1. Introduction
Endometriosis is a common problem in women. Its name comes from the word "endometrium", meaning the thin inner lining of the uterus [
1]. It is the growth of tissue usually found in the inner lining of the uterus (endometrium) in a location outside the uterine cavity. With the changing hormone levels during the menstrual cycle, the tissue can grow and/or break down, leading to the development of pathological tissue - referred to as scarring - and eventually abnormally high levels of pain. It can occur in the ovaries, fallopian tubes, behind the uterus, in the tissues that hold the uterus in place, in the intestine, abdominal wall, or other organs, i.e. in other areas of the body where it does not normally belong [
2]. Other locations where endometriosis can develop are the vagina, cervix, vulva, or even the bladder [
1,
2]. Endometriosis has multiple appearances, and the lesions may be confused with other non-endometriotic lesions, as well as endometriotic lesions that are nonendometriotic by appearance or deep infiltrating ones that may be missed on visual diagnosis. How often endometriosis occurs in women cannot be accurately determined, as the diagnosis is usually made only by direct visualization of the endometrial tissue (which requires a surgical procedure, typically a laparoscopy) [
3]. It is estimated, however, that approximately 6 to 10% of all women suffer from endometriosis. The percentage of women who have endometriosis is higher among women who are infertile (25 to 50%) and women who have pelvic pain (75 to 80%). The average age at diagnosis is 27, but it can also develop in adolescence.
According to studies, in the United States of America, more than 5 million women are faced with the problem of endometriosis. In a survey that was conducted, on 96 women it was observed that using histopathology as the gold standard, sensitivity for laparoscopic visualization was 90.1% (95% CI 81.0-95.1), while specificity was 40.0% (95% CI 23.4-59.3). Positive and negative predictive values were 81.0% (95% CI 71.0-88.1) and 58.8% (95% CI 36.0-78.4), respectively; and the accuracy was 77.1% (95% CI 67.7-84.4) [
4]. Laparoscopy is the minimally invasive method a doctor can use to see if there are areas of endometriosis. The endoscope is inserted into the abdominal cavity through a small incision most often made just above or below the belly button. It is the only safe way to know for sure that you have endometriosis. If it is not clear whether the detected tissue is normal or endometrial, a sample of the tissue is taken for biopsy. Depending on the location of the tissue, taking a sample for biopsy can be done endoscopically through the anus (sigmoidoscopy) or the bladder (cystoscopy) [
1,
2,
3,
5]. Based on the findings of laparoscopy, 4 stages of severity of the disease can be distinguished. Staging is judged according to the number of foci, their size and depth, the presence of adhesions, and the presence of endometriomas. The most severe stages require radical surgical treatment [
6]. For most women with moderate to severe endometriosis, the most effective treatment is surgical removal or destruction of the endometrial tissue. In the past, endometriosis was treated with open surgery, which involved a large incision. Now, however, it has prevailed that the surgery is performed with laparoscopic surgery or with its evolution, robotic surgery, to achieve the optimal medical result with the least possible tissue injury. Laparoscopic endometriosis repair is a minimally invasive procedure that involves not a large incision, like traditional open surgeries, but 3-4 very small holes in the patient's abdomen, through which laparoscopic instruments are inserted. Among them is the laparoscope, which includes a camera that offers the surgeon an extremely sharp image. This allows him to investigate all foci of endometriosis, minimizing injury to neighboring tissues and organs [
1,
7,
8,
9]. Through laparoscopy, medical video files can be created, for post-operative analysis. This enables physicians to review interventions at any time to gain useful insights or improve treatment planning and medical education [
10]. Computer-aided automatic content analysis can be employed for creating systems that highlight potentially relevant content to physicians during patient case inspections (
Figure 1). However, although obvious irrelevant video segments such as overly blurry frames or camera testing screens can reasonably be identified via video analysis, more sophisticated approaches are required for identifying very specific content such as scenes showing endometriosis lesions [
11].
The best-known classification system for endometriosis is the revised American Society for Reproductive Medicine (rASRM) score and the Enzian classification scheme [
12,
13]. The rASRM score describes superficial lesions of the peritoneum and ovarian endometriosis in four stages, whereas the Enzian classification categorizes deep endometriosis. Alongside these different possible locations, endometriotic lesions also strongly vary in their visual appearance, both intra and interpersonal. The rASRM score describes superficial lesions of the peritoneum and ovarian endometriosis in four stages, whereas the Enzian classification categorizes deep endometriosis. In direct comparison and without a specific medical background, the differences between normal and pathological tissue are very difficult to discern, which evidently holds true for laymen but even inexperienced medical practitioners. Consequently, with the current successful application of deep learning in many medical fields, attempting to solve this problem via computer-aided analysis seems reasonable. Moreover, being able to classify and potentially locate endometriosis can not only be helpful during interventions but also in treatment planning and particularly in teaching/training.
The purpose of this research is the development and evaluation of an intelligent assistive classification system for the recognition of endometriosis and the improvement of the accuracy of laparoscopic imaging in diagnosis. The research was developed with the use of Deep Learning approaches. By revising the labeling strategy of the publicly available endometriosis dataset GLENDA towards visual similarity, we discover a large improvement in lesion segmentation performance. Data from 4448 laparoscopy images were used [
14]. Based on simple clinical and imaging information and criteria such as the diagnosis of endometriosis (included in an open source dataset Glenda of repository Kaggle), data mining algorithms were used to improve laparoscopic imaging accuracy. The final developed computer system based on the ResNet50 algorithm predicted the best outcome for all participants who had laparoscopic surgical therapy. The Keras tool was used and the generated code was implemented in Python language.
