1. Introduction
The interleukin-1 (IL-1) family is a group of 11 cytokines and 10 receptors that regulate inflammatory responses with various roles. Some of them, such as IL-1ɑ, IL-33, IL-36, and IL-1β, mediate a pro-inflammatory response [
1], whereas others like IL-37 and IL-38 are anti-inflammatory [
1]. When the pro- inflammatory cytokines of the IL-1 family bind to the cognate receptor IL-1R1, they recruit the secondary receptor IL-1RacP [
2], thus initiating IL-1 signaling through two mediators, myeloid differentiation primary response 88 (MyD88) and interleukin-1 receptor associated kinases 4 (IRAK 4), to eventually activate nuclear factor κB (NFκB) and activator protein 1 (AP1). Both transcription factors will regulate target gene expression to mediate IL-1 functions. From the given signal transduction pathway, tight regulation is required to prevent detrimental effects that could lead to chronic inflammatory disorders.
Among IL-1 family, IL-1ɑ plays pleiotropic roles in both inflammation and hematopoiesis; it is widely expressed in various cells including epithelial, endothelial, stromal cells, neutrophils, and activated macrophages [
3]. Under the condition of cellular damage, necrotic cells release IL-1ɑ into the extracellular environment as an alarmin [
4]. In addition, IL-1ɑ is also induced in hematopoietic and non-hematopoietic cells in response to inflammatory stimuli. There exists IL-1RA as an antagonist to compete with IL-1 for binding to IL-1R1. The balance between IL-1RA and IL-1 becomes very crucial for the development of a variety of inflammatory diseases such as Deficiency of IL-1 Receptor Antagonist (DIRA), Rheumatoid Arthritis (RA), Gastric Cancer, and Osteoporosis.
DIRA is a rare autoinflammatory disorder characterized by marked skin and bone involvement, and elevation of acute phase reactants. DIRA is caused by a loss-of-function mutation occurring in the
IL1RN gene that prevents the expression of active IL-1RA, causing unchecked pro-inflammatory signaling and subsequent systemic inflammation [
5]. DIRA has a challenging feature with very early onset. As early as the first week of life, DIRA manifesting symptoms can be developed, including pustular rash, widening of ribs, periosteal reaction, multifocal osteolytic reactions, cervical vertebral fusion, hepatosplenomegaly, multifocal osteomyelitis [
5]. The primary drug choice for DIRA is Anakinra.
Anakinra is a recombinant form of the human IL-1RA protein. The primary difference between the recombinant and human form is that the recombinant contains an additional methionine residue in the amino terminus [
5]. Anakinra was approved by the U.S. Food and Drug Administration (FDA) in December 2020 for the treatment of DIRA, but had been previously approved and utilized for other disorders such as RA and Cryopyrin Associated Periodic Syndromes [
6]. Currently, Anakinra is the primary IL-1 therapeutic due to its short half-life, safety records, and subcutaneous route of delivery.
Even though Anakinra has been approved by the FDA for DIRA therapy, there is no previous review to evaluate Anakinra’s clinical efficacy and safety as a recombinant analog of IL-1RA in treatment of this autoinflammatory disease. Here, through review of current literature, we identified that Anakinra decreased acute phase reactants levels and improved anemia of patients in treatment of this autoimmune disease.
2. Materials and Methods
2.1. Literature Search Strategy
A literature search was conducted through PubMed, EMBASE, and Google Scholar, including articles from inception: keywords “DIRA” or “Deficiency of IL-1RA” and “Anakinra” were used to filter the studies evaluating the efficacy of Anakinra in DIRA therapy. Language was restricted to English only.
