Submitted:
07 December 2023
Posted:
07 December 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Antibiotics sales
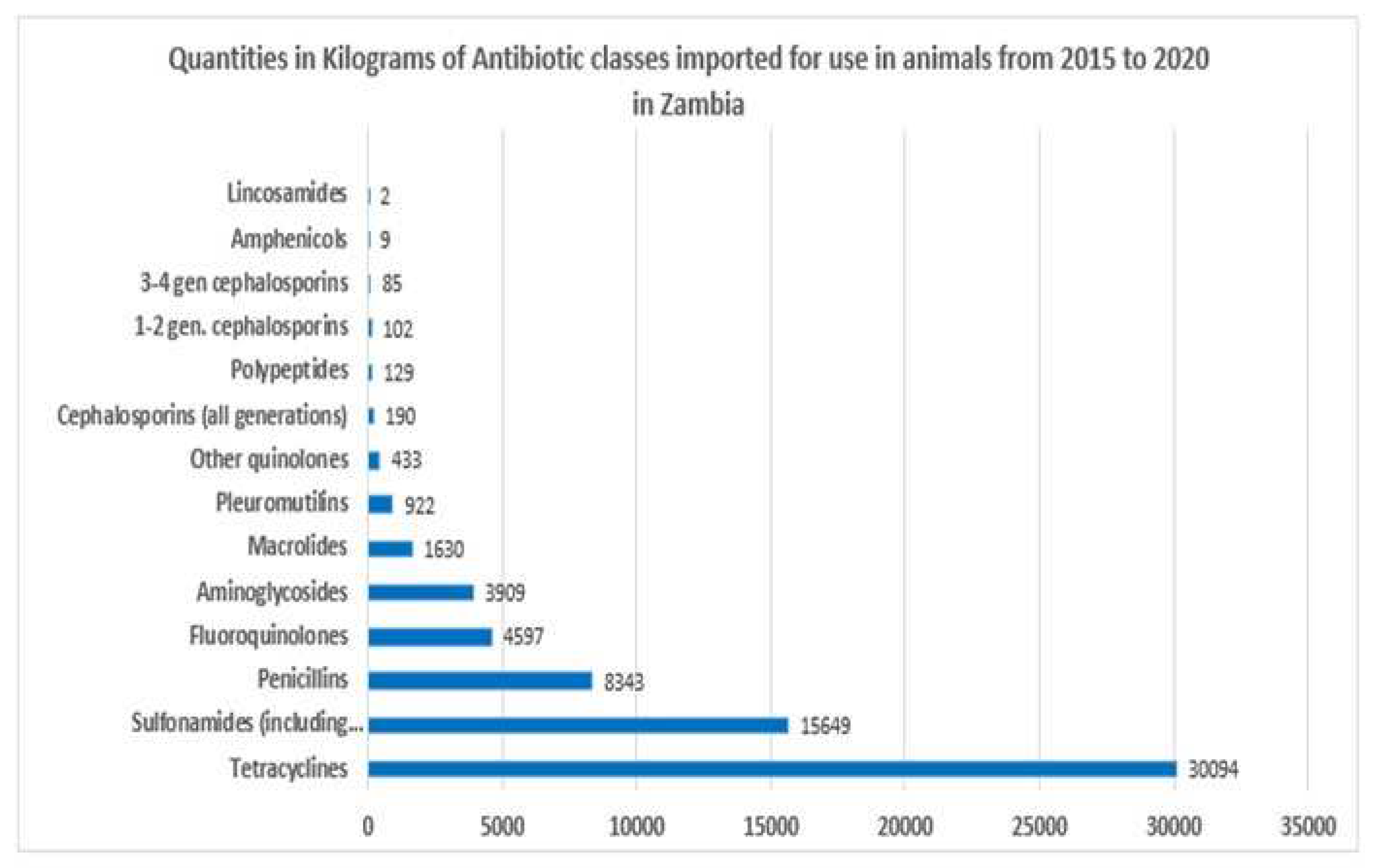
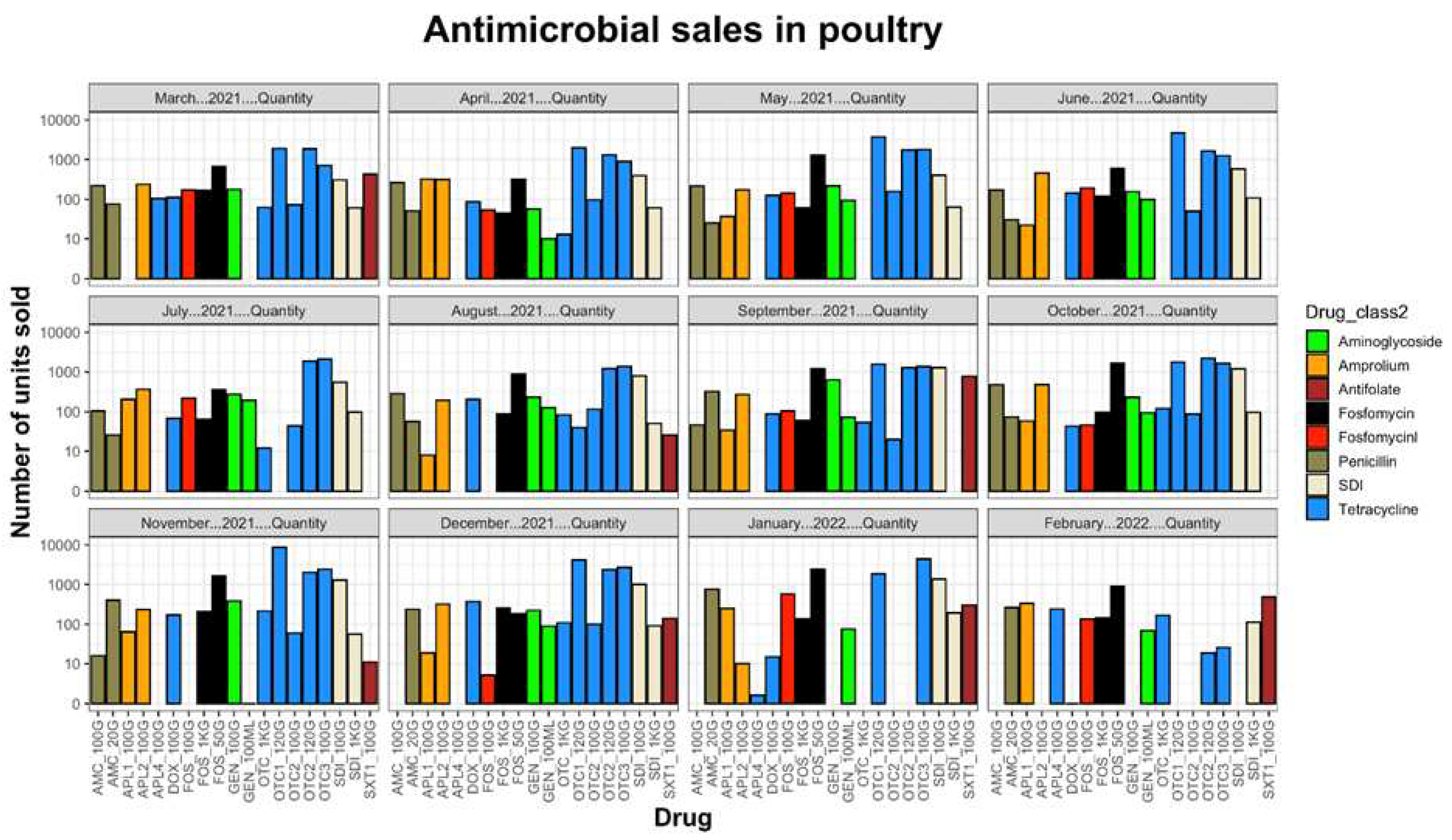
2.2. Prevalence of Enterobacteriaceae
2.3. Antimicrobial sensitivity showed multidrug resistance (MDR) among Enterobacteriaceae
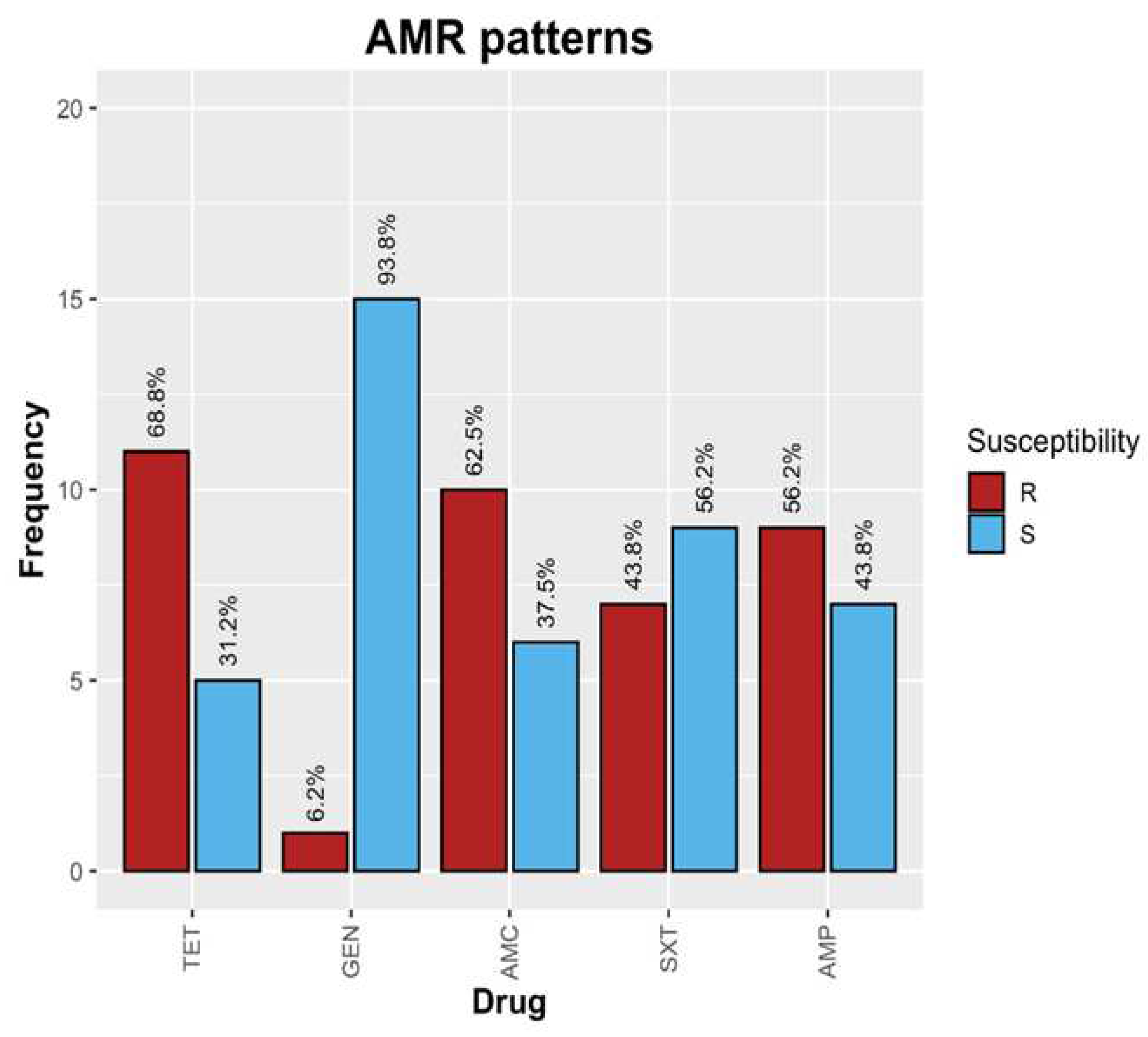
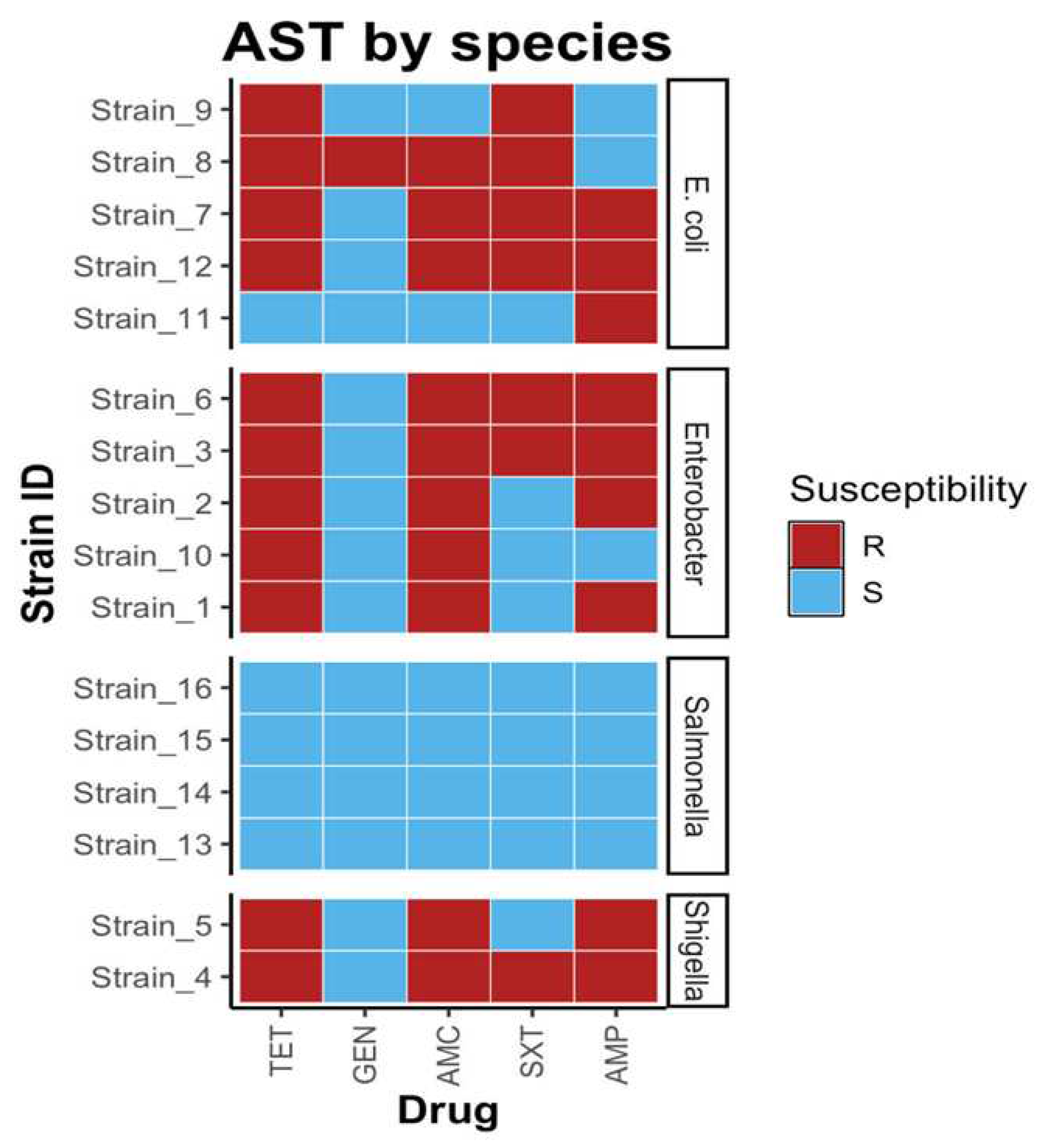
2.4. Association between cefotaxime (CTX) resistance and ESBL genes
3. Discussion
Study limitations
4. Materials and Methods
4.1. Study area and sampling.
4.2. Antimicrobial usage data.
4.3. Bacterial isolation.
4.4. Antibiotic sensitivity testing.
4.5. Cefotaxime (CTX) minimum inhibitory concentration (MIC).
4.6. Detection of ESBL genes by PCR
4.6. PCR product purification and Cycle sequencing
4.7. Data analysis
4.7.1. AMR data analysis
4.7.2. ESBL gene sequence analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Central Statistical Office Zambia 2010 Census of Population and Housing: Agriculture Analytical Report. 2014, 1–50.
- Muonga, E.M.; Mainda, G.; Mukuma, M.; Kwenda, G.; Hang’ombe, B.; Phiri, N.; Mwansa, M.; Munyeme, M.; Muma, J.B. Antimicrobial Resistance of Escherichia Coli and Salmonella Isolated from Raw Retail Broiler Chickens in Zambia. Res Sq 2019, 1–18. [Google Scholar]
