1. Introduction
Placenta accreta spectrum (PAS) is a serious obstetric complication characterized by excessive invasive growth of chorionic villi into adjacent tissue structures. This term encompasses both abnormal adherence (placenta creta, where villi adhere to the myometrium) and abnormal invasion (placenta increta, where villi invade the myometrium; placenta percreta, where villi invade the full thickness of the myometrium) [
1]. According to D.A. Carusi, the prevalence of PAS is reported as 1 in 1000 deliveries [
2], with an increasing incidence observed over time. For instance, in a tertiary south Italian center, the incidence of PAS tripled from 0.12% to 0.31% between 1970 and 2000 [
3]. Other authors report a 100-fold increase in PAS frequency since the 1950s [
4] [
5], which is attributed to the rising rate of cesarean section procedures [
6] [
7]. In turn, the risk of placenta previa significantly increases with the frequency of cesarean sections, representing an additional risk factor for PAS, constituting 50% [
8]. This is associated with the increased tropism of trophoblast cells of the blastocyst to the altered scar tissue area, leading to myofiber disarray, inflammatory processes, and dystrophy of elastic and collagen fibers.
Among the etiological factors of PAS, besides changes in scar tissue, curettage, myomectomy, uterine anomalies, endometriosis, and endometritis are noted, all of which may lead to endometrial fibrosis and poor decidualization [
8]. Several theories have been proposed to explain the origin of PAS One theory involves disorders in the coordinated regulation of extravillous trophoblast differentiation from progenitor cytotrophoblasts, resulting in excessive invasion into the myometrium to remodel the uterine vascular system, causing hypervascularity and vascular dysfunction [
9][
10][
11]. This trophoblast behavior resembles cancer-like progression [
11]. Single-cell transcriptome analysis of PAS and normally detached placenta tissues revealed close communication between excessive numbers of two cytotrophoblast cell types (LAMB4+ and KRT6A+) and maternal stromal cell subtypes (ADIRF+ and DES+), supporting trophoblast cell migration and invasion, as well as interactions with vascular endothelial cells through FLT1-VEGFA and JAG1-NOTCH2 cell‒cell interactions inducing abnormal blood vessels in the myometrium [
12]. Another hypothesis suggests that abnormal vascularization with local hypoxia in the uterine scar area impacts decidualization, causing a defect in the regulatory properties of the decidua. This defect allows trophoblast cells to be more aggressive and penetrative at the sites of the endometrial-myometrial interface and the myometrium itself [
13] [
14]. The main complications of PAS include massive blood loss, disseminated intravascular coagulation, hysterectomy, and preterm birth, leading to increased maternal and fetal morbidity and mortality [
15]. Despite improvements in ultrasound protocols [
1][
16][
17], PAS remains undiagnosed before delivery because of the different ultrasound equipment used by ultrasound examinators, subjective quality of ultrasound sings of PAS, lack of clear evaluation criteria for each of the three grades of PAS. Therefore, the timely and accurate antenatal diagnosis of PAS is essential to formulate the correct patient management algorithm and plan delivery by a multidisciplinary team to reduce the frequency of postpartum complications as also claimed by Pavón-Gomez N. et al [
18].
Circulating biomolecules in maternal blood were examined for their potential use in diagnosing PAS [
19][
20] [
21]. The sensitivity and specificity of maternal serum alpha-fetoprotein in the diagnosis of placenta previa complicated by PAS were only 71% and 46%, respectively [
22]. According to a meta-analysis, pregnant women with PAS have a high serum PAPP-A level in the first trimester [
23][
24], suggesting that this biomarker can be recommended for identifying the risk group for developing PAS. Several studies have shown that the level of β-hCG in maternal blood serum increases in the first and second trimesters of pregnancy during PAS, compared to a normal pregnancy [
25] [
22], however, blood serum hCG levels are also associated with miscarriage, ectopic pregnancy, and fetal abnormalities [
20]. PlGF levels are significantly higher in subgroups with pathological placental invasion compared to the group with normal placental implantation, while sFlt-1 levels and the sFlt-1/PlGF ratio are lower [
26][
27] [
28]. IL-8 promotes migration and invasion of extravillous trophoblast cells during pregnancy, and its elevation in blood serum may serve as a biomarker for PAS [
29]. The level of IL-33 is significantly higher in patients with PAS than in healthy pregnant women [
30]. Despite the identified correlations between PAS and the levels of these circulating biomolecules in maternal blood, it is necessary to prove their specificity for PAS and their ability to distinct from other pregnancy complications.
Due to the epigenetic regulation of trophoblast differentiation, migration, and invasion [
31][
32], miRNAs, acting as master regulators of the human genome at the transcriptional and post-transcriptional levels, were analyzed in various biological samples to associate their levels with PAS [
20] [
33] [
34][
35]. However, miRNA markers for PAS have been identified in the third trimester of pregnancy in studies conducted to date. To individualize the management tactics of pregnant women, preparing for qualified surgical assistance at the time of delivery with the possibility of blood transfusion, it is optimal to conduct screening of women in the first trimester of pregnancy for the content of miRNA markers of PAS in blood serum. Therefore, the aim of this study was to identify extracellular miRNAs circulating in the peripheral blood of women in the first trimester of pregnancy, specific to placental and/or myometrial tissue, and differentiating PAS from other pregnancy complications, such as preeclampsia.
2. Results
To identify circulating exosomal microRNA (miRNA) markers indicative of placental invasion during the first trimester of pregnancy, the study was conducted in four stages:
I) Generation of miRNA expression patterns using deep sequencing in both placental and myometrial tissues within the region of pathological trophoblast invasion and beyond at the time of delivery in women with Placenta Accreta Spectrum (PAS) conditions, including creta, increta, and percreta.
II) Identification of placenta- and myometrium-specific miRNAs in the blood plasma of pregnant women using deep sequencing and quantitative real-time polymerase chain reaction (qRT-PCR) on the day of delivery, enabling the diagnosis of PAS.
III) Qualitative and quantitative analysis of exosomal miRNA composition in the serum of women at 11-14 weeks of pregnancy, facilitating the diagnosis of PAS through deep sequencing with subsequent validation of the obtained data using qRT-PCR.
IV) Quantitative analysis of exosomal miRNA markers for PAS in the native serum of women at 11-14 weeks of pregnancy with either a normal course or complications such as the onset of pre-eclampsia symptoms after 20 weeks of gestation or complications involving placental invasion. The goal is to construct a logistic regression model for the accurate diagnosis of PAS.
2.1. Analysis of miRNA Expression Patterns Using Deep Sequencing in Placental and Myometrial Tissues from Patients with Placental Invasion at the Time of Delivery
In the initial stage of the study, deep sequencing of small non-coding RNAs was employed to obtain and compare the expression profiles of microRNAs (miRNAs) in various regions of the placenta and myometrium. Tissues were collected during cesarean section procedures from women in the first cohort (
Table 1) with diagnoses of placenta creta (7 patients), placenta increta (6 patients), and placenta percreta (4 patients).
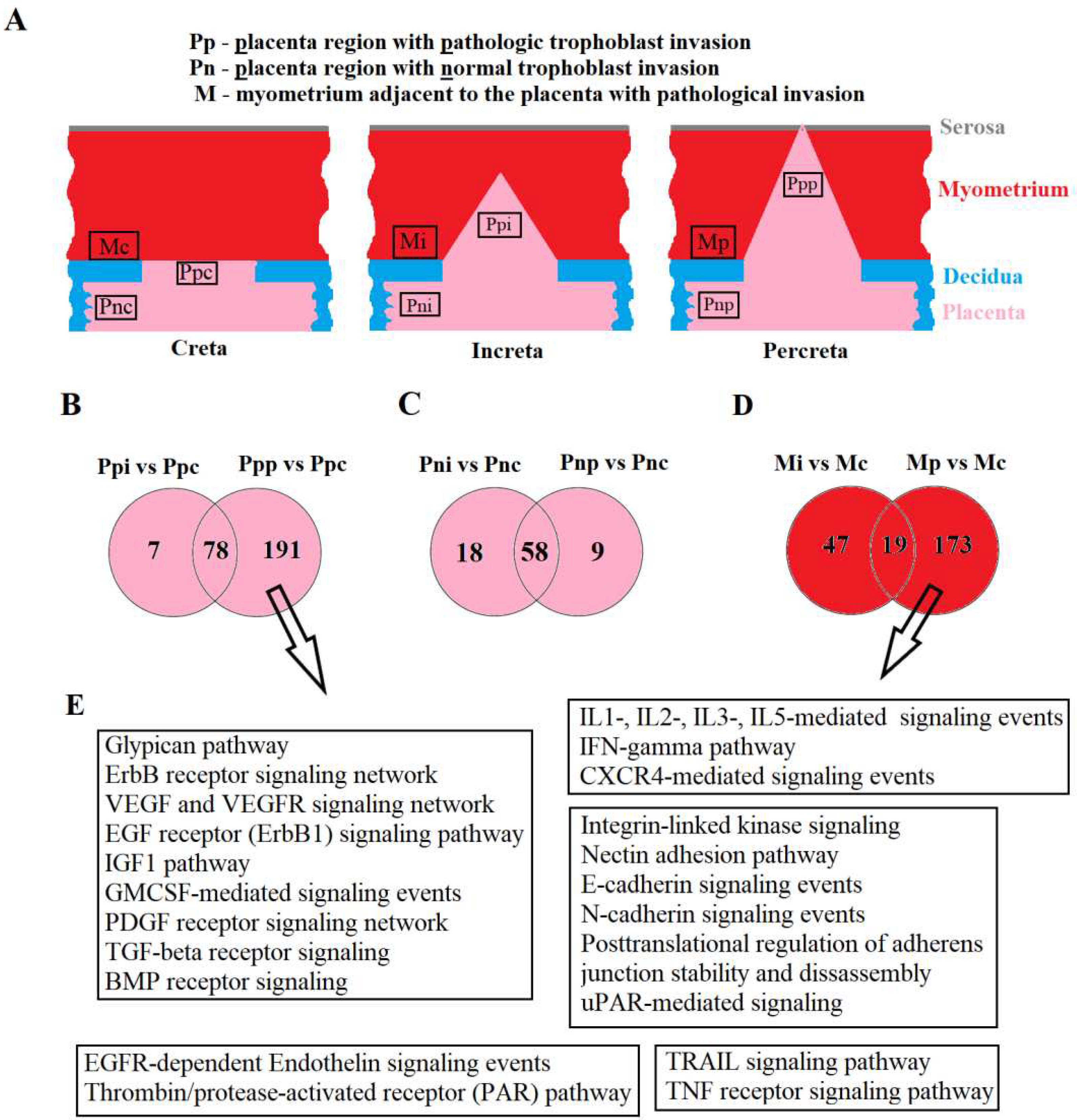
Placental samples were collected from areas of pathological trophoblast invasion and from regions outside this area. Myometrial samples were obtained from areas adjacent to the placental invasion site. The schematic localization of placental and myometrial sample collection and the designation of corresponding groups are illustrated in
Figure 1A.
During the analysis of miRNA expression profiles in placental tissue, two types of comparisons were performed: 1) comparison of placental areas with pathological trophoblast invasion in women with creta (7 samples), increta (6 samples), and percreta (4 samples): Ppp vs Ppc, Ppi vs Ppc; 2) comparison of placental areas outside pathological trophoblast invasion sites in women with creta (3 samples), increta (4 samples), and percreta (3 samples): Pnp vs Pnc, Pni vs Pnc.
In the first type of comparison, expression profiles of miRNAs were obtained, significantly differentiating samples of Ppi from Ppc in terms of expression levels for 85 miRNAs (
Table S1, Sheet1), and Ppp from Ppc for 269 miRNAs (
Table S2, Sheet1). Among them, altered expression levels in both types of invasions (increta and percreta) were identified for 78 miRNAs (
Figure 1B), constituting 92% of all differentially expressed miRNAs in the case of increta and 29% in the case of percreta. These data indicate similarities in molecular and biological changes in the placenta within the invasion area and the myometrial tissue in cases of increta and percreta, with an exacerbation of these changes in the case of percreta, involving an additional 191 miRNAs.
