SWAT4HCLS 2024: The 15th International Conference on Semantic Web Applications and Tools for Health Care and Life Sciences
Introduction
In the pursuit of advancing personalized healthcare and fostering collaborative research, the Swiss Personalized Health Network (SPHN) [
1] has undertaken the development of a national framework for semantic interoperability which strives to address two main challenges: 1) standardizing real-world health data to make it understandable and of good quality for research purposes and 2) bringing together the various stakeholders of the initiative who have distinct professional backgrounds (e.g. clinical and medical specialists, data engineers, computer/data scientists, researchers) to communicate and understand each other without ambiguity on the different aspects of health knowledge. The SPHN Semantic Interoperability Framework, developed by the SPHN Data Coordination Center (DCC), focuses on the semantic representation of health data within a Knowledge Graph [
1], aligning with the FAIR principles (Findable, Accessible, Interoperable, Reusable) [
2], with the aim of facilitating seamless sharing and integration of diverse health-related data. At the core is the semantic layer, which builds on standards or terminologies existing in the clinics e.g. for billing like International Statistical Classification Of Diseases And Related Health Problems, 10th revision, German Modification (ICD-10-GM) [
4], which is the German modification of the World Health Organization's ICD-10 classification and Swiss Classification of Operations (CHOP) [
5] as well as well-known international standards like Systematized Nomenclature of Medicine Clinical Terms (SNOMED CT) [
6], Logical Observation Identifiers Names and Codes (LOINC) [
7], Genotype Ontology (GENO) [
8] and Sequence Ontology (SO) [
9]. Over the last few years, the five Swiss University hospitals have established the necessary infrastructure to generate project-specific knowledge graphs that conform to the SPHN Semantic Interoperability Framework. These graphs are constructed by extending the SPHN RDF Schema with project-specific concepts. The resulting data, which comprises codes from standard terminologies, is made available to researchers in a secure and trusted research environment called BioMedIT [
10], facilitating their work with sensitive health data. A first challenge has been the incorporation of terminologies which are widely used in the clinics, such as Anatomical Therapeutic Chemical Classification System (ATC) [
11], CHOP, and ICD-10-GM, into the framework to enhance the interoperability and quality of health data. One challenge we faced was that ATC, CHOP and ICD-10-GM, when compared to other terminologies, do not exist in FAIR and machine-readable forms such as Resource Description Framework
(RDF). As a result, these terminologies need to be FAIRified and converted to RDF using World Wide Web Consortium (W3C) standards. The DCC Terminology Service transforms and provides current (and historical versions) of terminologies, relevant to SPHN, in RDF in a FAIR and SPHN-compatible manner [
12]. A second challenge revolved around the use of different versions of terminologies and how to integrate them over time. In hospitals, data has historically been coded in various versions of terminologies, as updating all codes to the newest version is time-consuming and often impractical. This is further complicated by the fact that the specific version of the code is often not documented within the system. Instead, it has to be inferred by the coding datetime. This issue would typically be inconsequential if the terminology itself remained inherently stable, ensuring codes maintain their unique identifier and do not change meanings. Unfortunately, this is not the case for ATC, CHOP, and ICD-10-GM.
To elaborate, there are many ways to represent concepts and terms that are used within a particular domain. These can be - with increasing complexity of representation - a collection of tags, a vocabulary, a data dictionary, a taxonomy, a schema, or an ontology. A terminology is a collection of terms that are of significance in a particular domain. Each term in a terminology has a fixed meaning and use. Terminologies can be of different flavors - some are collections of terms while others have terms organized in a hierarchy and resemble a taxonomy. These terminologies can be either a monohierarchy or a polyhierarchy. On the other hand, an ontology is a formal specification of a shared conceptualization that describes a domain. It is defined using a conventional knowledge representation language, typically the Web Ontology Language (OWL) [
13]. One common collection of ontologies is in the biological and biomedical domain where each ontology focuses on a specific aspect of biology, be it gene function (e.g. Gene Ontology [
14]), disease (e.g Disease Ontology [
15]), phenotype (e.g. Human Phenotype Ontology [
16]), or anatomy (e.g. Uberon [
17]). But there are also areas in biology for which there are no reliable or well used ontologies. Instead, we have terminologies that are less formally defined (i.e. collection of terms of significance in a particular domain) but have sufficient breadth and depth. These terminologies are available in simple formats such as comma separated values (CSV) or just plain text. To effectively use these terminologies within the context of linked data, one has to transform these terminologies to an RDF representation using W3C standards - like RDFS (RDF Schema), SKOS (Simple Knowledge Organization System), and OWL - as building blocks. The success and the quality of the transformation depends on the information provided by the original terminology providers.
