1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of coronavirus disease 2019 (COVID-19), poses a significant threat to global public health [
1,
2]. As November 22
rd 2023, there are over 772 million confirmed cases worldwide, resulting in over 6.97 million deaths (
https://covid19.who.int/?mapFilter=cases). SARS-CoV-2 is an enveloped, single-strand, positive-sense RNA virus that belongs to
Sarbecovirus lineage of betacoronavirus genus in the Coronaviridae family, which also includes the highly pathogenic SARS-CoV [
3]. Although there are approved vaccines for SARS-CoV-2, as the viruses continuously evolving, new variants emerge, such as the omicron variants, show markedly increased immune evasion to COVID-19 convalescent and vaccinated sera, indicating that the novel vaccine designs are urgently needed [
4]. A better understanding of the structure and function of viral glycoproteins would aid in the development of novel antiviral therapeutic targets.
The coronavirus spike (S) glycoproteins form homotrimeric spikes on the virion surface and mediate viral entry into host cells [
5]. The monomeric S protein consists of two subunits, designated as S1 and S2, which are responsible for receptor binding and membrane fusion, respectively. In the case of SARS-CoV-2, the S protein is cleaved into S1 and S2 by furin in Golgi during S protein maturation process [
4]. The native trimeric S protein is metastable, upon receptor binding and proteolytic cleavage, the S protein undergoes a cascade of conformational changes, from initial metastable prefusion conformation to a stable postfusion conformation, driving the fusion of viral and cellular membranes [
6,
7].
Similar to the human immunodeficiency virus (HIV) envelope protein, influenza virus hemagglutinin, and Ebola virus glycoprotein, S proteins of coronaviruses are also heavily glycosylated [
8,
9]. The trimeric S proteins of SARS-CoV, SARS-CoV-2 and MERS-CoV contain 69, 66, and 69 putative N-linked glycosylation sites, respectively [
8,
9]. The S protein is synthesized in the endoplasmic reticulum (ER) and transported to Golgi apparatus for further modifications. N-linked glycosylation is a common post-translational modification found in enveloped viruses, and the asparagine of the N-linked glycosylation sequon (Asn-X-Thr/Ser, where X is not Pro) is glycosylated [
10]. Mass spectrometry (MS) analysis revealed extensive heterogeneity of the N-glycans on the S protein, ranging from high-mannose-type glycans to complex-type glycans [
9,
11]. N-glycans cover approximately 3/4 of the surface of the trimeric S protein, shielding epitopes on the S protein from the host immune response and attenuating viral immunogenicity [
8,
12]. In parallel with epitope masking, glycan-dependent epitopes of viral glycoproteins can induce neutralizing antibodies [
13]. The N-glycans of SARS-CoV S protein are also involved in virus attachment to its attachment factors, such as DC/L-SIGN, and the N-glycosites N227 and N699 on SARS-CoV S protein are critical for binding to DC/L-SIGN [
14]. It has been reported that N-glycans at positions N165, N234, and N343 of the SARS-CoV-2 S protein facilitate the RBD in an open state and thus contribute to an increase in infectivity [
15,
16,
17], whereas N-glycans at position N370 stabilize the RBD in a closed state, resulting in decreased infectivity [
18,
19]. A recent study revealed N-glycans on the stalk hinge region of HCoV-NL63 S protein modulated protein bending [
20]. Although SARS-CoV-2 evolves continuously and new variants emerge and disappear, most of S protein N-linked glycosylation sites are highly conserved among variants, except for the deletion of N17 glycosite in the Delta variant and the introduction of N20 and N188 glycosites in the Gamma variant, indicating the important roles of N-glycosylation in the S proteins [
21]. In the present study, we investigated the roles of N-glycans on SARS-CoV-2 S glycoprotein in protein expression, stability, membrane fusion, and viral entry, and found some N-glycans on the SARS-CoV and SARS-CoV-2 S2 subunits are important for maintaining the structure of S protein in prefusion state.
2. Materials and Methods
2.1. Plasmid constructions
The codon-optimized SARS-CoV-2 S gene (GenBank: QHU36824.1) with a deletion of the last 19 amino acids was synthesized by GenScript (Nanjing, China) and cloned into pcDNA3.1(+) between the
Hind III and
BamH I sites. Plasmids encoding the SARS-CoV S protein with a deletion of the last 19 amino acids have been previously described [
22]. All mutagenesis experiments were performed using the Q5 mutagenesis kit (NEB, MA, USA). After the entire coding sequence was verified, the coding regions were subcloned into pcDNA3.1(+). Human ACE2-targeting gRNA (5ʹ-GAAAGCTGGAGATCTGAGGT-3ʹ) was synthesized and cloned into the plentiCRISPRv2 vector to generate the recombinant plasmid pCRISPRv2-hACE2.
2.2. Cell lines and culture conditions
Human embryonic kidney (HEK) 293T cells, HEK293 cells, and HEK293 cells stably expressing human angiotensin-converting enzyme 2 (293/hACE2) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin-fungizone (PSF) at 37 °C with 5% CO2. The endogenous ACE2 knockout HEK293T cell line (HEK293T/hACE2-KO) was generated using the CRISPR-Cas9 system.
2.3. Production and transduction of S-pseudotyped lentiviruses
Plasmids encoding different S coronavirus proteins were cotransfected with psPAX2 and pLenti-Luc-GFP into HEK293T cells. After 40 h of incubation, the pseudovirion-containing supernatants were centrifuged at 1000 g for 10 min to remove cell debris. To quantify the transduction efficiency of the S-pseudotyped lentiviruses, HEK293/hACE2 cells were seeded in a 24-well plate, and after overnight incubation, the cells were inoculated with pseudovirions. At 40 h post-inoculation, cells were lysed using Steady-Glo Reagent (Promega, Madison, WI, USA). Transduction efficiency was monitored by quantifying luciferase activity using a PerkinElmer EnVision® Multilabel Plate Reader.
2.4. Detection of viral spike glycoproteins using western blotting
To evaluate S protein expression in the cell lysates, HEK293T cells were transfected with plasmids encoding different S proteins. At 40 h post-transfection, cells were lysed using RIPA lysis buffer containing protease inhibitors. To determine the incorporation of S proteins into pseudovirions, the pseudovirus-containing supernatant was pelleted through a 20% sucrose cushion at 25000 rpm at 4 °C for 2 h. Viral pellets were resuspended in 1×loading buffer. Cell lysates and pseudovirion pellets were separated using 10% sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes. The SARS-CoV-2 S proteins were detected with a rabbit polyclonal anti-S2 antibody (Sinobiological Inc., Beijing, China), and the blot was further stained with horseradish peroxidase-conjugated goat anti-rabbit IgG and then visualized with Clarity Western ECL substrate (Bio-Rad, Hercules, CA, USA). β-actin and HIV capsid protein (p24) were detected using a mouse monoclonal anti-β-actin antibody (Sigma, St Louis, MO, USA) and a rabbit polyclonal anti-p24 antibody (Sinobiological Inc., Beijing, China), respectively.
2.5. Flow cytometric analysis of cell surface S protein expression
HEK293T cells transiently expressing different S proteins were digested with 1 mM ethylenediaminetetraacetic acid (EDTA). After washing, cells were incubated with polyclonal rabbit anti-SARS-CoV-2 S2 antibodies (Sinobiological Inc., Beijing, China) for 1 h on ice, followed by FITC-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch, PA, USA) for 1 h. After washing, the cells were analyzed using a FACSCalibur flow cytometer (Becton Dickinson, CA, USA).
2.6. Soluble hACE2 binding assay
HEK293T cells transiently expressing different S proteins were digested with 1 mM EDTA. After washing, cells were incubated with 5 μg/mL soluble hACE2 for 1 h on ice. Subsequently, the cells were washed and incubated with 1 µg/mL polyclonal goat anti-human ACE2 antibody (R&D Systems, Minneapolis, MN, USA) for 1 h, followed by incubation with FITC-conjugated rabbit anti-goat secondary antibody (Jackson ImmunoResearch, PA, USA) for 1 h. After washing, the cells were analyzed using a FACSCalibur flow cytometer (Becton Dickinson, CA, USA).
