1. Introduction
‘’Taxonomy (the science of classification) is often undervalued as a glorified form of filing-with each species in its folder, like a stamp in its prescribed place in an album; but taxonomy is a fundamental and dynamic science, dedicated to exploring the causes of relationships and similarities among organisms. Classifications are theories about the basis of natural order, not dull catalogues compiled only to avoid chaos’’. As the famous paleontologist, evolutionary biologist, and historian of science Stephen Jay Gould says.
Biological diversity or biodiversity is used to describe the variation within and between species as well as between ecosystems. Species is the basic unit of biodiversity and intraspecific genetic variation is the foundation for biodiversity [
1].
According to data collected from the International Union for Conservation of Nature (IUCN) Red List for 2022, more than 420,000 plant species exist around the world, yet a subset of them can be identified by traditional taxonomy [
2] based on morphological structures, namely on the phenotypic characteristics of individual organisms. The traditional taxonomy system has several limitations; morphological criteria, for instance vegetative characteristics that can be influenced by environmental factors, or cryptic plant species that look identical, but represent distinct evolutionary lineages, proving traditional taxonomy methods inadequate for plant identification [
3,
4,
5].
In the past 20 years a novel approach based on the use of DNA markers has been developed. DNA barcoding provides a reliable and rapid method for the identification of organisms [
6,
7]. As DNA is not affected by external or other environmental factors, it is stable and can be found in all tissues and thus, this method can be very useful [
8].
The term ‘DNA barcode’ was firstly introduced by Paul Hebert of University of Guelph in 2003 [
9]. DNA barcodes are standardized sequences of DNA, ideally unique, from the genome either of the organism or from its organelles, with a length between 400 and 800 base pairs- that is used to identify/classify an organismal group following amplification, sequencing and comparison with a reference database containing the relevant sequences from different species [
9]. By combining the strengths of two molecular biology methods, PCR amplification and sequencing, DNA barcoding offers a quick and accurate approach to identify different groups. Interestingly at the species level DNA barcoding can be used to characterize novel, unknown species, or cryptic species [
10] two or more species that are classified as a single, as they are superficially morphologically similar and hence, cannot be identified using classical taxonomy [
11,
12]. Notably, the highest cryptic diversity so far has been found in animals, particularly in invertebrates [
10,
13,
14,
15].
DNA barcoding can also be used for the preservation of rare endemic and endangered species [
6] and in general for the research of evolution, ecology, and conservation of plants, especially since biodiversity has been threatened by anthropogenic activity, pollution, deforestation, and resource extraction [
7,
10]. Notably, the US Food and Drug Administration (FDA) encourages the use of DNA-based technology for herbal products quality assessment, among other innovative analytical techniques [
16]. DNA barcoding is an effective approach used in food authentication and traceability, for instance in processed foods and nutritional supplements [
17,
18]. Additionally, it can be used to identify local or autochthonous varieties adding value to the crops or products promoting at the same time the consumption of locally grown vegetables, fruits, and aromatic plants [
18]. DNA barcoding is also used in forensics to link biological specimens to crime scenes. Analyzing both human and non-human DNA is becoming increasingly important for crime investigations. Above all, plant evidence found at the crime scene, such as the transportation of a corpse or the suspect's path, the identification of a narcotic plant etc., may have a crucial role in resolving a case [
4,
19]. Finally, a major application of DNA Barcoding is related with environmental and ecological genomic studies [
4].
A prerequisite for effective DNA barcoding is the establishment of database of the sequences of DNA barcodes. DNA barcoding has been successful used in identifying species from many taxa [
14,
20]; GenBank and the Barcode of Life Datasystem (BOLD) provide a repository for DNA sequences. In BOLD database, projects can be managed, files traced, herbarium specimen scanned, and pictures can be maintained along with DNA sequences. Other well-established databases are FISHBOL and the Chinese herbal medicine DNA barcode identification system. Finally, a DNA Barcode research center has been established in Canada (CCDB) [
18,
21,
22,
23].
The aim of the present review study was to summarize the work has been done on DNA barcoding in plants and discuss several different approaches that has been used.
2. DNA Barcodes
Standardization, minimalism, and scalability are the three oilers of DNA barcoding. This technique has been successfully used for species identification in animals; a 648-base pair (bp) fragment near the 5’-end of the mitochondrial gene cytochrome c oxidase subunit I (
COI) has been selected as the standard barcode [
24]: a) there is a large copy number per cell resulting in easier amplification from smaller or degraded samples, b) it is maternally inherited, c) there is no possibility of recombination with paternal copies, and d) it rapidly accumulates mutations [
25,
26]. While
COI is a suitable target for animals, does not discriminate most plants because of a much slower mutation rate. This has led to the search of alternative barcoding regions [
27,
28].
The fundamental idea behind DNA barcoding is that during species evolution, highly conserved DNA lengths, such as coding or non-coding regions, experienced minor changes. The cytoplasmic mitochondrial DNA, as well as chloroplast DNA and parts from the nuclear DNA include such sequences and have been suggested for use in DNA barcoding [
29]. The suitability of such loci or the combination of, is under discussion for plant species where there is not one easily applied solution [
30]. The design of universal primers and enabling efficient PCR amplification that following sequencing and bioinformatic analysis will identify, ideally all the known species. Unfortunately, so far the ideal DNA barcode does not exist in plants [
20,
21,
31,
32]. Several barcodes, single or multiple have been used and are presented below.
