Submitted:
11 December 2023
Posted:
12 December 2023
You are already at the latest version
Abstract
Keywords:
1. Hypotheses on the origin of foxtail millet and recent advances in foxtail millet genomics
2. Variation in morphological characteristics
3. Genetic differentiation of foxtail millet landraces, according to biochemical and genetic markers and intraspecific hybrid pollen sterility
4. Evolution of some genes (Waxy, PPO, HD1, PRR37, Svles1, and C) under human and/or natural selection
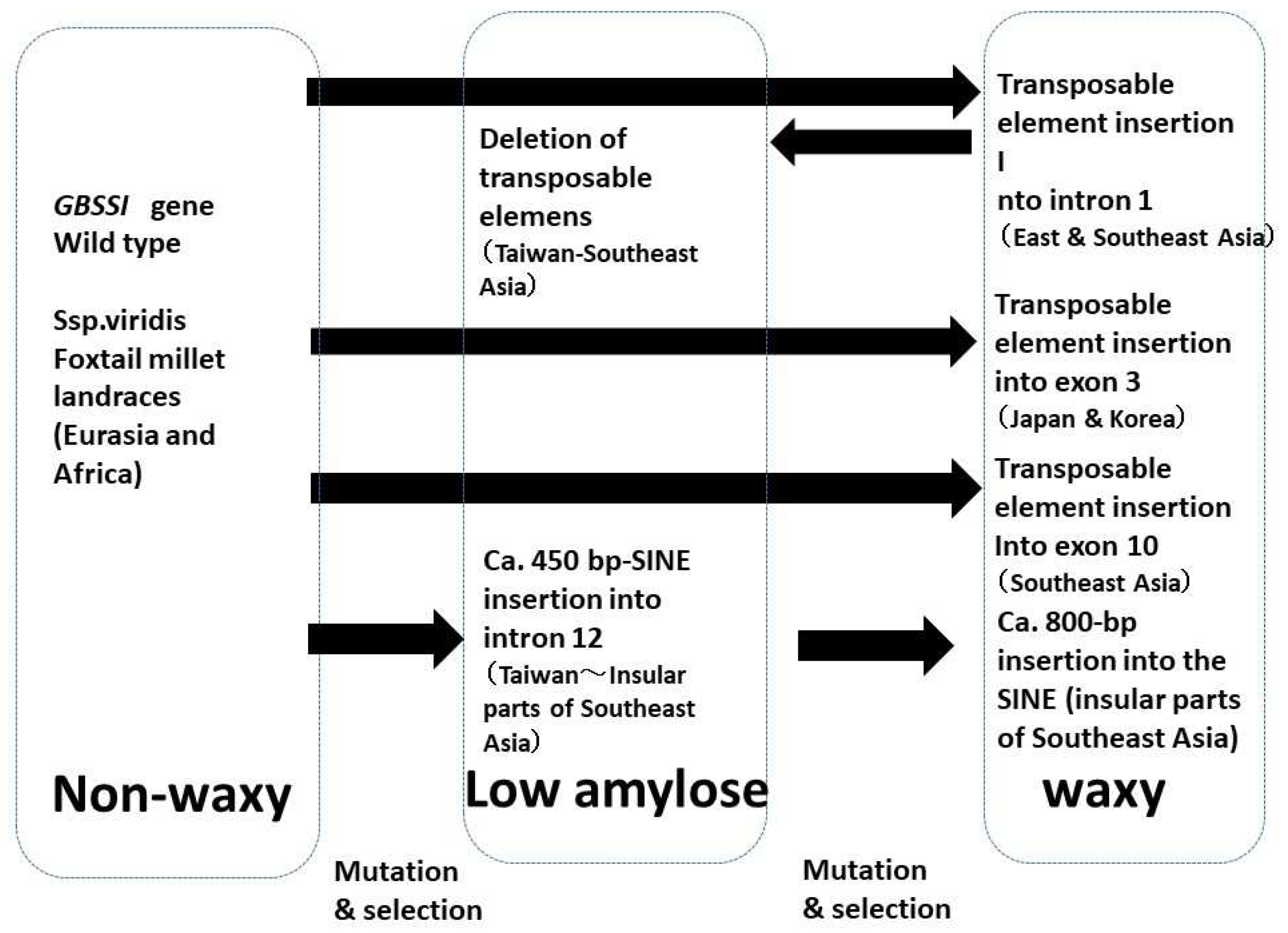
4.1. Waxy
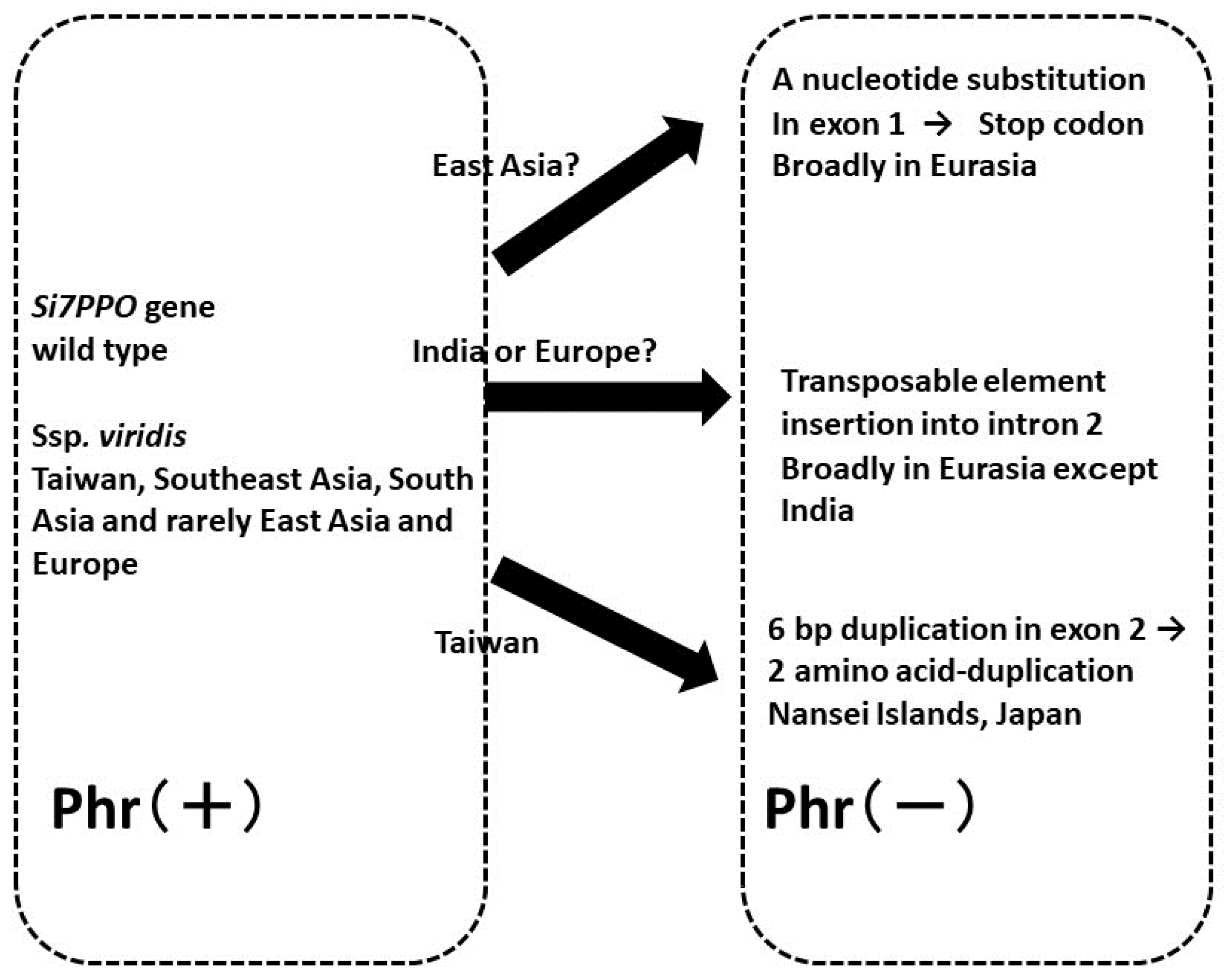
4.2. Variations in Phr and evolution of the polyphenol oxidase (Si7PPO) gene
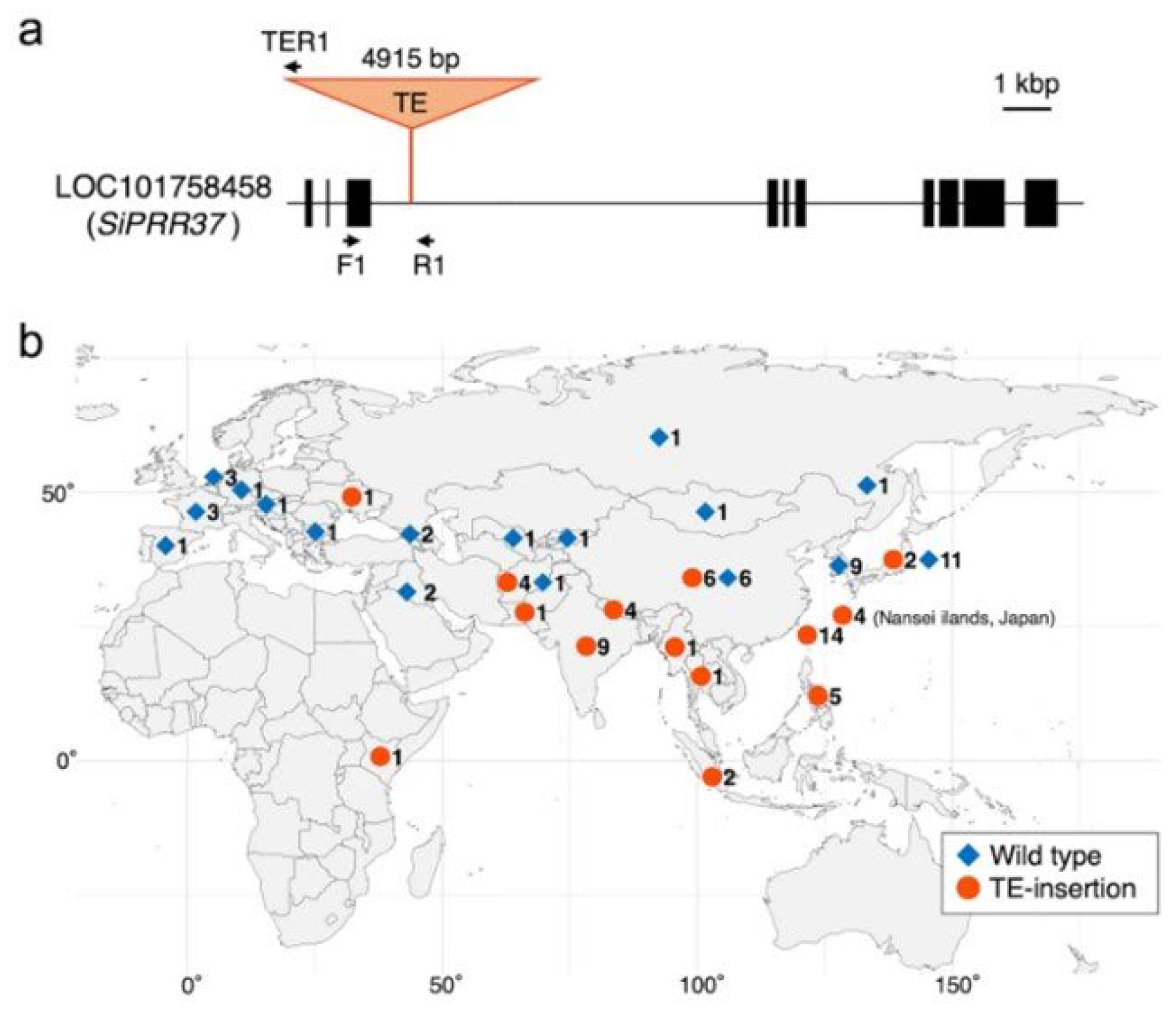
4.3. Heading time genes Heading Date1 (HD1) and SiPRR37
4.4. Shattering genes
4.5. The C gene involved in leaf sheath pigmentation
5. Perspective
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hunt, H.V.; Linden, M.V.; Liu, X.; Motuzaite-Matuzeviciute, G.; Colledge, S.; Jones, M.K. Millets across Eurasia: chronology and context of early records of the genera Panicum and Setaria from archaeological sites in the Old World. Veg. Hist. Archaeobot. 2008, 17, 5–18. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, S. Origin and dispersal of common millet and foxtail millet. Japan Agr Res Quart. 1987, 21, 84–89. [Google Scholar]
- Fukunaga, K. Genetic Differentiation and Crop Evolution of Foxtail Millet. In Genetics and Genomics of Setaria; Doust, A., Diao, X., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 115–131. [Google Scholar]
- Kihara, H.; Kishimoto, E. Bastarde zwischen Setaria italica und S.viridis. Bot Mag. 1942, 20, 63–67, (in Japanese with German summary). [Google Scholar]
- Li, H.W.; Li, C.H.; Pao, W.K. Cytological and genetical studies of the interspecific cross of the cultivated foxtail millet, Setaria italica (L.) Beauv., and the green foxtail millet, S. viridis. J Am Soc Agron. 1945, 37, 32–54. [Google Scholar] [CrossRef]
- Vavilov, N.I. Studies on the Origin of Cultivated Plants. Inst Appl Bot Plant Breed. 1926, pp. 1-248.
