Submitted:
19 December 2023
Posted:
20 December 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
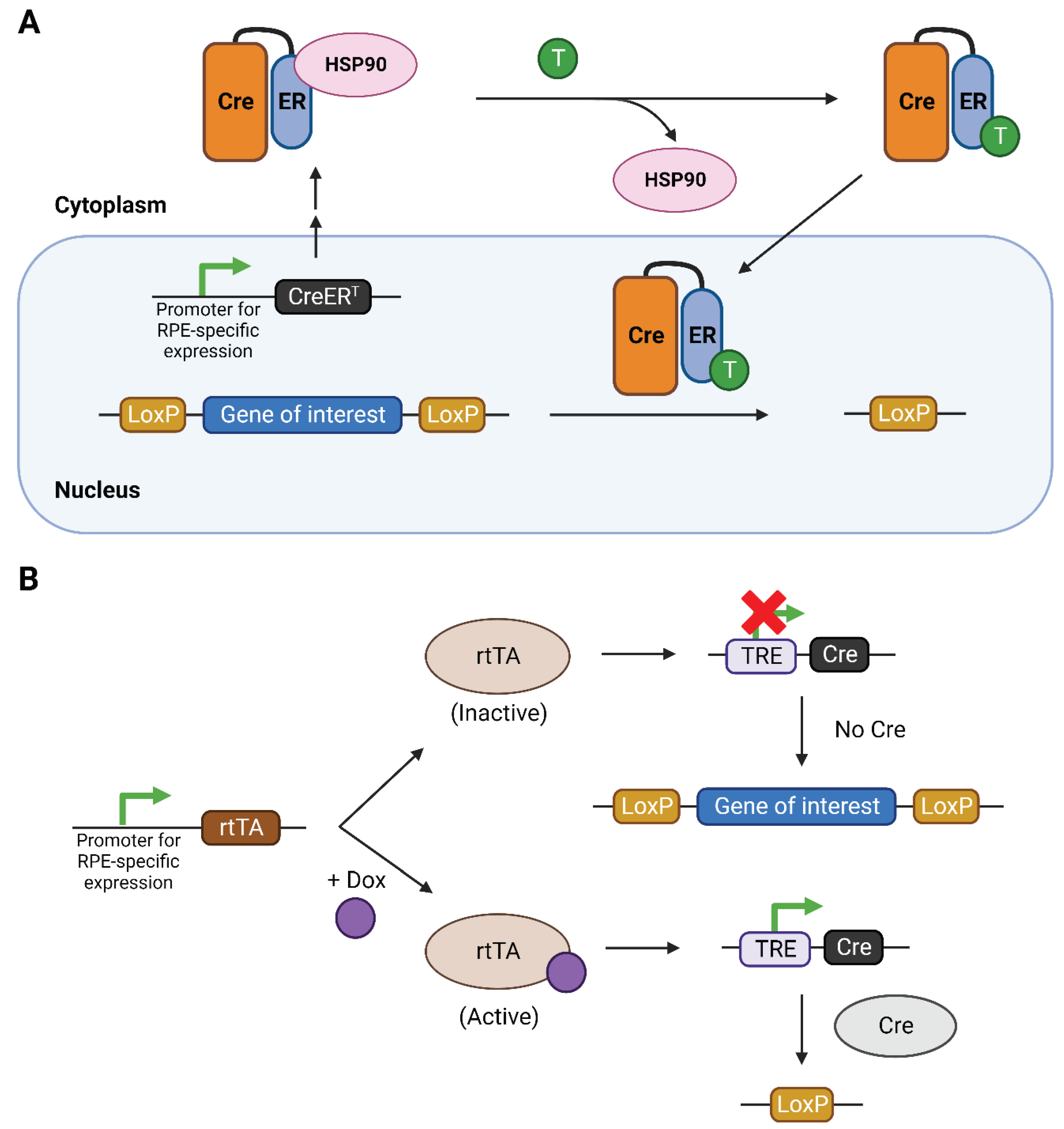
2. Regulation of the Cre-loxP system
3. Non-inducible Cre mice targeting the RPE
Tyrosinase gene family promoters
MART-1 promoter
Bestrophin 1 promoter
4. Inducible Cre mice targeting the RPE
Tetracycline/doxycycline-inducible VMD2 promoter
Tamoxifen-inducible MCT3 promoter
Tamoxifen-inducible Trp1 promoter
Tamoxifen-inducible hsp70 promoter with RPE-specific Cns-2 enhancer
Tamoxifen-inducible Best1 promoter in Rosa26 locus
Tamoxifen-inducible Cre recombinase gene conjugated to Rpe65 gene
Tetracycline/doxycycline-inducible Pmel promoter
5. Conclusion
Author Contributions
Funding
Conflicts of Interest
References
- Lakkaraju, A.; Umapathy, A.; Tan, L.X.; Daniele, L.; Philp, N.J.; Boesze-Battaglia, K.; Williams, D.S. The cell biology of the retinal pigment epithelium. Prog Retin Eye Res 2020, 100846. [Google Scholar] [CrossRef]
- Strauss, O. The retinal pigment epithelium in visual function. Physiol Rev 2005, 85, 845–881. [Google Scholar] [CrossRef]
- Yang, S.; Zhou, J.; Li, D. Functions and Diseases of the Retinal Pigment Epithelium. Front Pharmacol 2021, 12, 727870. [Google Scholar] [CrossRef]
- Longbottom, R.; Fruttiger, M.; Douglas, R.H.; Martinez-Barbera, J.P.; Greenwood, J.; Moss, S.E. Genetic ablation of retinal pigment epithelial cells reveals the adaptive response of the epithelium and impact on photoreceptors. Proc Natl Acad Sci U S A 2009, 106, 18728–18733. [Google Scholar] [CrossRef]
- Zhao, C.; Yasumura, D.; Li, X.; Matthes, M.; Lloyd, M.; Nielsen, G.; Ahern, K.; Snyder, M.; Bok, D.; Dunaief, J.L.; et al. mTOR-mediated dedifferentiation of the retinal pigment epithelium initiates photoreceptor degeneration in mice. J Clin Invest 2011, 121, 369–383. [Google Scholar] [CrossRef] [PubMed]
- Kocherlakota, S.; Das, Y.; Swinkels, D.; Vanmunster, M.; Callens, M.; Vinckier, S.; Vaz, F.M.; Sinha, D.; Van Veldhoven, P.P.; Fransen, M.; et al. The murine retinal pigment epithelium requires peroxisomal beta-oxidation to maintain lysosomal function and prevent dedifferentiation. Proc Natl Acad Sci U S A 2023, 120, e2301733120. [Google Scholar] [CrossRef] [PubMed]
- Guymer, R.H.; Campbell, T.G. Age-related macular degeneration. Lancet 2023, 401, 1459–1472. [Google Scholar] [CrossRef] [PubMed]
- Tsang, S.H.; Sharma, T. Leber Congenital Amaurosis. Adv Exp Med Biol 2018, 1085, 131–137. [Google Scholar] [CrossRef]
- Lenis, T.L.; Hu, J.; Ng, S.Y.; Jiang, Z.; Sarfare, S.; Lloyd, M.B.; Esposito, N.J.; Samuel, W.; Jaworski, C.; Bok, D.; et al. Expression of ABCA4 in the retinal pigment epithelium and its implications for Stargardt macular degeneration. Proc Natl Acad Sci U S A 2018, 115, E11120–E11127. [Google Scholar] [CrossRef]