3. Nuclear Genome DNA Barcodes
3.1. ITS (Internal Transcribed Spacer)
The nuclear ribosomal DNA (rDNA) genes have been widely used in plant molecular taxonomy; more specifically the rDNA cistron contains a multigene family that consist of the
18S,
26S and 5.8S coding regions that encode the rRNA core of the ribosome. These genes are separated by the internal transcribed spacer (
ITS) consisting of two subregions namely
ITS1 and
ITS2. So far the
ITS has been used as DNA barcode to identify more than 21,000 plant species [
6,
29,
33].
ITS is a powerful phylogenetic marker because of its widespread distribution and greater evolutionary rate it presents, which enables comparison of relative divergent taxa. Moreover, as it is a non-coding region it exhibits more variation because of a presumed reduction of functional constraints [
6,
20,
29,
32,
34].
ITS can be easily PCR-amplified with conserved primers; alternatively,
ITS1 and
ITS2 with the joining of
5.8S loci can be used as barcodes [
6,
29]. CBOL-Plant Working Group does not suggest its use as universal plant DNA barcode, but as a supplementary locus for taxonomic groupings [
2,
32]. Several limitations hinder the use of
ITS as a universal DNA barcode; gene paralogues, and the existence of secondary structural problems that result in difficulties of amplification and sequencing are major drawbacks [
27,
35,
36]. Despite these problems,
ITS has been used to identify flowers, parasitic plants, and algae very well [
21,
32,
35,
37]. Interestingly, a study of
ITS region as candidate for plant DNA barcoding, suggested that, in general is superior to chloroplast DNA barcodes having better discriminatory power and universality [
4,
29,
35].
3.2. ITS2
ITS2 can be used as an alternative DNA barcode for taxa [
38,
39,
40]; 92.7% of species have been correctly identified in more than 6,500 samples from 4,800 species [
41,
42]. Notably its secondary structure is also informative enabling species identification [
43].
ITS2 may also be used as a complementary locus for
COI to identify animal species [
4,
29,
41,
42].
ITS2 is easy to amplify, conserved, so it is easy to design universal primers, shorter, therefore easier to sequence and, due to its high diversity can be used to distinguish even closely related species [
41,
42]. Although
ITS2 has many strengths, it is not ideal for identifying every plant due to the presence of multiple copies with high levels of intraspecific variation as well as heterogeneity as a result of concerted evolution [
44].
4. Chloroplast DNA Barcodes
Chloroplast DNA is a circular molecule with size between 120-220 kb and consist of a large and a small single-copy region (LSC and SSC) intervened by two copies of a large, inverted repeat (Ira and Irb). There are about 100 functional genes that can be used for species identification and, according to some researchers, besides single-locus markers, whole plastid genome could be used for DNA Barcoding besides single-locus markers. DNA barcodes from chloroplast genes are extensively used in plant phylogenetic studies; the design of primers is easy, gene order in the genome of the organelle is conserved and amplification is much easier due to the high copy number per cell. Nevertheless, compared with the nuclear genome genes of the chloroplast genome are characterized by low evolutionary rate [
4,
23,
29]. Among the chloroplast markers the following have been successfully used:
4.1. matK
matK (Maturase K) is one of the most rapidly evolving, chloroplast genes, that has been used for identification at the family, the genus and even the species level.
matK exhibits interspecific divergence and low transition/ transversion rate. It is approximately 1550 bp long and encodes maturase K, an enzyme involved in the splicing of type-II introns [
35,
40,
45,
46]. However, its use as a universal DNA barcode is hampered by technical problems, mainly the design of the universal primer sets, due to the high substitution rate [
37,
47,
48]. However,
matK constitutes a suitable marker for angiosperms, flowering plants, bryophytes, lycophytes, gymnosperms and monilophytes identification [
46,
48].
4.2. rbcL
rbcL (Ribulose-bisphosphate carboxylase/oxygenase large subunit) is a candidate locus for comparing at the levels of family and genus, however it is not suitable for species identification as it has modest discriminatory power. This marker has been one of the most studied among the plastid genome, with wide representation from all major groups and many available sequences in GenBank [
20,
21,
28,
40]. It was the first gene sequenced from plant chloroplast genome and encodes the large subunit of rubilose-1,5-bisphosphate carboxylase/ oxygenase (RUBISCO), a critical photosynthetic enzyme [
49].
rbcL is easy to be amplified and sequenced, but has slow evolutionary rate [
8,
40]. Its length is approximately 1430 bp and thus at least two sets of primers are needed to sequence of the entire coding sequence [
21,
35,
50].
rbcL meets most of the desired criteria and can be used in conjunction with other markers [
28,
37]. It is also widely used for algae, peptidophytes and angiosperms identification [
7,
32].
4.3. trnH-psbA
trnH-psbA is one of the most variable non-coding plastid loci with inter-genic spacer suitable to offer high level of species discrimination [
35,
37]. It is easily amplified with universal primers but as it has high rates of insertion/ deletion and alignment can be difficult. Moreover, its length varies among different families, containing in some cases this region copies of
rps19, as well as a pseudogene that is located between
trnH and
psbA; this causes a problem as despite obtaining high-quality bidirectional sequences the alignment is difficult due to the high length variation. Most researchers proposed that
trnH-psbA should be used in combination with one or more loci to provide adequate resolution [
8,
20,
28,
37,
48]. Nevertheless, is has been shown that it is a suitable marker for flowering plants and peptidophytes [
7,
21].
4.4. rpoB and rpoC1
rpoB (RNA polymerase subunit B) and
rpoC1 (RNA polymerase subunit C1) are plastid genes, encoding subunits of the plastid-encoded plastid RNA polymerase that have been used for the identification at the family level but, due to their slow evolution rate they cannot be used for species discrimination in many plant families [
37,
46]. Both can be efficiently amplified with limited range of PCR conditions and primer sets [
37].
rpoB,
rpoC1,
rboC2 encode three out of four subunits of the chloroplast RNA polymerase [
51] and are suitable markers for bryophytes identification [
52].