In the second type of comparison, expression profiles of miRNAs significantly differentiating samples of Pni from Pnc in terms of expression levels for 76 miRNAs (
Table S3, Sheet1), and Pnp from Pnc for 67 miRNAs (
Table S4, Sheet1) were obtained. Among them, altered expression levels in both types of invasions (increta and percreta) were identified for 58 miRNAs (
Figure 1C), constituting 76% of all differentially expressed miRNAs in the case of increta and 87% in the case of percreta. These data suggest commonalities in molecular and biological changes throughout the entire placenta in women with increta and percreta, possibly formed during the embryo implantation stage due to the interaction of trophoblasts with a pathologically altered deciduous layer of the endometrium. Cells of the cytotrophoblast in certain areas of such a placenta acquire a phenotype with excessive invasive activity under predisposing conditions, such as structural changes in the myometrium. Therefore, deep sequencing analysis was performed on miRNA expression levels in myometrial tissue adjacent to the site of pathological trophoblast invasion for percreta (3 samples) and increta (4 samples) compared to creta (3 samples). Expression profiles of miRNAs significantly differentiating samples Mi from Mc (
Table S5, Sheet 1) and Mp from Mc (
Table S6, Sheet 1) were obtained. Among them, altered expression levels in myometrial tissue for both types of PAS (increta and percreta) were identified for only 19 miRNAs (
Figure 1D), constituting 29% of all differentially expressed miRNAs in the case of increta and 10% in the case of percreta. It is noteworthy that more pronounced changes in the qualitative and quantitative composition of miRNAs in myometrial tissue adjacent to placenta percreta were observed compared to that adjacent to placenta increta, possibly contributing to a greater depth of trophoblast cell invasion in the case of placenta percreta.
For each miRNA profile in a specific type of comparison (see above), the use of the MirWalk program (
http://mirwalk.umm.uni-heidelberg.de/search_mirnas/) allowed the identification of experimentally proven target genes according to the MiRTarBase algorithm (Sheet2 in the
Tables S1, S2, S5, S6). Additionally, using the FunRich program (
http://www.funrich.org/), signaling pathways regulated by differentially expressed miRNAs in placental and myometrial tissues in different types of PAS (Sheet3 in the
Tables S1, S2, S5, S6) were revealed. Since the most significant molecular and biological changes were identified in the placenta within the pathological trophoblast invasion site for percreta and in adjacent myometrial tissue compared to other types of PAS, for clarity, we presented in
Figure 1E common signaling pathways regulated by differentially expressed miRNAs in the comparison of Ppp vs Ppc and Mp vs Mc. Imbalances in the activity of these signaling pathways may account for local inflammatory processes, structural rearrangements in the extracellular matrix, neovascularization, apoptosis, and changes in the proliferative and invasive properties of cells, such as cytotrophoblast cells.
2.2. Identification of Placenta- and Myometrium-Specific miRNAs Circulating in the Blood Plasma of Pregnant Women on the Day of Delivery Using Deep Sequencing and Real-time Quantitative PCR
With the aim of developing a non-invasive diagnostic test for PAS in the third trimester of pregnancy, expression profiles of miRNAs were obtained in the peripheral blood plasma of 24 women from the first cohort (
Table 1) with various types of PAS using deep sequencing. Lists of differentially expressed (DE) miRNAs were generated for creta (
Table S7, Sheet 1), increta (
Tables S7, Sheet 2), and percreta (
Tables S7, Sheet 3) relative to the group with a normal pregnancy. These lists of DE miRNAs were compared by constructing a Venn-Euler diagram (
Figure 2A), indicating that most miRNAs differentially expressed in the blood plasma of women with creta and increta had significant altered expression levels in the blood plasma of women with percreta. As expression profiles of miRNAs in placental, myometrial and blood plasma samples were analyzed in the same cohort of patients using deep sequencing, the tissue specificity of circulating blood miRNA markers for PAS was evaluated.
The common DE list of 165 miRNAs in the blood plasma (BPi/p,
Figure 2A) for increta and percreta was compared with the lists of DE miRNAs in Ppi (
Table S1, Sheet 1), Ppp (
Table S2, Sheet 1), Mi (
Table S5, Sheet 1), and Mp (
Table S6, Sheet 1). Intersections were found between BPi/p and Ppi for 38 miRNAs, BPi/p and Ppp for 128 miRNAs, BPi/p and Mi for 33 miRNAs, BPi/p and Mp for 90 miRNAs (
Figure 2B). For further validation of the obtained data using real-time quantitative PCR, 40 miRNAs were selected that were differentially expressed in the blood plasma of women with invasive types of PAS (increta and percreta), as well as in placenta and/or myometrium in both invasive PAS types. The list of the selected 40 miRNAs is presented in
Figure 2C.
To validate the sequencing data, the peripheral blood plasma samples from the second cohort of patients (n = 46,
Table 2) were used.
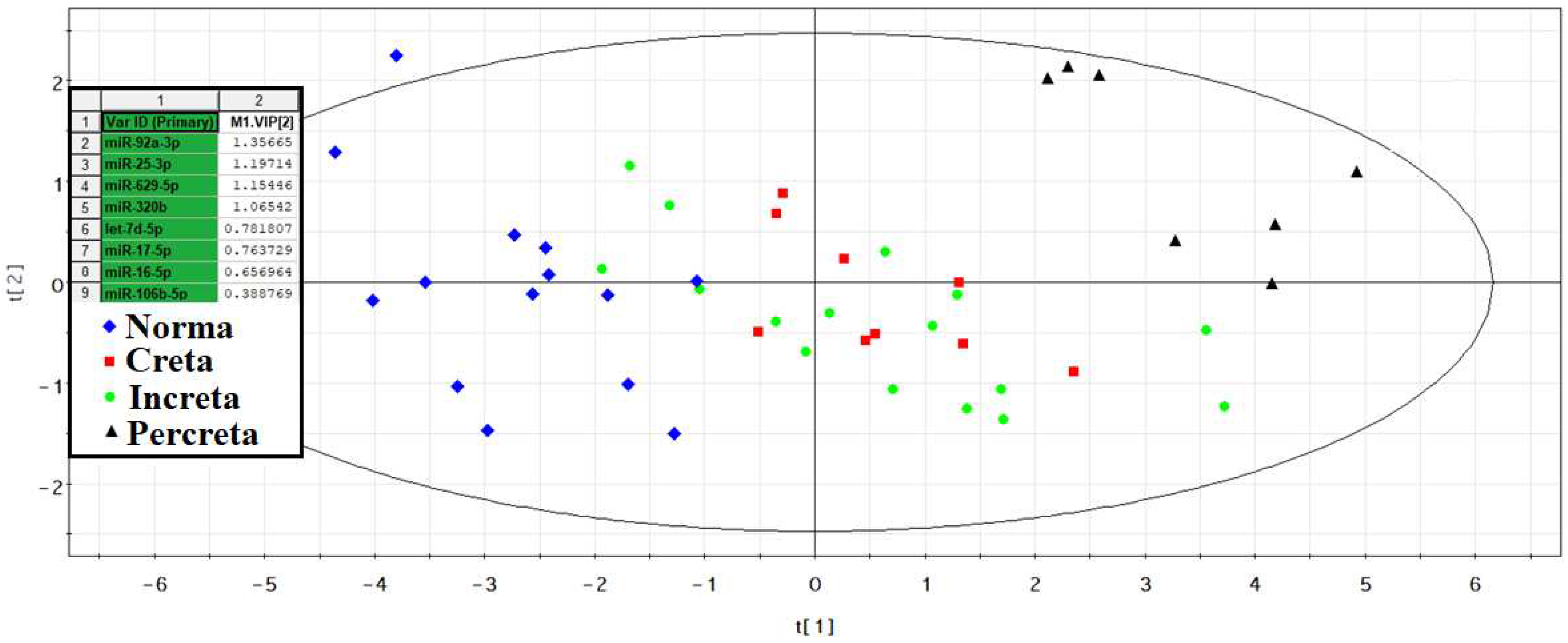
The -ΔCt values were calculated based on the difference between the Ct values of the analyzed 40 miRNAs and the Ct value of the exogenous RNA UniSp6 (see Materials and Methods) in each sample. Using the partial least squares regression (PLS-A) method, the greatest contribution to the clustering of blood plasma samples from women with different types of PAS was made by hsa-miR-92a-3p, hsa-miR-25-3p, hsa-miR-629-5p, hsa-miR-320b, hsa-let-7d-5p, hsa-miR-17-5p, hsa-miR-16-5p, hsa-miR-106b-5p (
Figure 3).
From
Figure 3, it is evident that all samples of peripheral blood plasma from women with PAS differ from control samples in the expression profile of 8 miRNAs, forming a distinct cluster. For this reason, it was decided to combine the Creta, Increta, and Percreta groups to develop logistic regression models for creating a non-invasive diagnostic test for PAS based on the quantitative assessment of miRNAs in blood plasma in the third trimester of pregnancy. Using RStudio, optimal combinations of miRNAs associated with the presence of PAS in pregnant women were found step by step, considering their contribution to building logistic regression models. In these models, the dependent variable (response variable) was the presence or absence of PAS in pregnant women (0 – absence of PAS; 1 – presence of PAS). The selected models, presented in
Figure 4, included statistically significant independent variables. The parameters of the models in
Figure 4 are listed in
Table 3. Formulas 1, 2 and 3, describing the models 1, 2 and 5 in
Figure 4, respectively, are provided below.
(1) Model 1 formula:
where х1 – «-∆Сt» value for hsa-miR-320b, х2 – «-∆Сt» value for hsa-miR-92a-3p;
(2) Model 2 formula:
where х1 – «-∆Сt» value for hsa-miR-320b, х2 – «-∆Сt» value for hsa-let-7d-5p.
(3) Model 5 formula:
where х1 – «-∆Сt» value for hsa-miR-629-5р, х2 – «-∆Сt» value for hsa-miR-17-5p.