2.7. Cell-cell fusion assay
HEK293T cells were co-transfected with plasmids encoding different S proteins and GFP in the absence or presence of 100 μg/mL castanospermine (#HY-N2022, MCE) or NB-DNJ (#HY-17020, MCE). After 40 h of transfection, the cells were detached using 0.25% trypsin for 2 min, and approximately 1/3 of these cells were overlaid on HEK293/hACE2 cells. After 4 h of incubation, images of syncytia were captured using a Nikon TE2000 epifluorescence microscope with the MetaMorph software (Molecular Devices).
2.8. Detection of S1 subunit shedding
HEK293T cells were transfected with plasmids encoding either wild-type or mutant S protein. After 40 h, the cell culture media were collected and centrifuged at 1000 g for 10 min to remove cell debris. The supernatants were then concentrated using Amicon Ultra-0.5 mL 3 kDa Centrifugal Filters (Millipore). Thereafter, the S1 subunits in the concentrated supernatants were detected using western blot analysis with antibodies against the SARS-CoV-2 RBD domain.
2.9. Statistic analysis
Data were presented as mean ± standard deviation of three experimental replicates using GraphPad Prism version 7. Comparisons between means were analyzed by the unpaired two-tailed Student’s t-test.
3. Results
3.1. α-Glucosidase inhibitors reduced the efficiency of SARS-CoV-2 S-mediated membrane fusion
To determine the effect of N-glycans on SARS-CoV-2 S protein expression, HEK293T cells transiently expressing SARS-CoV-2 S protein were treated with α-glycosidase inhibitors, castanospermine and NB-DNJ, and the levels of S protein in cell lysates was analyzed using western blot analysis. Consistent with our previous reports [
23], both cleaved and full-length (FL) uncleaved forms of the S protein were detected using antibodies against the SARS-CoV-2 S2 (
Figure 1A). There were two close bands corresponding to FL S proteins. The higher faint band might represent the fully glycosylated FL S proteins that would be further cleaved to S1 and S2 in Golgi and plasma membrane during S protein maturation, whereas the lower FL band likely reflects the nascent, unglycosylated FL S proteins. Treatment with α-glycosidase inhibitors almost had no effect on the levels of FL S proteins, but the levels of cleaved S proteins were significantly decreased when compared to the dimethyl sulfoxide (DMSO)-treated group (
Figure 1A), indicating that inhibition of N-linked glycosylation might greatly affect S protein folding and maturation process. Previously we and others showed that co-culturing SARS-CoV-2 S protein expressing HEK293T cells with HEK293/hACE2 cells in the presence of trypsin would induce large cell-cell fusion or syncytium formation. As shown in
Figure 1B, significant syncytium formation was observed when WT SARS-CoV-2 S expressing cells were mixed with HEK293/hACE2 cells in the presence of trypsin. However, the size and frequency of syncytium formation was significantly reduced in
Figure 1B when SARS-CoV-2 S protein-expressing HEK293T cells were pre-treated with castanospermine or NB-DNJ, consistent with the lower expression levels of S proteins in cells when treated with α-glycosidase inhibitors (
Figure 1A). We also evaluated the effects of glycosidase inhibitors on S-mediated viral entry by SARS-CoV-2 S-pseudotyped lentiviruses. Treatment of cells with α-glycosidase inhibitors also greatly decreased the levels of SARS-CoV-2 S protein incorporated into pseudovirions, resulting in significant reduction in transduction efficiency of SARS-CoV-2 S pseudovirons on HEK293/hACE2 (
Figure 1C,D). Together, these results indicate that inhibition of N-linked glycosylation in the SARS-CoV-2 S protein might greatly affect S protein maturation, S-mediated membrane fusion, and virus entry.
3.2. Effect of individual N-linked glycosylation on levels of expression, surface presentation, and receptor binding of SARS-CoV-2 S proteins
The SARS-CoV-2 S protein contains 22 putative N-linked glycosylation sites with 13 in the S1 subunit and 9 in the S2 subunit (
Figure 2A). We then determined which glycosylation site(s) might affect the S protein folding and maturation. The asparagine (N) residue in each glycosylation sites (NxT/S) was substituted with a glutamine (Q) to remove the N-linked glycosylation. Individual single mutation almost had no effect on the expression levels of the FL S proteins, whereas 9 out of 22 mutants gave marked decrease in the levels of cleaved S proteins when compared to WT S protein (
Figure 2B). The N61Q, N343Q, and N801Q mutants showed only slightly above background levels of cleaved S proteins, and N122Q, N165Q, N331Q, N717Q, N1098Q, and N1173Q also gave marked reduction in the cleaved S proteins, indicating that these N-linked glycosylation might have significant effect on S protein folding and maturation.
Next, we evaluated the levels of individual mutant S proteins on cell surface and their capabilities of binding to their cognate receptor, hACE2, using flow cytometry assay. As shown in
Figure 2C,D, the levels of each mutant S protein on cell surface and their receptor binding were largely in agreement with the amounts of the cleaved S proteins in cell lysate.
3.3. Effect of individual N-linked glycans on SARS-CoV-2 S-mediated cell-cell fusion and pseudovirus entry.
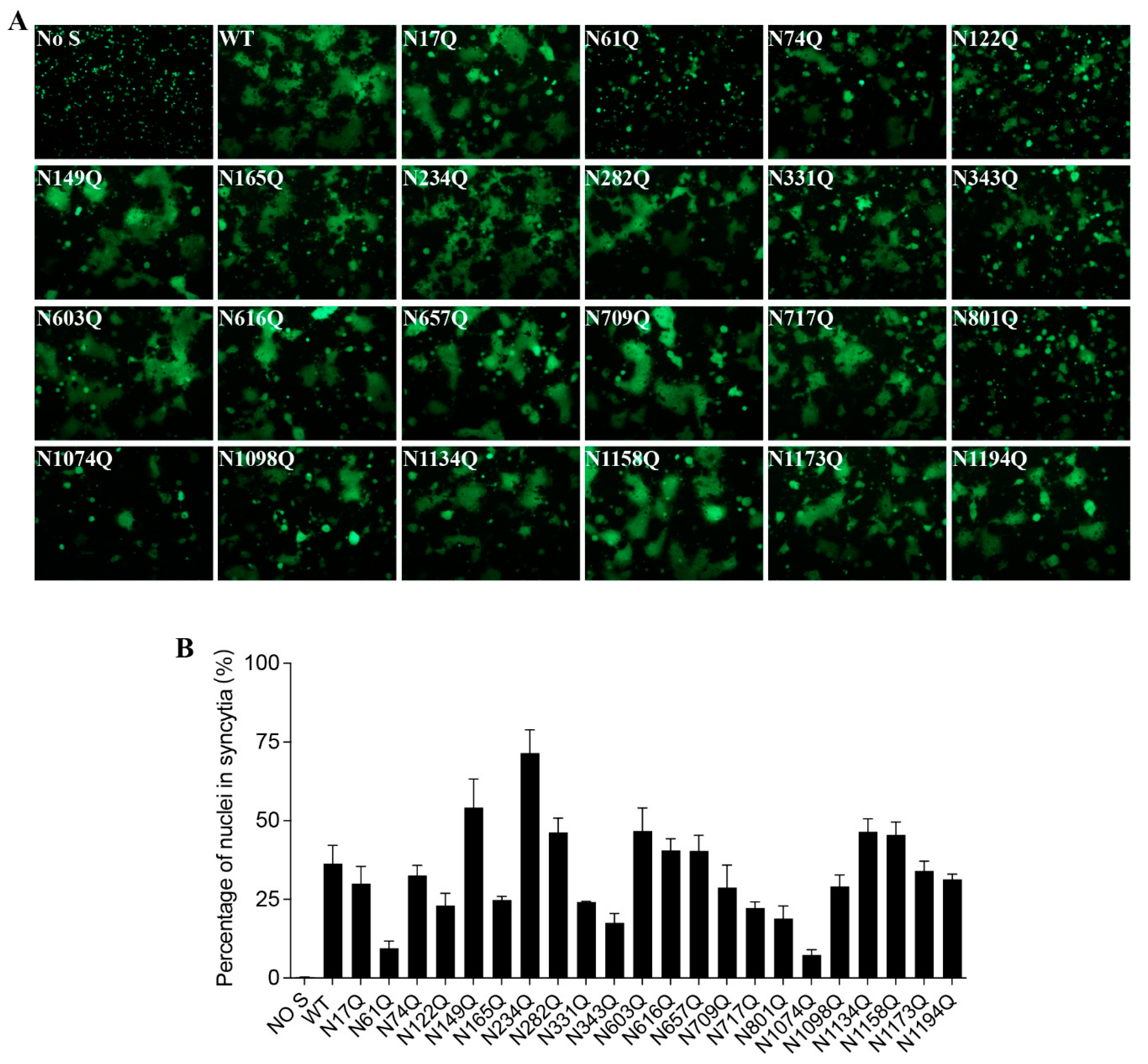
To investigate whether each individual N-glycosite affects S protein-mediated membrane fusion, we measured the levels of mutant S protein-mediated cell-cell fusion. Although most mutants induced comparable cell-cell fusion with the wild-type S protein, mutants N61Q, N122Q, N165Q, N331Q, N343Q, N801Q, and N1074Q-mediated cell-cell fusion were significantly impaired, consistent with the notably lower cell surface expression (
Figure 3A,B).