4.5. trnL-trnF (Genic, Intron and Intergenic Spacer)
trnL-trnF intergenic spacer has been proposed as universal plastid amplicon and is widely used in plant systematics and plylogeography since the 1990s [
35,
48]. This region located in the large single copy region of the chloroplast genome [
19]. Despite its slow rate of molecular evolution, the plastid
trnL intron is suggested as a possible marker, because of its conserved sites hence, it could be a useful tool for evolutionary studies at higher taxonomic levels, [
21,
37]. Taberlet et al., [
53] established primers that work for 19 species tested including algae, bryophytes, pteridophytes, gymnosperms and angiosperms.
4.6. psbK-psbI (Intergenic Spacer)
The
psbK and
psbI loci encode two low molecular weight polypeptides, K and I, of the photosystem II [
54]. The non-coding
psbK-psbI intergenic spacer is conserved and can be easily amplified with PCR, sequenced, and aligned [
55,
56]. It also, demonstrates high discriminatory power but low sequence quality and universality [
35]. Despite its discriminatory power the CBOL-Plant Working Group, propose its use as a supplementary locus due to the inconsistency in getting bidirectional unambiguous sequences [
28]. Nevertheless, it constitutes a suitable marker for bryophytes, lycophytes and monilophytes identification [
48].
4.7. atpF-atpH (Intergenic Spacer)
The non-coding, plastid region
atpF-atpH could be used as universal DNA barcoding marker for species level identification but its discriminatory power is medium. The genes
atpF and
atpH encode ATP synthase subunits CFO I and III. The length of
atpF-atpH sequences vary from 598 to 613 bp and the alignment of these sequences is difficult despite easy PCR amplification. For this reason, it could be useful only as supplementary marker in plants DNA Barcoding providing better resolution on specific projects and taxonomic groups [
35,
48,
57,
58,
59]. According to Wang W. et al., [
57] it is a suitable marker for duck seeds identification.
5. Single-Locus, Multi-Locus, and Viable Gene Combinations
The main idea of DNA barcoding is to use a DNA sequence present ideally in every organism to identify it; a prerequisite for this approach is the generation of a reference database containing the relevant sequences from different species [
4,
52]. As no single locus meets CBOLs data standards and guidelines for locus selection, and for this reason the use of combination of barcodes has been proposed [
28].
According to CBOL Plant Working Group, to identify unknown samples, the most common marker combinations are
matK,
rbcL,
rpoB,
rpoC1,
atpF-atpH,
psbK-psbI and
trnH-psbA sequence [
28]. These markers are suggested as they can be amplified with universal primer sets and contain enough sequence diversity - information, both individually and combined, to discriminate across species. Besides these genes, other candidates such as
ITS,
ITS2,
ycf5 and
trnL and combinations of them, that can improve a lot species discrimination have been proposed [
4,
6,
35]. As Newmaster and his colleagues [
20] suggested, to identify unknown species two sequences are needed, the first one would help organism classification into family or genus level and the second one would enable species identification within the higher systematic unit to which an object has been included by means of ‘primary’ sequence. This concept was expanded further by Kress and Erickson [
21], who looked at prospective plant DNA barcode sequences through the lenses of two criteria: universality of amplification and gene differentiation.
The Consortium for the Barcode of Life (CBOL) Plant Working Group, following evaluation of the seven candidate genes referred above, proposed the combination of
matK and
rbcL as the best plant barcode [
28].
rbcL offers high universality but lower species resolution, whereas
matK offers higher resolution but lower universality. This combination offers the best discrimination between species. However, even this combination cannot be used to discriminate between closely related species, so
trnH-psbA intergenic spacer has been proposed as a supplementary locus. To get the highest identification rates, even between closely related species, the China Plant BOL Group suggested adding nuclear
ITS to the
matK +
rbcL combination [
30,
60]. Additional combinations of non-coding and coding regions have also been proposed are
matK +
rbcL +
trnH-psbA,
matK +
atpF-atpH +
trnH-psbA,
matK +
atpF-atpH +
psbK-psbI [
48].
Kress et al. [
27] recommended that two non-coding areas, the nuclear
ITS region and the plastid
trnH-psbA intergenic spacer, could serve as universal plant barcodes. This combination predicted to have some difficulties at species level identification, because
ITS has variable lengths and
trnH-psbA provides insufficient variation, especially in groups with low divergence. However, they later advocated the pairing of
rbcL and
trnH-psbA [
21] and according to Ferri et al. [
61] and Tnah et al. [
8] this resolution is provided by well tested and robust universal primers for both loci that facilitate the reproducibility of results and the implementation of the method in forensics.
Chen et al. [
41] recommended
ITS2 as the best potential marker, which discriminate 92.7% plants at the species level in more than 6,600 plant samples. It has been used as the main DNA barcode for medicine plants and in combination with
trnH-psbA for herbal substrates. Additionally, two other combinations of plastid locus proposed from Chase et al. [
37] are
rpoC1 +
rpoB +
matK or
rpoC1 +
matK +
trnH-psbA. They are useful as a barcoding system, but only for identification of broad groups of species.
In general, different research groups have tested and recommended different combination using different taxa while attempting to select a universal barcode, nevertheless universal agreement is yet to be reached.