1.1. Related work
Laparoscopy is a minimally invasive procedure often alongside histopathological confirmation for diagnosing all types of pelvic endometriosis (ovarian endometriomas, DE, SE) since surgeons can directly visualize the pelvic and abdominal cavity. In 2022, ESHRE no longer considered laparoscopy as the "gold standard" and recommends its use if initial imaging results are negative and/or patients are not suitable/unresponsive to empirical treatment [
15]. Depending on the goal, during the same operation, surgeons could also aim for complete treatment of endometriotic lesions to provide symptomatic control and reduce the number of laparoscopies (which carries its own risks). Herein lies the issue with surgery as the diagnostic test of choice – many patients exhibit endometriosis that cannot be appropriately managed at a surgery that is simultaneously diagnostic. Laparoscopy for endometriosis should always involve a comprehensive exploration of the abdominal and pelvic contents. To identify subtle lesions, the laparoscope must be brought right up to the surfaces being inspected. In addition, the gastrointestinal and genitourinary systems need to be assessed as well, including the appendix. The diaphragm should routinely be inspected, especially if the patient describes right upper quadrant symptoms. If the patient underwent a preoperative ultrasound/MRI where endometriosis was visualized, these areas should be closely inspected surgically. In some situations, endometriosis may not be obviously visible at laparoscopy despite clear identification with imaging [
16].
SE has been described to have a black (“powder burn”) or dark bluish appearance from the accumulation of blood pigments [
17]. However, subtle forms can appear as white opacifications, red flamelike lesions, or yellow-brown patches in earlier, active stages of the disease [
18]. Ovarian endometriomas have a distinct morphology classically described as a "chocolate cyst" containing old menstrual blood giving it a dark brown appearance. Adhesions are often found in association with endometriomas and consist of fibrous scar tissue because of chronic inflammation. In many cases, there is endometriosis at the site of ovarian fixation [
19,
20]. Like an imaging-based assessment following the IDEA protocol, the posterior and anterior compartments should be assessed carefully for DE. Oftentimes, DE appears as multifocal nodules and may infiltrate the surrounding viscera and peritoneal tissue [
21]. As mentioned earlier, depending on the extent of POD obliteration, posterior compartment (USL, bowel, PVF) DE may be difficult to assess and diagnose surgically, with evidence supporting a better diagnostic test performance using non-invasive imaging-based assessment for this specific type of DE [
22].
When combining diagnostic laparoscopy with operative laparoscopy, the surgeon must consider the importance of a biopsy to ensure their visual diagnosis is correct. There is ample evidence that surgeons overcall endometriosis at surgery [
23,
24,
25]. When surgeons perform endometriosis excision, all specimens should be sent to pathology for analysis. In some cases, it is inappropriate to use biopsy as a diagnostic test. For example, in a patient who undergoes diagnostic laparoscopy and the surgeon suspects bowel DE, this area should only be biopsied/excised if the patient explicitly provided informed consent following a discussion about the benefits and risks of a “bowel surgery” component and the patient was adequately prepared with bowel preparation and antibiotic prophylaxis. Surgeons should also consider concurrent appendectomy during excision of endometriosis since women with DE have a high risk of appendiceal endometriosis [
26]. Often, appendectomy is not within the skill set of gynecologists, which again creates a unique challenge with surgery being used as a combined diagnostic test and treatment modality.
Deep convolutional networks such as GoogLeNet [
27] have been successfully applied in countless domains. Such networks represent valuable backbones in many deep architectures for image and video analysis. One such family of architectures that heavily use CNNs as a backbone for region-of-interest (ROI) prediction and labeling is called region-based convolutional neural networks, or R-CNNs. These R-CNNs after sufficient training are capable of detecting, classifying, and even segmenting objects in images through intelligent arrangement and use of CNN elements. The increasing performance improvements of deep learning in medicine entail an ever-expanding range of applications aimed at providing digital assistance to medical personnel in treating patients. Medical imaging also varies greatly according to its purpose: monochrome images obtained from computed tomography (CT) or ultrasound are very different compared to open surgery or endoscopic recordings. This generally makes scientific research on medical image classification more difficult to compare than traditional multimedia analysis. Surprisingly, endoscopic images have so far not been analyzed as digitally as many other technologies such as magnetic resonance imaging (MRI) [
28]. However, they offer a wide variety of research topics for the application of deep learning. For example, there are many studies on the classification or detection of content such as anatomy and surgical tasks [
29,
30,
31].
Ιn a recent research which is very close to our approach, Visalaxi et al 2021 have provided a system for automatic diagnosis of endometriosis using deep learning techniques [
32]. The trained model was verified based on the input images used and then model was used to classify the category as pathological and non-pathological images. The architecture of neural network model was designed where input consist of laparoscopic images are split as training, test and validation group independent of each other. The obtained image is in BGR format which once again converted into RGB format i.e. images are annotated and then converted into Numpy array format [
33]. The training and test image dataset in array format are split as features and labels. Since it is binary class, only two labels are mentioned in the model. The trained and tested accuracy were found to be 91% and 90% respectively. The proposed model identifies the endometriosis by providing the laparoscopic images alone as parameters for recognising the presence. An effective and efficient approach of OpenCV for pre-processing the data and ResNet50 based architecture for training and testing the model of large datasets with raw laparoscopic images were used. The prognostic model yields high accuracy (1) and throughputs as laparoscopic images were given as input.
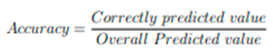
(1)
Accuracy is the quantity to measure the unambiguous value [
34]. The accuracy and loss were calculated by fitting the model in the architecture. The trained and tested model yielded an accuracy of 92% and 90% respectively. The predicted model yielded an accuracy of 90%.
2. Methodoly
Our approach aimed to detect and segment endometriosis. We start by using the published GLENDA dataset. Next, we thoroughly describe all the data augmentation techniques used. We detail the model training strategies and finally list our evaluation results.