In the case of ontologies, one key part of their design is the adoption of concept identifiers that are opaque i.e. concept meaning is not ascribed to the identifiers. One way to achieve this is by the use of numerical (and in some cases sequential) identifiers as concept identifiers. These numerical identifiers, along with the ontology namespace, uniquely identifies a concept within that ontology. The meaning of the concept is then applied to this numerical concept identifier (ID) via rdfs:label, skos:definition and other standardized predicates. The semantics of these concepts are further described using OWL axioms. Typically when working with ontologies, certain assumptions are made, within reason:
Concepts are unique throughout the lifetime of the ontology, regardless of the ontology version. Using numerical IDs as concept IDs help in ensuring concept uniqueness.
Concepts are never reused across successive versions of an ontology. Instead a new concept ID is generated when adding a new concept. This ensures that the meaning of the concept stays the same. If the meaning changes then the concept is treated as new and a new concept is added to the ontology, while deprecating the old concept.
Concepts are never removed throughout the lifetime of the ontology, regardless of its status. If a concept is deemed unusable then it is deprecated but never removed from the ontology. This ensures that the concept remains in the ontology and the fate of the concept is communicated in successive versions of the ontology.
These non-exhaustive assumptions can be made for ontologies that follow certain best practices. The Open Biological and Biomedical Ontologies (OBO) Foundry [
18] puts forth a set of principles for ontology design that improves the quality and interoperability of ontologies. But these principles and best practices are not applied when terminology providers deliver their terminologies to the broader community, due to, among other things, lack of sufficient metadata provided in the terminology. Thus, the assumptions made earlier cannot be applied - without careful consideration - to terminologies or RDF versions of these terminologies. This challenge is further complicated when dealing with the integration of different versions of a terminology. Key issues that need to be addressed include:
Semantic drift: This involves addressing the changes in meanings of the same code across different versions, which can lead to biases in research.
Managing retired codes: Identifying and distinguishing between codes that are no longer in use (retired) is crucial for maintaining data integrity.
Versioned and unversioned codes: Balancing the use of both versioned and unversioned codes in data, especially when the unversioned codes have historically undergone semantic changes.
The prevalence of these issues emphasize the need for standardized, version-controlled terminology management. Such standardization is critical for achieving interoperability and fostering a unified, patient-centric healthcare system. To address this challenge, our manuscript introduces an RDF-based versioning strategy focusing on terminologies susceptible to semantic drift, specifically ATC, CHOP, and ICD-10-GM. This strategy, aligned with the FAIR principles, represents a crucial step toward enhancing data quality, ensuring the reliability of data for research, and instilling confidence in contributions from hospitals and other data providers. Applying the versioning strategy to machine readable representation of ATC, CHOP and ICD-10-GM allowed us to leverage information about valid and deprecated codes in our quality assurance framework, based on Shapes Constraint Language (SHACL), and further allow the researcher to use this knowledge when querying the data with SPARQL Protocol and RDF Query Language (SPARQL).
Method and Results1.1. Label comparison to assess semantic drift
To illustrate the breakdown of assumptions we analyze codes across versions of a terminology. We adopt a simple approach where we compare the labels of the codes between successive versions and if the label of a code has changed then we assume that the code itself has changed. We start by first comparing labels for codes from ATC, CHOP and ICD-10-GM. The analysis is done such that we compare all codes from the current version with the prior version and then repeat where the prior version is the current version, and so on. To quantify the difference in labels we compute Jaro-Winkler Similarity measures [
19], a string similarity measure that provides a way of calculating how similar two given strings are, with a score of 1.0 indicating that the two strings are identical. We explain the analysis by taking the example of ATC where we compare the english labels of ATC codes. We approach the analysis in 3 parts: First, we consider a code different from its prior year if the labels are not identical (i.e. similarity score is less than 1.0).