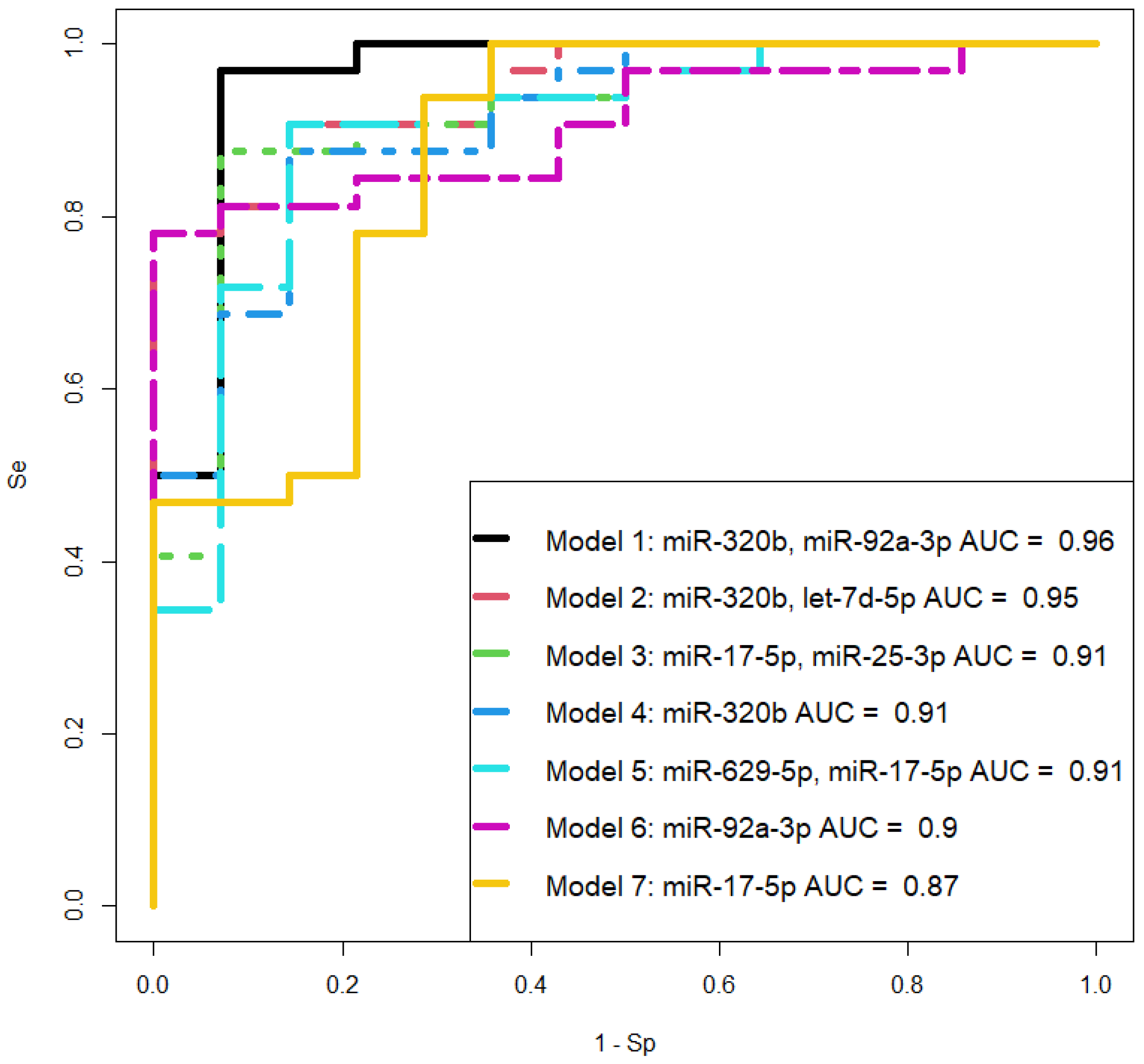
Model 1 demonstrates the highest sensitivity (96.9%) compared to Models 2 (Se = 78.1%) and 5 (Se = 78.1%), indicating a superior diagnostic value in identifying PAS in the third trimester of pregnancy. On the other hand, Models 2 and 5 exhibit 100% specificity, unlike Model 1 (Sp = 93%), ensuring 100% accuracy in detecting pregnancies without PAS. Simultaneous utilization of all three models on an extended training and testing dataset is essential for understanding their diagnostic value and selecting one for further consideration.
2.3. Analysis of Qualitative and Quantitative Composition of Exosome microRNA (miRNA) in the Blood Serum of Women at 11-14 Weeks of Pregnancy
One of the means of intercellular communication is the directed delivery of molecules, including miRNA, contained within exosomes. We hypothesized that morphofunctionally altered tissues of the placenta and myometrium in the area of pathological trophoblast invasion secrete exosomes with a modified miRNA profile into the blood. This profile could potentially diagnose the presence of PAS in early pregnancy stages, such as the 11-14 weeks of gestation.
Exosomes were isolated from blood serum samples of 48 women undergoing first-trimester screening (
Table 4) and analyzed for miRNA expression profiles using deep sequencing.
Four main groups were formed for the study: normal pregnancy (N, 10 women); high risk of developing pre-eclampsia according to the Astraia program without clinical manifestations after 20 weeks of pregnancy (Nhr, 7 women); development of clinical symptoms of early- or late-onset pre-eclampsia after 20 weeks of pregnancy (PE, 21 women); placenta creta, increta, or percreta in pregnant women (PAS, 10 women). The comparison of read numbers in the "PAS" group with those in the combined "N + PE" group is presented in
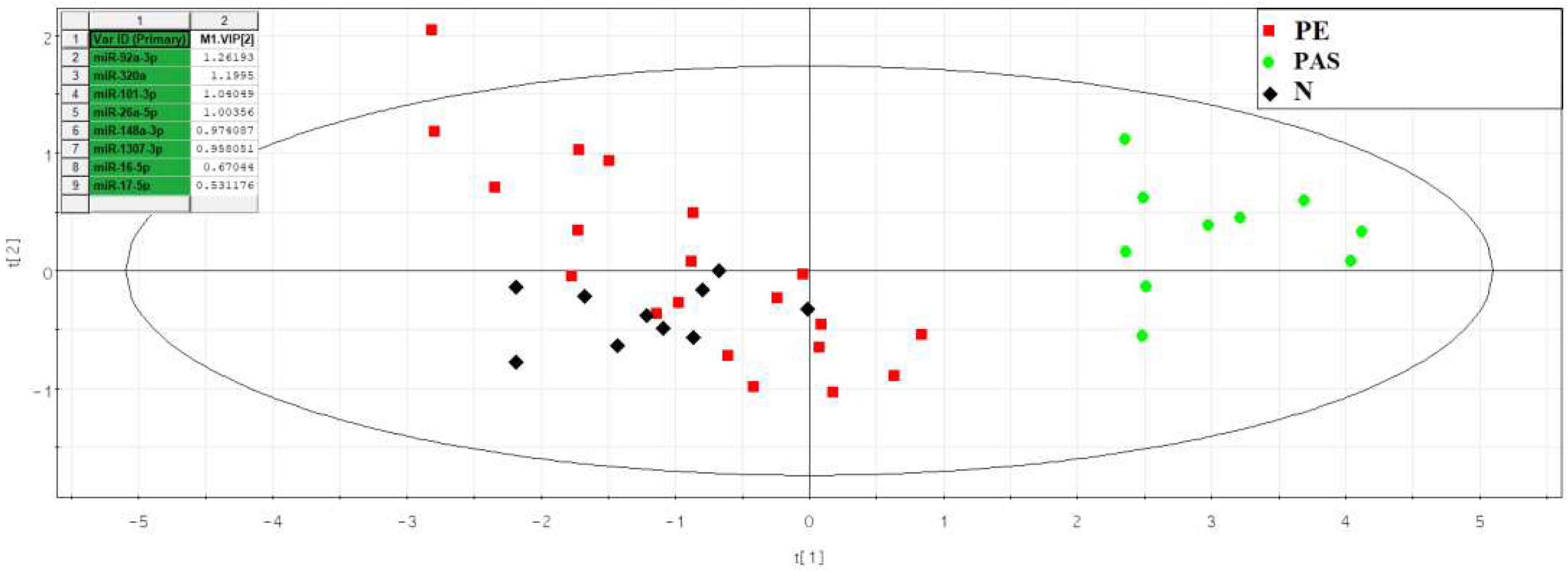
Table S8. The partial least squares regression (PLS) method was used to assess the contribution of each identified miRNA to the separation of samples from the analyzed groups (
Figure 5).
The distribution of samples in
Figure 5 shows that blood serum samples from women with PAS are distant from all other samples and form a separate cluster. The molecules hsa-miR-92a-3p, hsa-miR-320a, hsa-miR-101-3p, hsa-miR-26a-5p, hsa-miR-148a-3p, hsa-miR-1307-3p, hsa-miR-16-5p, hsa-miR-17-5p contributed most to this distribution. The levels of these molecules were evaluated by quantitative PCR in samples from the N and PAS groups (
Figure 6). The “-ΔCt” values were calculated based on the difference between the Ct value of the analyzed miRNA and the Ct value of the endogenous hsa-let-7a-5p, given its stable expression in all analyzed samples by deep sequencing (coefficient of variation was equal to 0.165, see
Table S8). The statistical significance of differences between the compared groups is presented in
Table 5.
From
Figure 6 and
Table 5, it can be inferred that the PAS group statistically significantly differed from the N group only in the elevated levels of hsa-miR-320a, hsa-miR-92a-3p, and hsa-miR-1307-3p among all the analyzed miRNAs. As miRNAs are secreted by cells not only as part of exosomes but also as part of lipoproteins [
36], we analyzed the levels of hsa-miR-92a-3p, hsa-miR-320a, hsa-miR-101-3p, hsa-miR-26a-5p, hsa-miR-148a-3p, hsa-miR-1307-3p, hsa-miR-16-5p, hsa-miR-17-5p in the native blood serum sample of the third patient cohort (
Table 4). Two types of normalization were used to calculate the “-ΔCt” values: normalization to the exogenous RNA UniSp6 (
Figure 7A,
Table 6) and endogenous hsa-let-7a-5p (
Figure 7B,
Table 6).
In comparison with the results of quantitative assessment of miRNA in the exosomal fraction of blood serum from women at 11-14 weeks of pregnancy, we observed a statistically significant increase in the levels of miR-101-3p, miR-26a-5p, miR-16-5p, and miR-17-5p in the native blood serum of women in the PAS group relative to the N group (
Table 6). We conclude that such increase in the levels of these miRNAs occurs in the non-exosomal fraction of blood serum. Notably, only miR-101-3p, miR-26a-5p, and miR-17-5p showed statistical significance in distinguishing the PAS group from all other groups (N, Nhr, PE) using two types of normalization: on UniSp6 or hsa-let-7a-5p. Hence, these miRNAs can be considered specific markers for PAS. The other three miRNAs—miR-92a-3p, miR-320a, and miR-16-5p—did not show statistically significant differences between the PAS group and the PE and/or Nhr groups (
Table 6), indicating that they cannot be considered specific markers for PAS.
The quantitative analysis of hsa-miR-92a-3p, hsa-miR-320a, hsa-miR-101-3p, hsa-miR-26a-5p, hsa-miR-148a-3p, hsa-miR-1307-3p, hsa-miR-16-5p, hsa-miR-17-5p in the native blood serum of women at 11-14 weeks of pregnancy, using endogenous hsa-let-7a-5p as a normalizing RNA, was employed to develop logistic regression models for creating a non-invasive diagnostic system for PAS. In RStudio, optimal combinations of miRNAs associated with the presence of PAS in pregnant women were identified by stepwise inclusion and exclusion of each molecule in logistic regression models, where the dependent variable (response variable) was the presence or absence of PAS in pregnant women (0 – combined groups N, Nhr, PE; 1 – PAS group). Models presented in
Figure 8 were selected, where all independent variables were statistically significant. The parameters of the models in
Figure 8 are indicated in
Table 7. Formulas 4 and 5, describing the models 2 and 3 in
Figure 8, respectively, are provided below.