2.1. Patient population and data collection
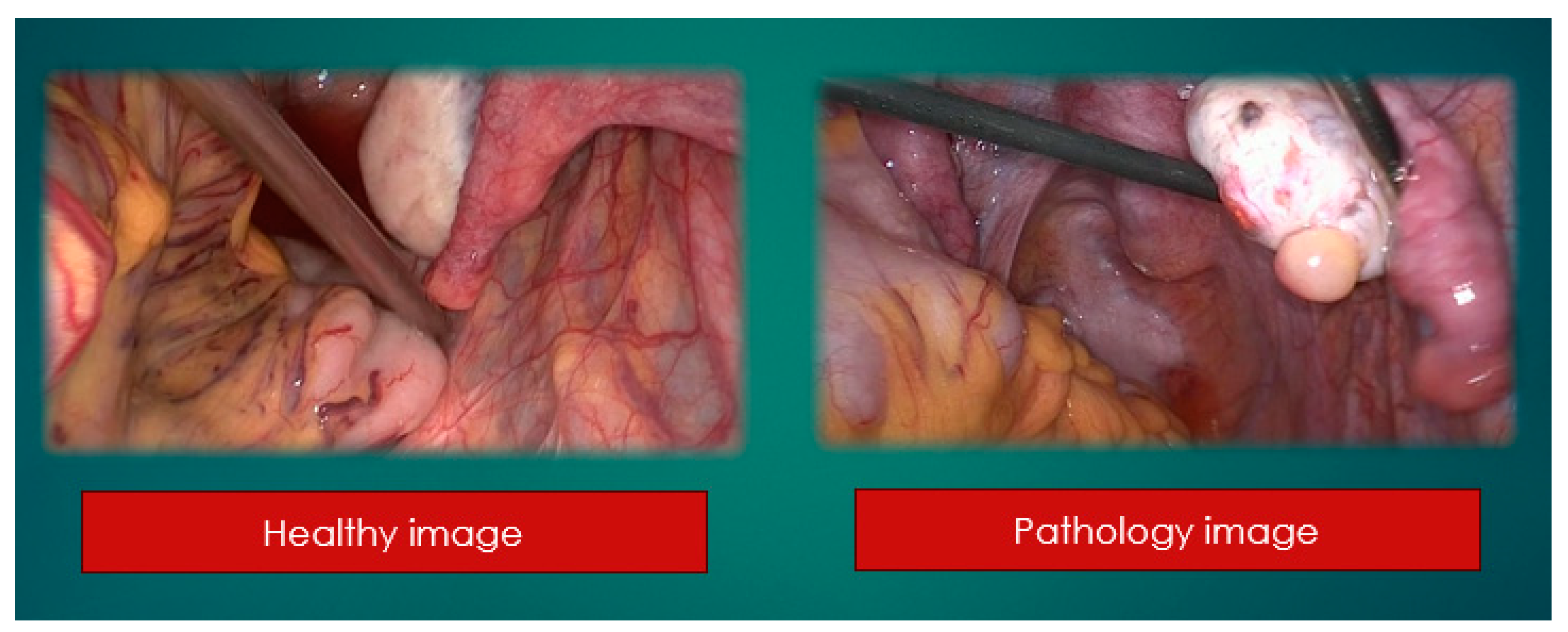
At the beginning a retrospective study of patients' health and pathology ones. 2157 healthy and 2291 pathological (
Figure 1). Based on the final results, we developed a novel assistant intelligent automated method to discriminate endometriosis with the improvement of laparoscopic images, as an additional tool to preoperative evaluation and improved planning of a minimally directed surgery. Glenda (Gynecologic Laparoscopy Endometriosis Dataset) was used. Gynecologic laparoscopy as a type of minimally invasive surgery (MIS) is performed via a live feed of a patient’s abdomen surveying the insertion and handling of various instruments for conducting treatment. Adopting this kind of surgical intervention not only facilitates a great variety of treatments, as well as it is also essential for numerous post-surgical activities, such as treatment planning, case documentation, and education. Nonetheless, the process of manually analyzing surgical images, as it is carried out in current practice, usually proves tediously time-consuming. In order to improve upon this situation, more sophisticated computer vision as well as machine learning approaches are actively developed. Since most such approaches heavily rely on sample data, which especially in the medical field is only sparsely available, with this work we publish the Gynecologic Laparoscopy ENdometriosis DAtaset (GLENDA) – an image dataset containing region-based annotations of a common medical condition named endometriosis, i.e. the dislocation of uterine-like tissue. The dataset is the first of its kind and it has been created in collaboration with leading medical experts in the field (
Figure 2) [
35].
2.2. Preoperative approach and final surgical procedure
Historically the only potential cure for endometriosis is laparoscopic.
2.2.1. Surgical Staging Methods
In 2017, the World Endometriosis Society consensus for the diagnosis of endometriosis recommended the use of the rASRM classification tool during surgery to classify endometriosis based on the size, extent, and location of lesions found (Figure 4) [
36]. There are four stages ranging from minimal (Stage 1), mild (Stage 2), moderate (Stage 3), and severe (Stage 4) [
36,
37]. However, a prospective analysis found considerable inter-variability among surgeons reducing its diagnostic accuracy. In 2021, the AAGL Special Interest Group in Endometriosis published the AAGL Classification Tool solely based on intraoperative findings to better quantify the surgical complexity of endometriosis cases (Figure 5). Like the rASRM system, there are four AAGL endometriosis stages. However, the recent validation study found higher reproducibility with the AAGL score (kappa = 0.621) compared to rASRM (kappa = 0.317) when discriminating surgical complexity [
38]. The rASRM system fails to properly incorporate DE, which often equates to surgical complexity, leading to this deficiency in the system. The Enzian classification tool has been introduced in recent years to better describe DE, and has recently introduced the updated system, #Enzian, which also incorporates SE and tubal pathology [
36,
39]. A recent study by Montanari and colleagues demonstrates that the #Enzian system score can be accurately predicted with ultrasound, increasing the utility of this model significantly [
40]. The Endometriosis Fertility Index (EFI), reliant on the rASRM staging system, was also recommended after surgical staging/treatment for women considering pregnancy in the future [
36,
40]. Although several staging tools exist, they mainly focus on the physical extent of the disease and do not correlate well with the pain symptoms and impact on quality of life for patients [
41]. Beyond accurately representing the experience of the patient, there should be functionality to prognosticate clinical outcomes, which currently only the EFI attempts to do. The utility of the newest system, the AAGL Classification Tool, remains to be seen but it must be validated [
42].