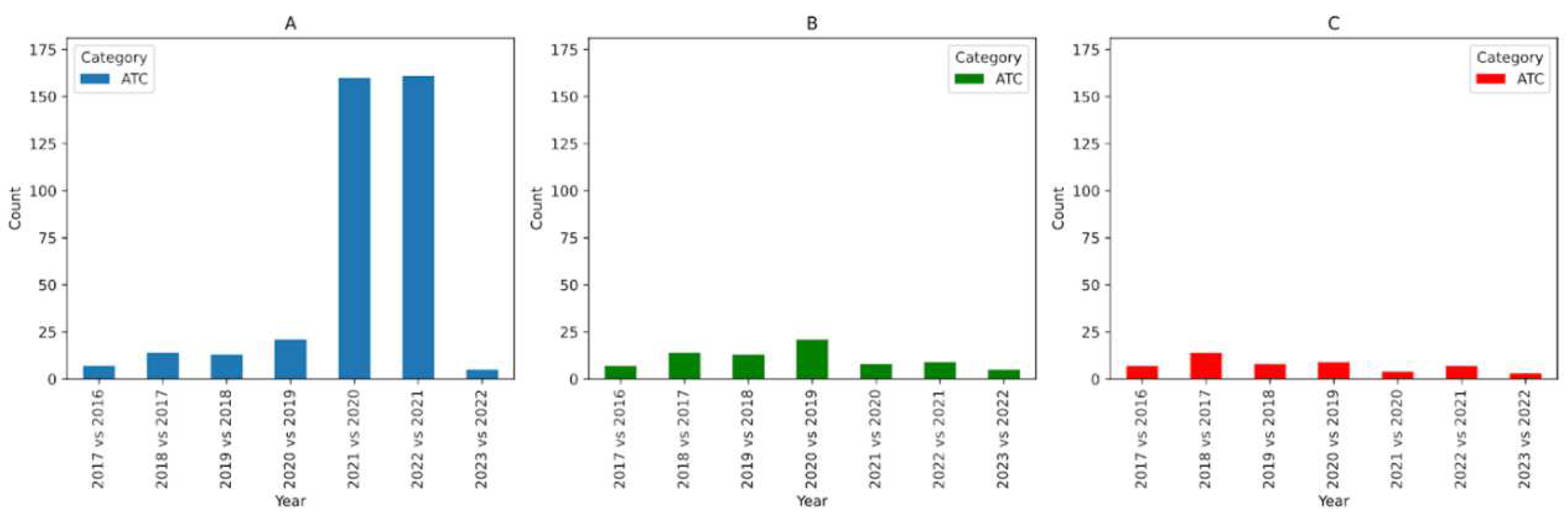
Figure 1.A illustrates the number of ATC codes that have changed for each year comparison. Second, we compare the labels after preprocessing. In this case, the following changes are ignored: introduction or removal of spaces, HTML tags, brackets, and character cases. After preprocessing, any change in labels is considered as a change in code meaning (i.e. similarity score is less than 1.0). As a result, the number of ATC codes that have changed is illustrated in
Figure 1.B. Finally, we compare the labels after preprocessing and then applying a Jaro-Winkler Similarity score cutoff of 0.85.
Figure 1.C illustrates the number of ATC codes that have changed for each year comparison. The choice of 0.85 as a cutoff was made after inspecting the codes manually and estimating a value that would maximize true positives while minimizing false negatives.
For ATC, to better explain the changes observed in
Figure 1, we consider the comparison between ATC 2017 and ATC 2016 where we see that 7 codes changed in their labels. Upon close inspection we can classify the changes according to 3 types:
No change in meaning: The ATC code 'L01XC05' in 2016 had a label of 'gemtuzumab' which changed in 2017 to 'gemtuzumab ozogamicin'. The label change here was for clarity and to better represent the name of the chemical compound. As a result there is no change in semantics for this code. In this scenario, we can state that L01XC05 is identical between 2016 and 2017.
Change in meaning leading to a split: The ATC code 'C07FB02' in 2016 had a label of 'metoprolol and other antihypertensives' which changed in 2017 to 'metoprolol and felodipine'. After inspection, it is evident that the 'C07FB02' code from 2016 was split where the scope of 'C07FB02' was narrowed and a new ATC code 'C07FB13' with label 'metoprolol and amlodipine' was introduced. In this scenario, we can say that C07FB02 in 2016 is different from C07FB02 in 2017 and treat the two codes as different.
Unclear change in meaning: The ATC code 'A12BA30' in 2016 had a label of 'combinations' which changed in 2017 to 'potassium (different salts in combination)'. This indicates that the label of the code was changed drastically to better scope and represent its value space. In this scenario, we treat A12BA30 in 2016 as different from A12BA30 in 2017.