6. Super Barcode
With the development of the NGS technology the use of the chloroplast (cp) genome for phylogenetic analysis has rapidly developed. Numerous analyses using the entire sequence of the cp genome have been conducted over the past ten years to solve phylogenetic problems at deep nodes [
62,
63]. The complete cp genome analysis generates the same amount of data as the
COI gene utilized in animals and it can provide molecular identification even between closely related species [
4,
64]. The cp genome ranging from 110-160 kb, offers more diversity enabling the discrimination between closely related plants [
65]. Additionally, PCR efficiency, potential problems with gene deletions and difficulties of sequence retrieval are avoided by using the cp-genome as a marker. Researchers have recommended the complete cp-genome as a super-barcode to differentiate closely related species, even though sequences from a single or many nuclear and chloroplast genes have proved useful for doing so [
4,
64].
The creation of a complete cp-genome database, the reduction of sequencing cost, and obtaining better DNA quality and quantity are the key to super barcoding [
66]. The first cp-genome was sequenced in 1986 [
67], since then the number of cp-genomes sequenced has significantly expanded duo to the development of next-generation sequencing (NGS) technology [
4].
7. Specific Barcode
Single locus barcodes do not offer enough variants, while fully annotated super barcodes are currently expensive and can be extremely complicated for laboratories without the specific technological knowledge. Thus, researchers proposed the use of “specific barcodes” which trade off single locus barcodes and super barcodes, to solve the above-mentioned problem. A specific barcode is a fragment of DNA sequence with high sufficient mutation rate to enable species identification within a specific taxonomic category. Universal primers for the target group can be simply designed since specific barcodes are selected directly from the plastid genome sequences of the target family or genus eliminating technical problems such as low PCR efficiency or extensive optimization that may be time and resource-consuming [
4,
37,
68].
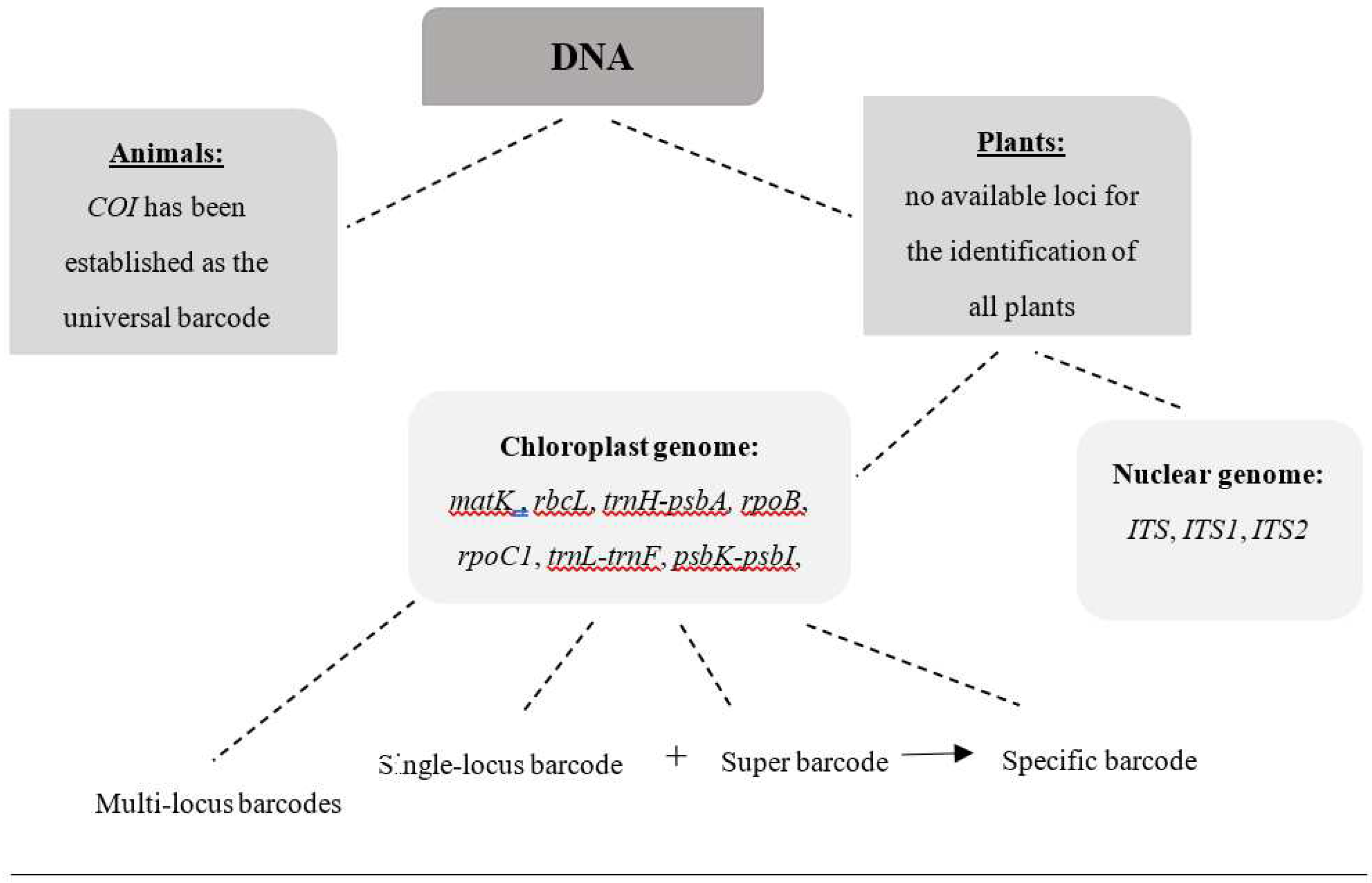
In
Figure 1, a schematic representation is presented, depicting the DNA Barcoding categories distinguished for both animals and plants as analyzed above.