- Harlan, J.R. Crops and Man. Soc Am Agron Crop Sci Soc America, Madison, WI, 1975; pp.1-284.
- de Wet, J.M.J.; Oestry-Stidd, L.L.; Cubero, J.I. Origins and evolution of foxtail millet (Setaria italica). Journ d’Agric et de Bot. 1979, 26, 53–64. [Google Scholar] [CrossRef]
- Jusuf, M.; Pernes, J. Genetic variability of foxtail millet (Setaria italica P. Beauv.). Theor. Appl. Genet. 1985, 71, 385–393. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Cao, Y.S.; Wu, S.Z.; Zhang, X.Z. A diversity analysis of foxtail millet (Setaria italica (L.) P. Beauv.) landraces of Chinese origin. Genet. Resour. Crop Evol. 1995, 45, 279–285. [Google Scholar] [CrossRef]
- Li, Y.; Wu, S.Z.; Cao, Y.S. Cluster analysis of an international collection of foxtail millet (Setaria italica (L.) P. Beauv). Euphytica 1995, 83, 79–85. [Google Scholar] [CrossRef]
- Fukunaga, K.; Kawase, M.; Sakamoto, S. Variation of caryopsis length and width among landraces of foxtail millet, Setaria italica (L.) P. Beauv. Jap J Trop. Agric. 1997, 41, 235–240. [Google Scholar]
- Nakayama, H.; Namai, H.; Okuno, K. Geographical variation of the alleles at the Two prolamin loci, Pro1 and Pro2, in foxtail millet, Setaria italica (L.) P. Beauv. Genes Genet Syst. 1999, 74, 293–297. [Google Scholar] [CrossRef]
- Fukunaga, K.; Domon, E.; Kawase, M. Ribosomal DNA variation in foxtail millet, Setaria italica (L.) P. Beauv. and a survey of variation from Europe and Asia. Theor. Appl. Genet. 1997, 97, 751–756. [Google Scholar] [CrossRef]
- Fukunaga, K.; Ichiatani, K.; Kawase, M. Phylogenetic analysis of rDNA intergenic spacer subrepeats and its implication for domestication history of foxtail millet, Setaria italica. Theor Appl Genet. 2006, 113, 261–269. [Google Scholar] [CrossRef] [PubMed]
- Schontz, D.; Rether, B. Genetic variability in foxtail millet, Setaria italica (L.) P. Beauv. - RFLP using a heterologous rDNA probe. Plant Breed 1998, 117, 231–234. [Google Scholar] [CrossRef]
- Eda, M.; Izumitani, A.; Ichitani, K.; Kawase, M.; Fukunaga, K. Geographical variation of foxtail millet, Setaria italica (L.) P.
