of Interest
The authors have no conflicts of interest to declare.
Abbreviations
| ER |
(Endoplasmic reticulum) |
| PEST |
(Proline, glutamic acid, serine, threonine) |
| RING |
(Really interesting new gene) |
| HOPS |
(Homotypic fusion and vacuole protein sorting) |
References
- Bretscher, A. Polarized growth and organelle segregation in yeast: The tracks, motors, and receptors. J Cell Biol 2003, 160, 811–816. [Google Scholar] [CrossRef] [PubMed]
- Pruyne, D.; Legesse-Miller, A.; Gao, L.; Dong, Y.; Bretscher, A. Mechanisms of polarized growth and organelle segregation in yeast. Annu Rev Cell Dev Biol 2004, 20, 559–591. [Google Scholar] [CrossRef] [PubMed]
- Estrada, P.; Kim, J.; Coleman, J.; Walker, L.; Dunn, B.; Takizawa, P.; Novick, P.; Ferro-Novick, S. Myo4p and She3p are required for cortical ER inheritance in Saccharomyces cerevisiae. J Cell Biol 2003, 163, 1255–1266. [Google Scholar] [CrossRef] [PubMed]
- Rossanese, O.W.; Reinke, C.A.; Bevis, B.J.; Hammond, A.T.; Sears, I.B.; O'Connor, J.; Glick, B.S. A role for actin, Cdc1p, and Myo2p in the inheritance of late Golgi elements in Saccharomyces cerevisiae. J Cell Biol 2001, 153, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, K.; Catlett, N.L.; Novak, J.L.; Tang, F.; Nau, J.J.; Weisman, L.S. Identification of an organelle-specific myosin V receptor. J Cell Biol 2003, 160, 887–897. [Google Scholar] [CrossRef] [PubMed]
- Tang, F.; Kauffman, E.J.; Novak, J.L.; Nau, J.J.; Catlett, N.L.; Weisman, L.S. Regulated degradation of a class V myosin receptor directs movement of the yeast vacuole. Nature 2003, 422, 87–92. [Google Scholar] [CrossRef] [PubMed]
- Hoepfner, D.; van den Berg, M.; Philippsen, P.; Tabak, H.F.; Hettema, E.H. A role for Vps1p, actin, and the Myo2p motor in peroxisome abundance and inheritance in Saccharomyces cerevisiae. J Cell Biol 2001, 155, 979–990. [Google Scholar] [CrossRef] [PubMed]
- Fagarasanu, A.; Mast, F.D.; Knoblach, B.; Jin, Y.; Brunner, M.J.; Logan, M.R.; Glover, J.N.; Eitzen, G.A.; Aitchison, J.D.; Weisman, L.S.; et al. Myosin-driven peroxisome partitioning in S. cerevisiae. J Cell Biol 2009, 186, 541–554. [Google Scholar] [CrossRef]
- Itoh, T.; Toh, E.A.; Matsui, Y. Mmr1p is a mitochondrial factor for Myo2p-dependent inheritance of mitochondria in the budding yeast. EMBO J 2004, 23, 2520–2530. [Google Scholar] [CrossRef]
- Itoh, T.; Watabe, A.; Toh, E.A.; Matsui, Y. Complex formation with Ypt11p, a rab-type small GTPase, is essential to facilitate the function of Myo2p, a class V myosin, in mitochondrial distribution in Saccharomyces cerevisiae. Mol Cell Biol 2002, 22, 7744–7757. [Google Scholar] [CrossRef]
- Altmann, K.; Frank, M.; Neumann, D.; Jakobs, S.; Westermann, B. The class V myosin motor protein, Myo2, plays a major role in mitochondrial motility in Saccharomyces cerevisiae. J Cell Biol 2008, 181, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Obara, K.; Yoshikawa, T.; Yamaguchi, R.; Kuwata, K.; Nakatsukasa, K.; Nishimura, K.; Kamura, T. Proteolysis of adaptor protein Mmr1 during budding is necessary for mitochondrial homeostasis in Saccharomyces cerevisiae. Nat Commun 2022, 13, 2005. [Google Scholar] [CrossRef] [PubMed]