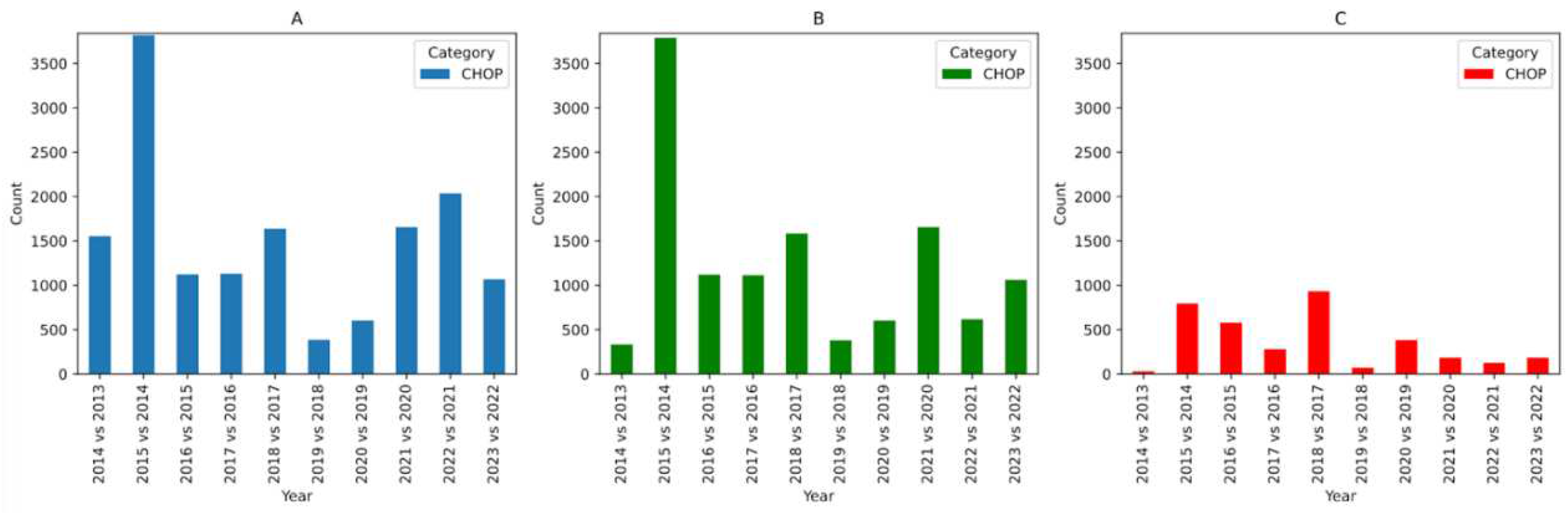
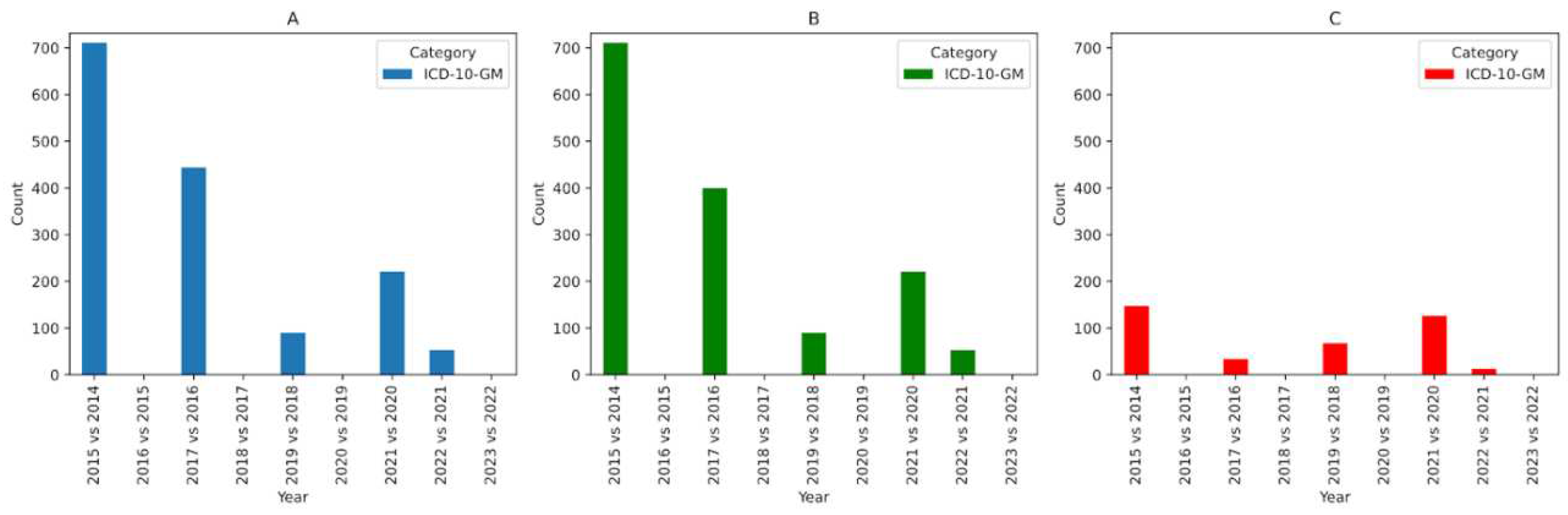
We apply the same analysis to CHOP (
Figure 2) and ICD-10-GM (
Figure 3). In the case of CHOP, we see a great deal of changes in codes between years, with the biggest change observed between 2014 and 2015 versions (as observed in
Figure 2.A and 2.B). After applying a more relaxed similarity score cutoff of 0.85, we see relatively fewer changes. Upon close inspection, one could attribute these changes to renaming or clarification of the code labels to aid their use. Whether or not these changes affect the semantics of the code is not always clear given that their interpretation is dependent on the existence of other codes.
In the case of ICD-10-GM, we see a great deal of changes between 2014 and 2015. Most of these changes are language-specific changes since labels in ICD-10-GM are in German. We also see that there are no changes between 2015-2016, 2017-2018, 2019-2020, and 2022-2023. This is because the codes from the years being compared are unchanged. i.e. 2015 version of ICD-10-GM is identical to the 2016 version due to the way the Bundesamt für Statistik (BFS) in Switzerland publishes ICD-10-GM. In Switzerland, a new version of the Bundesinstitut für Arzneimittel und Medizinprodukte (BfArM) release of ICD-10-GM is adopted every two years. In SPHN, we make use of the BFS release of ICD-10-GM. As a result, in our analysis we see no changes for every other year since the versions being compared are identical.
1.1. Identification and Reconciliation
Based on the analysis described in the previous section, it is clear that there are codes that change in their labels (and thus their meaning). To carefully verify all the changes is a manual task and requires subject matter experts to review each code that is flagged as different between versions. Furthermore, providing a list of codes that are different to hospitals and data providers is not sufficient for raising awareness within the community and improving data quality. Instead, we take the identified code label changes into consideration when preparing the terminologies as part of the DCC Terminology Service.