- Phiri, N.; Mainda, G.; Mukuma, M.; Ntazana, S.; Luke, J.; Banda, K.; Muligisa-Muonga, E.; Bumbangi, N.F.; Mwansa, M.; Yamba, K.; et al. Determination of Antimicrobial Resistant of Salmonella Species and Escherichia Coli in Broiler Chickens Slaughtered in Commercial Abattoirs in Lusaka Province, Zambia. 2019.
- Apata, D.F. Antibiotic Resistance in Poultry. Int J Poult Sci 2009, 8, 404–408. [Google Scholar] [CrossRef]
- Kiambi, S.; Mwanza, R.; Sirma, A.; Czerniak, C.; Kimani, T.; Kabali, E.; Dorado-Garcia, A.; Eckford, S.; Price, C.; Gikonyo, S.; et al. Understanding Antimicrobial Use Contexts in the Poultry Sector: Challenges for Small-Scale Layer Farms in Kenya. Antibiotics 2021, 10, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Lipsitch, M. The Rise and Fall of Antimicrobial Resistance. Trends Microbiol 2001, 9, 438–444. [Google Scholar] [CrossRef]
- Saliu, E.M.; Vahjen, W.; Zentek, J. Types and Prevalence of Extended-Spectrum Beta-Lactamase Producing Enterobacteriaceae in Poultry. Anim Health Res Rev 2017, 18, 46–57. [Google Scholar] [CrossRef] [PubMed]
- Smillie, C.; Garcillán-Barcia, M.P.; Francia, M.V.; Rocha, E.P.C.; de la Cruz, F. Mobility of Plasmids. Microbiology and Molecular Biology Reviews 2010, 74, 434–452. [Google Scholar] [CrossRef] [PubMed]