3. Model Development/Results
Keras is an open-source library that provides a Python interface for artificial neural networks. Keras acts as an interface for the TensorFlow library.
Up until version 2.3, Keras supported multiple backends, including TensorFlow, Microsoft Cognitive Toolkit, Theano, and PlaidML [
44,
45,
46]. As of version 2.4, only TensorFlow is supported. However, starting with version 3.0 (including its preview version, Keras Core), Keras will become multi-backend again, supporting TensorFlow, JAX, and PyTorch [
47]. Designed to enable fast experimentation with deep neural networks, it focuses on being user-friendly, modular, and extensible. It was developed as part of the research effort of project ONEIROS (Open-ended Neuro-Electronic Intelligent Robot Operating System) and its primary author and maintainer is François Chollet, a Google engineer. Chollet is also the author of the Xception deep neural network model [
48,
49].
Keras contains numerous implementations of commonly used neural network building blocks such as layers, objectives, activation functions, optimizers, and a host of tools for working with image and text data to simplify programming in deep neural network areas. The code is hosted on GitHub, and community support forums include the GitHub issues page and a Slack channel.
In addition to standard neural networks, Keras has support for convolutional and recurrent neural networks. It supports other common utility layers like dropout, batch normalization, and pooling [
50].
Keras allows users to produce deep models on smartphones (iOS and Android), on the web, or on the Java Virtual Machine [
45]. It also allows the use of distributed training of deep-learning models on clusters of graphics processing units (GPU) and tensor processing units (TPU) [
51]. Bibliographic research indicated that ResNet-50 is the best option as a base model for medical image data.
Using ResNet-50 for creating a medical image model is a popular choice due to its deep architecture and effectiveness in handling complex features in images. ResNet-50 is a variant of the ResNet (Residual Network) model, which introduced the concept of residual learning [
52,
53,
54]. This approach allows the model to learn residual functions, making it easier to train very deep neural networks without encountering the vanishing gradient problem. In the context of medical image analysis, where images can be highly detailed and intricate, the depth of the network and its ability to capture subtle patterns and features become crucial.
Here are a few reasons why ResNet-50 might be chosen for medical image analysis:
3.1. Handling Deep Networks
ResNet-50's architecture with residual blocks enables the training of very deep networks (50 layers in this case) without vanishing gradient issues. Deeper networks can learn more complex features, which is valuable in medical image analysis where identifying intricate patterns is essential [
54].
3.2. Feature Extraction
ResNet-50 excels at feature extraction. Medical images often contain hierarchical and multi-scale features. ResNet-50's design allows it to automatically learn and extract features at various levels of abstraction, making it suitable for diverse medical imaging tasks [
52,
53,
54].
3.3. Pretrained Models and Transfer Learning
ResNet-50 models pre-trained on large datasets (like ImageNet) are readily available. Transfer learning, where a pre-trained model is fine-tuned on a specific dataset, can be incredibly effective in medical image analysis where labeled datasets are limited. By using pre-trained weights, the model has already learned generic features from a massive dataset, which can boost its performance on smaller, specialized datasets [
52,
53,
54].
3.4. Regularisation and Generalization
The skip connections (residual connections) in ResNet-50 act as implicit regularizers. They help prevent overfitting, which is crucial when dealing with medical image datasets that are often small and can be prone to overfitting [
52,
53,
54].
3.5. Research and Benchmarking
ResNet-50 has been extensively studied and used in various research papers and competitions. Its performance and capabilities are well-documented in the literature, making it a reliable choice for medical image analysis tasks [
52,
53,
54].
By leveraging ResNet-50, researchers and practitioners in the field of medical image analysis can benefit from the model's depth, feature extraction capabilities, and the advantages of transfer learning, ultimately leading to more accurate and robust medical image analysis systems.
3.6. Pathological Class Processing
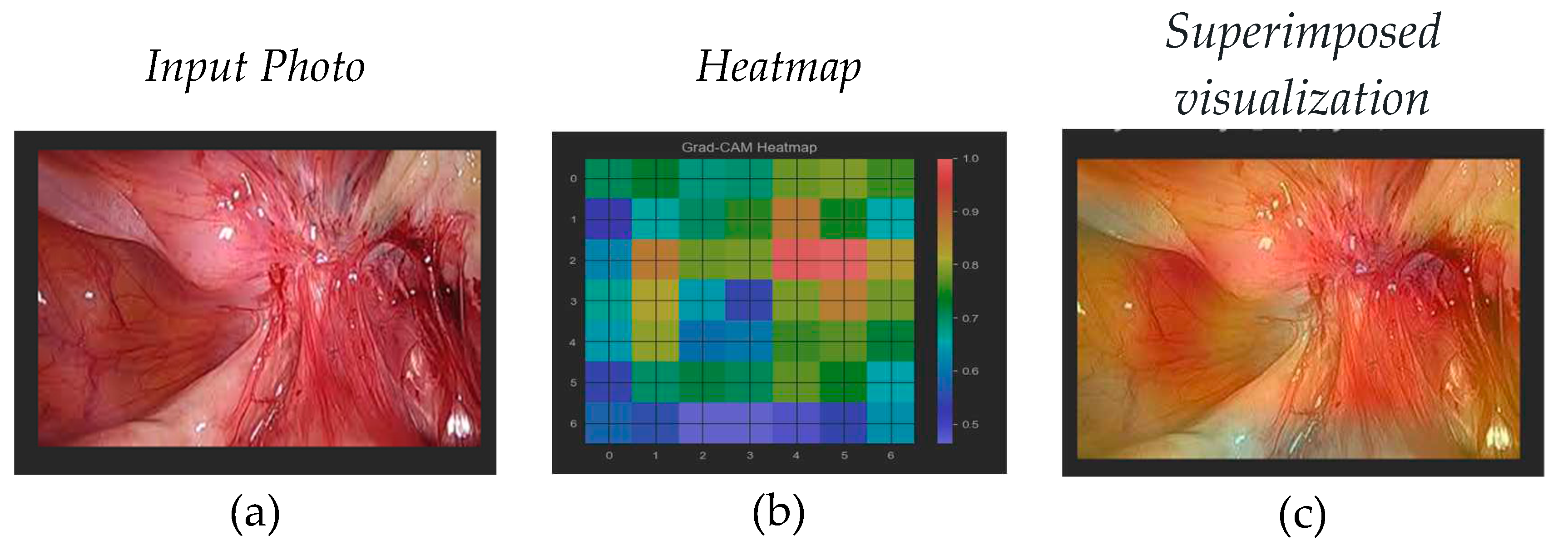
Visualizing the specific features that the network has learned to recognize as indicative of the pathological class can be challenging due to the complexity of deep neural networks. Techniques like activation maximization or gradient-based visualization methods can be used to gain some insights, but interpreting deep learning models in detail remains an ongoing area of research. In order to access the layer that can produce meaningful images from the understanding of the classes and the features used for classification we need to identify the latest 4-dimensional layer (
Figure 3).
