Submitted:
03 December 2023
Posted:
05 December 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction: Data analytics in modern cardiology
2. Fundamentals of Topological Data Analysis
2.1. Persistent Homology
- Every face of the complex must be a simplex, that is, a triangle or a higher-dimensional analogue of a triangle.
- Every face of the complex must be a subset of one of the vertices of the complex.
- If a face of the complex is a subset of another face, then the larger face must be a subset of one of the vertices of the complex.
2.1.1. Building the FSH
2.1.2. Calculating the PH
-
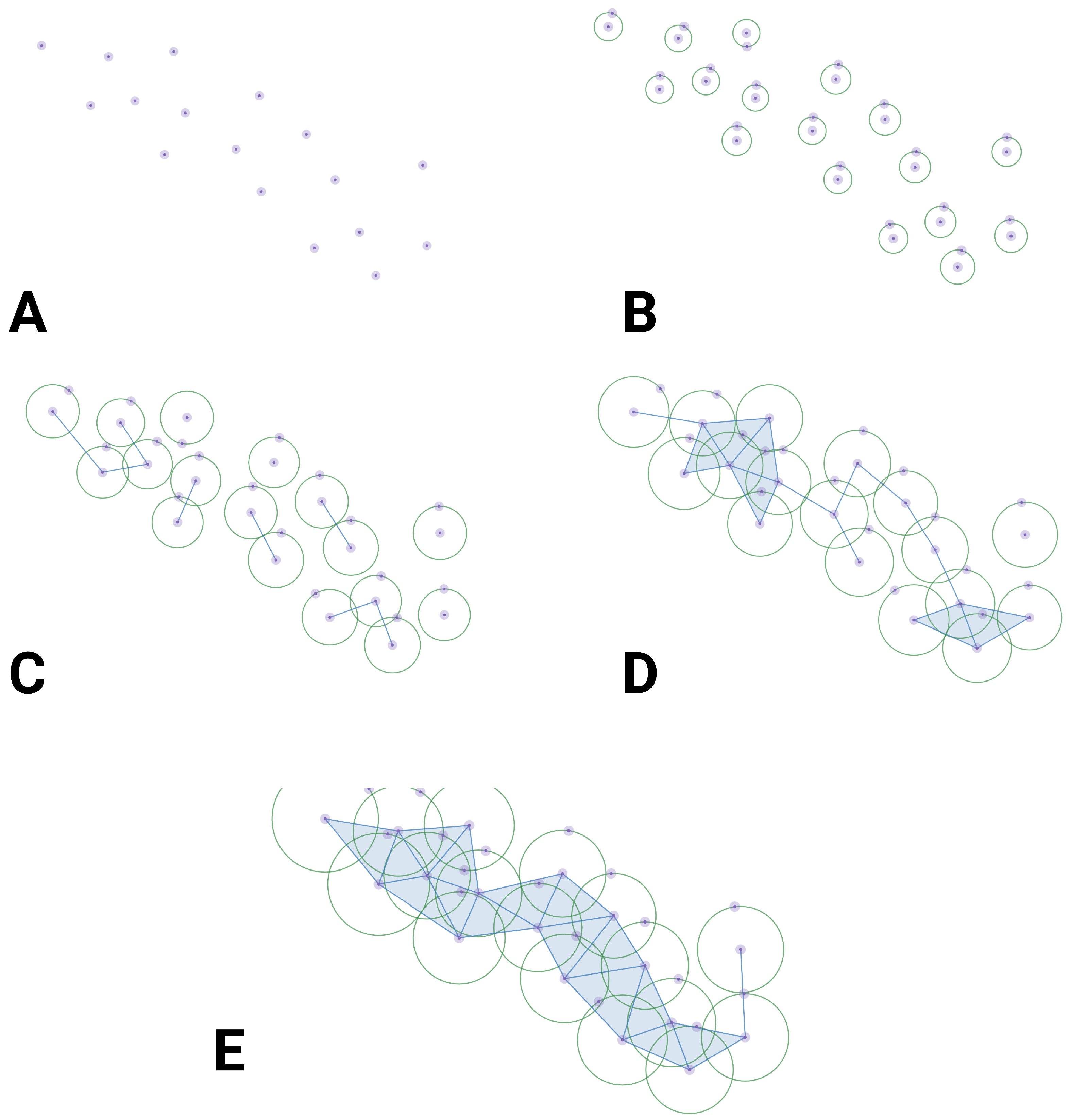
Simplicial Complex Construction: - Begin by constructing a simplicial complex from your data. This complex can be based on various covering maps, such as the Vietoris-Rips complex, Čech complex, or alpha complex, depending on the chosen strategy (See section 2.4 and section 2.5, as well as Table 1 below).- The simplicial complex consists of vertices (0-simplices), edges (1-simplices), triangles (2-simplices), and higher-dimensional simplices. The choice of the complex depends on your data and the topological features of interest.