3.4. Effect of SARS-CoV-2 S protein N-glycans on S protein-mediated viral entry
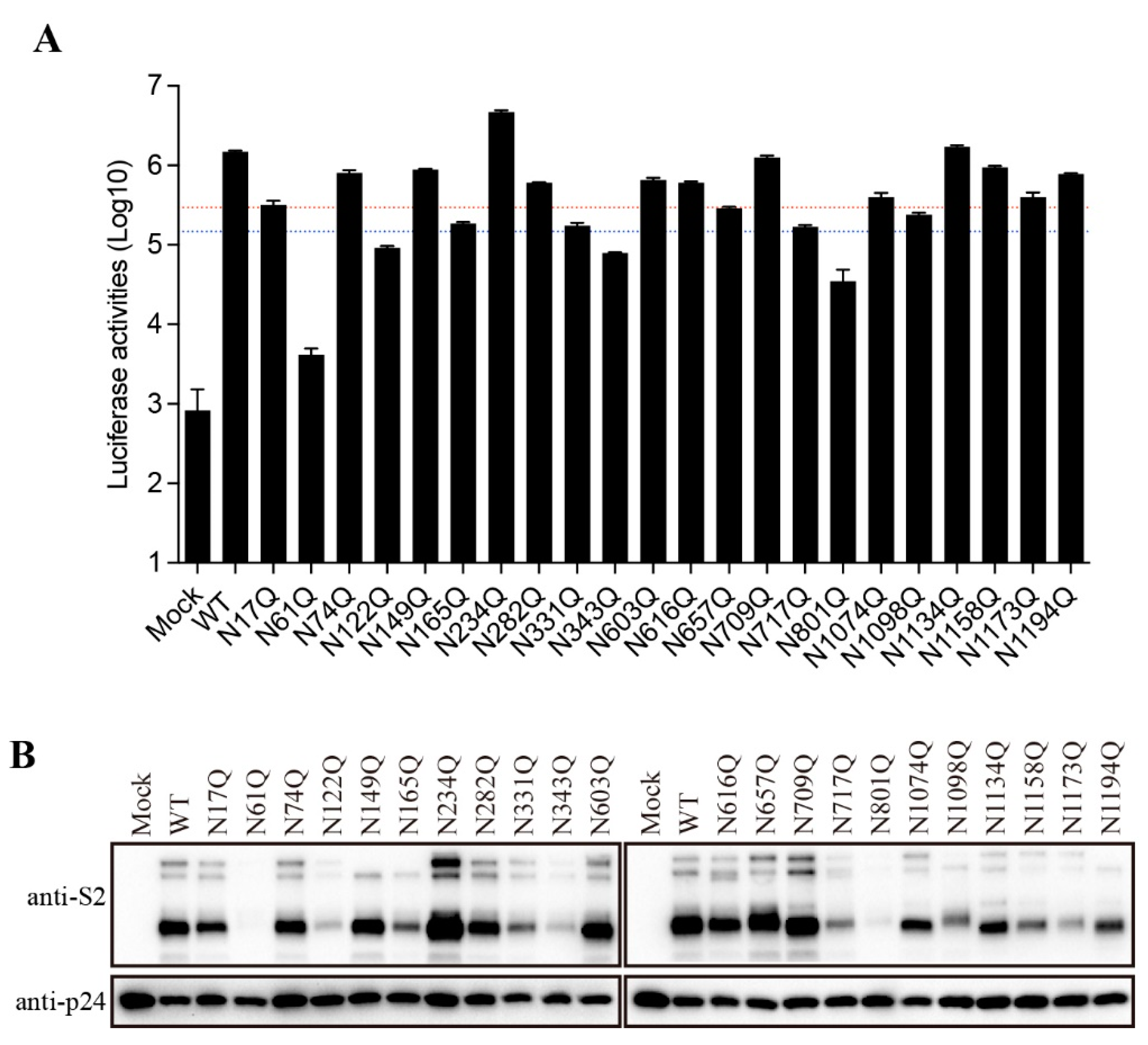
To further characterize the role of individual N-glycosites of the SARS-CoV-2 S protein in S-mediated membrane fusion, we used a lentiviral pseudotype system to evaluate mutant S protein-mediated viral entry. Majority of mutant S proteins showed reduction in transduction efficiency at various degree (
Figure 4A). Compared to WT S protein, the transduction efficiencies of N61Q, N122Q, N343Q, and N801Q were reduced by more than 10-fold, whereas N165Q, N331Q,N657Q, N717Q, and N1098Q pseudoviruses show more than 5-fold but less than 10-fold reduction in infectivity. Overall, the transduction efficiency of mutant S proteins is largely in agreement with the levels of S protein incorporation into pseudovirions (
Figure 4B). N61Q, N122Q, N165Q, N331Q, N343Q, N717Q, N801Q, N1098Q, and N1173Q showed marked reduction in S protein incorporation into pseudovirions at various degree (
Figure 4B), consistent to their relative low expression in cell lysates (
Figure 2B).
3.5. Removal of N-glycans at N1074 reduced the stability of SARS-CoV-2 S protein and induced receptor-independent cell-cell fusion in HEK293T cells
Surprisingly, when HEK293T cells were transfected with N1074Q, significant number of syncytia were observed even without expression of exogenous ACE2 (
Figure 5A). To further determine whether removal of N-glycans at N1074 was responsible for the syncytium formation in HEK293T cells, we also substituted the threonine residue at position 1076 with an alanine residue to abolish the N-link glycosylation sequon around N1074. The mutant T1076A show a phenotype similar to N1074Q, which further confirmed that the loss of N-linked glycosylation at position 1074 on the SARS-CoV-2 S protein caused syncytium formation in HEK293T cells (
Figure 5A). To investigate whether mutants N1074Q and T1076A-induced syncytia formation in HEK293T cells is ACE2-dependent or not, these two mutants-induced syncytia formation were also evaluated in endogenous ACE2 knockout HEK293T cells. We knocked out endogenous ACE2 in HEK293T cells using the CRISPR-Cas9 system, and the knockout was confirmed by sequencing (
Figure 5B). Next, mutants N1074Q and T1076A were transiently expressed in endogenous ACE2 knockout HEK293T cells, both mutants also induced syncytia formation in endogenous ACE2 knockout HEK293T cells. Thus, removal of N-glycans at N1074 of SARS-CoV-2 induced receptor-independent syncytia formation in HEK293T cells.
We hypothesized that mutants N1074Q and T1076A might loosen association between S1 subunit with the rest of S protein, resulting in an increase of S1 shedding and spontaneously conformational changes leading to syncytium formation. To test our hypothesis, we determined the amount of S1 in the supernatants of HEK293T cells transiently expressing either the wild-type or mutant S proteins. Consistent to our hypothesis, both N1074Q and T1076A mutants showed higher amount of S1 shedding than WT S proteins (
Figure 5C). Collectively, these data indicate that the glycans at N1074 on SARS-CoV-2 S protein is important for maintaining the stability of S protein, sheds the S1 subunit, and triggers cell-cell fusion in HEK293T cells.