- Yau, R.G.; Peng, Y.; Valiathan, R.R.; Birkeland, S.R.; Wilson, T.E.; Weisman, L.S. Release from myosin V via regulated recruitment of an E3 ubiquitin ligase controls organelle localization. Dev Cell 2014, 28, 520–533. [Google Scholar] [CrossRef]
- Weisman, L.S. Organelles on the move: Insights from yeast vacuole inheritance. Nat Rev Mol Cell Biol 2006, 7, 243–252. [Google Scholar] [CrossRef]
- Peng, Y.; Weisman, L.S. The cyclin-dependent kinase Cdk1 directly regulates vacuole inheritance. Dev Cell 2008, 15, 478–485. [Google Scholar] [CrossRef]
- Wang, Y.X.; Catlett, N.L.; Weisman, L.S. Vac8p, a vacuolar protein with armadillo repeats, functions in both vacuole inheritance and protein targeting from the cytoplasm to vacuole. J Cell Biol 1998, 140, 1063–1074. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.X.; Zhao, H.; Harding, T.M.; Gomes de Mesquita, D.S.; Woldringh, C.L.; Klionsky, D.J.; Munn, A.L.; Weisman, L.S. Multiple classes of yeast mutants are defective in vacuole partitioning yet target vacuole proteins correctly. Mol Biol Cell 1996, 7, 1375–1389. [Google Scholar] [CrossRef]
- Rogers, S.; Wells, R.; Rechsteiner, M. Amino acid sequences common to rapidly degraded proteins: The PEST hypothesis. Science 1986, 234, 364–368. [Google Scholar] [CrossRef] [PubMed]
- Fraschini, R.; Bilotta, D.; Lucchini, G.; Piatti, S. Functional characterization of Dma1 and Dma2, the budding yeast homologues of Schizosaccharomyces pombe Dma1 and human Chfr. Mol Biol Cell 2004, 15, 3796–3810. [Google Scholar] [CrossRef]
- Yau, R.G.; Wong, S.; Weisman, L.S. Spatial regulation of organelle release from myosin V transport by p21-activated kinases. J Cell Biol 2017, 216, 1557–1566. [Google Scholar] [CrossRef]
- Wong, S.; Hepowit, N.L.; Port, S.A.; Yau, R.G.; Peng, Y.; Azad, N.; Habib, A.; Harpaz, N.; Schuldiner, M.; Hughson, F.M.; et al. Cargo Release from Myosin V Requires the Convergence of Parallel Pathways that Phosphorylate and Ubiquitylate the Cargo Adaptor. Curr Biol 2020, 30, 4399–4412 e4397. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.; Weisman, L.S. Let it go: Mechanisms that detach myosin V from the yeast vacuole. Curr Genet 2021, 67, 865–869. [Google Scholar] [CrossRef] [PubMed]
- Seals, D.F.; Eitzen, G.; Margolis, N.; Wickner, W.T.; Price, A. A Ypt/Rab effector complex containing the Sec1 homolog Vps33p is required for homotypic vacuole fusion. Proc Natl Acad Sci U S A 2000, 97, 9402–9407. [Google Scholar] [CrossRef]
- Wurmser, A.E.; Sato, T.K.; Emr, S.D. New component of the vacuolar class C-Vps complex couples nucleotide exchange on the Ypt7 GTPase to SNARE-dependent docking and fusion. J Cell Biol 2000, 151, 551–562. [Google Scholar] [CrossRef] [PubMed]
- Chernyakov, I.; Santiago-Tirado, F.; Bretscher, A. Active segregation of yeast mitochondria by Myo2 is essential and mediated by Mmr1 and Ypt11. Curr Biol 2013, 23, 1818–1824. [Google Scholar] [CrossRef]
- Chen, W.; Ping, H.A.; Lackner, L.L. Direct membrane binding and self-interaction contribute to Mmr1 function in mitochondrial inheritance. Mol Biol Cell 2018, 29, 2346–2357. [Google Scholar] [CrossRef]
- McFaline-Figueroa, J.R.; Vevea, J.; Swayne, T.C.; Zhou, C.; Liu, C.; Leung, G.; Boldogh, I.R.; Pon, L.A. Mitochondrial quality control during inheritance is associated with lifespan and mother-daughter age asymmetry in budding yeast. Aging Cell 2011, 10, 885–895. [Google Scholar] [CrossRef]