-
Filtration: - Introduce a filtration parameter (often denoted as ) that varies over a range of values. This parameter controls which simplices are included in the complex based on some criterion (e.g., distance threshold).- As increases, more simplices are added to the complex, and the complex evolves. The filtration process captures the topological changes as varies.
-
Boundary Matrix: - For each value of in the filtration, compute the boundary matrix (also called the boundary operator) of the simplicial complex. This matrix encodes the relations between simplices.- Each row of the boundary matrix corresponds to a (k-1)-dimensional simplex, and each column corresponds to a k-dimensional simplex. The entries indicate how many times a (k-1)-dimensional simplex is a face of a k-dimensional simplex.
-
Persistent Homology Calculation: - Perform a sequence of matrix reductions (e.g., Gaussian elimination) to identify the cycles and boundaries in the boundary matrix.- A cycle is a collection of simplices whose boundaries sum to zero, while a boundary is the boundary of another simplex.- Persistent homology focuses on tracking the birth and death of cycles across different values of . These births and deaths are recorded in a persistence diagram or barcode.
-
Persistence Diagram or Barcode: - The persistence diagram is a graphical representation of the births and deaths of topological features (connected components, loops, voids) as varies.- Each point in the diagram represents a topological feature and is plotted at birth (x-coordinate) and death (y-coordinate).- Interpretation:- A point in the upper-left quadrant represents a long-lived feature that persists across a wide range of values.- A point in the lower-right quadrant represents a short-lived feature that exists only for a narrow range of values.- The diagonal represents features that are consistently present throughout the entire range of values.- The distance between the birth and death of a point in the diagram quantifies the feature’s persistence or lifetime. Longer persistence indicates a more stable and significant feature.
-