- Chishimba, K.; Hang’Ombe, B.M.; Muzandu, K.; Mshana, S.E.; Matee, M.I.; Nakajima, C.; Suzuki, Y. Detection of Extended-Spectrum Beta-Lactamase-Producing Escherichia Coli in Market-Ready Chickens in Zambia. Int J Microbiol 2016, 2016. [Google Scholar] [CrossRef] [PubMed]
- Azabo, R.; Mshana, S.; Matee, M.; Kimera, S.I. Antimicrobial Usage in Cattle and Poultry Production in Dar Es Salaam, Tanzania: Pattern and Quantity. BMC Vet Res 2022, 18, 1–12. [Google Scholar] [CrossRef]
- Kamini, M.G.; Keutchatang, F.T.; Mafo, H.Y.; Kansci, G.; Nama, G.M. Antimicrobial Usage in the Chicken Farming in Yaoundé, Cameroon: A Cross-Sectional Study. Int J Food Contam 2016, 3. [Google Scholar] [CrossRef]
- Caudell, M.A.; Dorado-Garcia, A.; Eckford, S.; Creese, C.; Byarugaba, D.K.; Afakye, K.; Chansa-Kabali, T.; Fasina, F.O.; Kabali, E.; Kiambi, S.; et al. Towards a Bottom-up Understanding of Antimicrobial Use and Resistance on the Farm: A Knowledge, Attitudes, and Practices Survey across Livestock Systems in Five African Countries. PLoS One 2020, 15, 1–26. [Google Scholar] [CrossRef]
- Mudenda, S.; Malama, S.; Munyeme, M.; Hang’ombe, B.M.; Mainda, G.; Kapona, O.; Mukosha, M.; Yamba, K.; Bumbangi, F.N.; Mfune, R.L.; et al. Awareness of Antimicrobial Resistance and Associated Factors among