8. Mini Barcoding
DNA mini barcoding has developed over the previous 10 years to either overcome the limitations of DNA barcoding or as an extension of it. DNA mini barcoding uses a DNA sequence shorter than conventional barcodes, usually equally to or less than 200 bp, which can easily and rapidly be amplified. Instead of achieving universality for most species, the major goal of creating a mini barcode is to identify specific target species of herbal plants to prevent adulteration of natural herbal goods. Nonetheless, in case of complex herbal mixtures made for instance from 10 different species mini barcoding is challenging. Considering the position and the length of the specific mini barcode sequence to distinguish between various species is very important, since certain DNA sequences may contain unstable mutation sites [
69,
70,
71].
9. Meta Barcoding
Meta barcoding is a method for identifying numerous species in a single environmental sample by using universal PCR primers to simultaneously amplify multiple DNA barcodes. The discovery and development of NGS (Next Generation Sequencing), which allows parallel reading of DNA sequences without the requirement of cloning, contributed to the development of meta barcoding techniques. In contrast to traditional DNA barcoding, which attempts to identify intact specimens up to the species level, metabarcoding aims to identify in various samples or in degraded DNA samples, like environmental samples (eDNA) up to the family level or higher. Metabarcoding represents a versatile and precise approach to analyze food matrices, to detect some adulterants or contaminants, in forensics and ecological analyses [
23,
29,
70,
71,
72].
10. Discussion
DNA barcoding developed approximately twenty years ago, is an approach that has significantly contributed to the development of molecular systematics. DNA barcodes are standardized sequences, ideally unique, coding or non-coding, either from the genome of the organism or from its organelles, that are used to identify/classify an organismal group; in short, the method includes amplification of the DNA barcode, sequencing and comparison with a reference database containing the relevant sequences from different species. This approach has been extremely successful in animals as COI has emerged as a universal DNA barcode that is used for taxa identification.
In plants, however, the use a universal DNA barcode has not been achieved so far. Several studies with the aim of characterizing plant DNA barcodes from chloroplast and nuclear genomes have been performed- ITS, rbcL, trnH-psbA, rpoB, and rpoC1, trnL-trnF, psbK-psbI, atpF-atp. Based on the data from single loci, the CBOL Plant Working Group has suggested the use of combinations of DNA barcodes, however, this approach also has not provided a universal combination for all plants; however, for specific organismal groups, specific combinations have been successfully used. As next generation sequencing becomes affordable, very long sequences- super barcodes, such as the chloroplast genome are being used successfully especially to distinguish between close species. Mini barcoding, meta barcoding and specific barcodes are approaches that have been successfully applied in various settings. Authors should discuss the results and how they can be interpreted from the perspective of previous studies and of the working hypotheses. The findings and their implications should be discussed in the broadest context possible. Future research directions may also be highlighted.
Funding
This research is part of the project, “Infrastructure of Microbiome Applications in Food Systems-FoodBiomes”, which is co-funded by the European Regional Development Fund (ERDF), under the Operational Program “Competitiveness, Entrepreneurship and Innovation-EPANEK 2014– 2020”, Call 111 “Support for Regional Excellence”.
Data Availability Statement
No new data were created.
Acknowledgments
We acknowledge the support of this work by the project “Infrastructure of Microbiome Applications in FoodSystems-FOODBIOMES” (MIS 5047291), which is implemented under the Action “Regional Excellence in R&D Infrastructures”, funded by the Operational Programme “Competitiveness, Entrepreneurship and Innovation” (NSRF 2014-2020) and co-financed by Greece and the European Union (European Regional Development Fund).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Mora, C.; Tittensor, D.P.; Adl, S.; Simpson, A.G.; Worm, B. How many species are there on Earth and in the ocean? PLoS biology 2011, 9, e1001127. [Google Scholar] [CrossRef]
- IUCN. The IUCN red list of threatened species, Version 2022-1. 2012. Available online: https://nc.iucnredlist.org.
- Erdtman, G. Pollen morphology and plant taxonomy. Svensk. Bot. Tidskr 1945, 39, 286–297. [Google Scholar]
- Li, X.; Yang, Y.; Henry, R.J.; Rossetto, M.; Wang, Y.; Chen, S. Plant DNA barcoding: From gene to genome. Biol. Rev. 2015, 90, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Sawarkar, A.D.; Shrimankar, D.D.; Kumar, M.; Kumar, P.; Kumar, S.; Singh, L. Traditional System Versus DNA Barcoding in Identification of Bamboo Species: A Systematic Review. Mol. Biotechnol. 2021, 63, 651–675. [Google Scholar] [CrossRef]
