Submitted:
19 December 2023
Posted:
20 December 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Ethical Aspects, target population and sampling
2.2. Capture of bats
2.3. Isolation and characterization of samples
2.4. Phenotypic antimicrobial sensitivity tests
2.5. Bacterial multiresistance index
2.6. Polymerase chain reaction (PCR): Staphylococcus aureus study
2.7. MecA gene determination
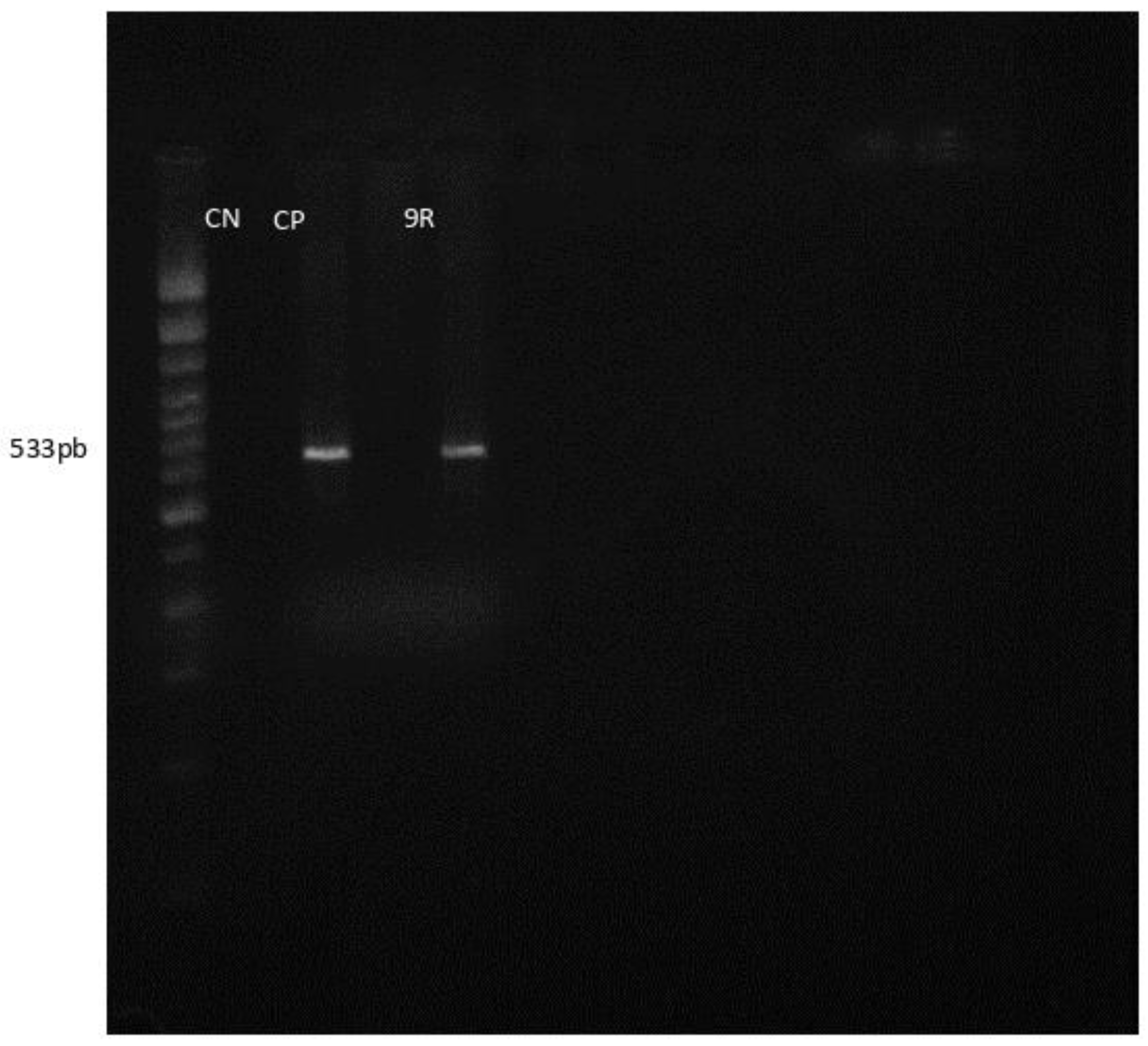
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pourramezan, N.; Ohadian Moghadam, S.; Pourmand, M. R. Methicillin-resistant Staphylococcus aureus tracking spread among health-care workers and hospitalized patients in critical wards at a university hospital, Tehran, Iran. New Microbes New Infect 2019, 27, 29–35. [CrossRef]
- Van Duijkeren, E.; Box, A.T.A.; Heck, M.E.O.C.; Wannet, W.J.B.; Fluit, A.C. Methicillin-resistant staphylococci isolated from animals. Vet. Microbiol. 2004, 103, 91–97. [CrossRef]
- Mrochen, D.M.; Grumann, D.; Schulz, D.; Gumz, J.; Trübe, P.; Pritchett-Corning, K.; Johnson, S.; Nicklas, W.; Kirsch, P.; Martelet, K. van den Brandt, J.; Berg, S.; Bröker, B.M.; Wiles, S.; Holtfreter, S. Global spread of mouse-adapted Staphylococcus aureus lineages CC1, CC15 , and CC88 among mouse breeding facilities. Int. J. Med. Microbiol. 2018, 308, 598–606. [CrossRef]
- Blodkamp, S.; Kadlec, K.; Gutsmann, T.; Naim, H.Y.; von Köckritz-Blickwede, M.; Schwarz, S. In vitro activity of human and animal cathelicidins against livestock-associated methicillin-resistant Staphylococcus aureus. Vet. Microbiol. 2015, 194, 107-111. [CrossRef]
- Akobi, B.; Aboderin, O.; Sasaki, T.; Shittu, A. Characterization of Staphylococcus aureus isolates from faecal samples of the straw-coloured fruit bat (Eidolon helvum) in Obafemi Awolowo University (OAU), Nigeria. BMC Microbiol. 2012, 12, 279. [CrossRef]
- Walther, B.; Wieler, L.H.; Friedrich, A.W.; Hanssen, A.M.; Kohn, B.; Brunnberg, L.; Lübke-Becker, A. Methicillin-resistant Staphylococcus aureus (MRSA) isolated from small and exotic animals at a university hospital during routine microbiological examinations. Vet. Microbiol. 2008, 127, 171-178. [CrossRef]