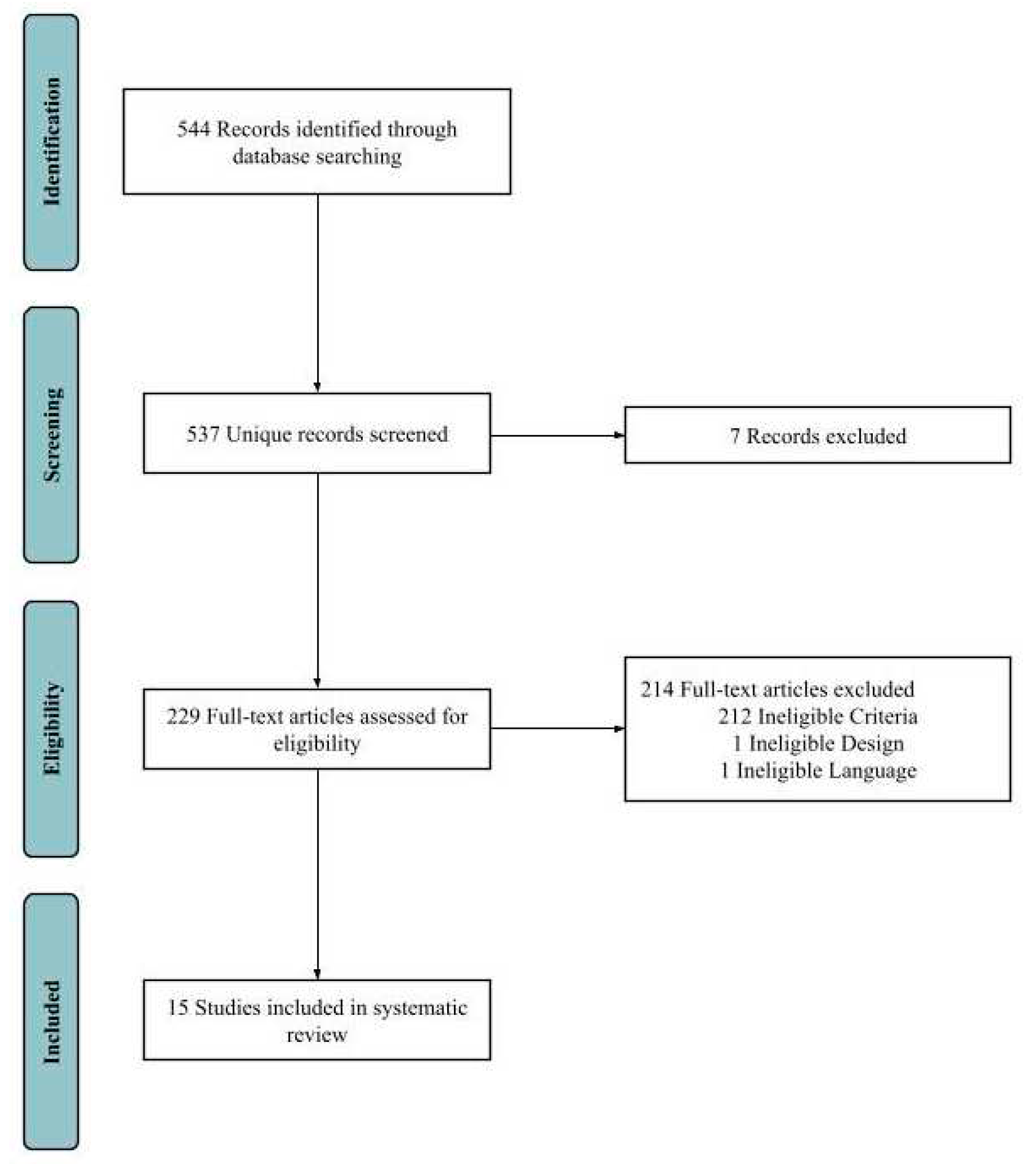
Gray literature was not included in the literature search. We included clinical trials, clinical studies, case reports, comparative study, original articles, meta-analysis, observational studies, and twin studies in the literature search. The detailed literature search and screening process was depicted below (
Figure 1).
2.2. Study Selection and Eligibility Criteria
Two paired reviewers independently conducted a title and abstract screening. We assessed the abstracts to identify articles that discussed Anakinra as the main treatment for DIRA and removed duplicate articles. Abstracts were also assessed to see if the efficacy of Anakinra was discussed with laboratory values or other means. Side effects of Anakinra were also considered. Abstracts that discussed Anakinra for the treatment of other rheumatological diseases but not DIRA were excluded from this study. Following screening, both reviewers independently evaluated the full-text of papers for eligibility.