Layer Poultry Farmers in Zambia: Implications for Surveillance and Antimicrobial Stewardship Programs. Antibiotics 2022, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Mudenda, S.; Mukosha, M.; Godman, B.; Fadare, J.; Malama, S.; Munyeme, M.; Hikaambo, C.N.; Kalungia, A.C.; Hamachila, A.; Kainga, H.; et al. Knowledge, Attitudes, and Practices of Community Pharmacy Professionals on Poultry Antibiotic Dispensing, Use, and Bacterial Antimicrobial Resistance in Zambia: Implications on Antibiotic Stewardship and WHO AWaRe Classification of Antibiotics. Antibiotics 2022, 11. [Google Scholar] [CrossRef]
- Mudenda, S.; Bumbangi, F.N.; Yamba, K.; Munyeme, M.; Malama, S.; Mukosha, M.; Hadunka, M.A.; Daka, V.; Matafwali, S.K.; Siluchali, G.; et al. Drivers of Antimicrobial Resistance in Layer Poultry Farming: Evidence from High Prevalence of Multidrug-Resistant Escherichia Coli and Enterococci in Zambia. Vet World 2023, 1803–1814. [Google Scholar] [CrossRef] [PubMed]
- Nhung, N.T.; Chansiripornchai, N.; Carrique-Mas, J.J. Antimicrobial Resistance in Bacterial Poultry Pathogens: A Review. Front Vet Sci 2017, 4, 1–17. [Google Scholar] [CrossRef]