- Russo, D.; Ancillotto, L. Sensitivity of bats to urbanization: A review. Mamm. Biol. 2015, 80, 205-212Buzón-Durán, L.; Capita, R.; Alonso-Calleja, C. Antibiotic susceptibility of methicillin-resistant staphylococci (MRS) of food origin: A comparison of agar disc diffusion method and a commercially available miniaturized test. Food Microbiol. 2018, 72, 220–224.
- Vandžurová, A.; Bačkor, P.; Javorský, P.; Pristaš, P. Staphylococcus nepalensis in the guano of bats (Mammalia). Vet. Microbiol. 2013, 164, 116-121. [CrossRef]
- Sens-Junior, H.; Trindade, W.A.; Oliveira, A.F.; Zaniolo, M.M.; Serenini, G.F.; Araujo-Ceranto, J.B.; Gonçalves, D.D.; Germano, R.M. Bacterial resistance in bats from the Phyllostomidae family and its relationship with unique health. Pesqui Vet Bras 2018, 38, 1207–1216. [CrossRef]
- Olatimehin, A.; Shittu, A.O.; Onwugamba, F.C.; Mellmann, A.; Becker, K.; Schaumburg, F. Staphylococcus aureus Complex in the Straw-Colored Fruit Bat ( Eidolon helvum ) in Nigeria. Front Microbiol 2018, 9, 1–7. [CrossRef]
- Liao, X.; Cullen, P.J.; Liu, D.; Muhammad, A.I.; Chen, S.; Ye, X.; Wang, J.; Ding, T. Combating Staphylococcus aureus and its methicillin resistance gene (mecA) with cold plasma. Science of the Total Environment 2018, 645, 1287–1295. [CrossRef]
- Alrabiah, K.; Al Alola, S.; Al Banyan, E.; Al Shaalan, M.; Al Johani, S. Characteristics and risk factors of hospital acquired–methicillin-resistant Staphylococcus aureus (HA-MRSA) infection of pediatric patients in a tertiary care hospital in Riyadh, Saudi Arabia. Int J Pediatr Adolesc Med 2016, 3, 71-77. [CrossRef]
- Shahbazian, J. H., Hahn, P. D., Ludwig, S., Ferguson, J., Baron, P., Christ, A., ... & Davis, M. F. (2017). Multidrug and mupirocin resistance in environmental methicillin-resistant Staphylococcus aureus (MRSA) isolates from homes of people diagnosed with community-onset MRSA infection. Applied and environmental microbiology, 83(22), e01369-17. [CrossRef]
- Kesharwani, A.K.; Mishra, J. Detection of β-lactamase and antibiotic susceptibility of clinical isolates of Staphylococcus aureus. Biocatal Agric Biotechnol 2019, 17, 720–725. [CrossRef]
- Greenhall, A. M.; Paradiso, J. L. Bats and bat banding. Bureau of Sport Fisheries and Wildlife Resource Publication 1968, 72, 47.