2.3. Data Extraction
The data were extracted from the selected papers on age of presentation of DIRA symptoms, sex, region of origin, reported cytogenetic abnormalities, all previous treatments, consanguinity status, side effects from anakinra treatment, and the reported common clinical presentation. Hemoglobin (HB), C-Reactive Protein (CRP), and Erythrocyte Sedimentation Rate (ESR) were also extracted from these papers. These markers were included to comprehensively assess DIRA as an autoinflammatory disease. The efficacy of Anakinra for the treatment of DIRA has been evaluated with the CRP and ESR laboratory values pre- and post-treatment. HB laboratory values have also reported pre- and post-treatment to evaluate anemia associated with chronic diseases.
2.4. Statistical Analysis
Statistical analysis in this work was performed using GraphPad Prism (version 9.5.1) with its embedded algorithms. The Wilcoxon rank-sum test and the Student’s t-test (two-tailed, paired) were applied to evaluate the significance of laboratory results pre- and post- treatment. Data unavailable either due to patient confidentiality or other reasons were excluded.
To optimize statistical testing, a quantile-quantile (QQ) plot was first plotted on all laboratory test data to evaluate skewness, central tendency, and dispersion of the data. This determined whether the data were in normal Gaussian distribution, and if further non-parametric tests were needed. Then, all laboratory data were plotted as a Violin plot using GraphPad Prism software to visualize the full distribution of the data. All data presented in this study were listed as the mean differences with 95% confidence intervals. Unless stated otherwise, a P-value less than 0.05 was considered significant.
3. Results
3.1. Overall Population
Of the 15 studies included in this review, a total of 25 patients were treated with Anakinra for DIRA [
7,
8,
9,
10,
11,
12,
13,
14,
15,
16,
17,
18,
19,
20,
21]. 19 patients (
n = 19, 76%) were 4 weeks old or younger, and a total of 23 patients (
n = 23, 92%) were under 17 weeks old (
Table 1). The sex of all patients was primarily male, making up 16 total (
n = 16, 64%) as shown in
Table 2. The most common ethnicity of patients was Dutch (
n = 6, 24%), Lebanese (
n = 3, 12%), Puerto Rican (
n = 4, 16%), and Brazilian (
n = 3, 12%) (
Table 3). 13 patients (
n = 13, 52%) were non-consanguineous and 9 patients (
n = 9, 36%) were consanguineous as shown in
Table 4.
3.2. Clinical Presentation and Cytogenetic Abnormalities
The common clinical manifestations of DIRA were pustular rashes (
n = 21, 84%), multifocal osteolytic lesions (
n = 17, 68%), widening of ribs (
n = 15, 60%), periosteal reaction (
n = 13, 52%), and multifocal osteomyelitis (
n = 12, 48%). All other clinical symptoms have been reported in
Table 5. The most prevalent nucleotide abnormalities leading to the clinical presentation of DIRA was c.229G→T (
n = 5, 20%), which led to a nonsense mutation on the amino acid found at position 77 (E77X). A 175-kb deletion mutation on chromosome 2q13 (
n = 5, 20%) was equally prevalent. All other mutations are listed in
Table 6.
3.3. Previous Treatments and Effects of Anakinra
Before treatment with Anakinra, 16 patients (
n = 16, 64%) were previously treated with Prednisolone/Methylprednisolone. 17 patients (
n = 17, 68%) with oral/intravenous antibiotics. Canakinumab, an IL-1β blocker, was also given to patients (
n = 2, 8%) before the administration of Anakinra. All other previous treatments utilized were listed in
Table 7. These previous treatments failed to reach clinical remission. With the treatment of Anakinra, most patients reported no side effects (
n = 22, 88%). All other side effects (
n = 3, 12%) we listed in
Table 8.