3.7. Data Preparation
Overview:
All data are separated into two folders, healthy and pathology ones.
2157 healthy
2291 pathology
80% for training, 20% for validation
Several preprocessing steps are applied to our dataset containing images of two classes: 'healthy' and 'pathology'.
Here's a breakdown of the preprocessing steps:
1.
Loading Images: Images from the directories specified in the `base_path` variable are loaded using the `load_img` function. This function loads an image file into a PIL (Python Imaging Library) object [
7,
55,
56].
2. Converting Images to Arrays: The loaded images are converted into numerical arrays using the `img_to_array` function from Keras. This function converts a PIL array instance to a Numpy array.
3. Resizing Images: The images are resized to fit the input size expected by the ResNet-50 model, which is 224x224 pixels. OpenCV's `cv2.resize` function is used for this purpose.
4. Building X (Feature) and y (Label) Arrays: The resized image arrays are appended to the list `X`, and the corresponding class labels ('healthy' or 'pathology') are appended to the list `y`. Vectorization also takes place in this step. Vectorization refers to the process of converting non-numeric data into a numerical format that can be processed by machine learning algorithms.
5. Converting y Labels to Numerical Format: The class labels in the list `y` are mapped to numerical values using a dictionary `y_dict`. 'healthy' is mapped to 0, and 'pathology' is mapped to 1.
6. Shuffling the Data: The data (both X and y) is shuffled using `np.random.permutation`. Shuffling the data is essential to ensure that the model does not learn any order-based patterns during training.
3.8. Choosing a basic model
Initially we decided to build a basic model and evaluate its efficiency for our dataset.
Therefore we chose a basic Sequential model with two additional layers:
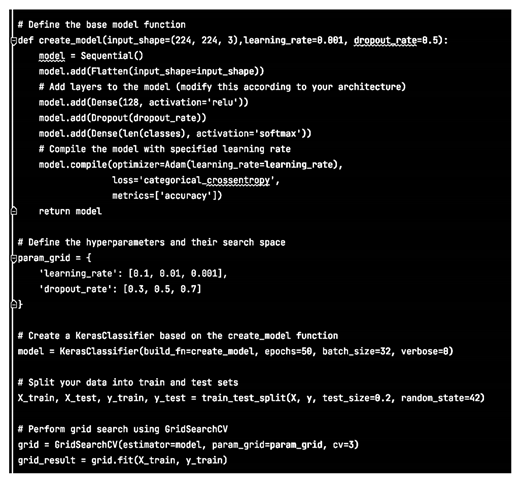
Consequently the results are extracted with:
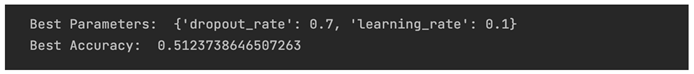
Based on the results, even after hyper-parameter tuning exploration, the accuracy could be described as equal to a randomly assigned result.
3.9. Model training with Resnet50
We chose as final model resnet50 with the same additional layers as in the base model. We used the same hyper-parameters as well.