- Mwansa, M.; Mukuma, M.; Mulilo, E.; Kwenda, G.; Mainda, G.; Yamba, K.; Bumbangi, F.N.; Muligisa-Muonga, E.; Phiri, N.; Silwamba, I.; et al. Determination of Antimicrobial Resistance Patterns of Escherichia Coli Isolates from Farm Workers in Broiler Poultry Production and Assessment of Antibiotic Resistance Awareness Levels among Poultry Farmers in Lusaka, Zambia. Front Public Health 2023, 10. [Google Scholar] [CrossRef]
- Nandi, S.P.; Sultana, M.; Hossain, M.A. Prevalence and Characterization of Multidrug-Resistant Zoonotic Enterobacter Spp. in Poultry of Bangladesh. Foodborne Pathog Dis 2013, 10, 420–427. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Munir, M.; Xagoraraki, I. Correlation of Tetracycline and Sulfonamide Antibiotics with Corresponding Resistance Genes and Resistant Bacteria in a Conventional Municipal Wastewater Treatment Plant. Science of the Total Environment 2012, 421–422, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Sreejith, S.; Shajahan, S.; Prathiush, P.R.; Anjana, V.M.; Viswanathan, A.; Chandran, V.; Ajith Kumar, G.S.; Jayachandran, R.; Mathew, J.; Radhakrishnan, E.K. Healthy Broilers Disseminate Antibiotic Resistance in Response to Tetracycline Input in Feed Concentrates. Microb Pathog 2020, 149. [Google Scholar] [CrossRef]
- Ameen-Ur-Rashid, S.S.A.S. Isolation of Escherichia Coli from Poultry Liver and Its Antibiogram. research journal for veterinary practitioners 2016, 25, 241–242. [Google Scholar] [CrossRef]