- Beauv. based on rDNA PCR–RFLP. Genet. Res. Crop Evol. 2013, 60, 265–274. [Google Scholar]
- Li, Y.; Jia, J.; Wang, W.; Wu, S. Intraspecific and interspecific variation in Setaria revealed by RAPD analysis. Genet Resour Crop Evol. 1998, 45, 279–285. [Google Scholar]
- Schontz, D.; Rether, B. Genetic variability in foxtail millet, Setaria italica (L.) P. Beauv.: Identification and classification of lines with RAPD markers. Plant Breed. 1999, 118, 190–192. [Google Scholar] [CrossRef]
- Le Thierry d’Ennequin, M.; Panaud, O.; Toupance, B.; Sarr, A. Assessment of genetic relationships between Setaria italica and its wild relative S. viridis using AFLP markers. Theor. Appl. Genet. 2000, 100, 1061–1066. [Google Scholar] [CrossRef]
- Fukunaga, K.; Wang, Z.M.; Kato, K.; Kawase, M. Geographical variation of nuclear genome RFLPs and genetic differentiation in foxtail millet, Setaria italica (L.) P. Beauv. Genet. Res. Crop Evol, 2002, 49, 95–101. [Google Scholar] [CrossRef]
- Fukunaga, K.; Kato, K. Mitochondrial DNA variation in foxtail millet, Setaria italica (L.)P.Beauv. Euphytica 2003, 129, 7–13. [Google Scholar] [CrossRef]
- Hirano, R.; Naito, K.; Fukunaga, K.; Watanabe, K.N.; Ohsawa, R.; Kawase, M. Genetic structure of landraces in foxtail millet (Setaria italica (L.) P. Beauv.) revealed with transposon display and interpretation to crop evolution of foxtail millet. Genome 2011, 54, 498–506. [Google Scholar] [CrossRef]
- Nasu, H.; Momohra, A.; Yasuda, Y.; He, J. The occurrence and identification of Setaria italica (L.) P. Beauv. (foxtail millet) grains from the Chengtoushan site (ca. 5800 cal B.P.) in central China, with reference to the domestication centre in Asia. Veget Hist Archaeobot. 2007, 16, 481–494. [Google Scholar] [CrossRef]
- Kawase, M.; Sakamoto, S. Geographical distribution of landrace groups classified by hybrid pollen sterility in foxtail millet, Setaria italica (L.) P. Beauv. J. Jpn Breed. 1987, 37, 1–9. [Google Scholar] [CrossRef]
- Bennetzen, J.L.; Schmutz, J.; Wang, H.; Percifield, R.; Hawkins, J.; Pontaroli, A.C.; Estep, M.; Feng, L.; Vaughn, J.N.; Grimwood, J.; et al. Reference genome sequence of the model plant Setaria. Nature Biotech. 2012, 30, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Liu, X.; Quan, Z.; Cheng, S.; Xu, X.; Pan, S.; Xie, M.; Zeng, P.; Yue, Z.; Wang, W.; et al. Genome sequence of foxtail millet (Setaria italica) provides insights into grass evolution and biofuel potential. Nat Biotechnol. 2012, 30, 549–554. [Google Scholar] [CrossRef] [PubMed]
- Mamidi, S.; Healey, A.; Huang, P.; Grimwood, J.; Jenkins, J.; Barry, K.; Sreedasyam, A.; Shu, S.; Lovell, J.T.; Feldman, M.; et al. A genome resource for green millet Setaria viridis enables discovery of agronomically valuable loci. Nat Biotechnol. 2020, 38, 1203–1210. [Google Scholar] [CrossRef] [PubMed]
- Dekaprelevich, L.L.; Kasparian, A.S. A contribution to the study of foxtail millet (Setaria italicaP.B. maxima. Alef.) cultivated in Georgia (western Transcaucasia). Bull. Appl Bot Plant Breed. 1928, 19, 533–572. [Google Scholar]
- Kawase, M. Genetic variation and landrace differentiation of foxtail millet, Setaria italica, in Eurasia. Ph. D. thesis. Kyoto University, Japan, 1986. 1-127.
- Takei, E; Sakamoto, S. Geographical variation of heading response to daylength in foxtail millet (Setaria italica P. Beauv.). Jpn. J. Breed. 1987, 37, 150–158. [Google Scholar] [CrossRef]
- Takei, E.; Sakamoto, S. Further analysis of geographical variation of heading response to daylength in foxtail millet (Setaria italica P.Beauv.). Jpn. J. Breed. 1989, 39, 285–298. [Google Scholar] [CrossRef]
- Prasada Rao, K.E.; de Wet, J.M.J; Brink, D.E.; Mengesha, M.H. Infraspecific variation and systematics of cultivated Setaria italica, foxtail millet (Poaceae). Econ Bot. 1987, 41, 108–116. [Google Scholar]
- Ochiai, Y.; Kawase, M.; Sakamoto, S. Variation and distribution of foxtail millet (Setaria italica P. Beauv.) in the mountainous areas of northern Pakistan. Breed Sci. 1994, 44, 413–418. [Google Scholar] [CrossRef]
- Ochiai, Y. Variation in tillering and geographical distribution of foxtail millet (Setaria italica P. Beauv.). Breed Sci. 1996, 46, 143–146. [Google Scholar] [CrossRef]
- Fukunaga, K. Differentiation of landraces of foxtail millet, Setaria italica (L.) P.Beauv. in RFLP and morphological characters. Doctor Thesis, Kyoto University, Kyoto, Japan, 1998; pp. 1–98. [Google Scholar]
- Hammer, K.; Khoshbakht, K. Foxtail millet (Setaria italica (L.) P. Beauv.) in Mazandaran/Northern Iran. Genet. Res. Crop Evol. 2007, 54, 907–911. [Google Scholar] [CrossRef]
- Nguyen Van, F.; Pernes, J. Genetic diversity of foxtail millet (Setaria italica). In: Genetic differentiation and dispersal in plants, P. Jacquard, Eds; NATO ASI Series, Vol. G5, Springer-Verlag, Berlin, 1985, pp. 113-128.