Topological Summaries: - By examining the persistence diagram or barcode, you can extract information about the prominent topological features in your data set.- Features with longer persistence are considered more robust and significant.- The number of connected components, loops, and voids can be quantified by counting points in specific regions of the diagram.
2.2. The Mapper algorithm
-
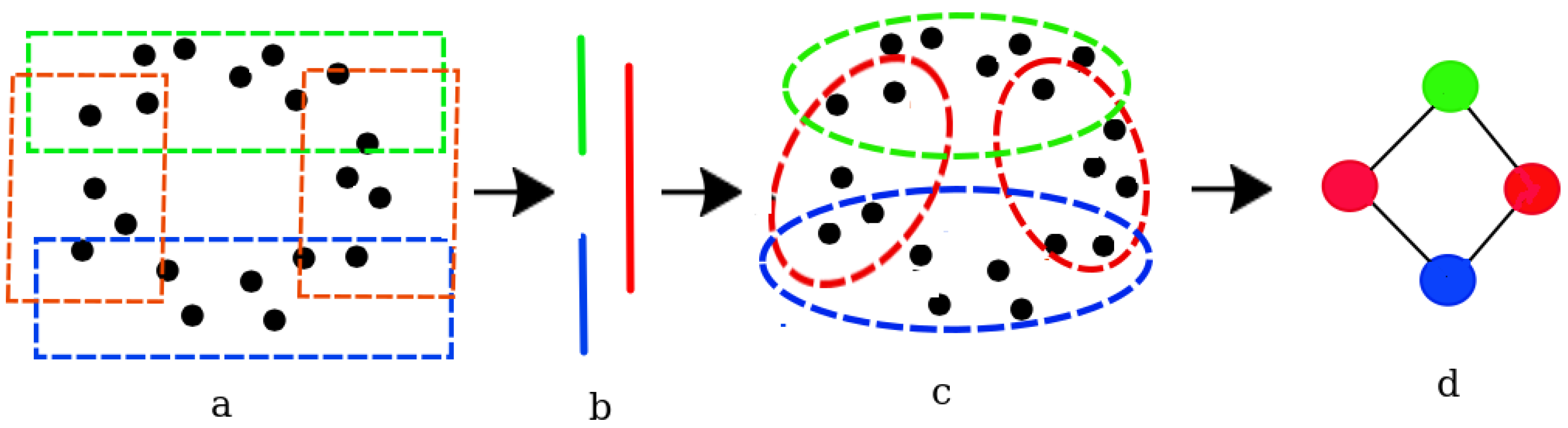
Covering the data set: The original data set (Figure 3 a) is partitioned into a number of overlapping subsets, called nodes (Figure 3 b). This is accomplished using a function called the covering map. The covering map assigns each point in the data set to a node. Since the nodes are allowed to overlap, every point potentially belongs to multiple nodes.There are several different ways to define a covering map. The choice of covering map, however, can significantly affect the resulting Mapper graph. Some common approaches to define a covering map include:
- (a)
- Filtering: The data set is partitioned based on the values of one or more variables. A data set may, for instance, be partitioned based on the values of a categorical variable, such as gender or race.
- (b)
- Projection: Data set partitioning is performed by calculating the distance between points in the data set and using it as a membership criteria. This can be done using a distance function, such as the Euclidean distance or the cosine similarity.
- (c)
- Overlapping intervals: The data set is partitioned into overlapping intervals, such as bins or quantiles. This can be useful for data sets that are evenly distributed or those having a known underlying distribution.
The choice of covering map depends on the characteristics of the data set and the research question being addressed. It is important to choose a covering map that is appropriate for the data set and that will yield meaningful results. - Clustering the nodes: The nodes are then clustered using a clustering algorithm, such as k-means or single-linkage clustering. The resulting clusters (Figure 3 c) represent the topological features of the data set, and the edges between the clusters represent the relationships between the features.
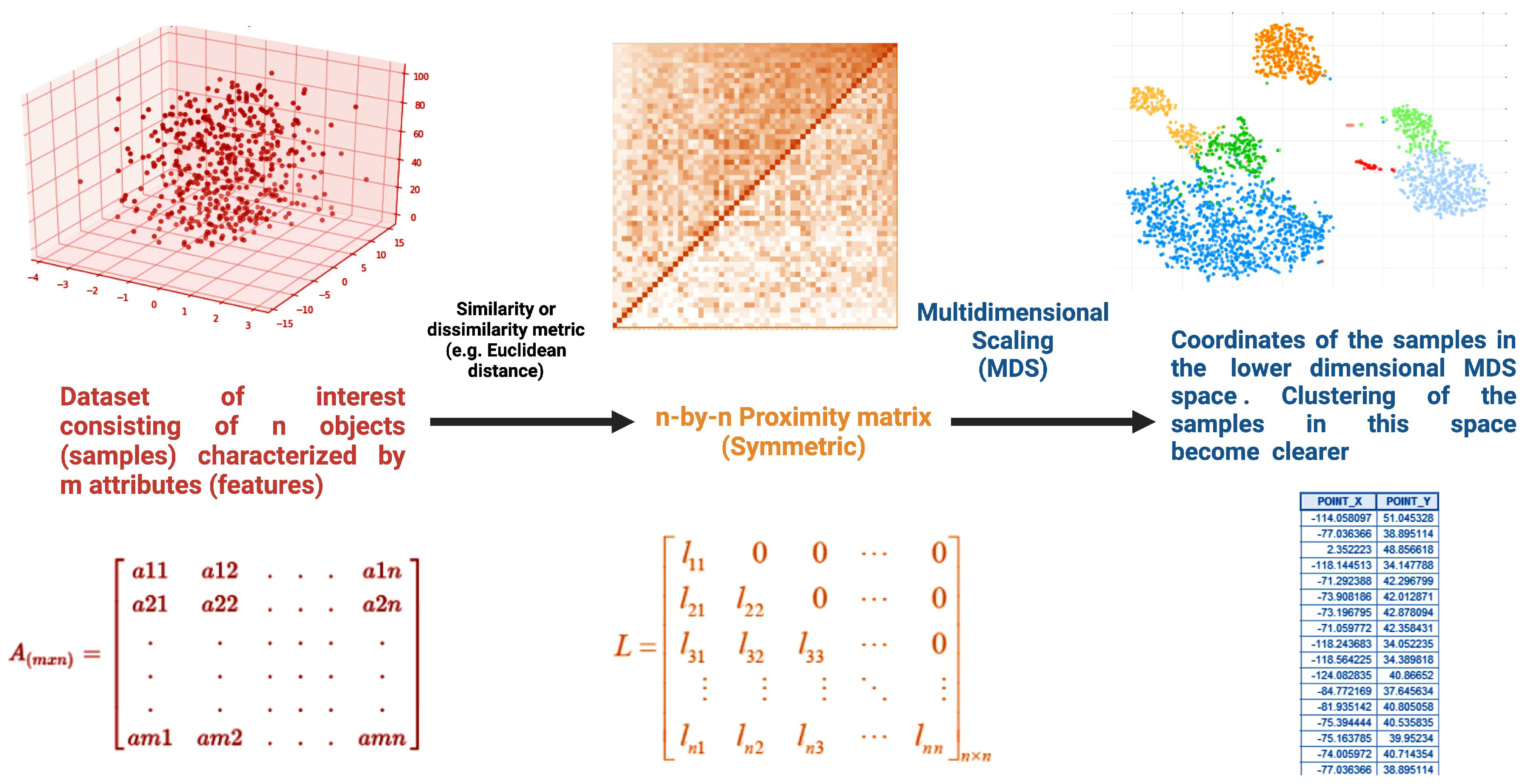
2.3. Multidimensional scaling
2.3.1. How to determine the scaling approach?
-