We adopt a simple lexical matching approach for establishing identity for concepts between successive versions of a terminology: any change in code labels is flagged as a change in meaning. Thus, in principle, any code between two versions is considered identical only if their label comparison yields a Jaro-Winkler Similarity score of 1.0. While this may yield a lot of false positives, we believe this is required to raise awareness within the community of the potential issues that one might face when working with ATC, CHOP, and ICD-10-GM. After establishing codes that are different, we use a lightweight approach to reconcile concepts that are identical across successive versions of a terminology. For this, we adopt a versioned URI for each version of the terminology allowing us to uniquely identify a code originating from one version of a terminology. With the versioned URIs in place, it is possible to make a statement about whether a code from one version of a terminology is the same or different in another version. Codes that are identical are annotated as equivalent via the
owl:equivalentClass axiom from the OWL specification (
http://www.w3.org/2002/07/owl#equivalentClass). For example, in the case of ATC, the code ‘A02BX05’ stays the same between 2016 and 2017 (
Figure 4). When coupled with a versioned URI, we can make the following statement:
The results from the analysis are leveraged to build a complete picture of how a code evolves across time. To facilitate the reconciliation we make use of versioned URIs to uniquely identify a code from a specific version of a terminology. This enables us to refer to a code with the same identifier but from different versions of a terminology and thus reducing ambiguity. Once we are able to uniquely refer to a code from a specific version of a terminology, we annotate codes that are identical via the
owl:equivalentClass (
http://www.w3.org/2002/07/owl#equivalentClass) predicate from the OWL specification. Conversely, the absence of an equivalent class axiom indicates that the codes are not identical between two versions and that the semantics of the code has changed.