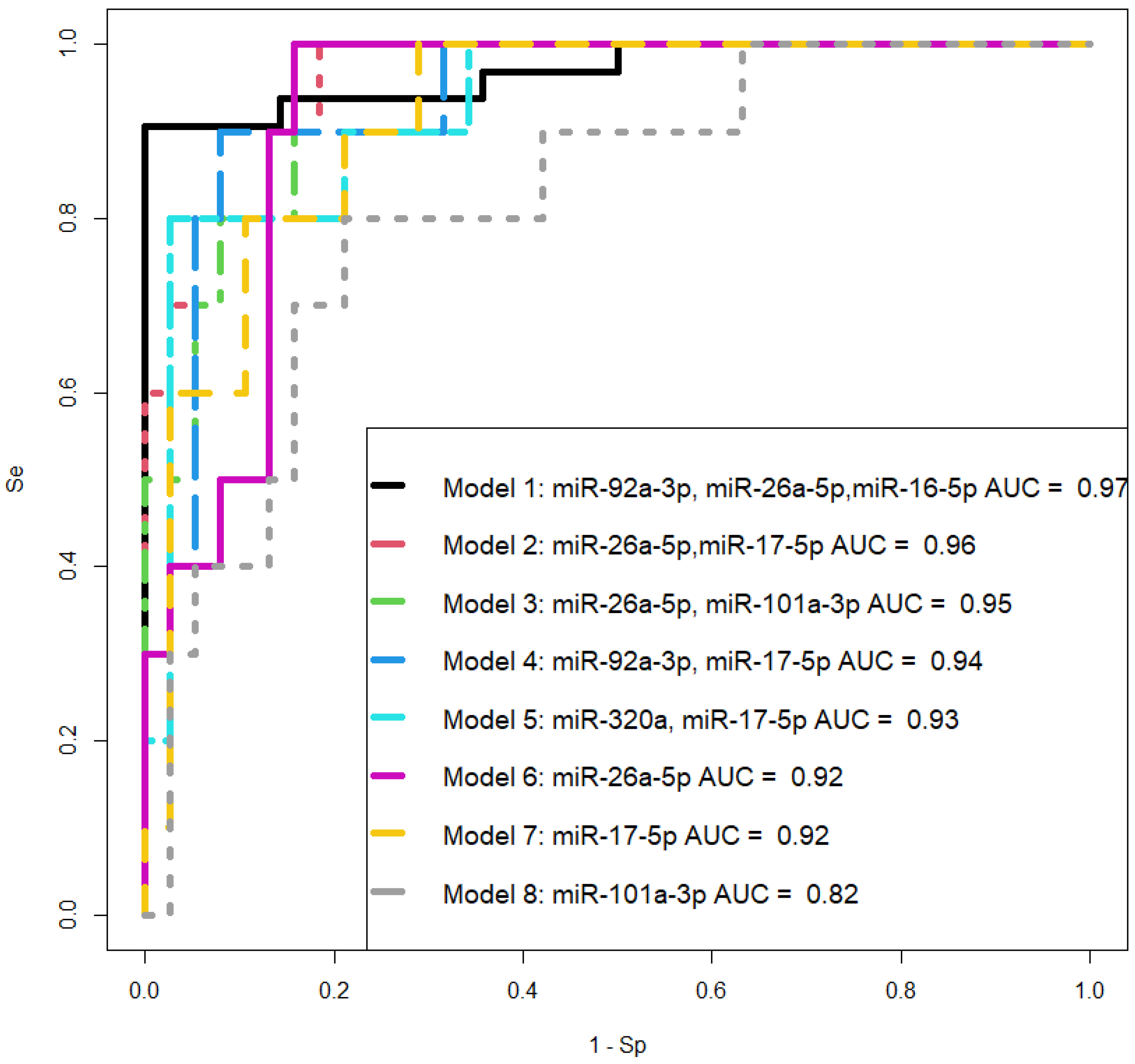
From
Table 7, it can be inferred that logistic regression models 1, 2, and 3 have 100% sensitivity in detecting PAS in women during the first trimester screening based on the quantitative analysis of the corresponding miRNAs in native blood serum. Since model 1 includes miR-92a-3p, which expression level was statistically significantly changed in both PE and PAS compared to normal pregnancy (
Table 6), it cannot be considered as a specific marker for PAS, and therefore model 1 was excluded from consideration.
(4) Formula for Model 2:
where х1 – «-∆Сt» value for hsa-miR-26a-5p, х2 – «-∆Сt» value for hsa-miR-17-5p;
(5) Formula for Model 3:
where х1 – «-∆Сt» value for hsa-miR-26a-5p, х2 – «-∆Сt» value for hsa-miR-101-3p.
Considering all 48 blood serum samples (
Table 4), a reliable correlation of miRNA level with biochemical and instrumental analysis data at 11-14 GW were found using the non-parametric Spearman rank correlation method: hsa-miR-101-3p with NT (r = 0.34, p = 0.0188), hsa-miR-17-5p with UA(PI)MoM (r = -0.3, p = 0.038), hsa-miR-26a-5p with PLGF (r = 0.49, p = 0.025), hsa-miR-17-5p with PLGF (r = 0.36, p = 0.0327). At the same time, statistically significant correlations were found for UA(PI)MoM with PAPP-A (r = -0.33, p = 0.0242) and with PAPP-A(MoM) (r = -0.29, p = 0.043), for PLGF with PAPP-A (r = 0.37, p = 0.0264).
3. Discussion
In this study, the miRNA signature in placental tissues was analyzed both within the invasive region and outside this area, in the adjacent myometrial tissues, and in the blood plasma from women using deep sequencing. The obtained data were validated through quantitative real-time PCR to construct logistic regression models as a non-invasive diagnostic approach for differentiating various types of placenta accreta spectrum (PAS) in the third trimester of pregnancy. It was observed that the quantitative assessment of hsa-miR-320b, hsa-let-7d-5p, hsa-miR-629-5p, and hsa-miR-17-5p in the blood plasma of women allows for the statistically significant identification of PAS cases with high specificity (93 – 100%) and sensitivity (78 – 97%). These molecules were found to be differentially expressed in both placental and myometrial tissues of women with different types of PAS. The constructed logistic regression models can be considered as an additional diagnostic method alongside commonly used instrumental diagnostic approaches such as ultrasound (US) and magnetic resonance imaging (MRI).
To formulate an individual management strategy for patients with PAS, involving the referral to a specialized hospital with a multidisciplinary team of surgeons, intensivists, neonatologists, and preparedness for blood transfusion in case of hemorrhage, it is imperative to diagnose this pregnancy complication in the first trimester. Due to the absence of precise biochemical and instrumental tests for detecting PAS during this gestational period, we conducted a retrospective study using deep sequencing and quantitative real-time PCR of cell-free miRNAs in the blood serum of women who underwent first-trimester screening and continued examination until the parturition with a clear diagnosis (physiological pregnancy or preeclampsia or PAS) at the Kulakov National Medical Research Center of Obstetrics, Gynecology, and Perinatology. The search for miRNA markers of placental invasion specifically in the exosomal fraction of blood serum was motivated by our previous findings [
37] indicating a decrease in the concentration of identified miRNA markers for preeclampsia upon repeated cycles of freezing/thawing of the analyzed blood serum sample. The question of the miRNA stability in the body's biological fluids has been thoroughly investigated by other researchers such as Coenen-Stass AML et al. [
38]. Exosomes, being membrane-containing structures, protect the encapsulated miRNAs from degradation by extracellular RNases and facilitate the targeted delivery of miRNAs to specific cells and tissues [
39]. We discovered that the most significant contributors to the differentiation of exosome fractions in the blood serum of women at 11-14 weeks of pregnancy with various types of PAS from combined groups of women with physiological pregnancy and preeclampsia, using principal component analysis, were miR-92a-3p, miR-320a, miR-101-3p, miR-26a-5p, miR-148a-3p, miR-1307-3p, miR-16-5p, miR-17-5p based on deep sequencing data. Among these, statistically significant differences between the PAS group and all other comparable groups were identified only for miR-320a, miR-92a-3p, and miR-1307-3p based on quantitative real-time PCR data.
However, the isolation of exosomes for identifying cases of placenta accreta spectrum (PAS) using marker mRNA is an additional time-consuming and costly method, which may not be feasible given the high patient volume during screening studies. Therefore, we decided to analyze exosome mRNA markers for PAS in the native blood serum of pregnant women at 11-14 weeks of gestation using quantitative real-time PCR. In comparison with the analysis of the exosome fraction of blood serum, we observed a significant increase in the levels of miR-101-3p, miR-26a-5p, miR-16-5p, and miR-17-5p, in addition to changes in miR-92a-3p and miR-320a, in the native blood serum of women from the PAS group compared to the group of women with physiological pregnancies. This increase in the levels of these mRNA markers occurs in the non-exosome fraction, possibly as part of very-low-density lipoproteins (VLDL), low-density lipoproteins (LDL), or high-density lipoproteins (HDL). For example, other researchers demonstrated the presence of some of the analyzed here miRNAs in blood lipoproteins, namely miR-16-5p in VLDL and HDL, miR-17-5p and miR-26a-5p in HDL [
36], dependent on miRNA sequence motifs. Mechanistically, it was found that HDL-miRNAs have roles in metabolic homeostasis and angiogenesis, whereas targets for LDL-miRNAs were enriched in pathways related to inflammation, immune system function, and different cardiomyopathies.
When calculating "-ΔCt" values for each analyzed miRNA, any of the RNA species was used as a reference: either UniSp6, introduced into the sample during the reverse transcription stage according to the Qiagen's recommendation, or the endogenous let-7a-5p, exhibiting a stable high expression level in the exosomal fraction of blood serum and native blood serum of women at 11-14 weeks of pregnancy according to our deep sequencing data. The use of UniSp6 allows for accounting for variations in reverse transcription and PCR efficiency but does not account for possible mRNA degradation due to extracellular RNases during repeated sample freezing/thawing cycles, unlike the use of endogenous let-7a-5p. The presence of let-7a-5p in exosomes, as identified in the current study, and in VLDL, as reported by Guido Rossi-Herring [
36], when used as an endogenous reference RNA, would consider detrimental processes affecting the concentration of analyzed miRNA in exosomes and the non-exosomal fraction of blood serum ex vivo.
It is important to note that miR-101-3p, miR-26a-5p, and miR-17-5p were found to be the most specific markers for PAS, as statistically significant differences in their expression levels were identified in the PAS group compared to all other comparison groups (physiological pregnancy, high risk of developing preeclampsia according to Astraia during the first pregnancy screening without clinical manifestations of preeclampsia after 20 weeks of pregnancy, development of early and late preeclampsia) using two types of normalization: on UniSp6 or let-7a-5p. Meanwhile, other miRNA associated with PAS (miR-92a-3p, miR-320a, miR-16-5p) did not significantly differentiate the PAS group from the preeclampsia and/or high-risk group for developing preeclampsia without clinical manifestations after 20 weeks of pregnancy. Therefore, the latter cannot be considered specific markers for PAS.
The potential role of miR-92a-3p, miR-320a, and miR-16-5p in the pathogenesis of preeclampsia (PE) has been studied in several works, revealing that i) intravascular inflammation occurs in PE as a sequence of Th1 polarization [
40][
41] through the targeting of GATA3 by up-regulated miR-92a-3p, contained in vesicles of activated NK cells [
42]; ii) miR-320a overexpression observed in PE inhibits trophoblast cell invasion and causes anomalous placentation by targeting estrogen-related receptor-gamma [
43], IL-4 [
44], and IGF-1R [
45]; iii) in a PE rat model, the up-regulation of miR-16-5p directly downregulates IGF-2 and provides inhibition of trophoblast cell viability and migration [
46]. Thus, the involvement of the same miRNA molecules in different diseases necessitates the search for a unique combination of marker miRNAs that can differentiate one pregnancy complication from another. Therefore, when selecting logistic regression models for diagnosing PAS at 11-14 weeks of gestation, we relied on the following criteria: the combination of molecules should include any or all the miRNAs miR-101-3p, miR-26a-5p, and miR-17-5p; all model parameters must be significant, and the model should have high specificity and sensitivity. These criteria are met by two models we developed: the combination of miR-26a-5p and miR-17-5p, and the combination of miR-26a-5p and miR-101-3p, both of which exhibit 100% sensitivity in detecting PAS in women during the first pregnancy screening through their quantitative analysis in native blood serum using real-time PCR.
It is important to note that the circulating miRNA markers for PAS identified in this study during the first trimester of pregnancy are differentially expressed in the placenta within the area of invasion and in the adjacent myometrium, specifically in the case of invasive forms (increta and percreta) relative to the adherent form (creta) in the third trimester of pregnancy at the time of delivery, as per deep sequencing data. Comparing areas of the placenta outside the invasive region led us to hypothesize that for placental invasion into the myometrium, molecular and biological changes in the myometrial tissue are necessary. This assumption is based on the observation that the intersection of differentially expressed miRNA lists in areas of the placenta outside the invasion site, in the case of increta and percreta relative to creta, is found for 70-80% of all miRNAs. And more pronounced quantitative and qualitative changes in the miRNA signature are observed in the area of trophoblast invasion in both placental tissue and adjacent myometrium from women with placenta percreta. In other words, for the invasive type of PAS, there are fundamental changes in the miRNA signature in the placenta, similar between increta and percreta but distinct from PAS adherent form (creta); the invasion of such altered placenta into the myometrial tissue occurs only if there are changes in the myometrium itself, most pronounced in the case of percreta. These findings are consistent with those of other researchers who adhere to the concept of a primary deciduomyometrium defect in PAS that impacts the formation of the migratory and invasive phenotype of interstitial and endovascular extravillous trophoblast cells [
13] [
14].