- Ibrahim, R.A.; Cryer, T.L.; Lafi, S.Q.; Basha, E.A.; Good, L.; Tarazi, Y.H. Identification of Escherichia Coli from Broiler Chickens in Jordan, Their Antimicrobial Resistance, Gene Characterization and the Associated Risk Factors. BMC Vet Res 2019, 15, 1–16. [Google Scholar] [CrossRef]
- Hamed, E.A.; Abdelaty, M.F.; Sorour, H.K.; Roshdy, H.; Abdelrahman, M.A.A.; Magdy, O.; Ibrahim, W.A.; Sayed, A.; Mohamed, H.; Youssef, M.I.; et al. Monitoring of Antimicrobial Susceptibility of Bacteria Isolated from Poultry Farms from 2014 to 2018. Vet Med Int 2021, 2021. [Google Scholar] [CrossRef] [PubMed]
- Kaonga, N.; Hang, B.M.; Lupindu, A.M.; Hoza, A.S. Detection of CTX-M-Type Extended Spectrum Beta-Lactamase Producing Salmonella Typhimurium in Commercial Poultry Farms in Copperbelt Province,. 2019.
- Collignon, P.; Aarestrup, F.M.; Irwin, R.; McEwen, S. Human Deaths and Third-Generation Cephalosporin Use in Poultry, Europe. Emerg Infect Dis 2013, 19, 1339–1340. [Google Scholar] [CrossRef] [PubMed]
- Simoneit, C.; Burow, E.; Tenhagen, B.A.; Käsbohrer, A. Oral Administration of Antimicrobials Increase Antimicrobial Resistance in E. Coli from Chicken - A Systematic Review. Prev Vet Med 2015, 118, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Shawa, M.; Furuta, Y.; Paudel, A.; Kabunda, O.; Mulenga, E.; Mubanga, M.; Kamboyi, H.; Zorigt, T.; Chambaro, H.; Simbotwe, M.; et al. Clonal Relationship between Multidrug-Resistant Escherichia Coli ST69 from Poultry and Humans in Lusaka, Zambia. FEMS Microbiol Lett 2021, 368. [Google Scholar] [CrossRef] [PubMed]
- Subramanya, S.H.; Bairy, I.; Nayak, N.; Amberpet, R.; Padukone, S.; Metok, Y.; Bhatta, D.R.; Sathian, B. Detection and Characterization of ESBLproducing Enterobacteriaceae from the Gut of Healthy Chickens, Gallus Gallus Domesticus in Rural Nepal: Dominance of CTX-M-15-Non-ST131 Escherichia Coli Clones. PLoS One 2020, 15, 1–15. [Google Scholar] [CrossRef]
- Ferreira, J.C.; Penha Filho, R.A.C.; Andrade, L.N.; Berchieri Junior, A.; Darini, A.L.C. Evaluation and Characterization of Plasmids Carrying CTX-M Genes in a Non-Clonal Population of Multidrug-Resistant Enterobacteriaceae Isolated from Poultry in Brazil. Diagn Microbiol Infect Dis 2016, 85, 444–448. [Google Scholar] [CrossRef] [PubMed]
- Zeynudin, A.; Pritsch, M.; Schubert, S.; Messerer, M.; Liegl, G.; Hoelscher, M.; Belachew, T.; Wieser, A. Prevalence and Antibiotic Susceptibility Pattern of CTX-M Type Extended-Spectrum β -Lactamases among Clinical Isolates of Gram-Negative Bacilli in Jimma , Ethiopia. 2018, 1–10.