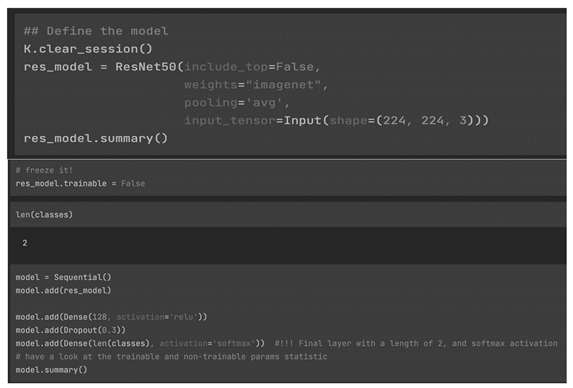
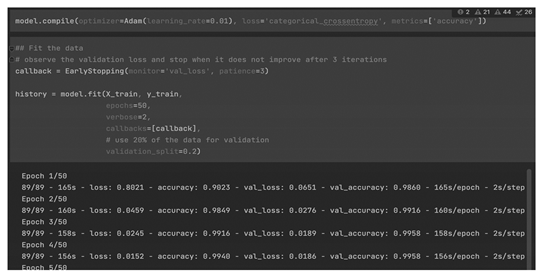
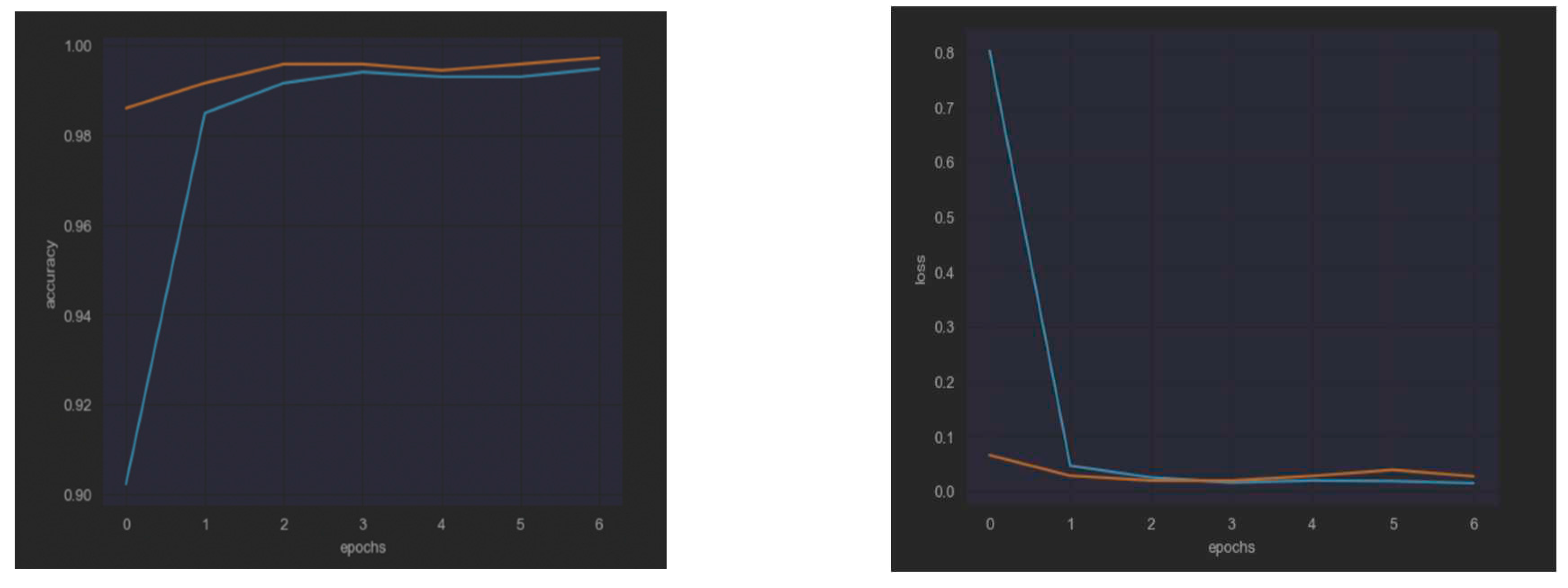
Checking the
accuracy and the
loss curves for both models we can see the results (
Figure 4):
Consequently, the best model is
ResNet50 because it provides High Accuracy in both validation and training data, with no over-fitting (
Table 1).
4. Discussion
In clinical practice the minimally invasive laparoscopy can be demonstrated beneficial as it could support health professionals in making critical and evidence-based medical decisions.
Closer to our approach Visalaxi et al 2021 have provided a system for automatic diagnosis of endometriosis using deep learning techniques by using ResNet50 algorithm and yielded an accuracy of 91%.
However, they reported that with a larger data set, they might have better accuracy rates. We developed our research using a much larger data set.
The final developed computer system in our research, based also on the ResNet50 algorithm, predicted the best outcome for all participants who had laparoscopic surgical therapy. The Keras tool was used and the generated code was implemented in Python language providing a mean accuracy of 99 %.
The presented approach revealed better performance than the commonly used imaging criteria in predicting endometriosis as it improves the time and the total accuracy of evidence-based diagnosis and consequently the following treatment. Additionally, using video during endoscopy the system could compare the input images with the internal reasoning model, even on-line accurate results.
References
- «Endometriosis: Overview». www.nichd.nih.gov. (. 18 May 2017.
- «Endometriosis: Condition Information». www.nichd.nih.gov (30 April 2017).
- «Endometriosis». womenshealth.gov. (). 13 February 2017.
- Gratton, S.-M.; Choudhry, A.J.; Vilos, G.A.; Vilos, A.; Baier, K.; Holubeshen, S.; Medor, M.C.; Mercier, S.; Nguyen, V.; Chen, I. Diagnosis of Endometriosis at Laparoscopy: A Validation Study Comparing Surgeon Visualization with Histologic Findings. J. Obstet. Gynaecol. Can. 2021, 44, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Bulletti, C.; Coccia, M.E.; Battistoni, S.; Borini, A. Endometriosis and infertility. J. Assist. Reprod. Genet. 2010, 27, 441–447. [Google Scholar] [CrossRef] [PubMed]
- Vercellini, P.; Eskenazi, B.; Consonni, D.; Somigliana, E.; Parazzini, F.; Abbiati, A.; Fedele, L. Oral contraceptives and risk of endometriosis: a systematic review and meta-analysis. Hum. Reprod. Updat. 2010, 17, 159–170. [Google Scholar] [CrossRef] [PubMed]
- GBD 2015 Disease and Injury Incidence and Prevalence Collaborators (8 October 2016). «Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990–2015: a systematic analysis for the Global Burden of Disease Study 2015». Lancet (London, England) 388 (10053): 1545–1602. . ISSN 0140-6736. PMID 27733282. PMC 5055577 (10 September 2017). [CrossRef]
- McGrath, Patrick J.· Stevens, Bonnie J.· Walker, Suellen M.· Zempsky, William T. (2013). Oxford Textbook of Paediatric Pain. (OUP Oxford. pp. 300. ISBN 9780199642656. (10 September 2017).