Classical MDS:- When to Use: Classical MDS is suitable when you have metric (distance) data that accurately represents the pairwise dissimilarities between objects. In classical MDS, the goal is to find a configuration of points in a lower-dimensional space (usually 2D or 3D) that best approximates the given distance matrix.- Pros: - It preserves the actual distances between data points in the lower-dimensional representation.- It provides a faithful representation when the input distances are accurate.- It is well-suited for situations where the metric properties of the data are crucial.- Cons: - It assumes that the input distances are accurate and may not work well with noisy or unreliable distance data.- It may not capture the underlying structure of the data if the metric assumption is violated.
-
Nonmetric MDS:- When to Use: Nonmetric MDS is appropriate when you have ordinal or rank-order data, where the exact distances between data points are not known, but their relative dissimilarities or rankings are available. Nonmetric MDS finds a configuration that best preserves the order of dissimilarities.- Pros: - It is more flexible than classical MDS and can be used with ordinal data.- It can handle situations where the exact distances are uncertain or difficult to obtain.- Cons: - It does not preserve the actual distances between data points, so the resulting configuration is only an ordinal representation.- The choice of a monotonic transformation function to convert ordinal data into dissimilarity values can affect the results.
-
Metric MDS:- When to Use: Metric MDS can be used when you have data that is inherently non-metric, but you believe that transforming it into a metric space could reveal meaningful patterns. Metric MDS aims to find a metric configuration that best approximates the non-metric dissimilarities.- Pros: - It provides a way to convert non-metric data into a metric representation for visualization or analysis.- It can help identify relationships in the data that may not be apparent in the original non-metric space.- Cons: - The success of metric MDS depends on the choice of the transformation function to convert non-metric data into metric distances.- It may not work well if the non-metric relationships in the data are too complex or cannot be adequately approximated by a metric space.