- Kawase, M.; Sakamoto, S. Variation, geographical distribution and genetical analysis of esterase isozymes in foxtail millet, Setaria italica (L.)P.Beauv. Theor Appl Genet. 1984, 67, 529–533. [Google Scholar] [CrossRef] [PubMed]
- Hunt, H.V.; Przelomska, N.A.S.; Campana, M.G.; Cockram, J.; Bligh, H.F.J.; Kneale, C.J.; Romanova, O.I.; Malinovskaya, E.V.; Jones, M.J. Population genomic structure of Eurasian and African foxtail millet landrace accessions inferred from genotyping-by-sequencing. Plant Genome, 2021, 14, e20081. [Google Scholar] [CrossRef] [PubMed]
- Konishi, S.; Izawa, T.; Lin, S.Y.; Ebana, K.; Fukuta, Y.; Sasaki, T.; Yano, M. An SNP caused loss of seed shattering during rice domestication. Science 2006, 312, 1392–1396. [Google Scholar] [CrossRef] [PubMed]
- Ishii, T.; Numaguchi, K.; Miura, K.; Yoshida, K.; Thanh, P.T.; Htun, T.M.; Yamasaki, M.; Komeda, N.; Matsumoto, T.; Terauchi, R.; et al. OsLG1 regulates a closed panicle trait in domesticated rice. Nat. Genet. 2013, 45, 462–465. [Google Scholar] [CrossRef] [PubMed]
- Doebley, J.; Stec, A.; Hubbard, L. The evolution of apical dominance in maize. Nature 1997, 386, 485–488. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Nussbaum-Wagler, T.; Li, B.; Zhao, Q.; Vigouroux, Y.; Faller, M.; Bomblies, K.; Lukens, L. Doebley, J.F. The origin of the naked grains of maize. Nature. 2005, 436, 714–719. [Google Scholar] [CrossRef] [PubMed]
- Komatsuda, T.; Pourkheirandish, M.; He, C.; Azhaguvel, P.; Kanamori, H.; Perovic, D.; Stein, N.; Graner, A.; Wicker, T.; Tagiri, A.; et al. Six-rowd barley originated from a mutation in a homeodomain-leucine zipper I-class homeobox gene. Proc Natl Acad Sci USA. 2007, 104, 1424–1429. [Google Scholar] [CrossRef]
- Taketa, S.; Amano, S.; Tsujino, Y.; Sato, T.; Saisho, D.; Kakeda, K.; Nomura, M.; Suzuki, T.; Matsumoto, T.; Sato, K.; et al. Barley grain with adhering hulls is controlled by an ERF family transcription factor gene regulating a lipid biosynthesis pathway. Proc. Natl. Acad. Sci. USA. 2008, 105, 4062–4067. [Google Scholar] [CrossRef]
- Sakamoto, S. Glutinous-endosperm starch food culture specific to Eastern and Southeastern Asia. In: Redefining Nature: ecology, culture and domestication; Ellen, R, Fukui, K. Eds.;. Berg Publishers, Oxford, UK, 1995, pp. 215–231.
- Yoshida, S. Wild food plants and vegeculture. In: Vegeculture in Eastern Asia and Oceania; Yoshida, S., Matthews, P.J., Ed.
- JCAS Symposium Series No. 16, National Museum of Ethnology, Osaka, Japan, 2002, pp. 31–44.
- Fogg, W.H. Swidden cultivation of foxtail millet by Taiwan aborigines: a cultural analogue of the domestication of Setaria italica in China. In: The Origins of Chinese Civilization. Keighty, D.N. Ed.;. Univ California Press, Berkeley, 1983, pp.95–115.
- Takei, E. Characteristics and ethnobotany of millets in the Southwestern (Nansei) Islands of Japan (in Japanese with English abstract).
- Ph.D thesis, Faculty of Agriculture, Kyoto University, Japan, 1994.
- Sano, Y. Differential regulation of gene expression in rice endosperm. Theor. Appl. Genet. 1984, 68, 467–473. [Google Scholar] [CrossRef]
- Collins, G.N. A new type of Indian corn from China. USDA Bur. Pl. Ind. Bull. 1909, 16, 1–30. [Google Scholar]
- Wessler, S.R.; Varagona, M.J. Molecular basis of mutations at Waxy locus of maize: Correlation with the fine structure genetic map. Proc Natl Acad Sci USA. 1985, 82, 4177–4181. [Google Scholar] [CrossRef]
- Hirano, H.Y.; Sano, Y. Molecular characterization of the waxy locus of rice (Oryza sativa). Plant Cell Physiol. 1991, 32, 989–997. [Google Scholar] [CrossRef]
- Hirano, H.Y.; Eighuchi, M.; Sano, Y. A single base change altered the regulation of the Waxy gene at the posttranscriptional level during the domestication of rice. Mol. Biol. Evol. 1998, 15, 978–987. [Google Scholar] [CrossRef]
- Isshiki, M.; Morino, K.; Nakajima, M.; Okagaki, R.J.; Wessler, S.R.; Izawa, T.; Shimamoto, K. A naturally occurring functional allele of the rice waxy locus has a GT to TT mutation at the 5’ splice site of the first intron. Plant J. 1998, 15, 133–138. [Google Scholar] [CrossRef]
- Wanchana, S.; Toojinda, T.; Tragoonrung, S.; Vanavichit, A. Duplicated coding sequence in the waxy allele of tropical glutinous rice (Oryza sativa L.). Plant Sci. 2003, 165, 1193–1199. [Google Scholar] [CrossRef]