- Kondo-Okamoto, N.; Ohkuni, K.; Kitagawa, K.; McCaffery, J.M.; Shaw, J.M.; Okamoto, K. The novel F-box protein Mfb1p regulates mitochondrial connectivity and exhibits asymmetric localization in yeast. Mol Biol Cell 2006, 17, 3756–3767. [Google Scholar] [CrossRef] [PubMed]
- Pernice, W.M.; Vevea, J.D.; Pon, L.A. A role for Mfb1p in region-specific anchorage of high-functioning mitochondria and lifespan in Saccharomyces cerevisiae. Nat Commun 2016, 7, 10595. [Google Scholar] [CrossRef]
- Christiano, R.; Nagaraj, N.; Frohlich, F.; Walther, T.C. Global proteome turnover analyses of the Yeasts S. cerevisiae and S. pombe. Cell Rep 2014, 9, 1959–1965. [Google Scholar] [CrossRef]
- Fagarasanu, A.; Mast, F.D.; Knoblach, B.; Rachubinski, R.A. Molecular mechanisms of organelle inheritance: Lessons from peroxisomes in yeast. Nat Rev Mol Cell Biol 2010, 11, 644–654. [Google Scholar] [CrossRef]
- Knoblach, B.; Rachubinski, R.A. Sharing with your children: Mechanisms of peroxisome inheritance. Biochim Biophys Acta 2016, 1863, 1014–1018. [Google Scholar] [CrossRef]
- Knoblach, B.; Sun, X.; Coquelle, N.; Fagarasanu, A.; Poirier, R.L.; Rachubinski, R.A. An ER-peroxisome tether exerts peroxisome population control in yeast. EMBO J 2013, 32, 2439–2453. [Google Scholar] [CrossRef] [PubMed]
- Fagarasanu, M.; Fagarasanu, A.; Tam, Y.Y.; Aitchison, J.D.; Rachubinski, R.A. Inp1p is a peroxisomal membrane protein required for peroxisome inheritance in Saccharomyces cerevisiae. J Cell Biol 2005, 169, 765–775. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Domenech, G.; Covill-Cooke, C.; Ivankovic, D.; Halff, E.F.; Sheehan, D.F.; Norkett, R.; Birsa, N.; Kittler, J.T. Miro proteins coordinate microtubule- and actin-dependent mitochondrial transport and distribution. EMBO J 2018, 37, 321–336. [Google Scholar] [CrossRef]
- Morris, R.L.; Hollenbeck, P.J. Axonal transport of mitochondria along microtubules and F-actin in living vertebrate neurons. J Cell Biol 1995, 131, 1315–1326. [Google Scholar] [CrossRef]
- Rohn, J.L.; Patel, J.V.; Neumann, B.; Bulkescher, J.; McHedlishvili, N.; McMullan, R.C.; Quintero, O.A.; Ellenberg, J.; Baum, B. Myo19 ensures symmetric partitioning of mitochondria and coupling of mitochondrial segregation to cell division. Curr Biol 2014, 24, 2598–2605. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.Y.; Steen, J.A.; Schwarz, T.L. Phosphorylation-Induced Motor Shedding Is Required at Mitosis for Proper Distribution and Passive Inheritance of Mitochondria. Cell Rep 2016, 16, 2142–2155. [Google Scholar] [CrossRef] [PubMed]
- Ohbayashi, N.; Fukuda, M. Recent advances in understanding the molecular basis of melanogenesis in melanocytes. F1000Res 2020, 9. [Google Scholar] [CrossRef]
- Higuchi-Sanabria, R.; Charalel, J.K.; Viana, M.P.; Garcia, E.J.; Sing, C.N.; Koenigsberg, A.; Swayne, T.C.; Vevea, J.D.; Boldogh, I.R.; Rafelski, S.M.; et al. Mitochondrial anchorage and fusion contribute to mitochondrial inheritance and quality control in the budding yeast Saccharomyces cerevisiae. Mol Biol Cell 2016, 27, 776–787. [Google Scholar] [CrossRef]
- Nakada, K.; Inoue, K.; Hayashi, J. Interaction theory of mammalian mitochondria. Biochem Biophys Res Commun 2001, 288, 743–746. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).