For the purpose of reconciliation, we will use examples from ATC to illustrate how we approach the task of annotating and cataloging the variations. We employ version-specific URIs to uniquely distinguish a code originating from a version of a terminology. For example, to represent code 'A02BX05' from ATC 2016, we make use of the URI '
https://www.whocc.no/atc_ddd_index/?year=2016&code=A02BX05' and to represent the same code from ATC 2017, we use the URI '
https://www.whocc.no/atc_ddd_index/?year=2017&code=A02BX05'. When the URIs are condensed into compact URIs (CURIEs), they are represented as ATC-2016:A02BX05 and ATC-2017:A02BX05. We add a triple that states that ATC-2016:A02BX05 is equivalent to ATC-2017:A02BX05 (i.e. using
owl:equivalentClass) in the ATC 2017 RDF serialization to indicate that A02BX05 in the current year (i.e. 2017) is identical to A02BX05 from the previous year (i.e. 2016). We do this for all codes where we have established that codes are identical between versions. Conversely, the lack of an equivalent class axiom suggests a disparity between the codes between versions, signaling a modification in the code.
The observation of possible semantic drift and coding changes in ATC, CHOP and ICD-10-GM, led to an upgrade of the SPHN DCC Terminology Service to take into consideration these changes. When a new version of these terminologies is released, the textual files are processed in the DCC Terminology Service. Each versioned RDF file generated for these terminologies contains all codes from the current release version and all codes from previous versions (back until 2016 for ATC, 2013 for CHOP and ICD-10-GM). The pipeline used by the DCC Terminology Service is available at
https://git.dcc.sib.swiss/dcc/biomedit-rdf. This implies that deprecated codes would now also be retrievable in a newer version of the terminology. The versioned terminology files are shared and made available via the DCC Terminology Service (
https://terminology.dcc.sib.swiss via a SWITCH Edu-ID). After which, the main value addition of this work is for a user to know if a code in a terminology has changed between version
n-1 and
n, where
n is any given version of a terminology.
Use of versioned codes for data validation
SPHN enhances data quality and consistency by utilizing SHACL shapes, which are derived from the SPHN RDF Schema. These shapes define a set of constraints to ensure data conforms to the SPHN specification. The generation of these SHACL shapes is automated using a specialized tool, the SHACLer (