3.4. Analysis of Laboratory Tests
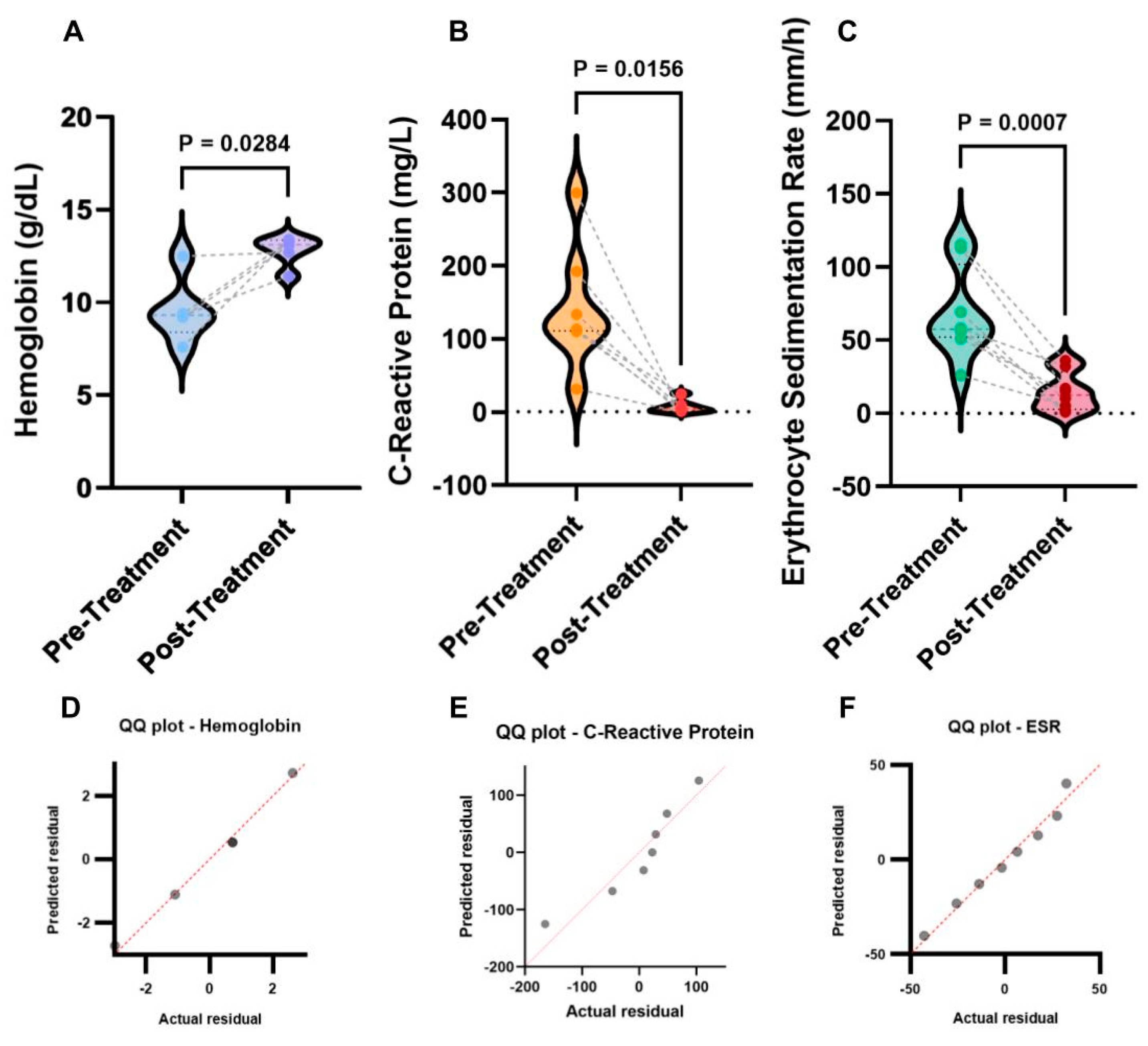
All laboratory testing results were plotted as Violin plots in
Figure 2A-C along with the QQ plots shown in
Figure 2D-F. From the QQ plots, the distance between the data and the predicted regression line, or the line of identity, displayed whether the laboratory test data was a normal Gaussian distribution. From
Figure 2D,F, the distribution of HB and ESR was confirmed to be a normal Gaussian distribution. However, the data listed on the QQ plot for CRP (
Figure 2E) appeared to be distant from the regression line, and therefore could not be assumed to be a normal Gaussian distribution. Furthermore, Student’s t-test (two-tailed, paired) was utilized to evaluate significance for HB and ESR. Since the distribution of CRP is not normal, the Wilcoxon rank-sum non-parametric test was utilized. The data were then plotted as Violin plots.