- Olsen, K.M.; Purugganan, M.D. Molecular evidence on the origin and evolution of glutinous rice. Genetics 2002, 162, 941–950. [Google Scholar] [CrossRef]
- Domon, E.; Fujita, M.; Ishikawa, N. The insertion/deletion polymorphisms in the Waxy gene of barley genetic resources from East Asia. Theor. Appl. Genet. 2002, 104, 132–138. [Google Scholar] [CrossRef]
- Domon, E.; Saito, A.; Takeda, K. Comparison of the waxy locus sequence from a non-waxy strain and two waxy mutants of spontaneous and artificial origins in barley. Genes Genet Syst. 2002, 77, 351–359. [Google Scholar] [CrossRef]
- Patron, N.J.; Smith, A.M.; Fahy, B.F.; Hylton, C.M.; Naldrett, M.J.; Rossnagel, B.G.; Denyer, K. The altered pattern of amylose accumulation in the endosperm of low-amylose barley cultivars is attributable to a single mutant allele of granule-bound starch synthase I with a deletion in the 5-non-coding region. Plant Physiol. 2002, 130, 190–198. [Google Scholar] [CrossRef]
- McIntyre, C.L.; Drenth, J.; Gonzalez, N.; Henzell, R.G; Jordan, D.R. Molecular characterization of the waxy locus in sorghum. Genome 2008, 51, 524–533. [Google Scholar] [CrossRef]
- Sattler, S.E.; Singh, J.; Haas, E.J.; Guo, L.; Sarath, G.; Pedersen, J.F. Two distinct waxy alleles impact the granule-bound starch synthase in sorghum. Mol Breed. 2009, 24, 349–359. [Google Scholar] [CrossRef]
- Kawahigashi, H.; Oshima, M.; Nishikawa, T.; Okuizumi, H.; Kasuga, S.; Yonemaru, J.-i. A novel waxy allele in sorghum land races in East Asia. Plant Breed. 2013, 132, 305–310. [Google Scholar] [CrossRef]
- Lu, Y.; Zhao, G.; Li, Y.; Fan, J.; Ding, G.; Zhao, J.; Ni, X.; Xu, Y.; Wang, W. Identification of two novel waxy alleles and development of their molecular markers in sorghum. Genome 2013, 56, 283–288. [Google Scholar] [CrossRef]
- Hachiken, T.; Masunaga, Y.; Ishii, Y.; Ohta, T.; Ichitani, K.; Fukunaga, K. Deletion commonly found in Waxy gene of Japanese and Korean cultivars of Job’s tears (Coix lacryma-jobi L.). Mol. Breed. 2012, 30, 1747–1756. [Google Scholar] [CrossRef]
- Fan, L.J.; Quan, L.Y.; Leng, X.D.; Guo, X.Y.; Hu, W.M.; Ruan, S.L.; Ma, H.S.; Zeng, M.Q. Molecular evidence for post- domestication selection in the Waxy gene of Chinese waxy maize. Mol. Breed. 2008, 22, 329–338. [Google Scholar] [CrossRef]
- Fan, LJ.; Bao, J.D.; Wang, Y.; Yao, J.Q.; Gui, Y.J.; Hu, W.M.; Zhu, J.Q.; Zeng, M.Q.; Li, Y.; Xu, Y.B. Post-domestication selection in the maize starch pathway. PLoS ONE 2009, 4, e7612. [Google Scholar] [CrossRef]
- Bao, J.D.; Yao, J.Q.; Zhu, J.Q.; Hu, W.M.; Cai, D.G.; Li, Y.; Shu, Q.Y.; Fan, L.J. Identification of glutinous maize landraces and inbred lines with altered transcription of waxy gene. Mol. Breed. 2012, 30, 1707–1714. [Google Scholar] [CrossRef]
- Zhang, L.L.; Chen, H.; Luo, M.; Zhang, X.W.; Deng, M.; Ma, J.; Qi, P.F.; Wang, J.R.; Chen, G.Y.; Liu, Y.X.; et al. Transposon insertion resulted in the silencing of Wx-B1n in Chinese wheat landraces. Theor. Appl. Genet. 2017, 130, 1331. [Google Scholar] [CrossRef]
- Wu, X.Y.; Chen, D.; Lu, Y.Q.; Liu, W.H.; Yang, X.M.; Li, X.Q.; Du, J.; Li, L.H. Molecular characteristics of two new waxy mutations in China waxy maize. Mol. Breed. 2017, 37, 1–7. [Google Scholar]
- Wu, X.; Wu, S.; Long, W.; Chen, D.; Zhou, G.; Du, J.; Cai, Q.; Huang, X. New Waxy allele wx-Reina found in Chinese waxy maize. Genet. Resour. Crop Evol. 2019, 66, 885–895. [Google Scholar] [CrossRef]
- Hunt, H.V.; Packman, L.C.; Jones, M.K.; Howe, C. J. Molecular basis of the Waxy endosperm starch phenotype in broomcorn millet (Panicum miliaceum L.). Mol. Biol. Evol. 2010, 27, 1478–1494. [Google Scholar] [CrossRef] [PubMed]
- Fukunaga, K. Origin of waxy cereals from a genetic point of view: From cultural history to genetic history of waxy cereals. Breed. Res. 2019, 21, 1–10, (in Japanese with English summary). [Google Scholar] [CrossRef]
- Gaur, V.S.; Sood, S.; Guzmán, C.; Olsen, K.M. Molecular insights on the origin and development of waxy genotypes in major crop plants. Briefings in Functional Genomics 2023, elad035. [Google Scholar] [CrossRef]
- Nakayama, H.; Afzal, M.; Okuno, K. Intraspecific differentiation and geographical distribution of Wx alleles for low amylase content in endosperm of foxtail millet, Setaria italica (L.)Beauv. Euphytica 1998, 102, 289–293. [Google Scholar] [CrossRef]