3.6. Removal of the furin-cleavage site from the SARS-CoV-2 S protein compensated for the stability of mutant N1074Q
The SARS-CoV-2 S protein harbors a furin-cleavage site at the S1/S2 boundary, which facilitates entry of SARS-CoV-2 into human respiratory cells. Removal of the furin-cleavage site significantly increases the overall stability of the S protein [
24,
25,
26]. Next, we determine whether removal of the furin-cleavage site might stabilize the N1074Q S proteins. Consistent with previous reports [
22], deletion of the furin-cleavage site reduced the cleavage of the S protein, as evidenced by the absence of the cleaved form of the S protein in the WT S without furin-cleavage site (WT-delFurin) and N1074Q mutant S without furin-cleavage site (N1074Q-delFurin) expressing cells (
Figure 6A). As shown in
Figure 6B, removal of the furin-cleavage site significantly stabilized the S protein and N1074Q-delFurin only gave minimal amount of syncytia formation in HEK293T cells, compared to extensive syncytia observed in N1074Q. As a control, receptor-dependent cell-cell fusion assay was also performed. To avoid the effect of syncytium formation in HEK293T cells expressing mutant S proteins on receptor-dependent cell-cell fusion assay, we performed the assay at 30 h post-transfection, when no obvious syncytia observed in HEK293T cells expressing the mutant N1074. We observed that all mutant S proteins induced similar levels of syncytia to WT in the presence of trypsin, indicating that removal of the furin cleavage site almost has no effect on overall function of S protein (
Figure 6C).
3.7. N-glycosylation of S2 subunit contributes to SARS-CoV S protein stability
There are nine N-linked glycosylation sites in the SARS-CoV-2 S2 subunit that are also conserved in SARS-CoV S2 (
Figure 7A). Then, we decided to determine whether these N-glycans in the SARS-CoV S2 subunit might have similar function. We mutated each of nine N-linked glycosylation sites in the SARS-CoV S2 subunit, and the expression levels of mutant S proteins were analyzed by western blot. All mutants were expressed as well as WT S protein (
Figure 7B). Strikingly, the N1056Q mutation in the SARS-CoV S protein, corresponding to the N1074Q mutation in the SARS-CoV-2 S protein, did not induce marked syncytia in HEK293T cells (
Figure 7C). In contrast, the mutants N699Q and N1080Q triggered extensive cell-cell fusion. The N1176Q mutant also induced minor syncytia in HEK293T cells at 40 h post-transfection. We further investigated whether N699Q and N1080Q mutants-induced syncytia formation were ACE2-dependent or not. Notably, syncytium formation induced by both mutants in HEK293T/hACE2-KO cells was comparable to that induced in HEK293T cells (
Figure 7D). Together, these results indicate that N-glycans on the S2 subunit are also involved in the stability of the SARS-CoV S protein.
4. Discussion
N-linked glycosylation is one of common glycosylation modifications found in enveloped viruses and contributes to proper protein structure and function. Similar to other type I viral fusion proteins, the SARS-CoV-2 S protein is heavily glycosylated [
8,
9]. In this study, we showed that N-linked glycosylation at residue N1074 of SARS-CoV-2 S2 subunit is crucial for S protein stability. Removal of the N-glycans at position N1074 of SARS-CoV-2 S protein by either N1074Q or T1076A mutation triggered an RIS formation in HEK293T/hACE2-KO cells. We previously reported RIS of the S protein of mouse hepatitis virus (MHV) A59 critical for virus fitness [
27]. However, MHV-A59 S-induced RIS was pH-dependent [
27], whereas the SARS-CoV-2 N1074Q or T1076A mutant S protein-mediated RIS was not pH-dependent (data not shown).
The SARS-CoV-2 D614G variant is an early human-adapted SARS-CoV-2 variant that rapidly becomes the dominant strain worldwide. The D614G mutation in S protein stabilized the S1/S2 complex and inhibited S1 shedding, suggesting modulation of S protein stability is critical in SARS-CoV-2 human adaptation [
27,
28]. N-glycans of viral fusion glycoproteins play a critical role in maintaining the conformation state of viral glycoproteins, therefore interfering viral infectivity and pathogenicity [
29,
30]. Avian influenza H5N6 promotes mammalian adaptation by acquiring an N-linked glycoprotein at the head of the HA protein that increases the stability of the HA trimeric structure [
31]. N-glycans at residues N165 and N234 of SARS-CoV-2 S protein modulates the conformational dynamics of the SARS-CoV-2 S protein by stabilizing the RBD in the ‘up’ conformation [
15,
16]. Here, we found that the glycosylation sites at position N1074 on the SARS-CoV-2 S protein and at positions N699 and N1080 on the SARS-CoV S protein are of great importance in maintaining the stability of the prefusion S trimers. The glycosylation site, N1074, of the SARS-CoV-2 S protein is located in the connector domain between heptad repeat 2 and the central helix [
9,
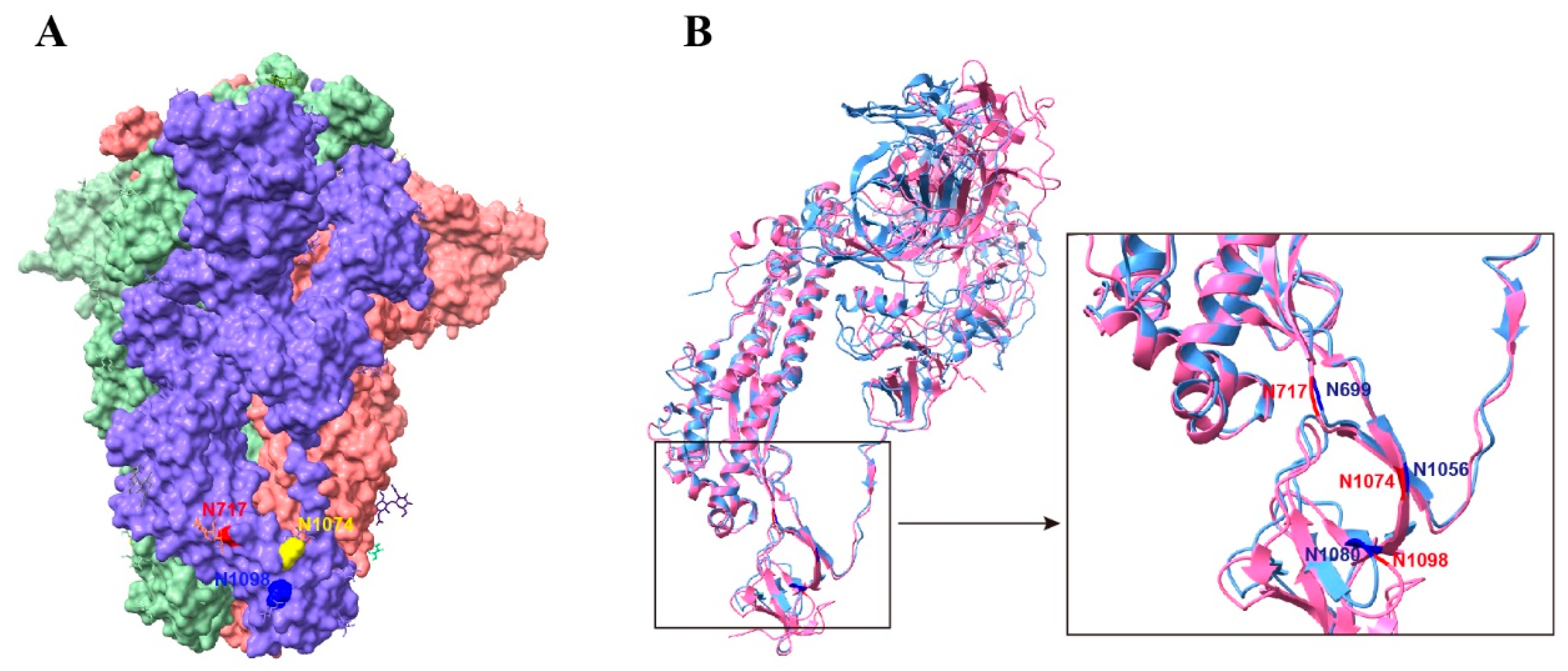
32]. In the prefusion S trimer, residue N1074 was located at the bottom of a cave formed by two adjacent S monomers (
Figure 8A). It is possible that the bulky hydrophilic carbohydrate chains at position N1074 interact with the side chains of the surrounding residues to fill the groove and aid in stabilizing the S trimer. Although the N-linked glycosylation sites of the S2 subunit are conserved between SARS-CoV-2 and SARS-CoV (
Figure 8B), the corresponding mutant in SARS-CoV S protein, the N1056Q mutant, had no obvious effect on the stability of the S trimer. However, N-glycans of two spatially close glycosylation sites N699 and N1080 are vital for the stability of the SARS-CoV S timer. Therefore, oligosaccharides on these glycosylation sites may function together to maintain the stability of the prefusion S trimer, although some sites may play a more important role. Further functional and structural analyses are required to confirm this hypothesis.