From these data analysis, it was found that before the treatment with Anakinra, CRP levels were 142.17 ± 62.20 mg/L, after treatment its levels were reduced to 6.72 ± 6.52 mg/L (Cohen’s d effect sizes = 2.27, Pearson’s r correlations = 0.75). HB levels were increased from 9.60 ± 1.56 g/dL (before the treatment with Anakinra) to 12.78 ± 0.71 g/dL (after the treatment; Cohen’s d effect sizes = 2.25, Pearson’s r correlations = 0.75). For ESR, its levels were reduced from 68.12 ± 21.40 mm/h (before Anakinra treatment) to 14.72 ± 9.18 mm/h (after the treatment; Cohen’s d effect sizes = -2.29, Pearson’s r correlations = -0.75).
4. Discussion
The majority of patients had an eruption of pustular rash within the first eight weeks of life. However, in the case report by Abdwani
et al. [
18] chronic diarrhea was reported as the initial unusual presentation of DIRA. Many patients also exhibited anemia. A small portion of patients also had extramedullary hematopoiesis leading to hepatomegaly and hepatosplenomegaly. With the administration of Anakinra, there was clinical remission of DIRA, however some patients experienced side effects. In the case report by Mendenco
et al. [
11], the patient experienced an urticarial rash within fifteen minutes of administration of Anakinra for the first time. This was well controlled by pre-medication with diphenhydramine, followed by the administration of Anakinra. However, in a case report by Ziaee
et al [
13], a patient showed signs of respiratory distress two weeks after administration of Anakinra. This patient also showed bone and skin complications. Then, Anakinra was discontinued, and Etanercept and prednisolone were started, and an improvement was observed. The patient’s respiratory involvement never went back to baseline, and the patient died eight months later. This may be a word of caution for clinicians who treat DIRA patients with bone, skin, and respiratory complications.
Given that the primary ethnicities of patients were Dutch, Puerto Rican, and Brazilian listed in
Table 3, their heterozygous relatives were not identified to exhibit any symptoms of DIRA shown in
Table 6. These cytogenetic abnormalities are first generation homozygous mutations and have been characterized in isolated geographical regions. The allele frequencies of the founder mutations in Puerto Rico (Arecibo) and Newfoundland have been estimated by Reddy
et al. [
17] and Aksentijevich
et al. [
21] to be 1.3% and 0.2%, respectively. In such isolated geographic regions, genetic counseling and prenatal screening is recommended for identification.
Recent studies such as Al-Herz
et al. [26] demonstrate that consanguineous marriages increase the risk of autoimmune diseases in offspring. However, in this study, shown in
Table 4, patients were primarily born to non-consanguineous parents. Due to the limited sample size of this review, further studies are required to precisely determine the correlation of consanguinity with DIRA.
In multiple studies, such as Kuemmerle-Deschner
et al. [
8] and Minkis
et al. [
12], DIRA patients were first diagnosed with pustular psoriasis based on skin biopsy and clinical presentation. However, genome-wide sequencing identified mutations in the
IL1RN gene and the diagnosis of DIRA. Across all studies, Sanger sequencing, SNP array, PCR,
In silico modeling, and Next-Generation Sequencing were utilized to identify DIRA to accurately differentiate DIRA from other similar systemic autoinflammatory disorders, such as chronic nonbacterial osteomyelitis. Thus, molecular diagnosis with the following genes has been incorporated:
IL1RN,
MEFV,
MVK,
TNFRSF1A,
NLRP12,
NLRP3,
NOD2,
LPIN2,
PSMB8,
PSTPIP1, provides a rapid and effective genetic screening for early treatment as the delayed treatment can cause major complications such as severe osteomyelitis.