- Vieira, C. O. C. Ensaio monográfico sobre os quirópteros do Brasil. Arquivo de Zoologia do Estado de São Paulo 1942, 3, 219-471.
- Vizoto, L. D.; Taddei, V. A. Chave para determinação de quirópteros brasileiros. Boletim de Ciências 1973, 1, 1-72.
- Jones, J. K.; Carter, D. C. Annotated checklist, with keys to subfamilies and genera. In: Biology of bats of the new world family Phyllostomidae, part I. Spec. Publ. Mus. Tex. Tech Univ 1976, 107-38.
- Quinn, P. J.; Carter, M. E.; Markey, B.; Carter, G. R. Clinical Veterinary Microbiology, 2 nd ed.; Wolfe publishing : London, England, 1994.
- CLINICAL AND LABORATORY STANDARDS INSTITUTE (CLSI). 2018. M100–S127. Performance Standards for Antimicrobial Susceptibility Testing: 28th Informational Supplement. Clinical and Laboratory Standards Institute, Wayne, PA.
- Krumperman, P.H. Multiple Antibiotic Resistance Indexing of Escherichia coli to Identify High-Risk Sources of Fecal Contamination of Foodst. Appl. Environ. Microbiol 1983, 46, 165–170.
- Martineau, F.; Picard, F. J.; Roy, P. H.; Ouellette, M.; Bergeron, M. G. Species-specific and ubiquitous-DNA-based assays for rapid identification of Staphylococcus aureus. J. Clin. Microbiol 1998, 36, 618-623. [CrossRef]
- Murakami, K.; Minamide, W.; Wada, K.; Nakamura, E.; Teraoka, H.; Watanabe, S. Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. J. Clin. Microbiol 1991, 29, 2240-2244. [CrossRef]
- Reis, N.R.; Peracchi, A.L.; Pedro, W.A.; Lima, I.P. Mamíferos do Brasil, 2nd ed.; Universidade Estadual de Londrina: Londrina, Brazil, 2011; pp. 437.
- Ortêncio-Filho, H.; Reis, N.R.; Pinto, D.; Anderson, R.; Testa, D.A.; Marques, M.A. Levantamento dos morcegos (Chiroptera, Mammalia) do parque municipal do Cinturão Verde de cianorte. Chiroptera Neotropical 2005 , 11, 211–215.
- Leandro, N.; Angelis, D.E.; Domingo, B.L. Parque Municipal Cinturão Verde De Cianorte – Módulo Mandhuy E Os Principais Impactos Da Área De Entorno. Rev GEOMAE 2011, 2, 51–70.
- Haddock, J.K.; Threlfall, C.G.; Law, B.; Hochuli, D.F. Light pollution at the urban forest edge negatively impacts insectivorous bats. Biological Conservation 2019, 236, 17–28. [CrossRef]
- Chaverri, G.; Kunz, T. H. Ecological determinants of social systems: perspectives on the functional role of roosting ecology in the social behavior of tent-roosting bats. Advances in the Study of Behavior 2010, 275-318.
- Witte, W.; Cuny, C.; Klare, I.; Nübel, U.; Strommenger, B.; Werner, G. Emergence and spread of antibiotic-resistant Gram-positive bacterial pathogens. Int J Med Microbiol 2008, 298, 365–377. [CrossRef]
- Gomes, G. P. L. A.; Souza, A. C. S.; Leão-Vasconcelos, L. N.O; Melo Costa, D.; Alves, S. B.; Neves, H. C. C.; Pereira, M. S. Manual resuscitators in successive use in the same patient: reservoir of multi- and extensively resistant bacteria. J. Hosp. Infect 2017, 95, 87–90. [CrossRef]
- Pristas, P.; Vandz, A. Staphylococcus nepalensis in the guano of bats (Mammalia). Vet. Microbiol 2013, 164, 116-121.