When analyzing experimentally validated target genes of identified here miRNAs differentially expressed in the placenta and myometrium within the PAS site, signaling pathways involving growth factors, glypicans, cell adhesion proteins, integrins, interleukins, and chemokines were identified. These pathways are responsible for processes such as cell adhesion, proliferation, migration, angiogenesis, inflammation, and apoptosis. These data align with discussions in published articles on pathways that stimulate trophoblast invasion [
12][
47].
A distinctive feature of invasive PAS, as observed through instrumental research methods and macroscopic examination of the uterine surface, is uteroplacental vascular changes in the accreta area resulting from both neovascularization and/or increased infiltration of deep uterine vessels (radial and even the arcuate arteries) by extravillous trophoblasts (EVT) [
1]. The role in angiogenesis of the identified here miRNAs as the markers of PAS in the first trimester of pregnancy has been demonstrated by numerous scientific teams. Hypoxia-responsive hsa-miR-101-3p known as angiomiR regulate angiogenesis by targeting cullin 3 thereby promoting Nrf2 nuclear accumulation and causing heme oxygenase-1 induction, VEGF expression, and nitric oxide production [
48], or by targeting c-Met [
49] – receptor for hepatocyte growth factor (HGF) which is one of the key molecule that stimulates endothelial cells to proliferate and migrate by upregulation of VEGF and its receptor KDR [
50] as well as metalloproteinases [
51] to degrade extracellular matrix for vascular growth. In addition, the inverse correlation between the expression of the miR-320a and the HGF gene was found [
49]. Different studies demonstrated the suppressor function of miR-320a in cell invasion and angiogenesis in ovary cancer [
52], hepatocellular carcinoma [
53][
54], endometrial cancer [
55]. The anti-inflammatory effect and promotion of the angiogenesis in the skeletal muscle injury model were found for miR-320a and miR-26a through reduction of the protein expression of their target genes – PTEN and TLR3, respectively [
56]. Enriched by miR-17-5p exosomes from endothelial progenitor cell decrease cell apoptosis, increase microvessel density and capillary angiogenesis as well as promote muscle structural integrity in a diabetic hind limb ischemia mode through increasing the levels of PI3K and phosphorylated Akt [
57]. The participation of miR-92a-3p in exosome-mediated angiogenesis was found in retinoblastoma by targeting transcription factor KLF2 [
58]which is able to modulate tumor proliferation and metastasis [
59]. The significant correlations of hsa-miR-17-5p expression level with uterine artery pulsatility index, and hsa-miR-26a-5p and hsa-miR-17-5p with PLGF revealed in the present study prove the important role of these PAS miRNA markers in angiogenesis.
Author Contributions
Conceptualization, A.V.T.; methodology, A.V.T., I.S.F. and T.Y.I.; software, I.S.F.; validation, A.V.T., I.S.F., T.M.Z., and T.Y.I.; investigation, A.V.T., I.S.F., Y.V.S., A.M.T., L.S.E., and T.Y.I.; data curation, A.V.T., O.N.V., and T.Y.I.; writing—original draft preparation, A.V.T.; writing—review and editing, A.V.T.; visualization, A.V.T. and I.S.F.; project administration, G.T.S.; funding acquisition, A.V.T. and G.T.S. All authors have read and agreed to the published version of the manuscript.
Figure 1.
Analysis of placental and myometrial tissues on the day of delivery in women with placenta creta, increta, and percreta using deep sequencing of miRNA. Schematic representation of sample collection locations for placenta and myometrium (a); Venn-Euler diagrams of differentially expressed miRNAs in the placenta within the pathological trophoblast invasion site for percreta and increta relative to creta (b), in the placenta outside areas of pathological trophoblast invasion for percreta and increta relative to creta (c), in myometrial tissues adjacent to areas of pathological trophoblast invasion for percreta and increta relative to creta (d), common signaling pathways regulated by differentially expressed miRNAs in the pathological trophoblast invasion site for percreta (Ppp vs Ppc) and in adjacent myometrial tissue (Mp vs Mc) according to MirTarbase and Funrich (e).
Figure 1.
Analysis of placental and myometrial tissues on the day of delivery in women with placenta creta, increta, and percreta using deep sequencing of miRNA. Schematic representation of sample collection locations for placenta and myometrium (a); Venn-Euler diagrams of differentially expressed miRNAs in the placenta within the pathological trophoblast invasion site for percreta and increta relative to creta (b), in the placenta outside areas of pathological trophoblast invasion for percreta and increta relative to creta (c), in myometrial tissues adjacent to areas of pathological trophoblast invasion for percreta and increta relative to creta (d), common signaling pathways regulated by differentially expressed miRNAs in the pathological trophoblast invasion site for percreta (Ppp vs Ppc) and in adjacent myometrial tissue (Mp vs Mc) according to MirTarbase and Funrich (e).
Figure 2.
Comparison of miRNA Expression Profiles in Placental Tissue, Adjacent Myometrium, and Peripheral Blood Plasma of Patients in Cohort 1. (a) Venn-Euler diagram for comparative analysis of lists of differentially expressed (DE) miRNAs in the blood plasma of women with creta, increta, and percreta relative to the group with a normal pregnancy using FunRich v. 3.1.3 (
http://www.funrich.org/) and Venny 2.1.0 (
https://bioinfogp.cnb.csic.es/tools/venny/).(b) Venn-Euler diagram for comparative analysis of the 165 DE miRNAs list in the blood plasma common for placenta increta and percreta cases (BPi/p, from
Figure 2A) with the DE miRNAs lists in placenta and myometrium for increta (Ppi and Mi, respectively) and percreta (Ppp and Mp, respectively) cases using FunRich v. 3.1.3 (
http://www.funrich.org/). (c) Venn-Euler diagram for comparative analysis of the DE miRNA list common to BPi/p and Ppi, BPi/p and Ppp, BPi/p and Mi, BPi/p and Mp (all obtained from
Figure 2B), constructed using Venny 2.1.0 (
https://bioinfogp.cnb.csic.es/tools/venny/).
Figure 2.
Comparison of miRNA Expression Profiles in Placental Tissue, Adjacent Myometrium, and Peripheral Blood Plasma of Patients in Cohort 1. (a) Venn-Euler diagram for comparative analysis of lists of differentially expressed (DE) miRNAs in the blood plasma of women with creta, increta, and percreta relative to the group with a normal pregnancy using FunRich v. 3.1.3 (
http://www.funrich.org/) and Venny 2.1.0 (
https://bioinfogp.cnb.csic.es/tools/venny/).(b) Venn-Euler diagram for comparative analysis of the 165 DE miRNAs list in the blood plasma common for placenta increta and percreta cases (BPi/p, from
Figure 2A) with the DE miRNAs lists in placenta and myometrium for increta (Ppi and Mi, respectively) and percreta (Ppp and Mp, respectively) cases using FunRich v. 3.1.3 (
http://www.funrich.org/). (c) Venn-Euler diagram for comparative analysis of the DE miRNA list common to BPi/p and Ppi, BPi/p and Ppp, BPi/p and Mi, BPi/p and Mp (all obtained from
Figure 2B), constructed using Venny 2.1.0 (
https://bioinfogp.cnb.csic.es/tools/venny/).
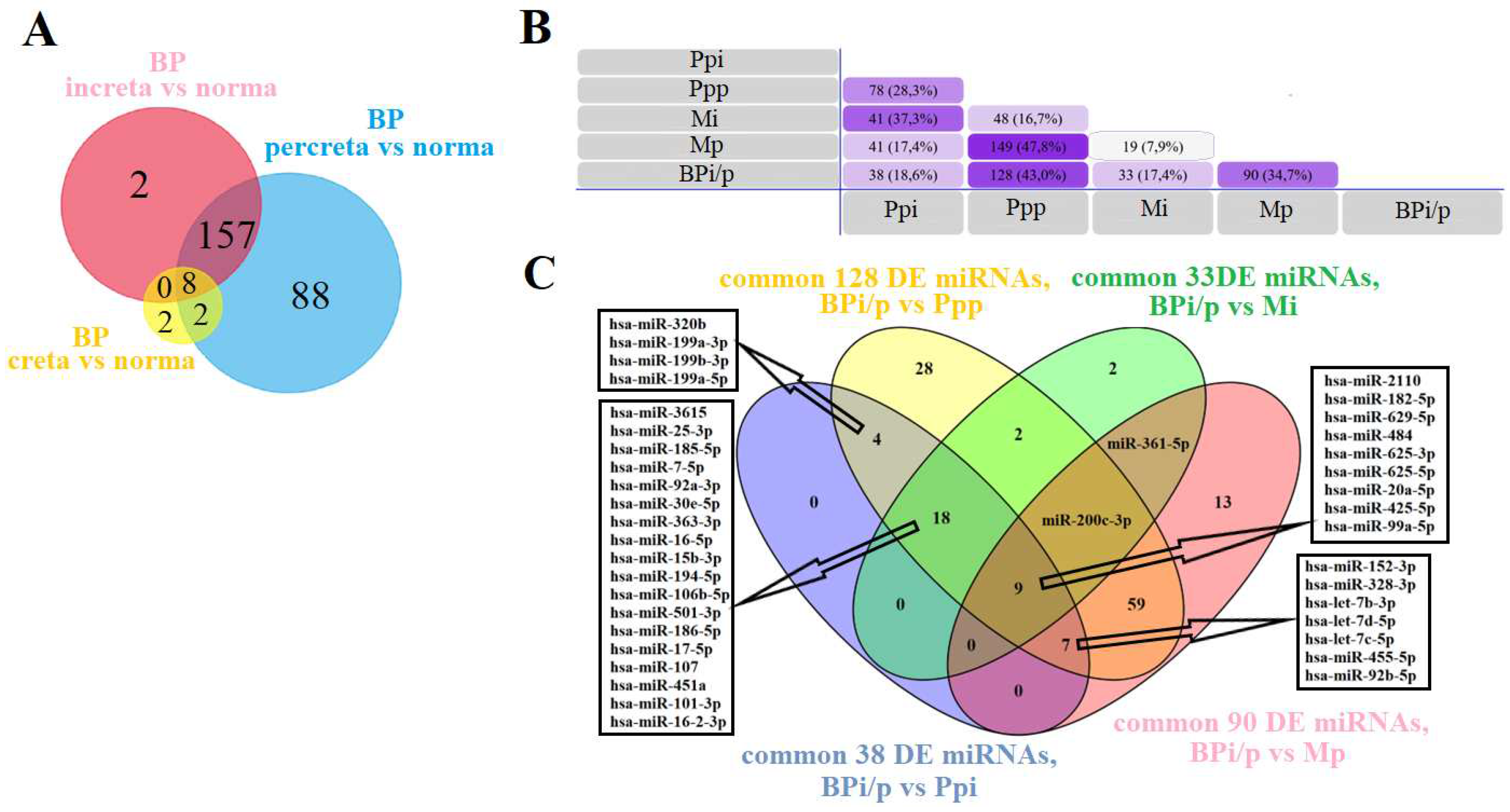
Figure 3.
PLS-A Analysis of Quantitative RT-PCR Data on the Expression Level of miRNAs in the Peripheral Blood Plasma of Pregnant Women with Physiological Pregnancy and PAS.
Figure 3.
PLS-A Analysis of Quantitative RT-PCR Data on the Expression Level of miRNAs in the Peripheral Blood Plasma of Pregnant Women with Physiological Pregnancy and PAS.
Figure 4.
Receiver Operating Characteristic (ROC) Curves of the Logistic Regression Models based on Real-Time Quantitative PCR Data when Comparing the Combined Groups "creta, increta, percreta" with the "norma" Group for the Content of miRNAs in the Blood Plasma of Pregnant Women in the Third Trimester. Se – Sensitivity, Sp – Specificity.
Figure 4.