- Fukunaga, K.; Kawase, M.; Kato, K. Structural variation in the Waxy gene and differentiation in foxtail millet [Setaria italica (L.) P. Beauv.]: implications for multiple origins of the waxy phenotype. Mol. Genet. Genomics 2002, 268, 214–222. [Google Scholar] [CrossRef] [PubMed]
- Kawase, M.; Fukunaga, K.; Kato, K. Diverse origins of waxy foxtail millet crops in East and Southeast Asia mediated by multiple transposable element insertions. Mol. Genet. Genomics 2005, 274, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Van, S.; Onoda, S.; Kim, M.Y.; Kim, K.D.; Lee, S.H. Allelic variation of the Waxy gene in foxtail millet [Setaria italica (L.) P. Beauv.] by single nucleotide polymorphisms. Mol. Genet. Genomics 2008, 279, 255–266. [Google Scholar] [CrossRef]
- Afzal, M.; Kawase, M.; Nakayama, H.; Okuno, K. Variation in electrophoregrams of total seed protein and Wx protein in foxtail millet. In: Progress in New Crops. Janick, J, Ed.,. ASHS Press, Alexandria, Va.,1996, pp.191–195.
- Okuno, K.; Fuwa, H.; Yano, M. A new mutant gene lowering amylose content in endosperm starch in rice, Oryza sativa L. Jpn. J. Breed. 1983, 33, 387–394. [Google Scholar] [CrossRef]
- Hachiken, T.; Sato, K.; Hasegawa, T.; Ichitani, K.; Kawase, M.; Fukunaga, K. Geographic distribution of Waxy gene SNPs and indels in foxtail millet, Setaria italica (L.)P.Beauv. Genet. Res. Crop Evol. 2013, 60, 1559–1570. [Google Scholar] [CrossRef]
- Oka, H.I. Phylogenetic differentiation of the cultivated rice plant. 1. Variations in respective characteristics and their combinations in rice cultivars. Jpn. J. Breed. 1953, 3, 33–43, (in Japanese with English summary). [Google Scholar] [CrossRef]
- Takeda, K.; Chang, C.L. Inheritance and geographical distribution of phenol reaction-less varieties of barley. Euphytica 1996, 90, 217–221. [Google Scholar] [CrossRef]
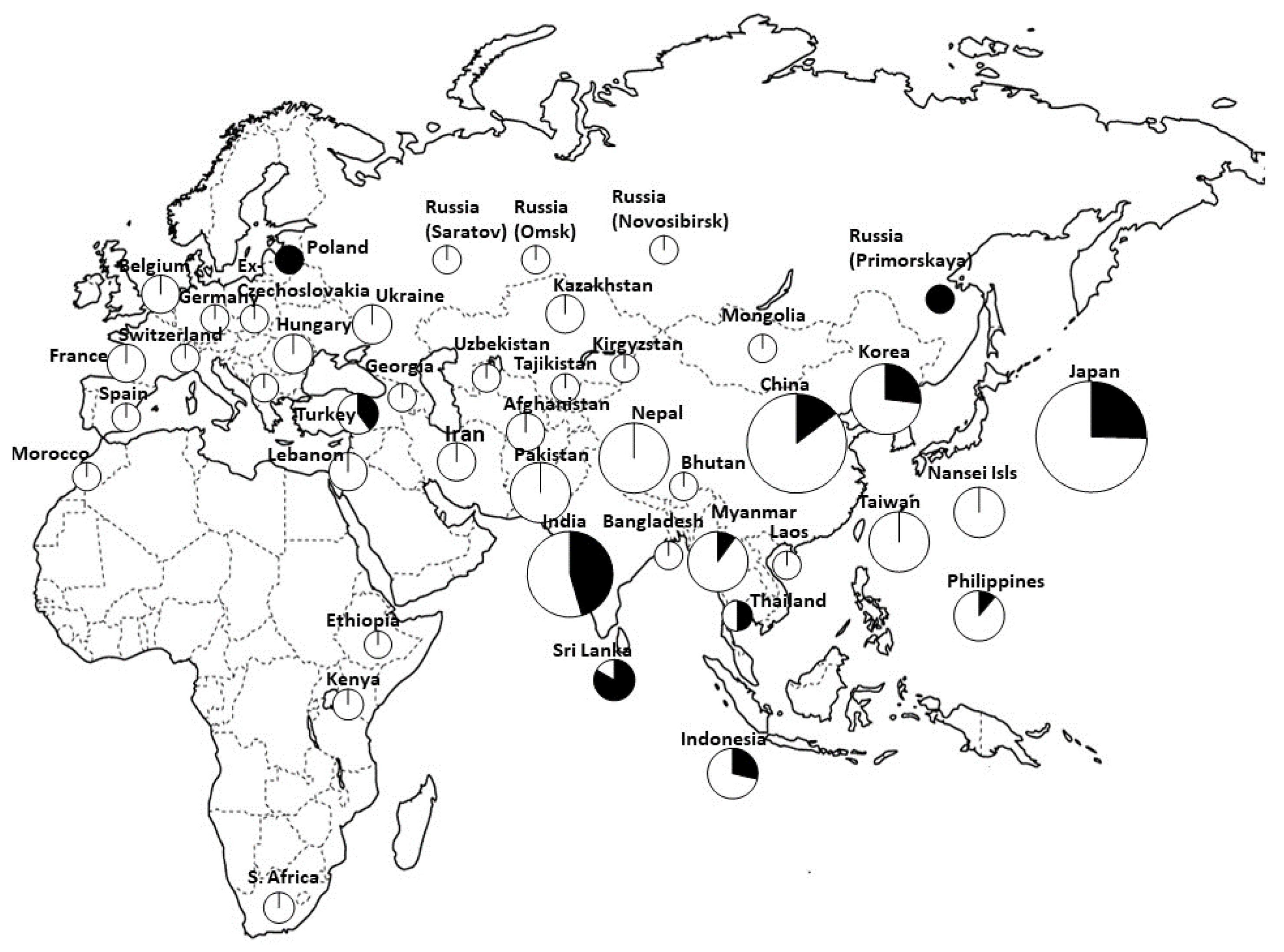
- Kawase, M.; Sakamoto, S. Geographical distribution and genetic analysis of phenol color reaction in foxtail millet, Setaria italica (L.) P. Beauv. Theor. Appl. Genet. 1982, 63, 117–119. [Google Scholar] [CrossRef] [PubMed]
- Inoue, T.; You, T.; Ohta, T.; Hitomi, E.; Ichitani, K.; Kawase, M.; Taketa, S.; Fukunaga, K. Multiple origins of the phenol reaction negative phenotype in foxtail millet, Setaria italica (L.)P. Beauv, were caused by independent loss-of-function mutations of the polyphenol oxidase (Si7PPO) gene during domestication. Mol. Genet. Genomics 2015, 290, 1563–1574. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Tang, T.; Qian, Q.; Wang, Y.; Yan, M.; Zeng, D.; Han, B.; Wu, C.I.; Shi, S.; Li, J. Independent losses of function in a polyphenol oxidase in rice: differentiation in grain discoloration between subspecies and the role of positive selection under domestication. Plant Cell 2008, 20, 2946–2959. [Google Scholar] [CrossRef] [PubMed]
- Taketa, S.; Matsuki, K.; Amano, S.; Saisho, D.; Himi, E.; Shitsukawa, N.; You, T.; Noda, K.; Takeda, K. Duplicate polyphenoloxidase genes on barley chromosome 2H and their functional differentiation in the phenol reaction of.
- spikes and grains. J Exp Bot. 2010, 61, 3983–3993.
- Fukunaga, K.; Nur, M.Z.; Inoue, T.; Taketa, S.; Ichitani, K. Phylogenetic analysis of the Si7PPO gene in foxtail millet, Setaria italica, provides further evidence for multiple origins of negative phenol color reaction phenotype. Genes Genet. Syst. 2020, 95, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Huang, X.; Zhi, H.; Zhao, Y.; Zhao, Q.; Li, W.; Chai, Y.; Yang, L.; Liu, K.; Lu, H.; et al. A haplotype map of genomic variations and genome-wide association studies of agronomic traits in foxtail millet (Setaria italica). Nat. Genet. 2013, 45, 957–961. [Google Scholar] [CrossRef]
- Koornneef, M.; Alonso-Blanco, C.; Vreugdenhil, D. Naturally occurring genetic variation in Arabidopsis thaliana. Annu. Rev. Plant Biol. 2004, 55, 141–172. [Google Scholar] [CrossRef]
- Yano, M.; Kojima, S.; Takahashi, Y.; Lin, H.; Sasaki, T. Genetic control of flowering time in rice, a short-day plant. Plant Physiol. 2001, 127, 1425–1429. [Google Scholar] [CrossRef]
- Izawa, T. Adaptation of flowering-time by natural and artificial selection in Arabidopsis and rice. J. Exp. Bot. 2007, 58, 3091–3097. [Google Scholar] [CrossRef]
- Ichitani, K.; Nagao, K.; Narita, Y.; Fujikawa, K.; Samejima, M.; Taura, S.; Sato, M. Genetic analysis of heading characters in foxtail millet (Setaria italica (L.) P. Beauv.) using the progeny from the cross between the two diverse strains, Gai 53 and Kuromochi. Mem. Fac. Agr. Kagoshima Univ. 2003, 38, 17–25. [Google Scholar]