- Hurley, J.B. Retina Metabolism and Metabolism in the Pigmented Epithelium: A Busy Intersection. Annu Rev Vis Sci 2021, 7, 665–692. [Google Scholar] [CrossRef]
- Liptak, N.; Gal, Z.; Biro, B.; Hiripi, L.; Hoffmann, O.I. Rescuing lethal phenotypes induced by disruption of genes in mice: a review of novel strategies. Physiol Res 2021, 70, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Baes, M.; Van Veldhoven, P.P. Mouse models for peroxisome biogenesis defects and beta-oxidation enzyme deficiencies. Biochim Biophys Acta 2012, 1822, 1489–1500. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Levine, B. Autophagy in mammalian development and differentiation. Nat Cell Biol 2010, 12, 823–830. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Kim, M.; Im, S.K.; Fang, S. Mouse Cre-LoxP system: general principles to determine tissue-specific roles of target genes. Lab Anim Res 2018, 34, 147–159. [Google Scholar] [CrossRef] [PubMed]
- Ye, R.; Wang, Q.A.; Tao, C.; Vishvanath, L.; Shao, M.; McDonald, J.G.; Gupta, R.K.; Scherer, P.E. Impact of tamoxifen on adipocyte lineage tracing: Inducer of adipogenesis and prolonged nuclear translocation of Cre recombinase. Mol Metab 2015, 4, 771–778. [Google Scholar] [CrossRef] [PubMed]
- Donocoff, R.S.; Teteloshvili, N.; Chung, H.; Shoulson, R.; Creusot, R.J. Optimization of tamoxifen-induced Cre activity and its effect on immune cell populations. Sci Rep 2020, 10, 15244. [Google Scholar] [CrossRef] [PubMed]
- Tewari, K.; Bonebrake, R.G.; Asrat, T.; Shanberg, A.M. Ambiguous genitalia in infant exposed to tamoxifen in utero. Lancet 1997, 350, 183. [Google Scholar] [CrossRef]
- Braems, G.; Denys, H.; De Wever, O.; Cocquyt, V.; Van den Broecke, R. Use of tamoxifen before and during pregnancy. Oncologist 2011, 16, 1547–1551. [Google Scholar] [CrossRef] [PubMed]
- Ved, N.; Curran, A.; Ashcroft, F.M.; Sparrow, D.B. Tamoxifen administration in pregnant mice can be deleterious to both mother and embryo. Lab Anim 2019, 53, 630–633. [Google Scholar] [CrossRef]
- Sun, M.R.; Steward, A.C.; Sweet, E.A.; Martin, A.A.; Lipinski, R.J. Developmental malformations resulting from high-dose maternal tamoxifen exposure in the mouse. PLoS One 2021, 16, e0256299. [Google Scholar] [CrossRef]
- Liu, Y.; Suckale, J.; Masjkur, J.; Magro, M.G.; Steffen, A.; Anastassiadis, K.; Solimena, M. Tamoxifen-independent recombination in the RIP-CreER mouse. PLoS One 2010, 5, e13533. [Google Scholar] [CrossRef]
- Lewis, K.T.; Oles, L.R.; MacDougald, O.A. Tetracycline response element driven Cre causes ectopic recombinase activity independent of transactivator element. Mol Metab 2022, 61, 101501. [Google Scholar] [CrossRef] [PubMed]
- Housset, M.; Samuel, A.; Ettaiche, M.; Bemelmans, A.; Beby, F.; Billon, N.; Lamonerie, T. Loss of Otx2 in the adult retina disrupts retinal pigment epithelium function, causing photoreceptor degeneration. J Neurosci 2013, 33, 9890–9904. [Google Scholar] [CrossRef] [PubMed]