Receiver Operating Characteristic (ROC) Curves of the Logistic Regression Models based on Real-Time Quantitative PCR Data when Comparing the Combined Groups "creta, increta, percreta" with the "norma" Group for the Content of miRNAs in the Blood Plasma of Pregnant Women in the Third Trimester. Se – Sensitivity, Sp – Specificity.
Figure 5.
PLS Analysis of Deep Sequencing Data of miRNAs in Women's Peripheral Blood Serum at 11-14 Weeks of Pregnancy with Physiological Course, Early- or Late-onset Pre-eclampsia, and PAS.
Figure 5.
PLS Analysis of Deep Sequencing Data of miRNAs in Women's Peripheral Blood Serum at 11-14 Weeks of Pregnancy with Physiological Course, Early- or Late-onset Pre-eclampsia, and PAS.
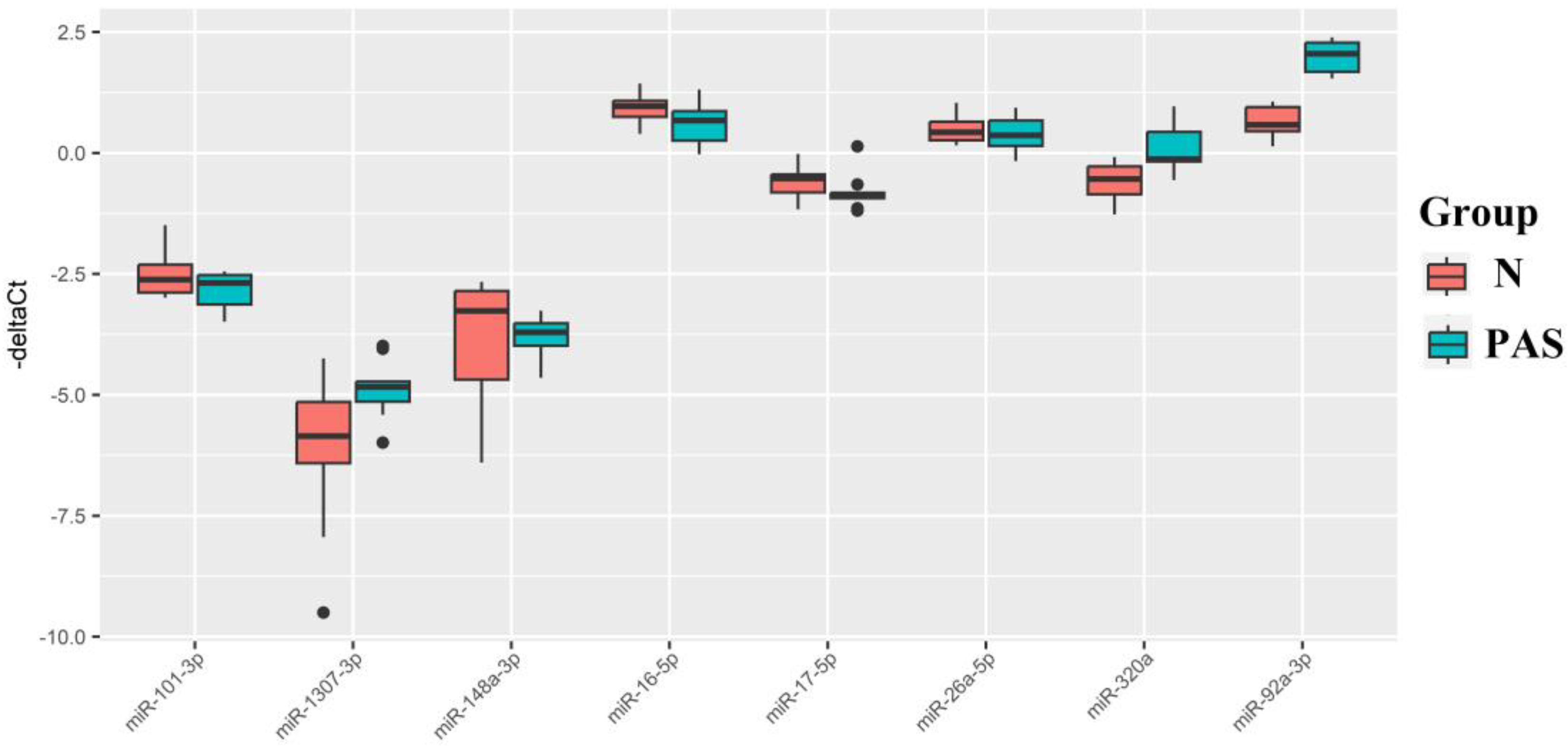
Figure 6.
Box plot of miRNA content in exosomes of blood serum from women at 11-14 weeks of pregnancy with a physiological course and PAS.
Figure 6.
Box plot of miRNA content in exosomes of blood serum from women at 11-14 weeks of pregnancy with a physiological course and PAS.
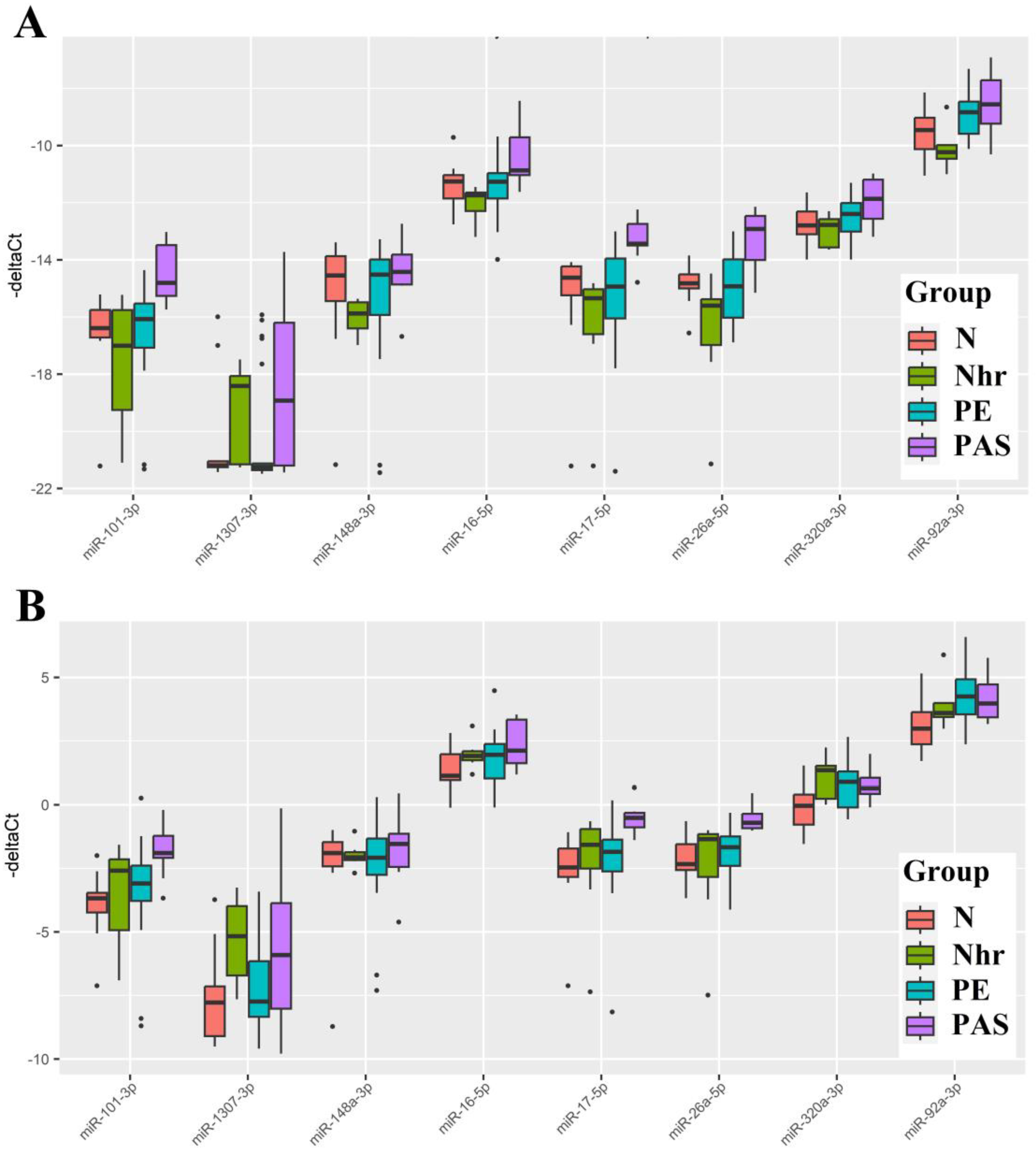
Figure 7.
Box Plot of miRNA Content in Native Blood Serum of Women at 11-14 Weeks of Pregnancy in Groups "N," "Nhr," "PE," "PAS." "-ΔCt" Values were Calculated Using Exogenous RNA UniSp6 (a). "-ΔCt" Values were calculated Relative to the Content of Endogenous miRNA hsa-let-7a-5p (b).
Figure 7.
Box Plot of miRNA Content in Native Blood Serum of Women at 11-14 Weeks of Pregnancy in Groups "N," "Nhr," "PE," "PAS." "-ΔCt" Values were Calculated Using Exogenous RNA UniSp6 (a). "-ΔCt" Values were calculated Relative to the Content of Endogenous miRNA hsa-let-7a-5p (b).
Figure 8.
ROC curves of logistic regression models based on real-time PCR data comparing the PAS group and combined groups "N + Nhr + PE" for the content of miRNA in native blood serum of women at 11-14 weeks of pregnancy. Se – sensitivity, Sp – specificity.
Figure 8.
ROC curves of logistic regression models based on real-time PCR data comparing the PAS group and combined groups "N + Nhr + PE" for the content of miRNA in native blood serum of women at 11-14 weeks of pregnancy. Se – sensitivity, Sp – specificity.
Table 1.
Clinical Characteristics of the First Cohort of Patients.
Table 2.
Clinical Characteristics of the Second Cohort of Patients.
Table 2.
Clinical Characteristics of the Second Cohort of Patients.
| Group name |
Group size |
Scar on the Uterus |
Placenta Previa |
Delivery time (weeks)* |
| Norma (pregnancy without complications) |
14 |
yes |
no |
37.5 (34.3; 39.2) |
| Creta |
9 |
yes |
yes |
36.4 (35; 37.4) |
| Increta |
16 |
yes |
yes |
34.6 (33.1; 36.3) |
| Percreta |
7 |
yes |
yes |
33 (32; 33) |
Table 3.
Parameters of the Logistic Regression Models in
Figure 4.
Table 3.