- Mauro-Herrera, M.; Wang, X.; Barbier, H.; Brutnell, T.P.; Devos, K.M; Doust, A.N. Genetic control and comparative genomic analysis of flowering time in Setaria (Poaceae). Genes, Genomes, and Genetics 2013, 3, 283–295. [Google Scholar] [CrossRef]
- Yoshitsu, Y.; Takakusagi, M.; Abe, A.; Takagi, H. Uemura, A.; Yaegashi, H.; Terauchi, R.; Takahata, Y.; Hatakeyama, K.; Yokoi, S. QTL-seq analysis identifies two genomic regions determining the heading date of foxtail millet, Setaria italica (L.) P.Beauv. Breed. Sci. 2017, 67, 518–527. [Google Scholar] [CrossRef]
- Fukunaga, K.; Izuka, N.; Hachiken, T.; Mizuguchi, S.; Ito, H.; Ichitani, K. A nucleotide substitution at the 5′splice site of intron 1 of rice HEADING DATE 1 (HD1) gene homolog in foxtail millet, broadly found in landraces from Europe and Asia. Crop J. 2015, 3, 481–488. [Google Scholar] [CrossRef]
- Liu, H.; Liu, H.; Zhou, L.; Zhang, Z.; Zhang, X.; Wang, M.; Li, H.; Lin, Z. Parallel domestication of the Heading Date 1 gene in cereals. Mol. Biol. Evol. 2015, 32, 2726–2737. [Google Scholar] [CrossRef]
- Li, C.; Wang, G.; Li, H.; Wang, G.; Ma, J.; Zhao, X.; Huo, L.; Zhang, L.; Jiang, Y.; Zhang, J.; et al. High-depth resequencing of 312 accessions reveals the local adaptation of foxtail millet. Theor. Appl. Genet. 2021, 134, 1303–1317. [Google Scholar] [CrossRef]
- Fukunaga, K.; Abe, A.; Mukainari, Y.; Komori, K.; Tanaka, K.; Fujihara, A.; Yaegashi, H.; Kobayashi, M.; Ito, K.; Ohsako, T.; Kawase, M. Recombinant inbred lines and next-generation sequencing enable rapid identification of candidate genes involved in morphological and agronomic traits in foxtail millet. Sci. Rep. 2020, 12, 218. [Google Scholar] [CrossRef]
- Koo, B.H.; Yoo, S.C.; Park, J.W.; Kwon, C.T.; Lee, B.D.; An, G.; Zhang, Z.; Li, J.; Li, Z.; Paek, N.C. Natural variation in OsPRR37 regulates heading date and contributes to rice cultivation at a wide range of latitudes. Mol. Plant 2013, 6, 1877–1888. [Google Scholar] [CrossRef]
- Murphy, R.L.; Klein, R.R.; Morishige, D.T.; Brady, J.A.; Rooney, W.L.; Miller, F.R.; Dugas, D.V.; Klein, P.E.; Mullet, J.E. Coincident light and clock regulation of pseudoresponse regulator protein 37 (PRR37) controls photoperiodic flowering in sorghum. Proc. Natl. Acad. Sci. USA. 2011, 108, 16469–16474. [Google Scholar] [CrossRef]
- Li, C.; Zhou, A.; Sang, T. Rice domestication by reducing shattering. Science 2006, 311, 1936–1939. [Google Scholar] [CrossRef]
- Onishi, K.; Takagi, K.; Kontani, M.; Tanaka, T.; Sano, Y. Different patterns of genealogical relationships found in the two major QTLs causing reduction of seed shattering during rice domestication. Genome 2007, 50, 757–766. [Google Scholar] [CrossRef]
- Lin, Z.; Li, X.; Shannon, L.M.; Yeh, C.T.; Wang, M.L.; Bai, G.; Peng, Z.; Li, J.; Trick, H.N.; Clemente, T.E.; et al. Parallel domestication of the Shattering1 genes in cereals. Nat. Genet. 2012, 44, 720–724. [Google Scholar] [CrossRef]
- Pourkheirandish, M.; Hensel, G.; Kilian, B.; Senthil, N.; Chen, G.; Sameri, M.; Azhaguvel, P.; Sakuma, S.; Dhanagond, S.; Sharma, R.; et al. Evolution of the grain dispersal system in barley. Cell 2015, 162, 527–539. [Google Scholar] [CrossRef]
- Odonkor, S.; Choi, S.; Chakraborty, D.; Martinez-Bello, L.; Wang, X.; Bahri, B.A.; Tenaillon, M.I.; Panaud, O.; Devos, K.M. QTL mapping combined with comparative analyses identified candidate genes for reduced shattering in Setaria italica. Front. Plant Sci. 2018, 9, 918. [Google Scholar] [CrossRef]
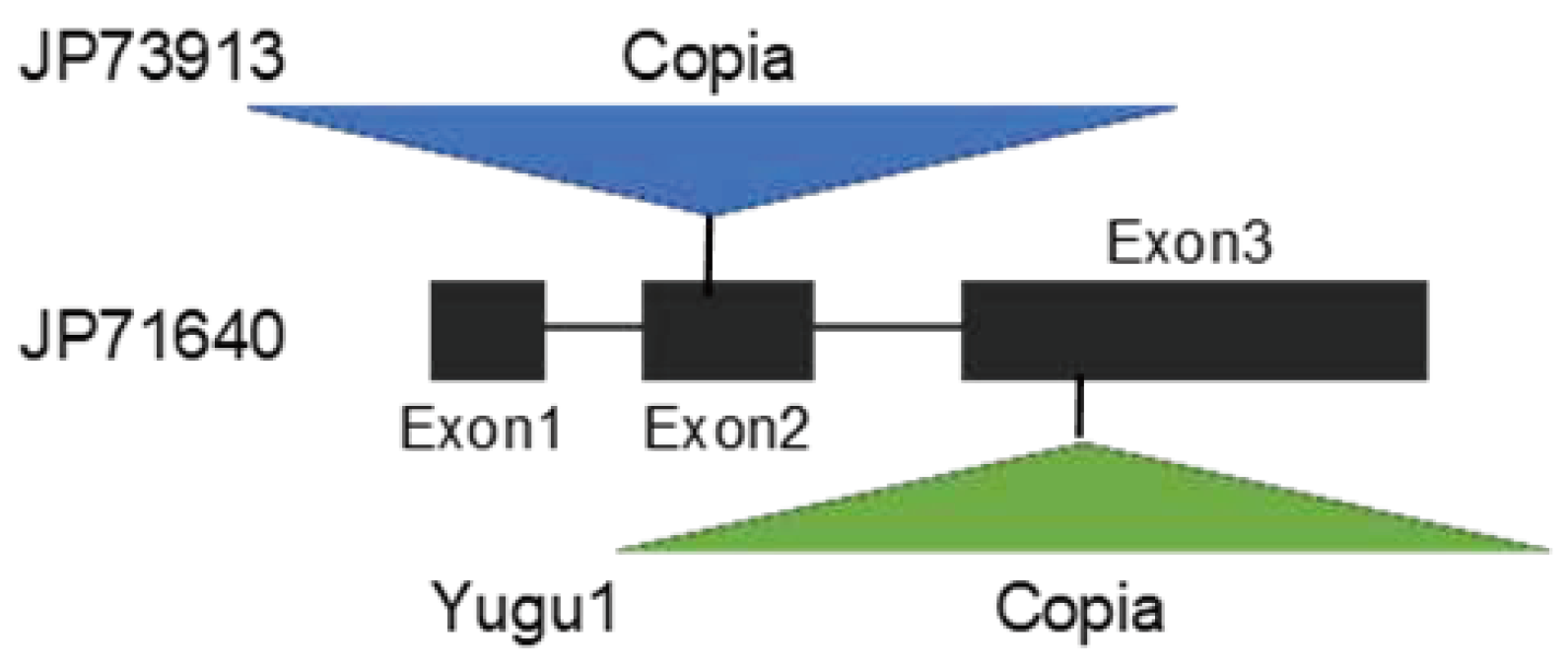
- Liu, H.; Fang, X.; Zhou, L.; Li, Y.; Zhu, C.; Liu, J.; Song, Y.; Jian, X.; Xu, M.; Dong, L.; Lin, Z. Transposon insertion drove the loss of natural seed shattering during foxtail millet domestication. Mol. Biol. Evol. 2022, 39, msac078. [Google Scholar] [CrossRef]
- Fukunaga, K.; Matsuyama, S.; Abe, A.; Kobayashi, M.; Ito, K. Insertion of a transposable element in Less Shattering1 (SvLes1)gene is not always involved in foxtail millet (Setaria italica) domestication. Genet. Resour. Crop Evol. 2021, 68, 2923–2930. [Google Scholar] [CrossRef]
- Diao, X.; Jia, G. Foxtail Millet Germplasm and Inheritance of Morphological Characteristics. In: Genetics and Genomics of Setaria Doust,A., Diao, X. Eds., Springer, Cham, Switzerland, 2017.