The pre-fusion state of the trimeric S protein is metastable and transforms into a stable post-fusion state upon receptor binding and protease cleavage or antibody binding, while mediating membrane fusion [
3,
6]. The presence of the furin-cleavage site in the S protein of SARS-CoV-2 facilitates the protease cleavage around S2’ sites and conformational changes in the S trimer during membrane fusion, but at the cost of protein stability [
25]. Although deletion of the furin-cleavage site on the mutant N1074Q did not affect S-mediated receptor-dependent membrane fusion, it no longer triggered cell-cell fusion in HEK293T cells, indicating that elimination of the furin-cleavage site of the S protein compensates for the instability of the mutant N1074Q. Unlike the SARS-CoV-2 S protein, the S protein of SARS-CoV does not harbor a furin-cleavage site at the boundary of the S1 and S2 subunits, but removal of N-glycans at positions N699 or N1080 of the SARS-CoV S protein still induced RIS formation in HEK293T cells, indicating the presence of subtle difference between two S proteins despite of high similarity in S2 subunits.
N-linked glycosylation of nascent polypeptides is initiated in the ER cotranslationally, which is thought to be critical for proper folding, and further modifications to glycans occur in the Golgi network [
33]. N-glycans at N61 and N801, structurally close to the S1/S2 and S2’ proteolytic sites, respectively, have been shown to regulate S protein maturation and viral infectivity [
34]. We found that removal of 9 out of the 22 N-linked glycosylation sites (N61, N122, N165, N331, N343, N717, N801Q, N1098, and N1173) impaired the cleavage of the S protein, which may result from improper protein folding and impaired trafficking, thus reducing S-mediated membrane fusion. This is in contrast to the SARS-CoV S protein, in which individual mutations of all putative N-linked glycosylation sites on the S1 subunit had no major effect on its expression [
14]. Therefore, N-glycans on the SARS-CoV-2 S protein are more crucial for protein maturation than those on the SARS-CoV S protein. A recent study has shown that N-linked glycosylation has a lesser effect on S protein expression in the SARS-CoV-2 Omicron variants compared to the D614G S protein [
35].
As a major surface protein, the coronavirus S protein is the focus of vaccine development. The ectodomain S trimer from MERS-CoV has been shown to elicit more potent neutralizing antibodies than the S1 monomer or RBD [
36]. However, the prefusion state of the S trimer from SARS-CoV-2 is unstable, as evidenced by the presence of the post-fusion state in SARS-CoV-2 intact virions and β-propiolactone-inactivated SARS-CoV-2 virions [
5,
37]. Cryo-EM structures of coronavirus S glycoproteins have revealed great differences between the pre-and post-fusion states, indicating that postfusion S immunogens may not efficiently induce neutralizing antibodies [
38]. Therefore, many studies have been conducted to stabilize the prefusion SARS-CoV-2 S timer, including removal of the furin-cleavage site, introduction of two consecutive proline substitutions (K986P and V987P) to the turn between heptad repeat 1 and the central helix, and introduction of six proline substitutions (F817P, A892P, A899P, A942P, K986P, and V987P) to the S2 subunit [
25,
39,
40]. Our study demonstrates that N-glycosylation on SARS-CoV-2 S2 subunit is involved in maintaining the stability of S trimer. The introduction of N-glycosylation to SARS-CoV-2 S2 subunit may be a novel approach to improve the stability of the prefusion SARS-CoV-2 S timer, which needs to be confirmed by further studies.
Author Contributions
Conceptualization, F.Z. and X.O.; methodology, F.Z., Y.Z. and T.C.; validation, F.Z., Y.Z. and T.C..; formal analysis, F.Z. and X.O.; investigation, F.Z., Y.Z., T.C., Y.C. and Z.M.; writing—original draft preparation, F.Z. and X.O.; writing—review and editing, Z.Q.; supervision, Z.Q. and X.O.; funding acquisition, Z.Q. and X.O. All authors have read and agreed to the published version of the manuscript.
Figure 1.
α-glycosidase inhibitors impaired SARS-CoV-2 S-mediated membrane fusion. (A) HEK293T cells transfected with plasmids expressing SARS-CoV-2 S protein were treated with 100 µg/mL castanospermine or NB-DNJ for 40 h and then the expression levels of S protein in cell lysates were detected using western blot analysis with antibodies against the S2 subunit of SARS-CoV-2 S protein. β-actin was used as a loading control. (B) HEK293T cells expressing SARS-CoV-2 S protein and GFP were treated with α-glycosidase inhibitors for 40 h and then trypsinized at 40 h post-transfection and co-cultured with HEK293/hACE2 for another 4 h, and then imaged using a fluorescence microscope. (C) Western blot analysis of SARS-CoV-2 S-pseudotyped lentiviral particles produced in HEK293T cells in the absence or presence of α-glycosidase inhibitors. p24 was used as a loading control. (D) The transduction efficiency of SARS-CoV-2 S-pseudotyped lentiviral particles produced in HEK293T cells in the absence or presence of α-glycosidase inhibitors. Unpaired two-tailed Student’s t-test was used for statistical analysis, **p<0.01.
Figure 1.
α-glycosidase inhibitors impaired SARS-CoV-2 S-mediated membrane fusion. (A) HEK293T cells transfected with plasmids expressing SARS-CoV-2 S protein were treated with 100 µg/mL castanospermine or NB-DNJ for 40 h and then the expression levels of S protein in cell lysates were detected using western blot analysis with antibodies against the S2 subunit of SARS-CoV-2 S protein. β-actin was used as a loading control. (B) HEK293T cells expressing SARS-CoV-2 S protein and GFP were treated with α-glycosidase inhibitors for 40 h and then trypsinized at 40 h post-transfection and co-cultured with HEK293/hACE2 for another 4 h, and then imaged using a fluorescence microscope. (C) Western blot analysis of SARS-CoV-2 S-pseudotyped lentiviral particles produced in HEK293T cells in the absence or presence of α-glycosidase inhibitors. p24 was used as a loading control. (D) The transduction efficiency of SARS-CoV-2 S-pseudotyped lentiviral particles produced in HEK293T cells in the absence or presence of α-glycosidase inhibitors. Unpaired two-tailed Student’s t-test was used for statistical analysis, **p<0.01.