- Vijayan, K.; Tsou, C.H. DNA barcoding in plants: Taxonomy in a new perspective. Curr. Sci. 2010, 1530–1541. [Google Scholar]
- Saddhe, A.A.; Kumar, K. DNA barcoding of plants: Selection of core markers for taxonomic groups. Plant Sci. Today 2018, 5, 9–13. [Google Scholar] [CrossRef]
- Tnah, L.; Lee, S.; Tan, A.; Lee, C.; Ng, K.; Ng, C.; Farhanah, Z.N. DNA barcode database of common herbal plants in the tropics: A resource for herbal product authentication. Food Control. 2019, 95, 318–326. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; Dewaard, J.R. Biological identifications through DNA barcodes. Proc. R. Soc. Lond. Ser. B Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef]
- Jiang, K.-W.; Zhang, R.; Zhang, Z.-F.; Pan, B.; Tian, B. DNA barcoding and molecular phylogeny of Dumasia (Fabaceae: Phaseoleae) reveals a cryptic lineage. Plant Divers. 2020, 42, 376–385. [Google Scholar] [CrossRef] [PubMed]
- Bickford, D.; Lohman, D.J.; Sodhi, N.S.; Ng, P.K.; Meier, R.; Winker, K.; Ingram, K.K.; Das, I. Cryptic species as a window on diversity and conservation. Trends Ecol. Evol. 2007, 22, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Struck, T.H.; Feder, J.L.; Bendiksby, M.; Birkeland, S.; Cerca, J.; Gusarov, V.I.; Kistenich, S.; Larsson, K.-H.; Liow, L.H.; Nowak, M.D.; et al. Finding Evolutionary Processes Hidden in Cryptic Species. Trends Ecol. Evol. 2018, 33, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, K.; Kumar, V.; Singha, D.; Chandra, K.; Laskar, B.A.; Kundu, S.; Chakraborty, R.; Chatterjee, S. DNA Barcoding studies on Thrips in India: Cryptic species and Species complexes. Sci. Rep. 2017, 7, 1–14. [Google Scholar]
- Hebert, P.D.N.; Penton, E.H.; Burns, J.M.; Janzen, D.H.; Hallwachs, W. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc. Natl. Acad. Sci. USA 2004, 101, 14812–14817. [Google Scholar] [CrossRef] [PubMed]
- Kanturski, M.; Lee, Y.; Choi, J.; Lee, S. DNA barcoding and a precise morphological comparison revealed a cryptic species in the Nippolachnus piri complex (Hemiptera: Aphididae: Lachninae). Sci. Rep. 2018, 8, 1–16. [Google Scholar]
- FDA. Botanical drug development guidance for industry. U.S. Department of Health and Human Services, Food and Drug Administration. 2016. Available online: https://www.fda.gov/ downloads/drugs/guidancecomplianceregulatoryinformation/guidances/ ucm458484.pdf.
- Uncu, A.O.; Uncu, A.T. A barcode-DNA analysis method for the identification of plant oil adulteration in milk and dairy products. Food Chem. 2020, 326, 126986. [Google Scholar] [CrossRef] [PubMed]
- Galimberti, A.; Casiraghi, M.; Bruni, I.; Guzzetti, L.; Cortis, P.; Berterame, N.M.; Labra, M. From DNA barcoding to personalized nutrition: The evolution of food traceability. Curr. Opin. Food Sci. 2019, 28, 41–48. [Google Scholar] [CrossRef]
- Park, E.; Kim, J.; Lee, H. Plant DNA barcoding system for forensic application. Forensic Sci. Int. Genet. Suppl. Ser. 2017, 6, e282–e283. [Google Scholar] [CrossRef]
- Newmaster, S.; Fazekas, A.; Ragupathy, S. DNA barcoding in land plants: Evaluation of rbcL in a multigene tiered approach. Can. J. Bot. 2006, 84, 335–341. [Google Scholar] [CrossRef]
- Kress, W.J.; Erickson, D.L. A two-locus global DNA barcode for land plants: The coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS ONE 2007, 2, e508. [Google Scholar] [CrossRef]
- Vere, N.D.; Rich, T.C.; Trinder, S.A.; Long, C. DNA barcoding for plants. In Plant Genotyping; Humana Press, New York, NY, USA, 2015; pp. 101–118.
- Yu, J.; Wu, X.; Liu, C.; Newmaster, S.; Ragupathy, S.; Kress, W.J. Progress in the use of DNA barcodes in the identification and classification of medicinal plants. Ecotoxicol. Environ. Saf. 2021, 208, 111691. [Google Scholar] [CrossRef] [PubMed]
- Coissac, E.; Hollingsworth, P.M.; Lavergne, S.; Taberlet, P. From barcodes to genomes: Extending the concept of DNA barcoding. Mol. Ecol. 2016, 25, 1423–1428. [Google Scholar] [CrossRef]
- Luo, A.; Zhang, A.; Ho, S.Y.; Xu, W.; Zhang, Y.; Shi, W.; Cameron, S.L.; Zhu, C. Potential efficacy of mitochondrial genes for animal DNA barcoding: A case study using eutherian mammals. BMC Genom. 2011, 12, 84. [Google Scholar] [CrossRef] [PubMed]
- Waugh, J. DNA barcoding in animal species: Progress, potential and pitfalls. BioEssays 2007, 29, 188–197. [Google Scholar] [CrossRef] [PubMed]
- Kress, W.J.; Wurdack, K.J.; Zimmer, E.A.; Weigt, L.A.; Janzen, D.H. Use of DNA barcodes to identify flowering plants. Proc. Natl. Acad. Sci. USA 2005, 102, 8369–8374. [Google Scholar] [CrossRef]
- CBOL Plant Working Group. A DNA barcode for land plants. Proc. Natl. Acad. Sci. USA 2009, 12794–12797. [Google Scholar]
- Ali, M.A.; Gyulai, G.; Hidvégi, N.; Kerti, B.; Al Hemaid, F.M.; Pandey, A.K.; Lee, J. The changing epitome of species identification – DNA barcoding. Saudi J. Biol. Sci. 2014, 21, 204–231. [Google Scholar]
- Techen, N.; Parveen, I.; Pan, Z.; A Khan, I. DNA barcoding of medicinal plant material for identification. Curr. Opin. Biotechnol. 2014, 25, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Sass, C.; Little, D.P.; Stevenson, D.W.; Specht, C.D. DNA Barcoding in the Cycadales: Testing the Potential of Proposed Barcoding Markers for Species Identification of Cycads. PLoS ONE 2007, 2, e1154. [Google Scholar] [CrossRef]