- Naghavi, M.; Wang, H.; Lozano, R.; Davis, A.; Liang, X.; Zhou, M.; Vollset, S.E.; Abbasoglu Ozgoren, A.; Abdalla, S.; Abd-Allah, F.; et al. Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990–2013: A systematic analysis for the Global Burden of Disease Study 2013. Lancet 2015, 385, 117–171. [Google Scholar] [CrossRef]
- Brosens I (2012). Endometriosis: Science and Practice. John Wiley & Sons. page. 3. ISBN 9781444398496.
- Leibetseder A, Petscharnig S, Primus MJ, Kletz S, Münzer B, Schoefmann K, Keckstein J (2018) Lapgyn4: a dataset for 4 automatic content analysis problems in the domain of laparoscopic gynecology. In: César P, Zink M, Murray N (eds) Proceedings of the 9th ACM multimedia systems conference, MMSys 2018, -15, 2018. ACM, Amsterdam, The Netherlands, pp 357–362. https:// doi.org/10.1145/3204949. 12 June 3208.
- Canis, M.; Donnez, J.G.; Guzick, D.S.; Halme, J.K.; Rock, J.A.; Schenken, R.S.; Vernon, M.W. Revised American Society for Reproductive Medicine classification of endometriosis: 1996. Fertil. Steril. 1997, 67, 817–821. [Google Scholar] [CrossRef]
- Keckstein J, Hudelist G. Classification of deep endometriosis (DE) including bowel endometriosis: From r-ASRM to# Enzian-classification. Best Pract. Res. Clin. Obstet. Gynaecol. 2021, 71, 27–37. [Google Scholar]
- Leibetseder A, Kletz S, Schoeffmann K, Keckstein S, Keckstein J (2020) GLENDA: gynecologic laparoscopy endometriosis dataset. In: Ro YM, Cheng W, Kim J, Chu W, Cui P, Choi J, Hu M, Neve WD (eds) MultiMedia Modeling - 26th International Conference, MMM 2020, Daejeon, South Korea, -8, 2020, Proceedings, Part II, Springer, Lecture Notes in Computer Science, vol 11962, pp 439–450. 5 January. [CrossRef]
- Becker, C.M.; Bokor, A.; Heikinheimo, O.; Horne, A.; Jansen, F.; Kiesel, L.; King, K.; Kvaskoff, M.; Nap, A.; Petersen, K.; et al. ESHRE guideline: endometriosis. Hum. Reprod. Open 2022, 2022, hoac009. [Google Scholar] [CrossRef]
- Goncalves, M.O.; Neto, J.S.; Andres, M.P.; Siufi, D.; de Mattos, L.A.; Abrao, M.S. Systematic evaluation of endometriosis by transvaginal ultrasound can accurately replace diagnostic laparoscopy, mainly for deep and ovarian endometriosis. Hum. Reprod. 2021, 36, 1492–1500. [Google Scholar] [CrossRef]
- Mettler, L.; Schollmeyer, T.; Lehmann-Willenbrock, E.; Schüppler, U.; Schmutzler, A.; Shukla, D.; Zavala, A.; Lewin, A. Accuracy of Laparoscopic Diagnosis of Endometriosis. JSLS 2003, 7, 15–18. [Google Scholar] [CrossRef]
- Jansen RP, Russell P. Nonpigmented endometriosis: clinical, laparoscopic, and pathologic definition. Am. J. Obstet. Gynecol. 1986, 155, 1154–1159. [Google Scholar] [CrossRef]
- Rao T, Condous G, Reid S. Ovarian Immobility at Transvaginal Ultrasound: An Important Sonographic Marker for Prediction of Need for Pelvic Sidewall Surgery in Women with Suspected Endometriosis. J. Ultrasound Med. 2021, 41, 1109–1113. [Google Scholar] [CrossRef]
- Reid S, Leonardi M, Lu C, et al. The association between ultrasound-based 'soft markers' and endometriosis type/location: A prospective observational study. Eur. J. Obstet. Gynecol. Reprod. Biol. 2019, 234, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Koninckx PR, Ussia A, Adamyan L, et al. Deep endometriosis: definition, diagnosis, and treatment. Fertil Steril 2012, 98, 564–571. [Google Scholar] [CrossRef] [PubMed]
- Leonardi M, Condous G. A pictorial guide to the ultrasound identification and assessment of uterosacral ligaments in women with potential endometriosis. Australas. J. Ultrasound Med. 2019, 22, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Gratton, S.-M.; Choudhry, A.J.; Vilos, G.A.; Vilos, A.; Baier, K.; Holubeshen, S.; Medor, M.C.; Mercier, S.; Nguyen, V.; Chen, I. Diagnosis of Endometriosis at Laparoscopy: A Validation Study Comparing Surgeon Visualization with Histologic Findings. J. Obstet. Gynaecol. Can. 2021, 44, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Fernando, S.; Soh, P.Q.; Cooper, M.; Evans, S.; Reid, G.; Tsaltas, J.; Rombauts, L. Reliability of Visual Diagnosis of Endometriosis. J. Minim. Invasive Gynecol. 2013, 20, 783–789. [Google Scholar] [CrossRef] [PubMed]
- Wykes CB, Clark TJ, Khan KS. Accuracy of laparoscopy in the diagnosis of endometriosis: a systematic quantitative review. BJOG Int. J. Obstet. Gynaecol. 2004, 111, 1204–1212. [Google Scholar] [CrossRef] [PubMed]
- Moulder JK, Siedhoff MT, Melvin KL, et al. Risk of appendiceal endometriosis among women with deep-infiltrating endometriosis. Int. J. Gynaecol. Obstet. 2017, 139, 149–154. [Google Scholar] [CrossRef]
- Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, Rabinovich A (2015) Going deeper with convolutions. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 1–9 ]. or ResNet [ He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: 2016 IEEE conference on computer vision and pattern recognition, CVPR 2016, Las Vegas, NV, USA, -30, 2016, IEEE Computer Society, pp 770–778. 27 June. [CrossRef]