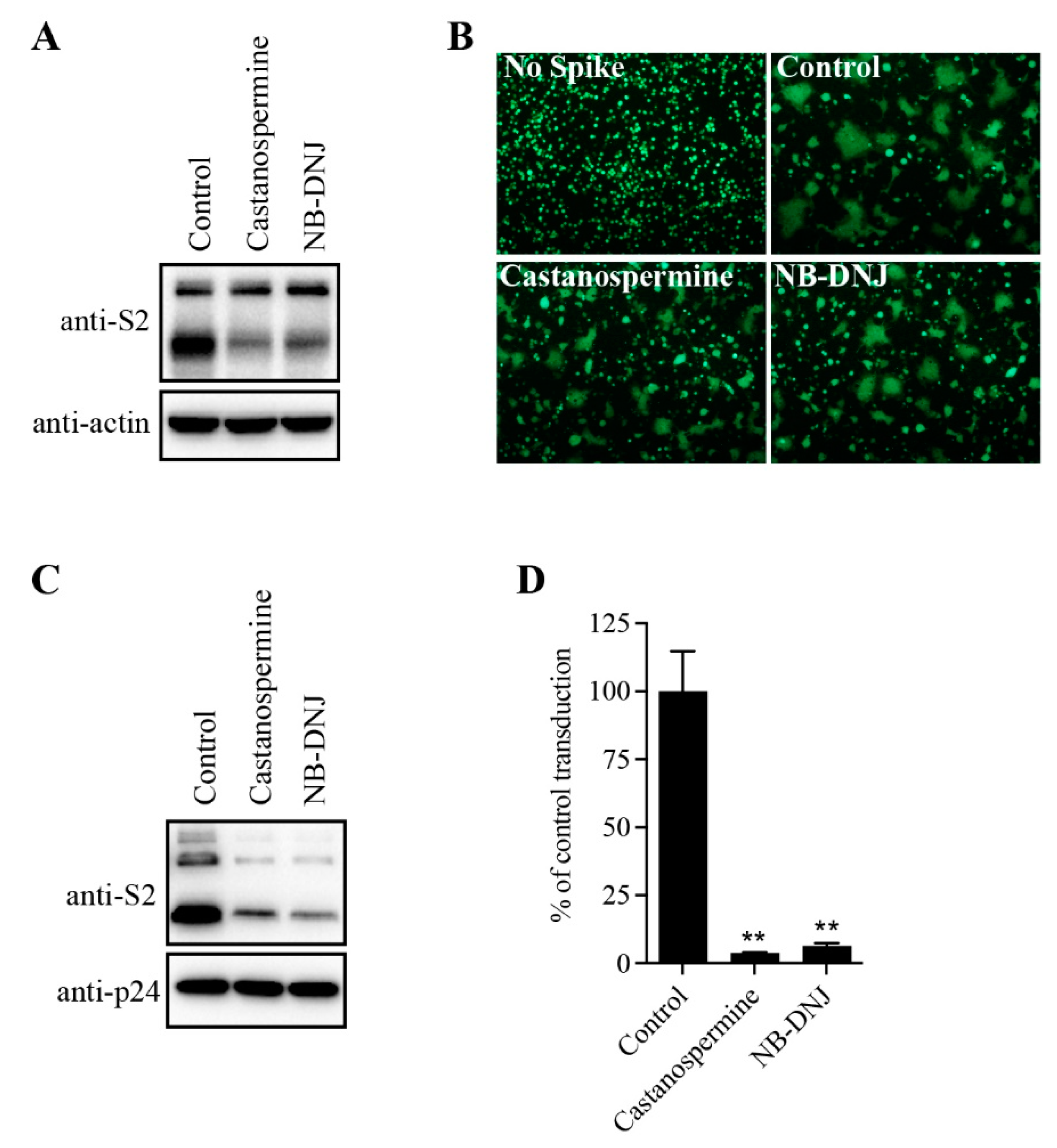
Figure 2.
The expression levels and receptor binding capacity of the N-glycosylation site mutants of the SARS-CoV-2 S protein. (A) Schematic representation of N-linked glycosylation sites in the SARS-CoV-2 S protein. (B) HEK293T cells were transfected with plasmids encoding wild-type S protein or N-glycosylation site mutants. After 40 h of transfection, the cells were lysed and analyzed using western blot analysis with antibodies against the SARS-CoV-2 S2 subunit. β-actin was used as a loading control. (C) Cell surface expression of wild-type S protein and different N-glycosylation site mutants using flow cytometry assay. (D) HEK293T cells were transfected with plasmids encoding wild-type S protein or different N-glycosylation mutants. After 40 h of transfection, the cells were detached with 1 mM EDTA, incubated with soluble hACE2 for 1 h on ice, incubated with primary antibodies against hACE2, and then incubated with FITC-conjugated secondary antibodies. The cells were then subjected to flow cytometry.
Figure 2.
The expression levels and receptor binding capacity of the N-glycosylation site mutants of the SARS-CoV-2 S protein. (A) Schematic representation of N-linked glycosylation sites in the SARS-CoV-2 S protein. (B) HEK293T cells were transfected with plasmids encoding wild-type S protein or N-glycosylation site mutants. After 40 h of transfection, the cells were lysed and analyzed using western blot analysis with antibodies against the SARS-CoV-2 S2 subunit. β-actin was used as a loading control. (C) Cell surface expression of wild-type S protein and different N-glycosylation site mutants using flow cytometry assay. (D) HEK293T cells were transfected with plasmids encoding wild-type S protein or different N-glycosylation mutants. After 40 h of transfection, the cells were detached with 1 mM EDTA, incubated with soluble hACE2 for 1 h on ice, incubated with primary antibodies against hACE2, and then incubated with FITC-conjugated secondary antibodies. The cells were then subjected to flow cytometry.
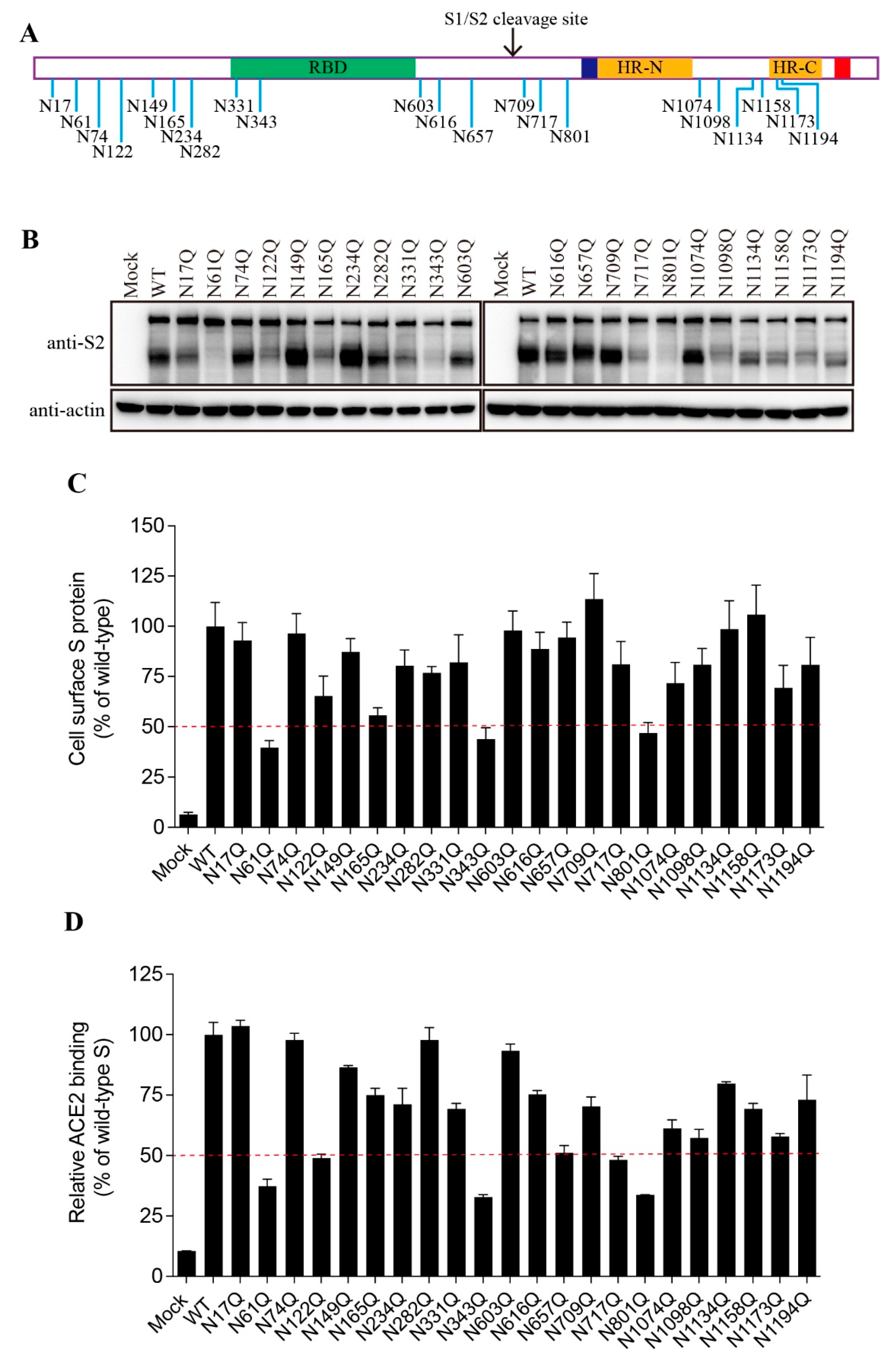
Figure 3.