- Nowakiewicz, A.; Ziółkowska, G.; Zięba, P.; Gnat, S.; Wojtanowicz-Markiewicz, K.; Trościańczyk, A. Comparative Immunology , Microbiology and Infectious Diseases Coagulase-positive Staphylococcus isolated from wildlife : Identification , molecular characterization and evaluation of resistance profiles with focus on a methicillin-resistant strain. Comp. Immunol. Microbiol. Infect. Dis 2016, 44, 21–28.
- Ben Yahia, H.; Chairat, S.; Hamdi, N.; Gharsa, H.; Sallem, R.B.; Ceballos, S.; Torres, C.; Slama, K. B.Antimicrobial resistance and genetic lineages of faecal enterococci of wild birds: Emergence of vanA and vanB2 harbouring Enterococcus faecalis. Innov Food Sci Emerg Technol 2018, 52, 936–941. [CrossRef]
- Garrido, A. M.; Gálvez, A.; Pulido, R. P. Antimicrobial resistance in enterococci. Journal of Infectious Diseases and Therapy 2014, 2.
- Kmeť, V.; Čuvalová, A.; Stanko, M. Small mammals as sentinels of antimicrobial-resistant staphylococci. Folia Microbiol 2018, 63, 665–668. [CrossRef]
- Watanabe, S.; Ito, T.; Sasaki, T.; Li, S.; Uchiyama, I.; Kishii, K.; Kikuchi, K.; Skov R.L.; Hiramatsu, K. Genetic diversity of staphylocoagulase genes (coa): insight into the evolution of variable chromosomal virulence factors in Staphylococcus aureus. PLoS One 2009, 4, 5714. [CrossRef]
- Becker, K.; Heilmann, C.; Peters, G. Coagulase-negative staphylococci. Clinical microbiology reviews 2014, 27, 870-926.
- Van Alen, S.; Ballhausen, B.; Peters, G.; Friedrich, A. W.; Mellmann, A.; Köck, R.; Becker, K. In the centre of an epidemic: Fifteen years of LA-MRSA CC398 at the University Hospital Münster. Vet. Microbiol 2017, 200, 19–24. [CrossRef]
- Bonsaglia, E. C. R.; Silva, N. C. C.; Rossi, B. F.; Camargo, C. H.; Dantas, S. T. A.; Langoni, H.;Guimarães, F.F.; Lima, F.S.; Fitzgerald, J.R.; Júnior, F.; Rall, V. L. M. Molecular epidemiology of methicillin-susceptible Staphylococcus aureus (MSSA) isolated from milk of cows with subclinical mastitis. Microbial Pathogenesis 2018, 124, 130–135. [CrossRef]
- Yang, F.; Liu, L.; Wang, L.; Wang, X.; Li, X.; Luo, J.; Zhang, Z.; Zhang, S.; Yan, Z.; Li, H. Penicillin-resistant characterization of Staphylococcus aureus isolated from bovine mastitis in Gansu, China. J Integr Agric 2017, 16, 1874–1878. [CrossRef]
- Miller, W. R.; Munita, J. M.; Arias, C. A. Mechanisms of antibiotic resistance in enterococci. Expert Rev Anti Infect Ther 2014, 12, 1221–1236. [CrossRef]
- Bouchiat, C.; El-Zeenni, N.; Chakrakodi, B.; Nagaraj, S.; Arakere, G.; Etienne, J. Epidemiology of Staphylococcus aureus in Bangalore, India: Emergence of the ST217 clone and high rate of resistance to erythromycin and ciprofloxacin in the community. New Microbes New Infect 2015, 7, 15–20. [CrossRef]
- Dadashi, M.; Hajikhani, B.; Darban-Sarokhalil, D.; van Belkum, A.; Goudarzi, M. Mupirocin Resistance in Staphylococcus aureus: A Systematic Review and Meta-Analysis. J Glob Antimicrob Resist 2019. [CrossRef]
- von Wintersdorff, C. J.; Penders, J.; Stobberingh, E. E.; Lashof, A. M. O.; Hoebe, C. J.; Savelkoul, P. H.; Wolffs, P. F. High rates of antimicrobial drug resistance gene acquisition after international travel, the Netherlands. Emerging Infectious Diseases 2014, 20, 649–657.