- Li, Y.; Fang, X.; Lin, Z. Convergent loss of anthocyanin pigments is controlled by the same MYB gene in cereals. J. Exp. Bot. 2022, 73, 6089–6102. [Google Scholar] [CrossRef]
- Bai, H.; Song, Z.; Zhang, Y.; Li, Z.; Wang, Y.; Liu, X.; Ma, J.; Quan, J.; Wu, X.; Liu, M.; et al. The bHLH transcription factor PPLS1 regulates the color of pulvinus and leaf sheath in foxtail millet (Setaria italica). Theor. Appl. Genet. 2020, 133, 1911–1926. [Google Scholar] [CrossRef]
- He, Q.; Tang, S.; Zhi, H.; Chen, J.; Zhang, J.; Liang, H.; Alam, O.; Li, H.; Zhang, H.; Xing, L.; et al. A graph-based genome and pan-genome variation of the model plant Setaria. Nat Genet. 2023, 55, 1232–1242. [Google Scholar] [CrossRef] [PubMed]
- Sato, K.; Mukainari, Y.; Naito, K.; Fukunaga, K. Construction of a foxtail millet linkage map and mapping spikelet-tipped bristles 1(stb1) by using transposon display markers and simple sequence repeat markers with genome sequence information. Mol. Breed. 2013, 31, 675–684. [Google Scholar] [CrossRef]
- Masumoto, H.; Takagi, H.; Mukainari, Y.; Terauchi, R.; Fukunaga, K. Genetic analysis of NEKODE1 gene involved in panicle branching of foxtail millet, Setaria italica (L.) P. Beauv., and mapping by using QTL-seq. Mol. Breed. 2016, 36, 59. [Google Scholar] [CrossRef]
- Doust, A.D.; Devos, K.M.; Gadberry, M.D.; Gale, M.D.; Kellogg, E.A. The genetic basis for inflorescence variation between foxtail and green millet (Poaceae) Genetics 2005, 169, 1659-1672.
- Mauro-Herrera, M.; Doust, A.N. Development and genetic control of plant architecture and biomass in the Panicoid grass, Setaria. PLoS One. 2016, 11, e0151346. [Google Scholar] [CrossRef] [PubMed]
- Doust, A.D.; Devos, K.M.; Gadberry, M.D.; Gale, M.D.; Kellogg, E.A. Genetic control of branching in foxtail millet. Proc. Nat. Acad. Sci. USA. 2004, 101, 9045–9050. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, J.; Peng, J.; Jiang, M.; Li, Y.; Han, F.; Du, G.; Yang, H.; Lian, S.; Yong, J.; et al. QTL mapping for 11 agronomic traits based on a genome-wide Bin-map in a large F2 population of foxtail millet (Setaria italica (L.) P. Beauv). Mol. Breed. 2019, 39, 18. [Google Scholar] [CrossRef]

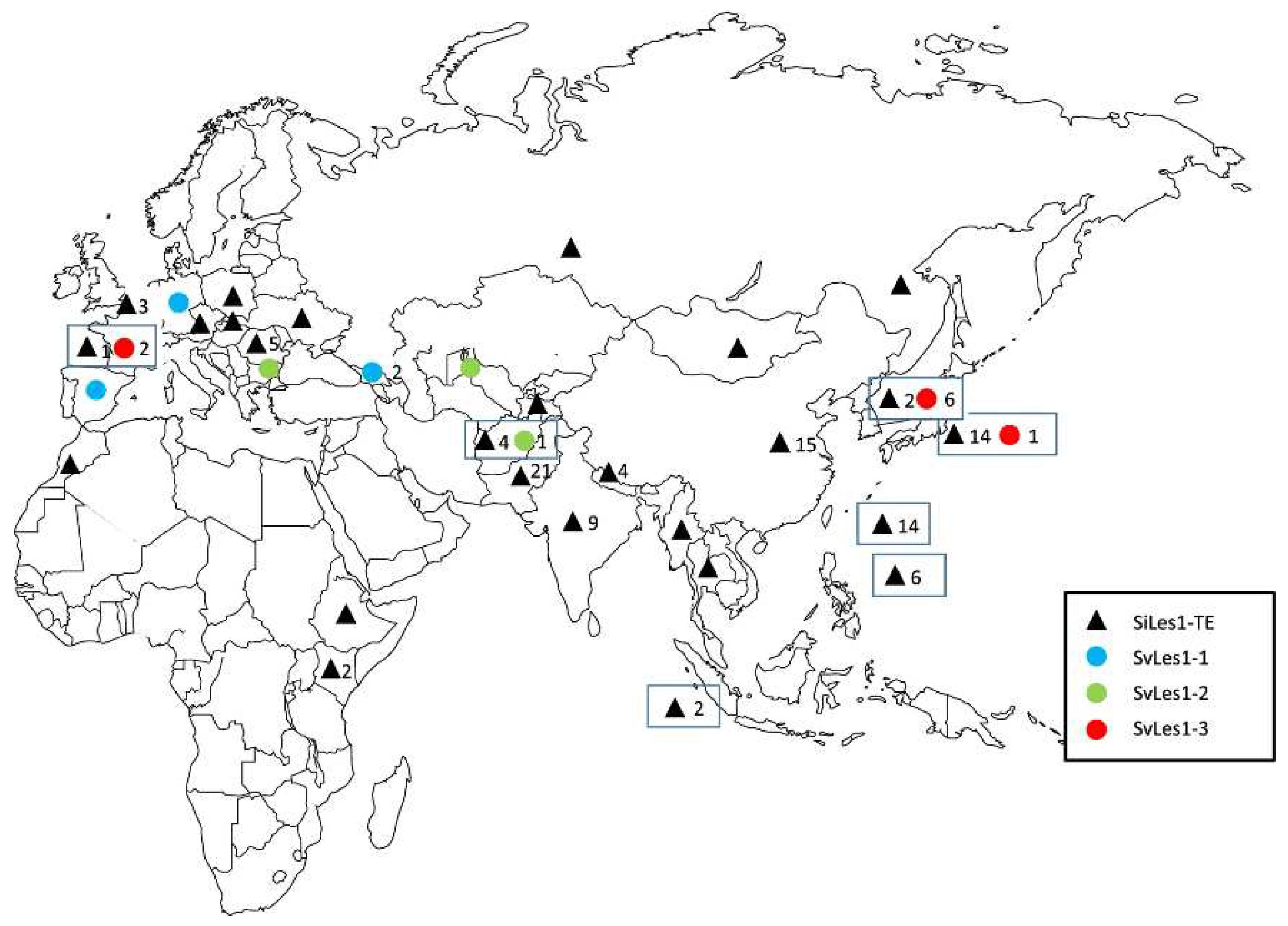
| Genetic markers/ intraspecific hybrid pollen sterility | Geographical groups | Center of diversity | References |
| esterase isozymes | East Asia vs Europe | East Asia | [39] |
| 10 isozymes | China-Korea-Japan, Okinawa (Nansei Islands of Japan)- Taiwan, India-Kenya, Europe | [9] | |
| prolamine | Europe, Tropical Groups | China | [13] |
| hybrid sterility | China-Korea-Japan, Okinawa (Nansei Islands of Japan)- Taiwan,Lan-Hsű-Batan Islands India-Afghanistan, Europe | [25] | |
| rDNA | China-Korea-Japan, Okinawa (Nansei Islands of Japan)-Taiwan-the Philippines, India, Afghanistan-northern Pakistan | China | [14,15,17] |
| nuclear RFLP | East Asia, Nansei Islands-Taiwan-the Philippines, India, Afghanistan-Central Asia-Europe | China | [21] |
| mtDNA | not clear | China | [22] |
| RAPD | central Europe and two Asiatic groups (north and south) | [19] | |
| AFLP | not clear | China | [20] |
| TD | East Asia, Nansei Islands-Taiwan-the Philippines, India, Central Asia, Europe | China | [23] |
| GBS | Four clusters 1. southern Asia (Nepal, Pakistan, Afghanistan, Iran, Turkey, and the Near East) +western and northern Europe, 2. Far East focus (including Japan, North and South Korea, and the region of Russia bordering the Sea of Japan)+ western China+ sporadically eastern Europe, 3. India, Sri Lanka, Bangladesh, and eastern and southern Africa, 4. widespread east-west distribution |
[40] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