- Rai HM, Chatterjee K, Gupta A, Dubey A (2020) A novel deep cnn model for classification of brain tumor from mr images. In: 2020 IEEE 1st international conference for convergence in engineering (ICCE), pp 134–138. [CrossRef]
- Leibetseder A, Petscharnig S, Primus MJ, Kletz S, Münzer B, Schoeffmann K, Keckstein J (2018) Lapgyn4: a dataset for 4 automatic content analysis problems in the domain of laparoscopic gynecology. In: César P, Zink M, Murray N (eds) Proceedings of the 9th ACM multimedia systems conference, MMSys 2018, -15, 2018. ACM, Amsterdam, The Netherlands, pp 357–362. 12 June. [CrossRef]
- Zadeh SM, Francois T, Calvet L, Chauvet P, Canis M, Bartoli A, Bourdel N. Surgai: deep learning for computerized laparoscopic image understanding in gynecology. Surg. Endosc. 2020, 34, 5377–5383. [Google Scholar] [CrossRef]
- Petscharnig S, Schöffmann K. Learning laparoscopic video shot classification for gynecological surgery. Multimed. Tools Appl. 2018, 77, 8061–8079. [Google Scholar] [CrossRef]
- Automated prediction of endometriosis using deep learning S. Visalaxia, T. Automated prediction of endometriosis using deep learning S. Visalaxia, T. Sudalai Muthua Int. J. Nonlinear Anal. Appl. 12 (2021) No. 2: 2, 2403-2416 ISSN, 2403; 2. [Google Scholar] [CrossRef]
- G. Bradski and A. Kaehler, Learning OpenCV: Computer Vision with the OpenCV Library, O’Reilly Media, Inc. 2008.
- Y. D. Wang, M. Shabaninejad, R.T. Armstrong and P. Mostaghimi, Deep neural networks for improving physical accuracy of 2D and 3D multi-mineral segmentation of rock micro-CT images. Appl. Soft Comput. 2021, 104, 107185. [Google Scholar] [CrossRef]
- GLENDA dataset https://dl.acm.org/doi/abs/10. 1007.
- Johnson NP, Hummelshoj L, Adamson GD, et al. World Endometriosis Society consensus on the classification of endometriosis. Hum. Reprod. 2017, 32, 315–324. [Google Scholar] [CrossRef]
- American Society for Reproductive, M. Revised American Society for Reproductive Medicine classification of endometriosis: 1996. Fertil. Steril. 1997, 67, 817–821. [Google Scholar] [CrossRef] [PubMed]
- Abrao MS, Andres MP, Miller CE, et al. AAGL 2021 Endometriosis Classification: An Anatomy-based Surgical Complexity Score. J. Minim. Invasive Gynecol. 2021, 28, 1941–1950. [Google Scholar] [CrossRef]
- Keckstein J, Saridogan E, Ulrich UA, et al. The #Enzian classification: A comprehensive non-invasive and surgical description system for endometriosis. Acta Obstet Gynecol Scand 2021, 100, 1165–1175. [Google Scholar]
- Adamson GD, Pasta DJ. Endometriosis fertility index: the new, validated endometriosis staging system. Fertil Steril 2010, 94, 1609–1615. [Google Scholar] [CrossRef] [PubMed]
- International working group of AAGL E, ESHRE, WES, et al. Endometriosis classification, staging and reporting systems: a review on the road to a universally accepted endometriosis classification†,‡. Hum. Reprod. Open 2021, 2021, hoab025. [Google Scholar] [CrossRef]
- Espada, M.; Leonardi, M.; Reid, S.; Condous, G. Regarding “AAGL 2021 Endometriosis Classification: An Anatomy-based Surgical Complexity Score”. J. Minim. Invasive Gynecol. 2021, 29, 449–450. [Google Scholar] [CrossRef]
- Montanari, E.; Bokor, A.; Szabó, G.; Kondo, W.; Trippia, C.H.; Malzoni, M.; Di Giovanni, A.; Tinneberg, H.R.; Oberstein, A.; Rocha, R.M.; et al. Accuracy of sonography for non-invasive detection of ovarian and deep endometriosis using #Enzian classification: prospective multicenter diagnostic accuracy study. Ultrasound Obstet. Gynecol. 2021, 59, 385–391. [Google Scholar] [CrossRef]
- "Keras backends". keras.io. Retrieved 2018-02-23.
- "Why use Keras?". keras.io. Retrieved 2020-03-22.
- "R interface to Keras". keras.rstudio.com. Retrieved 2020-03-22.
- "Introducing Keras Core: Keras for TensorFlow, JAX, and PyTorch". Keras.io. Retrieved 11 July 2023.
- "Keras Documentation". keras.io. Retrieved 2016-09-18.
- Chollet, François (2016). "Xception: Deep Learning with Depthwise Separable Convolutions". arXiv:1610.02357.
- "Core - Keras Documentation". keras.io. Retrieved 2018-11-14.
- "Using TPUs | TensorFlow". TensorFlow. Archived from the original on 2019-06-04. Retrieved 2018-11-14.
- Deep Learning in Image Classification using Residual Network (ResNet) Variants for Detection of Colorectal Cancer - ScienceDirect.
- ResNet and its application to medical image processing: Research progress and challenges - ScienceDirect.
- Transfer learning with fine-tuned deep CNN ResNet50 model for classifying COVID-19 from chest X-ray images - PMC.
- [1512.03385]. Deep Residual Learning for Image Recognition.
- Convolutional Neural Networks for Medical Image Analysis: Fine Tuning or Full Training?
- A survey on deep learning in medical image analysis - ScienceDirect.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
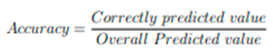 (1)
(1)