- Mühldorfer, K. Bats and Bacterial Pathogens: A Review. Zoonoses Public Health 2013, 60, 93–103. [CrossRef]
- Ito, T.; Hiramatsu, K.; Tomasz, A.; De Lencastre, H.; Perreten, V.; Holden, M. T.; Coleman, D. C.; Goering, R.; Giffard, P. M.; Skov, R. L.; Zhang, K.; Westh, H.; O'Brien, F.; Tenover, F. C.; Oliveira, D. C.; Boyle-Vavra, S.; Laurent, F.; Kearns, A. M.; Kreiswirth, B.; Ko, K. S.; Grundmann, H.; Sollidu, J. E.; John Jr., J. F.; Daum, R.; Soderquist, B.; Buist, G. Guidelines for reporting novel mecA gene homologues. Antimicrob Agents Chemother 2012, 56, 4997-4999.
- Koukos, G.; Sakellari, D.; Arsenakis, M.; Tsalikis, L.; Slini, T.; Konstantinidis, A. Prevalence of Staphylococcus aureus and methicillin resistant Staphylococcus aureus ( MRSA ) in the oral cavity. Arch. Oral Biol 2015, 60, 1410–1415. [CrossRef]
- Wendlandt, S.; Feßler, A. T.; Monecke, S.; Ehricht, R.; Schwarz, S.; Kadlec, K. The diversity of antimicrobial resistance genes among staphylococci of animal origin. Int. J. Med. Microbiol, Jena 2013, 303, 338–349. [CrossRef]
- Resman, F.; Thegerström, J.; Månsson, F.; Ahl, J.; Tham, J.; Riesbeck, K. The prevalence, population structure and screening test specificity of penicillin-susceptible Staphylococcus aureus bacteremia isolates in Malmö, Sweden. Journal of Infection 2016, 73, 129–135.
- Cheatham, S.; Thapaliya, D.; Taha, M.; Milliken, K.; Dalman, M. R.; Kadariya, J.; Grenier, D.; Smith, T. C. Prevalence of Staphylococcus aureus and methicillin-resistant S aureus on environmental surfaces in Ohio nursing homes. American Journal of Infection Control 2019, 000, 1–5. [CrossRef]
- Peterson, L. R.; Boehm, S.; Beaumont, J. L.; Patel, P. A.; Schora, D. M.; Peterson, K. E.; Burdsall, D.; Hines, C.; Fausone, M.; Robicsek, A.; Smith, B. A. Reduction of methicillin-resistant Staphylococcus aureus infection in long-term care is possible while maintaining patient socialization: A prospective randomized clinical trial. American Journal of Infection Control 2016, 44, 1622-1627. [CrossRef]
- Banskar, S.; Mourya, D.T.; Shouche, Y.S. Bacterial diversity indicates dietary overlap among bats of different feeding habits. Microbiol Res 2016, 182, 99–108. [CrossRef]
- Feßler, A. T.; Schuenemann, R.; Kadlec, K.; Hensel, V.; Brombach, J.; Murugaiyan, J.; Oechtering, G.; Burgener, I.A.; Schwarz, S. Methicillin-resistant Staphylococcus aureus (MRSA) and methicillin-resistant Staphylococcus pseudintermedius (MRSP) among employees and in the environment of a small animal hospital. Vet. Microbiol 2018, 221, 153–158. [CrossRef]
- Baptista, I.; Rocha, S.M.; Cunha, Â.; Saraiva, J.A.; Almeida, A. Inactivation of Staphylococcus aureus by high pressure processing : An overview. Innov Food Sci Emerg Technol 2016, 36, 128–149. [CrossRef]

| Bacterial Resistance | |||||
|---|---|---|---|---|---|
| Antimicrobials | Feces | ||||
| R | Total | % | |||
| Ampicillin Cephalothin Oxacillin Amoxicillin+clavulanic acid Norfloxacin Enrofloxacin Gentamicin |
5 5 5 4 1 0 0 |
6 6 6 6 6 6 6 |
83.33 83.33 83.33 66.66 16.66 0% 0% |
||
| TOTAL | 20 | ||||
| Sample | Antimicrobials and Resistance | Antimicrobial Resistance Index | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMC | AMP | CFL | ENO | GEN | NOR | OXA | |||||||||||
| 12 | S | S | S | I | S | R | S | 0.14 | |||||||||
| 13* | R | R | R | I | S | S | R | 0.57 | |||||||||
| 14 | S | R | R | S | S | S | R | 0.42 | |||||||||
| 15* | R | R | R | S | I | S | R | 0.57 | |||||||||
| 16* | R | R | R | S | S | S | R | 0.57 | |||||||||
| 19* | R | R | R | S | I | S | R | 0.57 | |||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