- Kowalska, Z.; Pniewski, F.; Latała, A. DNA barcoding–A new device in phycolog’st's toolbox. Ecohydrol. Hydrobiol. 2019, 19, 417–427. [Google Scholar] [CrossRef]
- Chen, S.; Pang, X.; Song, J.; Shi, L.; Yao, H.; Han, J.; Leon, C. A renaissance in herbal medicine identification: From morphology to DNA. Biotechnol. Adv. 2014, 32, 1237–1244. [Google Scholar] [CrossRef] [PubMed]
- Álvarez IJ, F.W.; Wendel, J.F. Ribosomal ITS sequences and plant phylogenetic inference. Mol. Phylogenetics Evol. 2003, 29, 417–434. [Google Scholar] [CrossRef] [PubMed]
- Hollingsworth, P.M.; Graham, S.W.; Little, D.P. Choosing and Using a Plant DNA Barcode. PLoS ONE 2011, 6, e19254. [Google Scholar] [CrossRef] [PubMed]
- De Mattia, F.; Bruni, I.; Galimberti, A.; Cattaneo, F.; Casiraghi, M.; Labra, M. A comparative study of different DNA barcoding markers for the identification of some members of Lamiacaea. Food Res. Int. 2011, 44, 693–702. [Google Scholar] [CrossRef]
- Chase, M.W.; Cowan, R.S.; Hollingsworth, P.M.; Van Den Berg, C.; Madriñán, S.; Petersen, G.; Wilkinson, M. A proposal for a standardised protocol to barcode all land plants. Taxon 2007, 56, 295–299. [Google Scholar] [CrossRef]
- Gao, T.; Yao, H.; Song, J.; Liu, C.; Zhu, Y.; Ma, X.; Pang, X.; Xu, H.; Chen, S. Identification of medicinal plants in the family Fabaceae using a potential DNA barcode ITS2. J. Ethnopharmacol. 2010, 130, 116–121. [Google Scholar] [CrossRef]
- Schultz, J.; Wolf, M. ITS2 sequence–structure analysis in phylogenetics: A how-to manual for molecular systematics. Mol. Phylogenetics Evol. 2009, 52, 520–523. [Google Scholar] [CrossRef] [PubMed]
- Miao, L.; Xi-Wen, L.; Bao-Sheng, L.; Lu, L.; Yue-Ying, R. Species identification of poisonous medicinal plant using DNA barcoding. Chin. J. Nat. Med. 2019, 17, 585–590. [Google Scholar]
- Chen, S.L.; Yao, H.; Han, J.P.; Liu, C.; Song, J.Y.; Shi, L.C.; Zhu, Y.J.; Ma, X.Y.; Gao, T.; Pang, X.H.; et al. Validation of the ITS2 Region as a Novel DNA Barcode for Identifying Medicinal Plant Species. PLoS ONE 2010, 5, e8613. [Google Scholar] [CrossRef]
- Yao, H.; Song, J.; Liu, C.; Luo, K.; Han, J.; Li, Y.; Pang, X.; Xu, H.; Zhu, Y.; Xiao, P.; et al. Use of ITS2 Region as the Universal DNA Barcode for Plants and Animals. PLoS ONE 2010, 5, e13102. [Google Scholar] [CrossRef]
- Wolf, M.; Chen, S.; Song, J.; Ankenbrand, M.; Müller, T. Compensatory Base Changes in ITS2 Secondary Structures Correlate with the Biological Species Concept Despite Intragenomic Variability in ITS2 Sequences – A Proof of Concept. PLoS ONE 2013, 8, e66726. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Chen, M.; Dong, X.; Lin, R.; Fan, J.; Chen, Z. Evaluation of Four Commonly Used DNA Barcoding Loci for Chinese Medicinal Plants of the Family Schisandraceae. PLoS ONE 2015, 10, e0125574. [Google Scholar] [CrossRef] [PubMed]
- Hilu, K.W.; Liang, G. The matK gene: Sequence variation and application in plant systematics. Am. J. Bot. 1997, 84, 830–839. [Google Scholar] [CrossRef]
- Dong, W.; Liu, J.; Yu, J.; Wang, L.; Zhou, S. Highly Variable Chloroplast Markers for Evaluating Plant Phylogeny at Low Taxonomic Levels and for DNA Barcoding. PLoS ONE 2012, 7, e35071. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Xue, J.; Zhou, S. New universal matK primers for DNA barcoding angiosperms. J. Syst. Evol. 2011, 49, 176–181. [Google Scholar] [CrossRef]
- Fazekas, A.J.; Burgess, K.S.; Kesanakurti, P.R.; Graham, S.W.; Newmaster, S.G.; Husband, B.C.; Percy, D.M.; Hajibabaei, M.; Barrett, S.C.H. Multiple Multilocus DNA Barcodes from the Plastid Genome Discriminate Plant Species Equally Well. PLoS ONE 2008, 3, e2802. [Google Scholar] [CrossRef]
- Zurawski, G.; Perrot, B.; Bottomley, W.; Whitfeld, P.R. The structure of the gene for the large subunit of ribulose 1,5-bisphosphate carboxylase from spinach chloroplast DNA. Nucleic Acids Res. 1981, 9, 3251–3270. [Google Scholar] [CrossRef] [PubMed]
- Blaxter, M.L. The promise of a DNA taxonomy. Philos. Trans. R. Soc. London. Ser. B: Biol. Sci. 2004, 359, 669–679. [Google Scholar] [CrossRef] [PubMed]
- Serino, G.; Maliga, P. RNA Polymerase Subunits Encoded by the Plastid rpoGenes Are Not Shared with the Nucleus-Encoded Plastid Enzyme1. Plant Physiol. 1998, 117, 1165–1170. [Google Scholar] [CrossRef]
- Liu, Y.; Yan, H.; Cao, T.; Ge, X. Evaluation of 10 plant barcodes in Bryophyta (Mosses). J. Syst. Evol. 2010, 48, 36–46. [Google Scholar] [CrossRef]
- Taberlet, P.; Gielly, L.; Pautou, G.; Bouvet, J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol. Biol. 1991, 17, 1105–1109. [Google Scholar] [CrossRef] [PubMed]
- Meng, B.Y.; Wakasugi, T.; Sugiura, M. Two promoters within the psbK-psbI-trnG gene cluster in tobacco chloroplast DNA. Curr. Genet. 1991, 20, 259–264. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.L.; Yi, D.K.; Kim, J.S.; Kim, K.J. Development of plant DNA barcoding markers from the variable noncoding regions of chloroplast genome. In Abstract presented at the second international barcode of life conference. Academia Sinica: Taipei, Taiwan, 2007.