Cell-cell fusion capacity of N-glycosylation mutants of the SARS-CoV-2 S protein. (A) HEK293T cells were co-transfected with plasmids encoding wild-type or different N-glycosylation site mutant S proteins and plasmids encoding GFP. After 40 h of transfection, the cells were trypsinized and co-cultured with HEK293/hACE2 cells for an additional 4 h and then imaged using a fluorescence microscope. (B) Quantification of cell-cell fusion. The total number of nuclei and the number of nuclei in fused cells for each image were counted. The fusion efficiency was calculated as the number of nuclei in syncytia/total number of nuclei ×100.
Figure 3.
Cell-cell fusion capacity of N-glycosylation mutants of the SARS-CoV-2 S protein. (A) HEK293T cells were co-transfected with plasmids encoding wild-type or different N-glycosylation site mutant S proteins and plasmids encoding GFP. After 40 h of transfection, the cells were trypsinized and co-cultured with HEK293/hACE2 cells for an additional 4 h and then imaged using a fluorescence microscope. (B) Quantification of cell-cell fusion. The total number of nuclei and the number of nuclei in fused cells for each image were counted. The fusion efficiency was calculated as the number of nuclei in syncytia/total number of nuclei ×100.
Figure 4.
The role of N-glycans on the SARS-CoV-2 S protein in viral entry. (A) The transduction efficiency of wild-type or N-glycosylation mutant S-pseudotyped lentiviral particles. The red and blue dashed lines represent the 1/5 and 1/10 of wild-type transduction values, respectively. (B) Western blot analysis of wild-type or N-glycosylation mutant S-pseudotyped lentiviral particles.
Figure 4.
The role of N-glycans on the SARS-CoV-2 S protein in viral entry. (A) The transduction efficiency of wild-type or N-glycosylation mutant S-pseudotyped lentiviral particles. The red and blue dashed lines represent the 1/5 and 1/10 of wild-type transduction values, respectively. (B) Western blot analysis of wild-type or N-glycosylation mutant S-pseudotyped lentiviral particles.
Figure 5.
Removal of N-glycans on the SARS-CoV-2 S protein destabilized the S protein. (A) HEK293T cells were co-transfected with plasmids encoding EGFP and plasmids encoding wild-type SARS-CoV-2 S protein, mutant N1074Q, or mutant T1076A. After 40 h of transfection, the cells were visualized using a fluorescence microscope. (B) Endogenous ACE2 knockout HEK293T cells were confirmed by DNA sequencing. A nucleotide insertion in the ACE2 gene causes a frameshift and leads to premature termination of ACE2 translation. (C) The endogenous ACE2 knockout HEK293T cells were co-transfected with plasmids encoding EGFP and plasmids encoding wild-type SARS-CoV-2 S protein, mutant N1074Q, or mutant T1076A. After 40 h of transfection, the cells were visualized using a fluorescence microscope. (D-E) HEK293T cells were transfected with plasmids encoding wild-type SARS-CoV-2 S protein, mutant N1074Q or mutant T1076A. After 40 h of transfection, the S protein in cell lysates (D) and supernatants (E) were analyzed using western blot analysis with antibodies against S2 subunit and RBD subunit of SARS-CoV-2 S protein, respectively.
Figure 5.
Removal of N-glycans on the SARS-CoV-2 S protein destabilized the S protein. (A) HEK293T cells were co-transfected with plasmids encoding EGFP and plasmids encoding wild-type SARS-CoV-2 S protein, mutant N1074Q, or mutant T1076A. After 40 h of transfection, the cells were visualized using a fluorescence microscope. (B) Endogenous ACE2 knockout HEK293T cells were confirmed by DNA sequencing. A nucleotide insertion in the ACE2 gene causes a frameshift and leads to premature termination of ACE2 translation. (C) The endogenous ACE2 knockout HEK293T cells were co-transfected with plasmids encoding EGFP and plasmids encoding wild-type SARS-CoV-2 S protein, mutant N1074Q, or mutant T1076A. After 40 h of transfection, the cells were visualized using a fluorescence microscope. (D-E) HEK293T cells were transfected with plasmids encoding wild-type SARS-CoV-2 S protein, mutant N1074Q or mutant T1076A. After 40 h of transfection, the S protein in cell lysates (D) and supernatants (E) were analyzed using western blot analysis with antibodies against S2 subunit and RBD subunit of SARS-CoV-2 S protein, respectively.
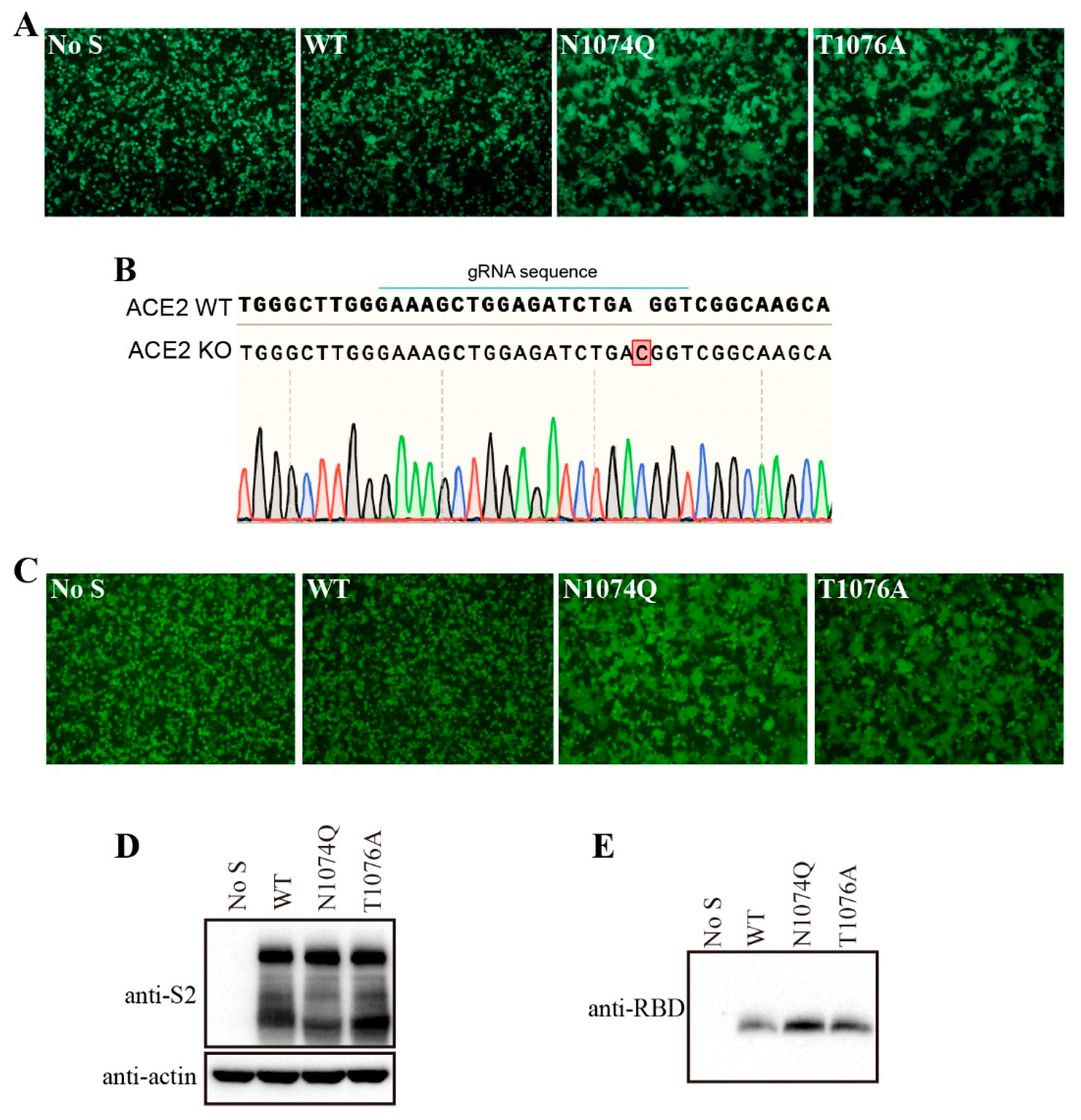
Figure 6.