One of the limitations of this study is a lack of clinical trials and animal models. Given the nature of DIRA as an ultra-rare disorder, clinical trials with enough participants would be very challenging. Randomized animal trials could be utilized to solve this problem for the better evaluation of the effectiveness of Anakinra as compared to multiple interventions.
From this review, we obtained more evidence on the effectiveness of Anakinra therapy for the treatment of DIRA. With a dose of 2-3 mg/kg/day Anakinra, complete remission of DIRA is possible. Although most patients have reported no side effects of Anakinra, some patients have reported adverse effects from Anakinra. This could be attributed to the different genetic predisposition of each individual patient. With proper desensitization to Anakinra with diphenhydramine, improvement of treatment and full clinical remission of DIRA can be reached.
5. Conclusions
DIRA is a rare autoinflammatory disorder with early onset (1-8 weeks) and poor prognosis (systemic inflammation, mortality, etc). Due to its similarity to other common heterozygous autoinflammatory disorders, it has been difficult to diagnose DIRA. In this review, we evaluated the efficacy of Anakinra in the treatment of DIRA and reported statistical analyses of laboratory data from the selected studies. The results indicated that most patients had no adverse effects to Anakinra. In addition, we also identified a decrease in acute phase reactants and an increase in hemoglobin levels from these laboratory data. In summary, our study further confirms that complete remission of DIRA is possible through Anakinra therapy with minimal or no side effects.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org. Table S1: Supplementary Data.
Author Contributions
Conceptualization, K.P., J.P. and J.L.; methodology, K.P.; software, J.P.; validation, K.P., J.P. and J.L.; formal analysis, K.P. and J.P.; investigation, K.P. and J.P.; resources, K.P. and J.P.; data curation, K.P. and J.P.; writing—original draft preparation, K.P. and J.P.; writing—review and editing, K.P., J.P. and J.L.; visualization, K.P., J.P. and J.L.; supervision, J.L.; project administration, K.P., J.P. and J.L.; funding acquisition, J.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Acknowledgments
We thank the internal research funds of California University of Science and Medicine to J.L. for the partial support of this study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Dinarello CA. Overview of the IL-1 family in innate inflammation and acquired immunity. Immunol Rev. 2018;281(1):8–27. [CrossRef]
- Brint E, Kamradt T, Doyle SL. Editorial: IL-1 family members in health and disease. Front Immunol. 2019;10. [CrossRef]
- Cavalli G, Colafrancesco S, Emmi G, Imazio M, Lopalco G, Maggio MC, et al. Interleukin 1α: a comprehensive review on the role of IL-1α in the pathogenesis and treatment of autoimmune and inflammatory diseases. Autoimmun Rev. 2021;20(3):102763. [CrossRef]
- Van Den Eeckhout B, Tavernier J, Gerlo S. Interleukin-1 as innate mediator of T cell immunity. Front Immunol. 2021;11. [CrossRef]
- Cvetkovic RS, Keating G. Anakinra. BioDrugs. 2002;16(4):303–11. [CrossRef]
- KINERET® (anakinra) official website. Kineretrx.com. [cited 2023 Jul 20]. Available from: https://www.kineretrx.com/.