2.4. Choosing the covering map
- Data Dimensionality: The dimensionality of the data under consideration is crucial. Covering maps should be chosen to preserve the relevant topological information in the data. For high-dimensional data, dimension reduction techniques may be applied before selecting a covering map.
- Noise and Outliers: The presence of noise and outliers in the data can affect the choice of a covering map. Robust covering maps can help mitigate the influence of noise and outliers on the topological analysis.
- Data Density: The distribution of data points in the feature space matters. A covering map should be chosen to account for variations in data density, especially if there are regions of high density and regions with sparse data.
- Topological Features of Interest: It is important to consider the specific topological features one is interested in analyzing. Different covering maps may emphasize different aspects of the data topology, such as connected components, loops, or voids. The election of a covering map should align with the particular research objectives.
- Computational Efficiency: The computational complexity of calculating the covering map should also be taken into account. Some covering maps may be computationally expensive, which can be a limiting factor for large data sets.
- Continuous vs. Discrete Data: Determine whether the data you are analyzing is continuous or discrete. The choice of a covering map may differ based on the nature of the data.
- Metric or Non-Metric Data: Some covering maps are designed for metric spaces, where distances between data points are well-defined, while others may work better for non-metric or qualitative data.
- Geometric and Topological Considerations: Think about the geometric and topological characteristics of your data. Certain covering maps may be more suitable for capturing specific geometric or topological properties, such as persistence diagrams or Betti numbers.
- Domain Knowledge: Incorporate domain-specific knowledge into your choice of a covering map. Understanding the underlying structure of the data can guide you in selecting an appropriate covering map.
- Robustness and Stability: Assess the robustness and stability of the chosen covering map. TDA techniques should ideally produce consistent results under small perturbations of the data or variations in sampling.
2.5. Different strategies for topological feature selection
2.5.1. Vietoris-Rips (VR) Strategy:
2.5.2. Witness Strategy (WS):
2.5.3. Lazy-Witness Strategy (LW):
3. Applications of TDA to analyze Cardiovascular signals
3.1. General features
3.2. ECG data and heart rate signals
3.3. Stenosis and vascular data
3.4. TDA in Echocardiography
4. Perspectives and Limitations
Appendix: Computational tools
References
- Seetharam, K.; Shrestha, S.; Sengupta, P.P. Artificial intelligence in cardiovascular medicine. Current treatment options in cardiovascular medicine 2019, 21, 1–14. [Google Scholar] [CrossRef]
- Silverio, A.; Cavallo, P.; De Rosa, R.; Galasso, G. Big health data and cardiovascular diseases: a challenge for research, an opportunity for clinical care. Frontiers in medicine 2019, 6, 36. [Google Scholar] [CrossRef] [PubMed]
- Kagiyama, N.; Shrestha, S.; Farjo, P.D.; Sengupta, P.P. Artificial intelligence: practical primer for clinical research in cardiovascular disease. Journal of the American Heart Association 2019, 8, e012788. [Google Scholar] [CrossRef] [PubMed]
- Shameer, K.; Johnson, K.W.; Glicksberg, B.S.; Dudley, J.T.; Sengupta, P.P. Machine learning in cardiovascular medicine: are we there yet? Heart 2018, 104, 1156–1164. [Google Scholar] [CrossRef] [PubMed]
- Aljanobi, F.A.; Lee, J. Topological Data Analysis for Classification of Heart Disease Data. In Proceedings of the 2021 IEEE International Conference on Big Data and Smart Computing (BigComp), 2021; pp. 210–213. [Google Scholar] [CrossRef]