- Holmgaard, A.; Askou, A.L.; Benckendorff, J.N.E.; Thomsen, E.A.; Cai, Y.; Bek, T.; Mikkelsen, J.G.; Corydon, T.J. In Vivo Knockout of the Vegfa Gene by Lentiviral Delivery of CRISPR/Cas9 in Mouse Retinal Pigment Epithelium Cells. Mol Ther Nucleic Acids 2017, 9, 89–99. [Google Scholar] [CrossRef]
- Muzumdar, M.D.; Tasic, B.; Miyamichi, K.; Li, L.; Luo, L. A global double-fluorescent Cre reporter mouse. Genesis 2007, 45, 593–605. [Google Scholar] [CrossRef]
- Soriano, P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet 1999, 21, 70–71. [Google Scholar] [CrossRef]
- Novak, A.; Guo, C.; Yang, W.; Nagy, A.; Lobe, C.G. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis 2000, 28, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Lobe, C.G.; Koop, K.E.; Kreppner, W.; Lomeli, H.; Gertsenstein, M.; Nagy, A. Z/AP, a double reporter for cre-mediated recombination. Dev Biol 1999, 208, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Mori, M.; Metzger, D.; Garnier, J.M.; Chambon, P.; Mark, M. Site-specific somatic mutagenesis in the retinal pigment epithelium. Invest Ophthalmol Vis Sci 2002, 43, 1384–1388. [Google Scholar] [PubMed]
- Thanos, A.; Morizane, Y.; Murakami, Y.; Giani, A.; Mantopoulos, D.; Kayama, M.; Roh, M.I.; Michaud, N.; Pawlyk, B.; Sandberg, M.; et al. Evidence for baseline retinal pigment epithelium pathology in the Trp1-Cre mouse. Am J Pathol 2012, 180, 1917–1927. [Google Scholar] [CrossRef]
- Guyonneau, L.; Rossier, A.; Richard, C.; Hummler, E.; Beermann, F. Expression of Cre recombinase in pigment cells. Pigment Cell Res 2002, 15, 305–309. [Google Scholar] [CrossRef]
- Aydin, I.T.; Beermann, F. A mart-1::Cre transgenic line induces recombination in melanocytes and retinal pigment epithelium. Genesis 2011, 49, 403–409. [Google Scholar] [CrossRef]
- Iacovelli, J.; Zhao, C.; Wolkow, N.; Veldman, P.; Gollomp, K.; Ojha, P.; Lukinova, N.; King, A.; Feiner, L.; Esumi, N.; et al. Generation of Cre transgenic mice with postnatal RPE-specific ocular expression. Invest Ophthalmol Vis Sci 2011, 52, 1378–1383. [Google Scholar] [CrossRef]
- He, L.; Marioutina, M.; Dunaief, J.L.; Marneros, A.G. Age- and gene-dosage-dependent cre-induced abnormalities in the retinal pigment epithelium. Am J Pathol 2014, 184, 1660–1667. [Google Scholar] [CrossRef]
- Zolghadri, S.; Bahrami, A.; Hassan Khan, M.T.; Munoz-Munoz, J.; Garcia-Molina, F.; Garcia-Canovas, F.; Saboury, A.A. A comprehensive review on tyrosinase inhibitors. J Enzyme Inhib Med Chem 2019, 34, 279–309. [Google Scholar] [CrossRef]
- Korner, A.M.; Pawelek, J. Dopachrome conversion: a possible control point in melanin biosynthesis. J Invest Dermatol 1980, 75, 192–195. [Google Scholar] [CrossRef]
- Lapedriza, A.; Petratou, K.; Kelsh, R.N. Chapter 14 - Neural Crest Cells and Pigmentation. In Neural Crest Cells, Trainor, P.A., Ed.; Academic Press: Boston, 2014; pp. 287–311. [Google Scholar]