- Aworh, M.K.; Kwaga, J.K.P.; Hendriksen, R.S.; Okolocha, E.C.; Thakur, S. Genetic Relatedness of Multidrug Resistant Escherichia Coli Isolated from Humans, Chickens and Poultry Environments. Antimicrob Resist Infect Control 2021, 10. [Google Scholar] [CrossRef]
- Shams, E.; Nateghi, B.; Eshaghiyan, A.; Behshood, P. TEM Gene Detection in Clinical Pseudomonas Aeruginosa and Escherichia Coli Samples. Research in Molecular Medicine 2019, 7, 43–51. [Google Scholar] [CrossRef]
- Magiorakos, A. Osobennosti Narusheni I Rechi Pri Dykhanii Pod Izbytochnym Davleniem. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance 2012, 25, 10–14.
- Pokharel, S.; Shrestha, P.; Adhikari, B. Antimicrobial Use in Food Animals and Human Health: Time to Implement ‘One Health’ Approach. Antimicrob Resist Infect Control 2020, 9, 1–5. [Google Scholar] [CrossRef]
- Wickham, H. Reshaping Data with the Reshape Package; 2006.
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J Open Source Softw 2019, 4, 1686. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; Mcgettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X Version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
| Species | Facility | Total | ||
| A | B | C | ||
| Enterobacter | 2 | 26 | 15 | 43 |
| E. coli | 5 | 11 | 4 | 20 |
| Salmonella | 2 | 8 | 0 | 10 |
| Klebsiella | 0 | 1 | 0 | 1 |
| Shigella | 0 | 5 | 3 | 8 |
| Yersinia | 0 | 2 | 0 | 2 |
| Citrobacter | 0 | 4 | 0 | 4 |
| Vibrio | 1 | 0 | 0 | 1 |
| Proteus | 4 | 5 | 1 | 10 |
| Morganella | 0 | 4 | 0 | 0 |
| Total | 14 | 66 | 23 | 103 |
| FACILITY | PATHOGENS | |||
| Enterobacter | E. coli | Salmonella | Shigella | |
| A | 3 | 3 | 3 | 1 |
| B | 1 | 1 | 1 | |
| C | 1 | 1 | 1 |
| SAMPLE ID | ORGANISM | CTX MIC | blaTEM | blaCTX-M |
| LS 1 | Enterobacter | 2 | + | - |
| LS 2 | E. coli | 4 | - | + |
| LS 3 | Enterobacter | 2 | + | - |
| UZ 1 | E. coli | ≥ 512 | + | + |
| LS 4 | Enterobacter | 2 | + | - |
| AV 1 | E. coli | 2 | + | + |
| LS 5 | E. coli | ≥ 512 | + | + |
| LS 6 | Enterobacter | 128 | + | - |
| AV 2 | Enterobacter | ≥ 512 | + | - |
| AV 3 | Enterobacter | 2 | + | - |
| UZ 2 | Enterobacter | 2 | + | - |
| LS 7 | E. coli | ≥ 512 | - | + |
| LS 8 | E. coli | 16 | + | + |
| LS 9 | Enterobacter | 128 | + | - |
| PRIMERS | TARGET GENE | SEQUENCE 5’-3’ | EXPECTED AMPLICON SIZE |
|---|---|---|---|
|
TEM1F TEM1R |
blaTEM | ATGAGTATTCAACATTTCCG CTGACAGTTACCAATGCTTA |
864 |
|
SHVF SHVR |
blaSHV | GGTTATGCGTTATATTCGCC TTAGCGTTGCCAGTGCTC |
865 |
|
CTX-MA1 CTX-MA2 |
blaCTX-M | *SCSATGTGCAG≠YACCAGTAA CCGC¥RATATGRTTGGTGGTG |
544 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