Parameters of the Logistic Regression Models in
Figure 4.
| Model |
Estimate (95% CI) |
Wald |
p_value |
OR (95% CI) |
Se |
Sp |
| 1 |
(Intercept) |
12.4342(5.8431;25.1735) |
2.6862 |
0.0072 |
251251.6477(344.8485;85646274941.2615) |
0.97 |
0.93 |
| miR-320b |
-2.6129(-5.2514;-0.9477) |
-2.504 |
0.0123 |
0.0733(0.0052;0.3876) |
| miR-92a-3p |
-2.1527(-4.8586;-0.5747) |
-2.0715 |
0.0383 |
0.1162(0.0078;0.5629) |
| 2 |
(Intercept) |
9.7122(4.807;18.8667) |
2.8577 |
0.0043 |
16518.5771(122.3664;156204305.6105) |
0.78 |
1 |
| miR-320b |
-4.0078(-7.9435;-1.83) |
-2.6992 |
0.007 |
0.0182(0.0004;0.1604) |
| let-7d-5p |
-0.6938(-1.4087;-0.1879) |
-2.3393 |
0.0193 |
0.4996(0.2445;0.8287) |
| 3 |
(Intercept) |
11.9713(5.7515;21.3935) |
3.0688 |
0.0021 |
158149.4506(314.6611;1954609961.5401) |
0.88 |
0.93 |
| miR-17-5p |
-0.7119(-1.3684;-0.2466) |
-2.5573 |
0.0105 |
0.4907(0.2545;0.7815) |
| miR-25-3p |
-0.5773(-1.0826;-0.2314) |
-2.7482 |
0.006 |
0.5614(0.3387;0.7934) |
| 4 |
(Intercept) |
5.4891(2.9334;9.5854) |
3.3713 |
<0.001 |
242.042(18.7919;14550.8746) |
0.88 |
0.86 |
| miR-320b |
-2.8885(-5.1368;-1.4507) |
-3.1781 |
0.0015 |
0.0557(0.0059;0.2344) |
| 5 |
(Intercept) |
9.5615(4.6005;16.6831) |
3.1352 |
0.0017 |
14206.4468(99.5329;17593816.9241) |
0.78 |
1 |
| miR-629-5p |
-0.4622(-0.8576;-0.1463) |
-2.61 |
0.0091 |
0.6299(0.4242;0.8639) |
| miR-17-5p |
-0.5663(-1.1491;-0.1557) |
-2.2973 |
0.0216 |
0.5676(0.3169;0.8558) |
| 6 |
(Intercept) |
8.6533(4.3535;15.0105) |
3.2633 |
0.0011 |
5729.094(77.7512;3303537.1587) |
0.91 |
0.86 |
| miR-92a-3p |
-2.2867(-3.9974;-1.1017) |
-3.1624 |
0.0016 |
0.1016(0.0184;0.3323) |
| 7 |
(Intercept) |
5.891(2.3393;11.7884) |
2.5906 |
0.0096 |
361.7674(10.3736;131711.3987) |
0.94 |
0.71 |
| miR-17-5p |
-0.5333(-1.0924;-0.1761) |
-2,4108 |
0,0159 |
0.5866(0.3354;0.8385) |
Table 4.
Clinical characteristics of the third cohort of patients.
Table 4.
Clinical characteristics of the third cohort of patients.
| Sample ID |
Diagnose at delivery, Group name |
Biochemical data |
Ultrasound data |
| 1st trimester screening, peripheral blood sampling, GW |
PAPP-A (0.7–6.0 IU/L) |
РАРР-А (0.5‒2.0 MoM) |
b-hCG (50.0–55.0 IU/ml) |
b-hCG (0.5‒2.0 MoM) |
PLGF (21,9-71,2 пг/мл) |
Сrown rump length, CRL (43.0 - 84.0 mm) |
Nuchal translucency thickness, NT (1.6‒1.7 mm) |
Uterine artery pulsatility index, UA (PI), MoM |
Uterine artery pulsatility index, UA (PI), 0.9‒2.6 (5th and 95th percentiles) |
| 1 |
late-onset preeclampsia, PE |
12.1 |
1.72 |
0.65 |
51.1 |
0.94 |
23.8 |
57.4 |
1.3 |
0.75 |
1.27 |
| 6 |
12.2 |
1 |
0.5 |
53.6 |
1.64 |
15.8 |
58 |
1.7 |
0.94 |
1.6 |
| 7 |
12.1 |
4.3 |
1.7 |
27.9 |
0.57 |
15.79 |
60.5 |
1.9 |
1.21 |
2.06 |
| 11 |
12.5 |
1.92 |
0.64 |
40.5 |
0.81 |
20.9 |
63.2 |
1.91 |
1.4 |
2.35 |
| 12 |
12 |
4.27 |
1.21 |
28.1 |
0.46 |
8.6 |
58.7 |
1.5 |
0.73 |
1.28 |
| 14 |
12.5 |
3.13 |
0.65 |
27.8 |
0.45 |
24.01 |
63.3 |
1.4 |
1.33 |
2.32 |
| 16 |
12.5 |
2.96 |
1.03 |
35.6 |
0.79 |
15.6 |
64 |
2 |
0.4 |
0.65 |
| 17 |
12.1 |
0.61 |
0.4 |
28.3 |
0.76 |
15.8 |
58.6 |
1.5 |
1.45 |
2.28 |
| 18 |
11.6 |
1.63 |
0.55 |
42.9 |
0.69 |
9.34 |
55.1 |
1.5 |
1.04 |
1.86 |
| 19 |
12.4 |
5.01 |
2.65 |
28.1 |
0.51 |
26.5 |
57.6 |
1.3 |
0.81 |
1.41 |
| 21 |
early-onset preeclampsia, PE |
12.1 |
1.86 |
0.9 |
54 |
1.09 |
|
57 |
1.6 |
2.07 |
3.5 |
| 22 |
12.1 |
0.8 |
0.46 |
36 |
0.6 |
|
50.6 |
1.76 |
1.18 |
2.09 |
| 23 |
12.2 |
3.36 |
1.17 |
45.9 |
0.92 |
11.7 |
60.2 |
2.9 |
1.22 |
2.04 |
| 24 |
11.2 |
1.96 |
1.04 |
94.3 |
1.66 |
|
50 |
1.07 |
1.2 |
2.12 |
| 25 |
11.6 |
6.2 |
2.96 |
43.8 |
0.76 |
10 |
56 |
1.45 |
1.03 |
1.83 |
| 26 |
12.1 |
2.06 |
0.79 |
48.8 |
1.05 |
9.2 |
60.3 |
1.7 |
1.56 |
2.6 |
| 27 |
12.3 |
0.86 |
0.45 |
15.6 |
0.36 |
|
59 |
1.9 |
1.2 |
2 |
| 28 |
11.6 |
2.47 |
2.27 |
34.3 |
0.98 |
12.84 |
57 |
1.3 |
1.01 |
1.59 |
| 29 |
12.4 |
2.9 |
1 |
56.8 |
1.1 |
18 |
62 |
1.3 |
0.79 |
1.31 |
| 30 |
12.4 |
1.71 |
0.64 |
31.3 |
0.7 |
6.3 |
62 |
1.5 |
0.98 |
1.68 |
| 31 |
12 |
2.09 |
0.83 |
16.9 |
0.33 |
16 |
57.7 |
1.7 |
0.33 |
1.686 |
| 33 |
physiological pregnancy, N |
12.5 |
2.28 |
0.86 |
89.7 |
2.17 |
19.7 |
63.9 |
2.2 |
0.7 |
1.12 |
| 34 |
12.1 |
2 |
0.71 |
89.8 |
2.25 |
16.39 |
66 |
1.6 |
1.09 |
1.7 |
| 35 |
13.4 |
2.57 |
0.83 |
62.2 |
1.81 |
32.7 |
74.7 |
1.8 |
1.19 |
1.77 |
| 39 |
12 |
3.02 |
1.52 |
66 |
1.32 |
16.11 |
54 |
1.1 |
1.028 |
1.75 |
| 41 |
13 |
3.1 |
0.88 |
61 |
1.26 |
26.68 |
67.5 |
1.6 |
0.28 |
0.44 |
| 43 |
12.2 |
1.62 |
0.52 |
86.7 |
1.77 |
14.7 |
58 |
1.2 |
1.14 |
1.95 |
| 44 |
13 |
3.42 |
1.3 |
56.6 |
1.44 |
15.97 |
66 |
1.21 |
1.02 |
1.6 |
| 46 |
12 |
3.011 |
1.23 |
64.2 |
1.15 |
15.49 |
54.5 |
1.2 |
1.002 |
1.77 |
| 47 |
12.6 |
2.9 |
0.61 |
52.3 |
1.06 |
32.52 |
65 |
1.17 |
1.3 |
2.15 |
| 49 |
12 |
6.9 |
3.15 |
58.8 |
1.105 |
26.2 |
55 |
1.3 |
0.88 |
1.49 |
| 51 |
high risk of preeeclampsia according to Astraia but no preeclampsia at delivery, Nhr |
11.2 |
1.05 |
0.41 |
25.5 |
0.36 |
|
50 |
1 |
1.33 |
2.48 |
| 52 |
13 |
3.38 |
0.68 |
39.8 |
0.74 |
|
68 |
2 |
1.15 |
1.92 |
| 53 |
11.6 |
1.55 |
1.32 |
23.7 |
0.717 |
12.7 |
59.7 |
1.9 |
1.4 |
2.17 |
| 54 |
12.1 |
2.1 |
1.07 |
89.8 |
2.5 |
|
56 |
1.1 |
1.02 |
1.68 |
| 55 |
12.2 |
2.25 |
0.97 |
23.2 |
0.48 |
19.99 |
57.6 |
1.6 |
1.19 |
2.02 |
| 56 |
11.6 |
2.27 |
1.23 |
114.7 |
2.3 |
|
53 |
1.29 |
1.05 |
1.8 |
| 58 |
12.1 |
4.18 |
1.8 |
51.7 |
1.02 |
|
57 |
1.9 |
0.81 |
1.36 |
| 110 |
Placenta percreta, PAS |
12.2 |
13.7 |
4.6 |
101.21 |
2.047 |
37.7 |
61.9 |
1.7 |
1.069 |
1.75 |
| 111 |
Placenta increta, PAS |
12.5 |
5.117 |
1.983 |
47 |
0.872 |
25.6 |
61 |
1.3 |
0.49 |
0.855 |
| 112 |
Placenta creta, PAS |
12 |
4.46 |
2.145 |
38.14 |
0.69 |
22.8 |
53 |
1.2 |
0.981 |
1.715 |
| 113 |
Placenta increta, PAS |
12.3 |
1.891 |
0.995 |
35 |
0.86 |
|
59.6 |
1.8 |
0.845 |
1.365 |
| 114 |
Placenta percreta, PAS |
12.6 |
3.257 |
1.043 |
65.17 |
1.428 |
23.3 |
65 |
1.9 |
0.552 |
0.89 |
| 115 |
Placenta percreta, PAS |
13.1 |
2.5 |
1.151 |
28.06 |
0.9 |
56.2 |
70 |
2 |
0.999 |
1.475 |
| 116 |
Placenta increta, PAS |
12.0 |
5.84 |
2.021 |
223.6 |
3.97 |
|
57.9 |
1.2 |
1.164 |
1.995 |
| 117 |
Placenta increta, PAS |
13.0 |
8.6 |
2.8 |
53 |
1.309 |
34.8 |
68 |
1.5 |
0.485 |
0.755 |
| 118 |
Placenta increta, PAS |
12.1 |
3.838 |
1.562 |
45.84 |
0.865 |
|
56.8 |
1.6 |
1.312 |
2.21 |
| 119 |
Placenta percreta, PAS |
12.4 |
2.252 |
0.989 |
86.85 |
2.06 |
|
61.7 |
1.9 |
0.753 |
1.22 |
Table 5.
Comparison of groups “N” and “PAS” by the miRNA “-ΔCt” value plotted as a box-diagram in
Figure 6.
Table 5.
Comparison of groups “N” and “PAS” by the miRNA “-ΔCt” value plotted as a box-diagram in
Figure 6.
| miRNA |
Group |
Me |
Q1 |
Q3 |
p-value |
| miR-101-3p |
N |
-2.62 |
-2.89 |
-2.31 |
0.25 |
| PAS |
-2.69 |
-3.13 |
-2.52 |
| miR-16-5p |
N |
0.96 |
0.75 |
1.08 |
0.09 |
| PAS |
0.67 |
0.26 |
0.86 |
| miR-17-5p |
N |
-0.53 |
-0.82 |
-0.44 |
0.08 |
| PAS |
-0.89 |
-0.93 |
-0.82 |
| miR-26a-5p |
N |
0.43 |
0.26 |
0.64 |
0.53 |
| PAS |
0.37 |
0.15 |
0.67 |
| miR-320a |
N |
-0.54 |
-0.85 |
-0.28 |
0.006 |
| PAS |
-0.13 |
-0.18 |
0.43 |
| miR-92a-3p |
N |
0.58 |
0.45 |
0.95 |
<0.001 |
| PAS |
2.05 |
1.68 |
2.28 |
| miR-1307-3p |
N |
-5.85 |
-6.41 |
-5.15 |
0.04 |
| PAS |
-4.84 |
-5.14 |
-4.72 |
| miR-148a-3p |
N |
-3.26 |
-4.69 |
-2.85 |
0.44 |
| PAS |
-3.71 |
-3.99 |
-3.52 |
Table 6.