- Delmas, V.; Martinozzi, S.; Bourgeois, Y.; Holzenberger, M.; Larue, L. Cre-mediated recombination in the skin melanocyte lineage. Genesis 2003, 36, 73–80. [Google Scholar] [CrossRef]
- Murisier, F.; Guichard, S.; Beermann, F. A conserved transcriptional enhancer that specifies Tyrp1 expression to melanocytes. Dev Biol 2006, 298, 644–655. [Google Scholar] [CrossRef]
- Murisier, F.; Beermann, F. Genetics of pigment cells: lessons from the tyrosinase gene family. Histol Histopathol 2006, 21, 567–578. [Google Scholar] [CrossRef]
- Murisier, F.; Guichard, S.; Beermann, F. Distinct distal regulatory elements control tyrosinase expression in melanocytes and the retinal pigment epithelium. Dev Biol 2007, 303, 838–847. [Google Scholar] [CrossRef]
- Hoashi, T.; Watabe, H.; Muller, J.; Yamaguchi, Y.; Vieira, W.D.; Hearing, V.J. MART-1 is required for the function of the melanosomal matrix protein PMEL17/GP100 and the maturation of melanosomes. J Biol Chem 2005, 280, 14006–14016. [Google Scholar] [CrossRef] [PubMed]
- Kunzelmann, K. TMEM16, LRRC8A, bestrophin: chloride channels controlled by Ca(2+) and cell volume. Trends Biochem Sci 2015, 40, 535–543. [Google Scholar] [CrossRef] [PubMed]
- Masuda, T.; Esumi, N. SOX9, through interaction with microphthalmia-associated transcription factor (MITF) and OTX2, regulates BEST1 expression in the retinal pigment epithelium. J Biol Chem 2010, 285, 26933–26944. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Kim, L.; Lu, C.W.; Zeng, H.; Vollrath, D. An efficient inducible RPE-Selective cre transgenic mouse line. Exp Eye Res 2021, 202, 108370. [Google Scholar] [CrossRef] [PubMed]
- Go, Y.M.; Zhang, J.; Fernandes, J.; Litwin, C.; Chen, R.; Wensel, T.G.; Jones, D.P.; Cai, J.; Chen, Y. MTOR-initiated metabolic switch and degeneration in the retinal pigment epithelium. FASEB J 2020, 34, 12502–12520. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Gu, S.; Chen, M.; Zhang, S.J.; Jiang, Z.; Chen, X.; Jiang, C.; Liu, G.; Radu, R.A.; Sun, X.; et al. Abnormal mTORC1 signaling leads to retinal pigment epithelium degeneration. Theranostics 2019, 9, 1170–1180. [Google Scholar] [CrossRef] [PubMed]
- Heffner, C.S.; Herbert Pratt, C.; Babiuk, R.P.; Sharma, Y.; Rockwood, S.F.; Donahue, L.R.; Eppig, J.T.; Murray, S.A. Supporting conditional mouse mutagenesis with a comprehensive cre characterization resource. Nat Commun 2012, 3, 1218. [Google Scholar] [CrossRef] [PubMed]
- Frenz, S.; Rak, K.; Volker, J.; Jurgens, L.; Scherzad, A.; Schendzielorz, P.; Radeloff, A.; Jablonka, S.; Hansen, S.; Mlynski, R.; et al. Mosaic pattern of Cre recombinase expression in cochlear outer hair cells of the Brn3.1 Cre mouse. Neuroreport 2015, 26, 309–313. [Google Scholar] [CrossRef] [PubMed]