- Phinyomark, A.; Ibáñez-Marcelo, E.; Petri, G. Topological Data analysis of Biomedical Big Data. In Signal Processing and Machine Learning for Biomedical Big Data; CRC Press, 2018; pp. 209–233. [Google Scholar]
- Carlsson, G. The shape of biomedical data. Current Opinion in Systems Biology 2017, 1, 109–113. [Google Scholar] [CrossRef]
- Skaf, Y.; Laubenbacher, R. Topological data analysis in biomedicine: A review. Journal of Biomedical Informatics 2022, 130, 104082. [Google Scholar] [CrossRef]
- Carlsson, G.; Vejdemo-Johansson, M. Topological Data Analysis with Applications; Cambridge University Press, 2021. [Google Scholar]
- Ristovska, D.; Sekuloski, P. Mapper algorithm and its applications. Mathematical Modeling 2019, 3, 79–82. [Google Scholar]
- Zhou, Y.; Chalapathi, N.; Rathore, A.; Zhao, Y.; Wang, B. Mapper Interactive: A scalable, extendable, and interactive toolbox for the visual exploration of high-dimensional data. In Proceedings of the 2021 IEEE 14th Pacific Visualization Symposium (PacificVis); IEEE, 2021; pp. 101–110. [Google Scholar]
- Brown, A.; Bobrowski, O.; Munch, E.; Wang, B. Probabilistic convergence and stability of random mapper graphs. Journal of Applied and Computational Topology 2021, 5, 99–140. [Google Scholar] [CrossRef]
- Wasserman, L. Topological data analysis. Annual Review of Statistics and Its Application 2018, 5, 501–532. [Google Scholar] [CrossRef]
- Chazal, F.; Michel, B. An introduction to topological data analysis: fundamental and practical aspects for data scientists. Frontiers in artificial intelligence 2021, 4. [Google Scholar] [CrossRef]
- Lopez, J.E.; Datta, E.; Ballal, A.; Izu, L.T. Topological Data Analysis of Electronic Health Record Features Predicts Major Cardiovascular Outcomes After Revascularization for Acute Myocardial Infarction. Circulation 2022, 146, A14875–A14875. [Google Scholar] [CrossRef]
- Yan, Y.; Ivanov, K.; Cen, J.; Liu, Q.H.; Wang, L. Persistence landscape based topological data analysis for personalized arrhythmia classification. 2019; Preprints (non-peer reviewed yet). [Google Scholar]
- Falsetti, L.; Rucco, M.; Proietti, M.; Viticchi, G.; Zaccone, V.; Scarponi, M.; Giovenali, L.; Moroncini, G.; Nitti, C.; Salvi, A. Risk prediction of clinical adverse outcomes with machine learning in a cohort of critically ill patients with atrial fibrillation. Scientific reports 2021, 11, 1–11. [Google Scholar] [CrossRef]
- Safarbali, B.; Hashemi Golpayegani, S.M.R. Nonlinear dynamic approaches to identify atrial fibrillation progression based on topological methods. Biomedical Signal Processing and Control 2019, 53, 101563. [Google Scholar] [CrossRef]
- Goldberger, A.L.; Amaral, L.A.; Glass, L.; Hausdorff, J.M.; Ivanov, P.C.; Mark, R.G.; Mietus, J.E.; Moody, G.B.; Peng, C.K.; Stanley, H.E. PhysioBank, PhysioToolkit, and PhysioNet: components of a new research resource for complex physiologic signals. circulation 2000, 101, e215–e220. [Google Scholar] [CrossRef] [PubMed]
- Graff, G.; Graff, B.; Pilarczyk, P.; Jabłoński, G.; Gąsecki, D.; Narkiewicz, K. Persistent homology as a new method of the assessment of heart rate variability. Plos one 2021, 16, e0253851. [Google Scholar] [CrossRef] [PubMed]
- Ling, T.; Zhu, Z.; Zhang, Y.; Jiang, F. Early Ventricular Fibrillation Prediction Based on Topological Data Analysis of ECG Signal. Applied Sciences 2022, 12. [Google Scholar] [CrossRef]
- Mjahad, A.; Frances-Villora, J.V.; Bataller-Mompean, M.; Rosado-Muñoz, A. Ventricular Fibrillation and Tachycardia Detection Using Features Derived from Topological Data Analysis. Applied Sciences 2022, 12. [Google Scholar] [CrossRef]
- Caffrey, S.L.; Willoughby, P.J.; Pepe, P.E.; Becker, L.B. Public use of automated external defibrillators. New England journal of medicine 2002, 347, 1242–1247. [Google Scholar] [CrossRef] [PubMed]