Removal of the furin-cleavage site compensated the stability of the N-glycosylation mutant N1074Q. (A) HEK293T cells were transfected with plasmids encoding wild-type SARS-CoV-2 S protein, furin-cleavage site mutant wild-type S protein (WT-delFurin), mutant N1074Q, or N1074Q furin-cleavage site mutant (N1074Q-delFurin). After 40 h of transfection, the cells were lysed and analyzed using western blot analysis with antibodies against the SARS-CoV-2 S2 subunit. β-actin was used as a loading control. (B) HEK293T cells were co-transfected with plasmids encoding GFP and plasmids encoding wild-type SARS-CoV-2 S protein, mutant WT-delFurin, mutant N1074Q, or mutant N1074Q-delFurin. After 40 h of transfection, the cells were visualized using a fluorescence microscope. (C) The cell-cell fusion assay was performed to assess the fusogenicity of wild-type SARS-CoV-2 S protein, mutant WT-delFurin, mutant N1074Q, and mutant N1074Q-delFurin. HEK293T cells were co-transfected with plasmids encoding wild-type or mutant S proteins and plasmids encoding GFP. After 30 h of transfection, the cells were trypsinized and co-cultured with HEK293/hACE2 cells for an additional 4 h and then imaged using a fluorescence microscope.
Figure 6.
Removal of the furin-cleavage site compensated the stability of the N-glycosylation mutant N1074Q. (A) HEK293T cells were transfected with plasmids encoding wild-type SARS-CoV-2 S protein, furin-cleavage site mutant wild-type S protein (WT-delFurin), mutant N1074Q, or N1074Q furin-cleavage site mutant (N1074Q-delFurin). After 40 h of transfection, the cells were lysed and analyzed using western blot analysis with antibodies against the SARS-CoV-2 S2 subunit. β-actin was used as a loading control. (B) HEK293T cells were co-transfected with plasmids encoding GFP and plasmids encoding wild-type SARS-CoV-2 S protein, mutant WT-delFurin, mutant N1074Q, or mutant N1074Q-delFurin. After 40 h of transfection, the cells were visualized using a fluorescence microscope. (C) The cell-cell fusion assay was performed to assess the fusogenicity of wild-type SARS-CoV-2 S protein, mutant WT-delFurin, mutant N1074Q, and mutant N1074Q-delFurin. HEK293T cells were co-transfected with plasmids encoding wild-type or mutant S proteins and plasmids encoding GFP. After 30 h of transfection, the cells were trypsinized and co-cultured with HEK293/hACE2 cells for an additional 4 h and then imaged using a fluorescence microscope.
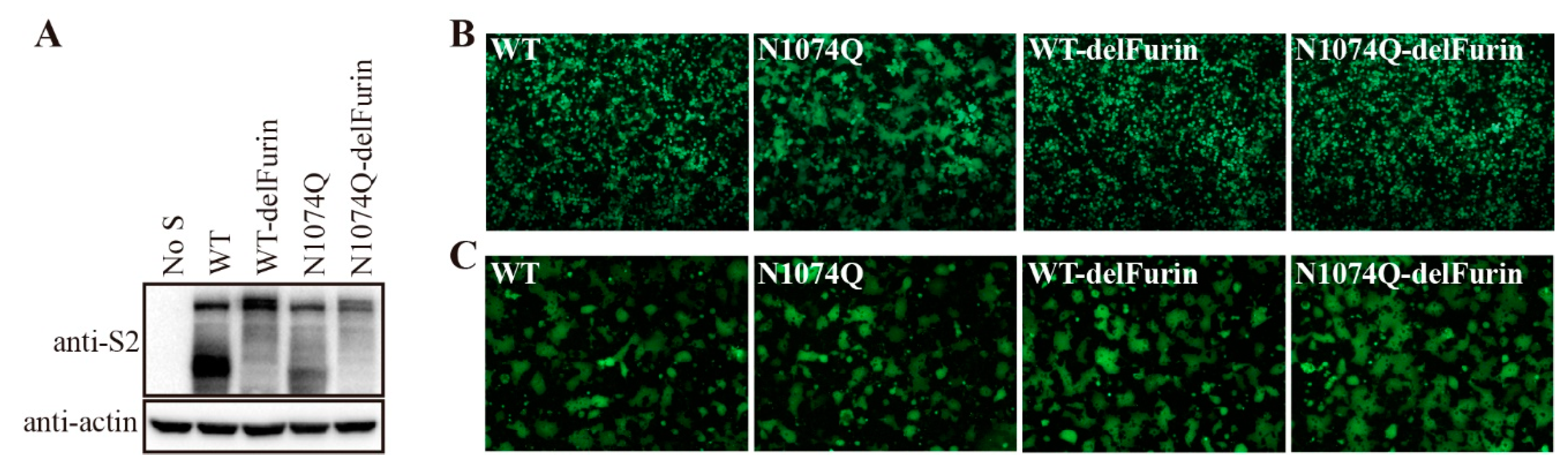
Figure 7.
Removal of N-glycans on the S2 subunit of SARS-CoV S protein destabilized the S protein. (A) Amino acid sequence alignment of S2 subunits from SARS-CoV and SARS-CoV-2, red asterisks indicate N-linked glycosylation sites. (B) HEK293T cells were transfected with an empty vector or plasmids encoding the wild-type SARS-CoV S protein or the different glycosylation mutants. After 40 h of transfection, the cells were lysed and analyzed with polyclonal rabbit anti-SARS S1 antibodies T62. β-actin was used as a loading control. (C) HEK293T cells were co-transfected with plasmids encoding EGFP and plasmids encoding wild-type SARS-CoV S protein or the different glycosylation mutants. After 40 h of transfection, the cells were visualized using a fluorescence microscope. (D) HEK293T/ACE2-KO cells were co-transfected with plasmids encoding EGFP and plasmids encoding wild-type SARS-CoV S protein, mutant N699Q, or mutant N1080Q. After 40 h of transfection, the cells were visualized using a fluorescence microscope.
Figure 7.
Removal of N-glycans on the S2 subunit of SARS-CoV S protein destabilized the S protein. (A) Amino acid sequence alignment of S2 subunits from SARS-CoV and SARS-CoV-2, red asterisks indicate N-linked glycosylation sites. (B) HEK293T cells were transfected with an empty vector or plasmids encoding the wild-type SARS-CoV S protein or the different glycosylation mutants. After 40 h of transfection, the cells were lysed and analyzed with polyclonal rabbit anti-SARS S1 antibodies T62. β-actin was used as a loading control. (C) HEK293T cells were co-transfected with plasmids encoding EGFP and plasmids encoding wild-type SARS-CoV S protein or the different glycosylation mutants. After 40 h of transfection, the cells were visualized using a fluorescence microscope. (D) HEK293T/ACE2-KO cells were co-transfected with plasmids encoding EGFP and plasmids encoding wild-type SARS-CoV S protein, mutant N699Q, or mutant N1080Q. After 40 h of transfection, the cells were visualized using a fluorescence microscope.
Figure 8.
Stabilizing glycosylation sites on the structure of SARS-CoV-2 S protein. (A) Prefusion structure of the trimeric SARS-CoV-2 S proteins with all RBDs in the closed state (PDB 7qus). (B) Structural comparison of the SARS-CoV-2 and SARS-CoV S monomers. SARS-CoV S protein PDB accession number: 5x58.
Figure 8.
Stabilizing glycosylation sites on the structure of SARS-CoV-2 S protein. (A) Prefusion structure of the trimeric SARS-CoV-2 S proteins with all RBDs in the closed state (PDB 7qus). (B) Structural comparison of the SARS-CoV-2 and SARS-CoV S monomers. SARS-CoV S protein PDB accession number: 5x58.