- Garrick, D.; Fiering, S.; Martin, D.I.; Whitelaw, E. Repeat-induced gene silencing in mammals. Nat Genet 1998, 18, 56–59. [Google Scholar] [CrossRef]
- Cain-Hom, C.; Splinter, E.; van Min, M.; Simonis, M.; van de Heijning, M.; Martinez, M.; Asghari, V.; Cox, J.C.; Warming, S. Efficient mapping of transgene integration sites and local structural changes in Cre transgenic mice using targeted locus amplification. Nucleic Acids Res 2017, 45, e62. [Google Scholar] [CrossRef]
- Choi, E.H.; Suh, S.; Einstein, D.E.; Leinonen, H.; Dong, Z.; Rao, S.R.; Fliesler, S.J.; Blackshaw, S.; Yu, M.; Peachey, N.S.; et al. An inducible Cre mouse for studying roles of the RPE in retinal physiology and disease. JCI Insight 2021, 6. [Google Scholar] [CrossRef] [PubMed]
- Swarup, A.; Samuels, I.S.; Bell, B.A.; Han, J.Y.S.; Du, J.; Massenzio, E.; Abel, E.D.; Boesze-Battaglia, K.; Peachey, N.S.; Philp, N.J. Modulating GLUT1 expression in retinal pigment epithelium decreases glucose levels in the retina: impact on photoreceptors and Muller glial cells. Am J Physiol Cell Physiol 2019, 316, C121–C133. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Jia, L.; Khan, N.; Lin, C.; Mitter, S.K.; Boulton, M.E.; Dunaief, J.L.; Klionsky, D.J.; Guan, J.L.; Thompson, D.A.; et al. Deletion of autophagy inducer RB1CC1 results in degeneration of the retinal pigment epithelium. Autophagy 2015, 11, 939–953. [Google Scholar] [CrossRef] [PubMed]
- Sethna, S.; Scott, P.A.; Giese, A.P.J.; Duncan, T.; Jian, X.; Riazuddin, S.; Randazzo, P.A.; Redmond, T.M.; Bernstein, S.L.; Riazuddin, S.; et al. CIB2 regulates mTORC1 signaling and is essential for autophagy and visual function. Nat Commun 2021, 12, 3906. [Google Scholar] [CrossRef] [PubMed]
- Bullock, J.; Polato, F.; Abu-Asab, M.; Bernardo-Colon, A.; Aflaki, E.; Agbaga, M.P.; Becerra, S.P. Degradation of Photoreceptor Outer Segments by the Retinal Pigment Epithelium Requires Pigment Epithelium-Derived Factor Receptor (PEDF-R). Invest Ophthalmol Vis Sci 2021, 62, 30. [Google Scholar] [CrossRef] [PubMed]
- Le, Y.Z.; Zheng, W.; Rao, P.C.; Zheng, L.; Anderson, R.E.; Esumi, N.; Zack, D.J.; Zhu, M. Inducible expression of cre recombinase in the retinal pigmented epithelium. Invest Ophthalmol Vis Sci 2008, 49, 1248–1253. [Google Scholar] [CrossRef] [PubMed]
- Ueki, Y.; Ash, J.D.; Zhu, M.; Zheng, L.; Le, Y.Z. Expression of Cre recombinase in retinal Muller cells. Vision Res 2009, 49, 615–621. [Google Scholar] [CrossRef]
- Fu, S.; Zhu, M.; Wang, C.; Le, Y.Z. Efficient induction of productive Cre-mediated recombination in retinal pigment epithelium. Mol Vis 2014, 20, 480–487. [Google Scholar] [PubMed]
- Mori, M.; Gargowitsch, L.; Bornert, J.M.; Garnier, J.M.; Mark, M.; Chambon, P.; Metzger, D. Temporally controlled targeted somatic mutagenesis in mouse eye pigment epithelium. Genesis 2012, 50, 828–832. [Google Scholar] [CrossRef]