- Delhomme, C.; Njeim, M.; Varlet, E.; Pechmajou, L.; Benameur, N.; Cassan, P.; Derkenne, C.; Jost, D.; Lamhaut, L.; Marijon, E.; et al. Automated external defibrillator use in out-of-hospital cardiac arrest: Current limitations and solutions. Archives of cardiovascular diseases 2019, 112, 217–222. [Google Scholar] [CrossRef] [PubMed]
- Kamp, N.J.; Al-Khatib, S.M. The subcutaneous implantable cardioverter-defibrillator in review. American heart journal 2019, 217, 131–139. [Google Scholar] [CrossRef]
- Friedman, P.; Murgatroyd, F.; Boersma, L.V.; Manlucu, J.; O’Donnell, D.; Knight, B.P.; Clémenty, N.; Leclercq, C.; Amin, A.; Merkely, B.P.; et al. Efficacy and safety of an extravascular implantable cardioverter–defibrillator. New England Journal of Medicine 2022, 387, 1292–1302. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Xu, B.; Zhu, Z.; Zhang, B. Topological Data Analysis Approach to Extract the Persistent Homology Features of Ballistocardiogram Signal in Unobstructive Atrial Fibrillation Detection. IEEE Sensors Journal 2022, 22, 6920–6930. [Google Scholar] [CrossRef]
- Ignacio, P.S.; Bulauan, J.A.; Manzanares, J.R. A Topology Informed Random Forest Classifier for ECG Classification. In Proceedings of the 2020 Computing in Cardiology; IEEE, 2020; pp. 1–4. [Google Scholar]
- Ignacio, P.S.; Dunstan, C.; Escobar, E.; Trujillo, L.; Uminsky, D. Classification of single-lead electrocardiograms: TDA informed machine learning. In Proceedings of the 2019 18th IEEE International Conference On Machine Learning And Applications (ICMLA); IEEE, 2019; pp. 1241–1246. [Google Scholar]
- Byers, M.; Hinkle, L.B.; Metsis, V. Topological Data Analysis of Time-Series as an Input Embedding for Deep Learning Models. In Proceedings of the IFIP International Conference on Artificial Intelligence Applications and Innovations; Springer, 2022; pp. 402–413. [Google Scholar]
- Seversky, L.M.; Davis, S.; Berger, M. On time-series topological data analysis: New data and opportunities. In Proceedings of the IEEE conference on computer vision and pattern recognition workshops; 2016; pp. 59–67. [Google Scholar]
- Karan, A.; Kaygun, A. Time series classification via topological data analysis. Expert Systems with Applications 2021, 183, 115326. [Google Scholar] [CrossRef]
- Sun, F.; Ni, Y.; Luo, Y.; Sun, H. ECG Classification Based on Wasserstein Scalar Curvature. Entropy 2022, 24, 1450. [Google Scholar] [CrossRef] [PubMed]
- Fraser, B.A.; Wachowiak, M.P.; Wachowiak-Smolíková, R. Time-delay lifts for physiological signal exploration: An application to ECG analysis. In Proceedings of the 2017 IEEE 30th Canadian Conference on Electrical and Computer Engineering (CCECE); IEEE, 2017; pp. 1–4. [Google Scholar]
- Dlugas, H. Electrocardiogram feature extraction and interval measurements using optimal representative cycles from persistent homology. bioRxiv 2022. [Google Scholar]
- Nicponski, J.; Jung, J.H. Topological data analysis of vascular disease: A theoretical framework. Frontiers in Applied Mathematics and Statistics 2020, 6, 34. [Google Scholar] [CrossRef]
- Bresten, C.L.; Kweon, J.; Chen, X.; Kim, Y.H.; Jung, J.H. Preprocessing of general stenotic vascular flow data for topological data analysis. bioRxiv 2021. Available online: https://www.biorxiv.org/content/early/2021/01/07/2021.01.07.425693.full.pdf. [CrossRef]
- Tokodi, M.; Shrestha, S.; Ashraf, M.; Casaclang-Verzosa, G.; Sengupta, P. Topological Data Analysis for quantifying inter-patient similarities in cardiac function. Journal of the American College of Cardiology 2019, 73, 751–751. [Google Scholar] [CrossRef]
- Tokodi, M.; Shrestha, S.; Bianco, C.; Kagiyama, N.; Casaclang-Verzosa, G.; Narula, J.; Sengupta, P.P. Interpatient similarities in cardiac function: a platform for personalized cardiovascular medicine. Cardiovascular Imaging 2020, 13, 1119–1132. [Google Scholar] [PubMed]