- Mendonca LO, Malle L, Donovan FX, Chandrasekharappa SC, Montealegre Sanchez GA, Garg M, et al. Deficiency of interleukin-1 receptor antagonist (DIRA): Report of the first Indian patient and a novel deletion affecting IL1RN. J Clin Immunol. 2017;37(5):445–51. [CrossRef]
- Kuemmerle-Deschner JB, Welzel T, Hoertnagel K, Tsiflikas I, Hospach A, Liu X, et al. New variant in the IL1RN-gene (DIRA) associated with late-onset, CRMO-like presentation. Rheumatology (Oxford). 2020;59(11):3259–63. [CrossRef]
- Sözeri B, Gerçeker-Türk B, Yıldız-Atıkan B, Mir S, Berdeli A. A novel mutation of interleukin-1 receptor antagonist (il1rn) in a dira patient from turkey: diagnosis and treatment. Turk J Pediatr. 2018;60(5):588. [CrossRef]
- Schnellbacher C, Ciocca G, Menendez R, Aksentijevich I, Goldbach-Mansky R, Duarte AM, et al. Deficiency of interleukin-1 receptor antagonist responsive to anakinra. Pediatr Dermatol. 2013;30(6):758–60. [CrossRef]
- Mendonça LO, Grossi A, Caroli F, de Oliveira RA, Kalil J, Castro FFM, et al. A case report of a novel compound heterozygous mutation in a Brazilian patient with deficiency of Interleukin-1 receptor antagonist (DIRA). Pediatr Rheumatol Online J. 2020;18(1). [CrossRef]
- Minkis K, Aksentijevich I, Goldbach-Mansky R, Magro C, Scott R, Davis JG, et al. Interleukin 1 receptor antagonist deficiency presenting as infantile pustulosis mimicking infantile pustular psoriasis. Arch Dermatol. 2012;148(6). [CrossRef]
- Ziaee V, Youssefian L, Faghankhani M, Jazayeri A, Saeidian AH, Vahidnezhad H, et al. Homozygous IL1RN mutation in siblings with deficiency of interleukin-1 receptor antagonist (DIRA). J Clin Immunol. 2020;40(4):637–42. [CrossRef]
- Sakran W, Shalev SA, Sakran W, Shalev SA, El-Shanti H, Uziel Y, et al. Chronic recurrent multifocal osteomyelitis and deficiency of interleukin-1–receptor antagonist. Pediatr Infect Dis J. 2013;32(1):94. [CrossRef]
- Jesus AA, Osman M, Silva CA, Kim PW, Pham T-H, Gadina M, et al. A novel mutation of IL1RN in the deficiency of interleukin-1 receptor antagonist syndrome: Description of two unrelated cases from Brazil. Arthritis Rheum. 2011;63(12):4007–17. [CrossRef]
- Brau-Javier CN. Chronic cutaneous pustulosis due to a 175-kb deletion on chromosome 2q13: Excellent response to anakinra. Arch Dermatol. 2012;148(3):301. [CrossRef]
- Reddy S, Jia S, Geoffrey R, Lorier R, Suchi M, Broeckel U, et al. An autoinflammatory disease due to homozygous deletion of theIL1RNlocus. N Engl J Med. 2009;360(23):2438–44. [CrossRef]
- Abdwani R, Abdalla E, Al Masilhi B, Shalaby A, Al-Maawali A. Novel mutation in interleukin 1 receptor antagonist associated with chronic diarrhoea in infancy. J Paediatr Child Health. 2022;58(1):186–8. [CrossRef]
- Stenerson M, Dufendach K, Aksentijevich I, Brady J, Austin J, Reed AM. The first reported case of compound heterozygous IL1RN mutations causing deficiency of the interleukin-1 receptor antagonist. Arthritis Rheum. 2011;63(12):4018–22. [CrossRef]
- Thacker PG, Binkovitz LA, Thomas KB. Deficiency of interleukin-1-receptor antagonist syndrome: a rare auto-inflammatory condition that mimics multiple classic radiographic findings. Pediatr Radiol. 2012;42(4):495–8. [CrossRef]
- Aksentijevich I, Masters SL, Ferguson PJ, Dancey P, Frenkel J, van Royen-Kerkhoff A, et al. An autoinflammatory disease with deficiency of the interleukin-1–receptor antagonist. N Engl J Med. 2009;360(23):2426–37. [CrossRef]
- Al-Herz W, Aldhekri H, Barbouche M-R, Rezaei N. Consanguinity and primary immunodeficiencies. Hum Hered. 2014;77(1–4):138–43. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).