https://git.dcc.sib.swiss/sphn-semantic-framework/sphn-shacl-generator). Over time, medical codes can become outdated or their meanings might evolve - as demonstrated in our analysis - and thus create challenges in maintaining data accuracy and relevance. The SHACL shapes address this issue by identifying instances where data might reference outdated or modified codes. One key aspect of the SPHN semantic interoperability strategy is its dual approach where on one hand, it maintains terminologies with versioned codes, ensuring that the most current and accurate codes are available. On the other hand, it empowers both data providers and consumers to recognize and handle instances where outdated or invalid codes are still in use. This is achieved through specific SHACL shapes designed for this purpose. A notable example is the SHACL shape called ‘constraints:OldVersionedCodeStillValid’ (
Figure 5) which checks whether a code used in the data is still valid. Additionally, it determines if the code is from an older version or aligns with the most recent terminology version.
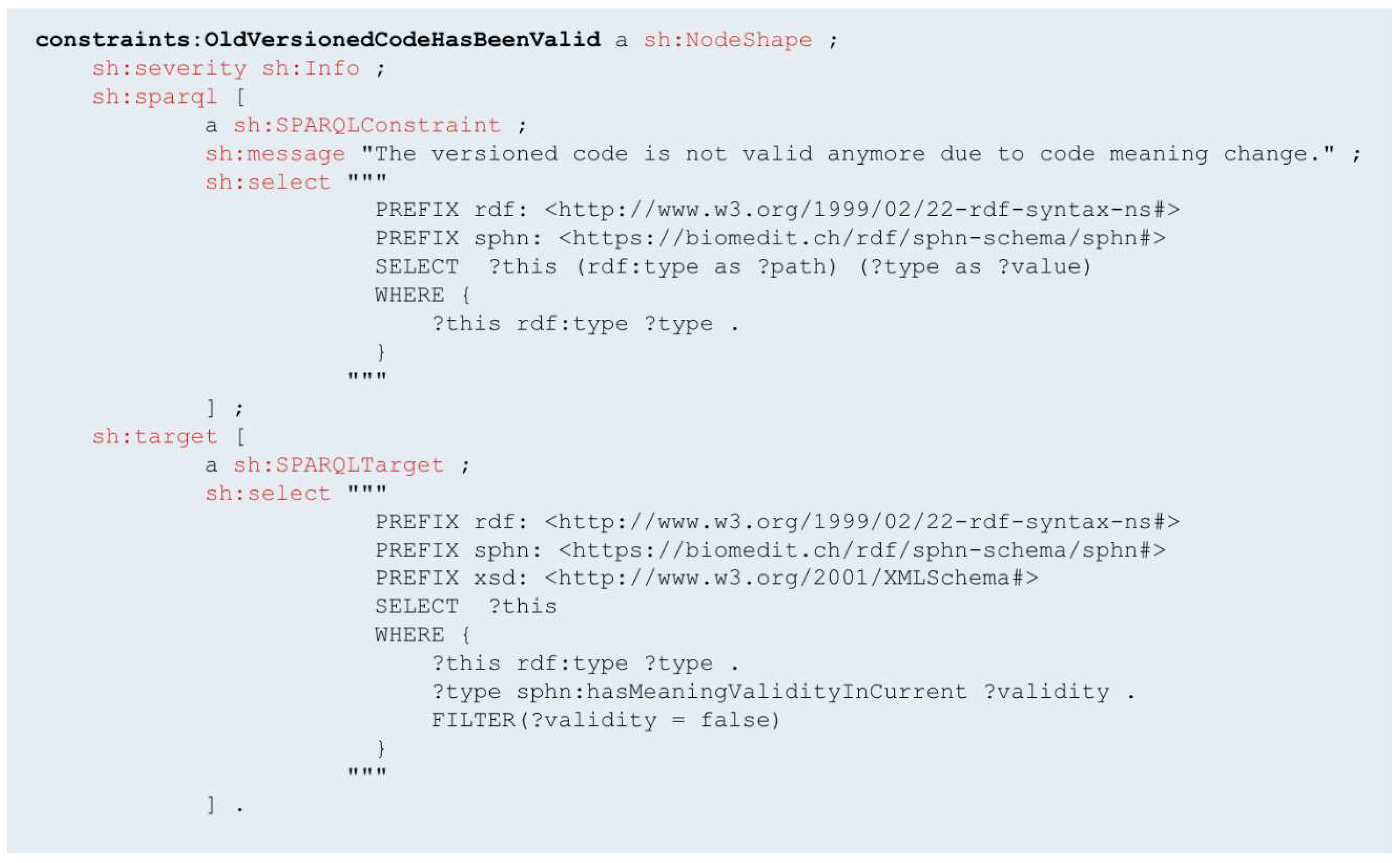
Another example is the SHACL shape called 'constraints:OldVersionedCodeHasBeenValid’ (
Figure 6) which is designed to identify and flag instances where a code, previously valid in the data, has become invalid due to changes in terminologies. This SHACL shape operates by examining the codes used in data against the updated versions of a terminology. It is specifically designed to detect discrepancies between the codes in use and the most current versions available. When a code that was once valid is found to be outdated or superseded, this shape flags it and raises a warning.
This ability to identify old and invalid codes is essential during data analysis and processing, especially when comparing historical and current data sets. Moreover, the shapes are useful for retrospective studies and longitudinal data analysis in healthcare. By identifying codes that were once valid but are now outdated, this shape helps maintain the integrity of such analyses, ensuring that conclusions drawn are based on accurate and relevant data. The implementation of such SHACL shapes significantly enhances the robustness and reliability of data management within the SPHN Semantic Interoperability Framework, ensuring that the health data remains current, valid, and in line with the latest medical standards and practices.