- Lahaye, R.; Savolainen, V.; Duthoit, S.; Maurin, O.; van der Bank, M. A test of psbK-psbI and atpF-atpH as potential plant DNA barcodes using the flora of the Kruger National Park (South Africa) as a model system. Nature Precedings 2008, 1. [Google Scholar] [CrossRef]
- Wang, W.; Wu, Y.; Yan, Y.; Ermakova, M.; Kerstetter, R.; Messing, J. DNA barcoding of the Lemnaceae, a family of aquatic monocots. BMC Plant Biol. 2010, 10, 1–11. [Google Scholar] [CrossRef]
- Drager, R.G.; Hallick, R.B. A novel Euglena gracilis chloroplast operon encoding four ATP synthase subunits and two ribosomal proteins contains 17 introns. Curr. Genet. 1993, 23, 271–280. [Google Scholar] [CrossRef]
- Neto, A.B.; Morais, M.B.; Dutra, E.D.; Junior, T.C. Biological diversity of Lemna aequinoctialis Welw. isolates influence biomass production and wastewater phytoremediation. Bioresour. Technol. Rep. 2019, 6, 251–259. [Google Scholar] [CrossRef]
- China Plant BOL Group 1; Li, D.Z.; Gao, L.M.; Li, H.T.; Wang, H.; Ge, X.J.; Duan, G.W. Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc. Natl. Acad. Sci. USA 2011, 108, 19641–19646. [Google Scholar]
- Ferri, G.; Corradini, B.; Ferrari, F.; Santunione, A.; Palazzoli, F.; Alu’, M. Forensic botany II, DNA barcode for land plants: Which markers after the international agreement? Forensic Sci. Int. Genet. 2015, 15, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Zhai, W.; Duan, X.; Zhang, R.; Guo, C.; Li, L.; Xu, G.; Shan, H.; Kong, H.; Ren, Y. Chloroplast genomic data provide new and robust insights into the phylogeny and evolution of the Ranunculaceae. Mol. Phylogenetics Evol. 2019, 135, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Xu, C.; Wu, P.; Cheng, T.; Yu, J.; Zhou, S.; Hong, D.-Y. Resolving the systematic positions of enigmatic taxa: Manipulating the chloroplast genome data of Saxifragales. Mol. Phylogenetics Evol. 2018, 126, 321–330. [Google Scholar] [CrossRef]
- Wu, L.; Wu, M.; Cui, N.; Xiang, L.; Li, Y.; Li, X.; Chen, S. Plant super-barcode: A case study on genome-based identification for closely related species of Fritillaria. Chin. Med. 2021, 16, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Parks, M.; Cronn, R.; Liston, A. Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biol. 2009, 7, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Kane, N.; Sveinsson, S.; Dempewolf, H.; Yang, J.Y.; Zhang, D.; Engels, J.M.M.; Cronk, Q. Ultra-barcoding in cacao (Theobroma spp.; Malvaceae) using whole chloroplast genomes and nuclear ribosomal DNA. Am. J. Bot. 2012, 99, 320–329. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, K.; Ohme, M.; Tanaka, M.; Wakasugi, T.; Hayashida, N.; Matsubayashi, T.; Zaita, N.; Chunwongse, J.; Obokata, J.; Yamaguchi-Shinozaki, K.; et al. The complete nucleotide sequence of the tobacco chloroplast genome: Its gene organization and expression. EMBO J. 1986, 5, 2043–2049. [Google Scholar] [CrossRef]
- Chen, Q.; Wu, X.; Zhang, D. Comparison of the abilities of universal, super, and specific DNA barcodes to discriminate among the original species of Fritillariae cirrhosae bulbus and its adulterants. PLoS ONE 2020, 15, e0229181. [Google Scholar] [CrossRef] [PubMed]
- Meusnier, I.; Singer, G.A.; Landry, J.-F.; Hickey, D.A.; Hebert, P.D.; Hajibabaei, M. A universal DNA mini-barcode for biodiversity analysis. BMC Genom. 2008, 9, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Liu, Y.; Wang, X.; Wei, X.; Han, J. DNA Mini-Barcoding: A Derived Barcoding Method for Herbal Molecular Identification. Front. Plant Sci. 2019, 10, 987. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.S. DNA barcoding in plants and animals: A critical review. 2022.
- Taberlet, P.; Prud’homme, S.M.; Campione, E.; Roy, J.; Miquel, C.; Shehzad, W.; Gielly, L.; Rioux, D.; Choler, P.; Clément, J.; et al. Soil sampling and isolation of extracellular DNA from large amount of starting material suitable for metabarcoding studies. Mol. Ecol. 2012, 21, 1816–1820. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).