Pairwise Comparison of 3rd Patient Cohort Groups Listed in
Table 5 by miRNA "-ΔCt" Value, Presented as a Box Diagram in
Figure 6.
Table 6.
Pairwise Comparison of 3rd Patient Cohort Groups Listed in
Table 5 by miRNA "-ΔCt" Value, Presented as a Box Diagram in
Figure 6.
| Normalization to UniSp6 |
| miRNA |
Group |
Me |
Q1 |
Q3 |
p-value |
| N |
Nhr |
PE |
PAS |
| miR-101-3p |
N |
-16.39 |
-16.72 |
-15.75 |
- |
0.53 |
0.54 |
<0.001 |
| Nhr |
-17 |
-19.25 |
-15.75 |
0.53 |
- |
0.43 |
<0.001 |
| PE |
-16.08 |
-17.07 |
-15.53 |
0.54 |
0.43 |
- |
<0.001 |
| PAS |
-14.81 |
-15.26 |
-13.48 |
<0.001 |
<0.001 |
<0.001 |
- |
| miR-16-5p |
N |
-11.26 |
-11.85 |
-11.04 |
- |
0.06 |
0.98 |
0.02 |
| Nhr |
-11.74 |
-12.29 |
-11.64 |
0.06 |
- |
0.08 |
<0.001 |
| PE |
-11.27 |
-11.85 |
-10.96 |
0.98 |
0.08 |
- |
0.01 |
| PAS |
-10.87 |
-11.03 |
-9.71 |
0.02 |
<0.001 |
0.01 |
- |
| miR-17-5p |
N |
-14.63 |
-15.24 |
-14.23 |
- |
0.07 |
0.85 |
<0.001 |
| Nhr |
-15.35 |
-16.6 |
-15.03 |
0.07 |
- |
0.17 |
<0.001 |
| PE |
-14.93 |
-16.05 |
-13.95 |
0.85 |
0.17 |
- |
<0.001 |
| PAS |
-13.44 |
-13.5 |
-12.75 |
<0.001 |
<0.001 |
<0.001 |
- |
| miR-26a-5p |
N |
-14.82 |
-15 |
-14.51 |
- |
0.02 |
0.88 |
0.002 |
| Nhr |
-15.6 |
-16.98 |
-15.38 |
0.02 |
- |
0.08 |
<0.001 |
| PE |
-14.93 |
-16.02 |
-13.98 |
0.88 |
0.08 |
- |
<0.001 |
| PAS |
-12.92 |
-14.01 |
-12.46 |
0.002 |
<0.001 |
<0.001 |
- |
| miR-320a-3p |
N |
-12.79 |
-13.11 |
-12.31 |
- |
0.88 |
0.39 |
0.02 |
| Nhr |
-12.78 |
-13.57 |
-12.58 |
0.88 |
- |
0.09 |
0.01 |
| PE |
-12.39 |
-13.01 |
-12.01 |
0.39 |
0.09 |
- |
0.07 |
| PAS |
-11.86 |
-12.56 |
-11.2 |
0.02 |
0.01 |
0.07 |
- |
| miR-92a-3p |
N |
-9.46 |
-10.13 |
-9.03 |
- |
0.13 |
0.12 |
0.05 |
| Nhr |
-10.23 |
-10.46 |
-9.98 |
0.13 |
- |
0.001 |
0.009 |
| PE |
-8.84 |
-9.59 |
-8.46 |
0.12 |
0.001 |
- |
0.24 |
| PAS |
-8.56 |
-9.23 |
-7.72 |
0.05 |
0.009 |
0.24 |
- |
| miR-1307-3p |
N |
-21.19 |
-21.25 |
-21.05 |
- |
0.31 |
0.34 |
0.31 |
| Nhr |
-18.41 |
-21.15 |
-18.06 |
0.31 |
- |
0.14 |
0.60 |
| PE |
-21.24 |
-21.35 |
-21.13 |
0.34 |
0.14 |
- |
0.09 |
| PAS |
-18.92 |
-21.2 |
-16.2 |
0.31 |
0.60 |
0.09 |
- |
| miR-148a-3p |
N |
-14.55 |
-15.44 |
-13.87 |
- |
0.07 |
0.63 |
0.73 |
| Nhr |
-15.88 |
-16.4 |
-15.48 |
0.07 |
- |
0.10 |
0.003 |
| PE |
-14.52 |
-15.93 |
-13.99 |
0.63 |
0.10 |
- |
0.30 |
| PAS |
-14.42 |
-14.86 |
-13.81 |
0.73 |
0.003 |
0.30 |
- |
| Normalization to hsa-let-7a-5p |
| miRNA |
Group |
Me |
Q1 |
Q3 |
p-value |
| N |
Nhr |
PE |
PAS |
| miR-101-3p |
N |
-3.68 |
-4.24 |
-3.46 |
- |
0.36 |
0.12 |
<0.001 |
| Nhr |
-2.59 |
-4.93 |
-2.14 |
0.36 |
- |
0.95 |
0.05 |
| PE |
-3.09 |
-3.78 |
-2.39 |
0.12 |
0.95 |
- |
0.01 |
| PAS |
-1.9 |
-2.09 |
-1.22 |
<0.001 |
0.05 |
0.01 |
- |
| miR-16-5p |
N |
1.13 |
0.96 |
1.98 |
- |
0.16 |
0.28 |
0.02 |
| Nhr |
1.91 |
1.74 |
2.09 |
0.16 |
- |
0.83 |
0.41 |
| PE |
1.96 |
1.03 |
2.38 |
0.28 |
0.83 |
- |
0.17 |
| PAS |
2.12 |
1.62 |
3.34 |
0.02 |
0.41 |
0.17 |
- |
| miR-17-5p |
N |
-2.46 |
-2.84 |
-1.72 |
- |
0.41 |
0.21 |
<0.001 |
| Nhr |
-1.58 |
-2.51 |
-0.95 |
0.41 |
- |
0.53 |
0.009 |
| PE |
-1.85 |
-2.62 |
-1.37 |
0.21 |
0.53 |
- |
<0.001 |
| PAS |
-0.51 |
-0.89 |
-0.31 |
<0.001 |
0.009 |
<0.001 |
- |
| miR-26a-5p |
N |
-2.33 |
-2.56 |
-1.55 |
- |
0.73 |
0.23 |
<0.001 |
| Nhr |
-1.35 |
-2.83 |
-1.15 |
0.73 |
- |
0.91 |
<0.001 |
| PE |
-1.67 |
-2.4 |
-1.24 |
0.23 |
0.91 |
- |
<0.001 |
| PAS |
-0.71 |
-0.92 |
-0.35 |
<0.001 |
<0.001 |
<0.001 |
- |
| miR-320a-3p |
N |
-0.04 |
-0.78 |
0.4 |
- |
0.05 |
0.11 |
0.05 |
| Nhr |
1.35 |
0.24 |
1.52 |
0.05 |
- |
0.29 |
0.60 |
| PE |
0.9 |
-0.1 |
1.3 |
0.11 |
0.29 |
- |
0.88 |
| PAS |
0.65 |
0.43 |
1.05 |
0.05 |
0.60 |
0.88 |
- |
| miR-92a-3p |
N |
2.99 |
2.37 |
3.64 |
- |
0.10 |
0.03 |
0.03 |
| Nhr |
3.6 |
3.44 |
3.99 |
0.10 |
- |
0.46 |
0.53 |
| PE |
4.25 |
3.55 |
4.93 |
0.03 |
0.46 |
- |
1 |
| PAS |
3.97 |
3.43 |
4.72 |
0.03 |
0.53 |
1 |
- |
| miR-1307-3p |
N |
-7.78 |
-9.1 |
-7.14 |
- |
0.04 |
0.41 |
0.19 |
| Nhr |
-5.17 |
-6.71 |
-3.98 |
0.04 |
- |
0.03 |
0.60 |
| PE |
-7.74 |
-8.34 |
-6.15 |
0.41 |
0.03 |
- |
0.30 |
| PAS |
-5.91 |
-8.02 |
-3.87 |
0.19 |
0.60 |
0.30 |
- |
| miR-148a-3p |
N |
-1.9 |
-2.42 |
-1.47 |
- |
0.96 |
0.95 |
0.48 |
| Nhr |
-2.07 |
-2.17 |
-1.86 |
0.96 |
- |
1 |
0.36 |
| PE |
-2.08 |
-2.76 |
-1.33 |
0.95 |
1 |
- |
0.41 |
| PAS |
-1.54 |
-2.44 |
-1.14 |
0.48 |
0.36 |
0.41 |
- |
Table 7.
Parameters of Logistic Regression Models in
Figure 8.
Table 7.
Parameters of Logistic Regression Models in
Figure 8.
| Models |
Estimate (95% CI) |
Wald |
p-value |
OR(95% CI) |
Se |
Sp |
| (Intercept) |
12.264(3.532;27.813) |
2.149 |
0.032 |
211919.348(34.208;1.2Е+12) |
Model 1 |
| miR-92a-3p |
-4.955(-10.911;-1.566) |
-2.246 |
0.025 |
0.007(0.00001;0.209) |
1 |
0.84 |
| miR-26a-5p |
5.093(2.068;10.91) |
2.355 |
0.019 |
162.828(7.905;54728.702) |
| miR-16-5p |
5.584(1.896;12.356) |
2.267 |
0.023 |
266.031(6.658;232425.187) |
| (Intercept) |
5.661(1.863;12.559) |
2.157 |
0.031 |
287.563(6.445;284717.908) |
Model 2 |
| miR-26a-5p |
4.189(1.259;9.276) |
2.157 |
0.031 |
65.939(3.523;10674.696) |
1 |
0.82 |
| miR-17-5p |
2.921(0.646;6.887) |
1.916 |
0.055 |
18.564(1.908;979.604) |
| (Intercept) |
8.049(2.747;17.86) |
2.193 |
0.028 |
3129.605(15.589;5.7Е+07) |
Model 3 |
| miR-26a-5p |
5.538(2.059;12.068) |
2.274 |
0.023 |
254.057(7.835;174165.021) |
1 |
0.84 |
| miR-101-3p |
1.989(0.502;4.585) |
2.012 |
0.044 |
7.308(1.652;98.023) |
| (Intercept) |
9.303(2.248;19.251) |
2.21 |
0.027 |
10967.88(9.473;2.3Е+08) |
Model 4 |
| miR-92a-3p |
-1.451(-3.193;-0.173) |
-1.941 |
0.052 |
0.234(0.041;0.841) |
0.9 |
0.9211 |
| miR-17-5p |
4.333(1.956;8.034) |
2.872 |
0.004 |
76.151(7.071;3084.714) |
| (Intercept) |
4.152(1.217;8.272) |
2.386 |
0.017 |
63.567(3.378;3911.935) |
Model 5 |
| miR-320a-3p |
-1.477(-3.168;-0.148) |
-1.973 |
0.048 |
0.228(0.042;0.862) |
0.8 |
0.9737 |
| miR-17-5p |
4.108(1.878;7.607) |
2.899 |
0.004 |
60.854(6.539;2013.226) |
| (Intercept) |
2.256(0.321;4.915) |
1.974 |
0.048 |
9.544(1.378;136.287) |
Model 6 |
| miR-26a-5p |
3.389(1.483;6.373) |
2.773 |
0.006 |
29.638(4.405;585.998) |
1 |
0.8421 |
| (Intercept) |
1.628(-0.009;3.742) |
1.742 |
0.082 |
5.093(0.991;42.181) |
Model 7 |
| miR-17-5p |
2.533(1.127;4.672) |
2.877 |
0.004 |
12.592(3.087;106.945) |
1 |
0.7105 |
| (Intercept) |
1.136(-0.568;3.188) |
1.218 |
0.223 |
3.115(0.566;24.245) |
Model 8 |
| miR-101-3p |
0.985(0.33;1.887) |
2.533 |
0.011 |
2.679(1.391;6.598) |
0.8 |
0.7895 |