1.1. Use of versioned codes for querying
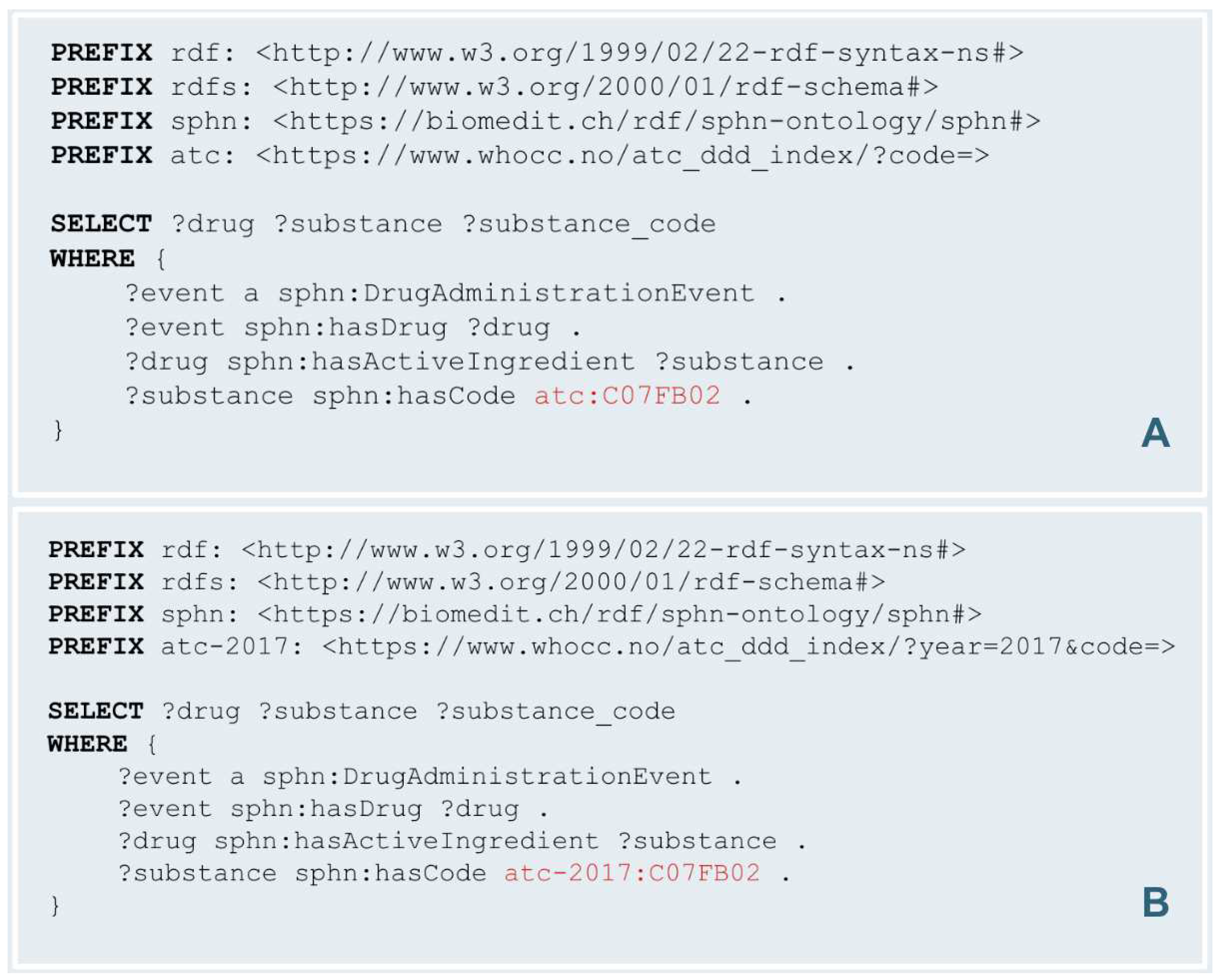
SPHN enhances data analysis capabilities through the use of SPARQL queries, derived from the SPHN RDF Schema. These queries are useful for querying and getting all relevant information associated with a concept. Versioned codes from a terminology can also be leveraged in SPARQL queries to get results that are relevant and appropriate. Let's consider the example of the ATC code 'C07FB02'. As explained in the previous section, this code underwent a change between 2016 and 2017, acquiring a different label in 2017. This evolution in the code's meaning poses a challenge for researchers seeking accurate data.
Consider a researcher interested in analyzing Drug Administration Events where the active ingredients are 'metoprolol and felodipine'. To achieve this, they would typically employ a SPARQL query (
Figure 9.A). However, if the data is coded according to the 2016 version of the ATC, the results would significantly differ. In 2016, the code ‘C07FB02’ was associated with 'metoprolol and other hypertensives', implying a broader range of drug combinations. This discrepancy underscores the importance of versioned URIs in SPARQL queries. By specifying the exact version of the ATC code, researchers can refine their search to retrieve data that accurately reflects their intended analysis.
For example, to focus exclusively on data from 2017, the researcher can modify their query to specifically target the 2017 version of ‘C07FB02’ (
Figure 9.B). This distinction is necessary for accurate data retrieval and analysis, particularly when studying treatment trends and outcomes over time. It enables researchers and healthcare professionals to conduct targeted analyses based on specific code versions, thereby ensuring that the data they work with is both relevant and accurate.