- Fasy, B.T.; Lecci, F.; Rinaldo, A.; Wasserman, L.; Balakrishnan, S.; Singh, A. Confidence sets for persistence diagrams. The Annals of Statistics 2014, 2301–2339. [Google Scholar] [CrossRef]
- Chazal, F.; Fasy, B.T.; Lecci, F.; Rinaldo, A.; Wasserman, L. Stochastic convergence of persistence landscapes and silhouettes. In Proceedings of the thirtieth annual symposium on Computational geometry; 2014; pp. 474–483. [Google Scholar]
- Wadhwa, R.R.; Williamson, D.F.; Dhawan, A.; Scott, J.G. TDAstats: R pipeline for computing persistent homology in topological data analysis. Journal of open source software 2018, 3, 860. [Google Scholar] [CrossRef]
- Bauer, U.; Kerber, M.; Reininghaus, J.; Wagner, H. Phat–persistent homology algorithms toolbox. Journal of symbolic computation 2017, 78, 76–90. [Google Scholar] [CrossRef]
- Bauer, U.; Kerber, M.; Reininghaus, J. Distributed computation of persistent homology. In Proceedings of the 2014 proceedings of the sixteenth workshop on algorithm engineering and experiments (ALENEX); SIAM, 2014; pp. 31–38. [Google Scholar]
- Zhang, S.; Xiao, M.; Wang, H. GPU-accelerated computation of Vietoris-Rips persistence barcodes. arXiv 2020, arXiv:2003.07989. [Google Scholar]
- Kerber, M.; Nigmetov, A. Efficient approximation of the matching distance for 2-parameter persistence. arXiv 2019, arXiv:1912.05826. [Google Scholar]
| Covering map | Type of data | Brief description |
|---|---|---|
| Vietoris-Rips Complex | Point cloud data, particularly when dealing with metric spaces. It is often used in applications like sensor networks, molecular chemistry, and computer graphics. |
The Vietoris-Rips complex connects points in the data if they are within a certain distance (the radius parameter) of each other, forming simplices (e.g., edges, triangles, tetrahedra) based on pairwise distances |
| Čech Complex | Similar to the Vietoris-Rips complex, Čech complexes are used for point cloud data in metric spaces |
The Čech complex connects points if they belong to the same open ball of a specified radius. It can capture similar topological features as the Vietoris-Rips complex but may have a different geometric structure |
| Alpha Complex | Alpha complexes are useful for point cloud data in metric spaces and provide an alternative representation of the topological structure |
The alpha complex connects points with a Delaunay triangulation, considering balls whose radii can vary at each point to ensure that the complex is a subcomplex of the Vietoris-Rips complex |
| Witness Complex | Witness complexes are used for point cloud data but are particularly useful when dealing with data that may not be uniformly sampled or when dealing with non-metric or qualitative data |
Witness complexes are constructed by selecting a subset of witness points from the data. Each witness point witnesses the presence of other data points within a specified distance. This can be used to capture topological features in a more robust way, especially when data is irregular |
| Mapper | Mapper is a flexible approach that can be applied to various types of data, including both metric and non-metric spaces |
Mapper is not a traditional covering map but rather a method for creating a topological summary of data by combining clustering and graph theory. It can be adapted to different data types and is useful for exploratory data analysis |
| Rips-Filtration and Čech-Filtration | These are extensions of the Vietoris-Rips and Čech complexes, respectively, that allow for the analysis of topological features at different scales |
By varying the radius parameter continuously, Rips-filtration and Čech-filtration produce a sequence of simplicial complexes. This can be useful for capturing topological features at different levels of detail and studying persistence diagrams |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