- Schneider, S.; Hotaling, N.; Campos, M.; Patnaik, S.R.; Bharti, K.; May-Simera, H.L. Generation of an inducible RPE-specific Cre transgenic-mouse line. PLoS One 2018, 13, e0207222. [Google Scholar] [CrossRef]
- Nasrin, M.; Ahmed, O.; Han, X.; Nojebuzzaman, M.; Abo-Ahmed, A.I.; Yazawa, S.; Osawa, M. Generation of Pmel-dependent conditional and inducible Cre-driver mouse line for melanocytic-targeted gene manipulation. Pigment Cell Melanoma Res 2023, 36, 53–70. [Google Scholar] [CrossRef] [PubMed]
- Philp, N.J.; Yoon, H.; Grollman, E.F. Monocarboxylate transporter MCT1 is located in the apical membrane and MCT3 in the basal membrane of rat RPE. Am J Physiol 1998, 274, R1824–1828. [Google Scholar] [CrossRef] [PubMed]
- Philp, N.J.; Yoon, H.; Lombardi, L. Mouse MCT3 gene is expressed preferentially in retinal pigment and choroid plexus epithelia. Am J Physiol Cell Physiol 2001, 280, C1319–1326. [Google Scholar] [CrossRef] [PubMed]
- Donnelly, M.L.L.; Luke, G.; Mehrotra, A.; Li, X.; Hughes, L.E.; Gani, D.; Ryan, M.D. Analysis of the aphthovirus 2A/2B polyprotein 'cleavage' mechanism indicates not a proteolytic reaction, but a novel translational effect: a putative ribosomal 'skip'. J Gen Virol 2001, 82, 1013–1025. [Google Scholar] [CrossRef]
- Kim, J.H.; Lee, S.R.; Li, L.H.; Park, H.J.; Park, J.H.; Lee, K.Y.; Kim, M.K.; Shin, B.A.; Choi, S.Y. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS One 2011, 6, e18556. [Google Scholar] [CrossRef]
- Mori, M.; Metzger, D.; Picaud, S.; Hindelang, C.; Simonutti, M.; Sahel, J.; Chambon, P.; Mark, M. Retinal dystrophy resulting from ablation of RXR alpha in the mouse retinal pigment epithelium. Am J Pathol 2004, 164, 701–710. [Google Scholar] [CrossRef]
| Model | Promoter | Cre expression start | Important features | References |
|---|---|---|---|---|
| Trp1-Cre | Tyrosinase-related protein 1 (Trp1) promoter | E10.5 | - Untargeted insertion of Cre - Cre toxicity to RPE - Ectopic Cre expression in some cells of neural retina, along with some other tissues |
[29,30] |
| Dct-Cre | Dopachrome tautamerase (Dct) promoter | E9.5 | - Untargeted insertion of Cre - Mosaic Cre expression - Cre expression also in melanocytes and in cells of telencephalon* - Ectopic Cre expression in caudal nerves and dorsal root ganglia |
[31] |
| MART-1-Cre | Melanoma associated antigen recognized by T-cells (MART-1) promoter | E12.5 | - Untargeted insertion of Cre - Uniform Cre expression - Cre expression also in all melanocytes* - Minimal ectopic Cre expression in some epidermal cells of the skin |
[32] |
| Best1-Cre | Bestrophin-1 (Best1) promoter | P10 | - Untargeted insertion of Cre - Mosaic Cre expression - Age and dosage-dependent Cre toxicity to RPE$ - Cre expression also in testis* |
[33,34] |
| Model | Promoter | Induction by | Important features | References |
|---|---|---|---|---|
| Inducible VMD2-Cre | Vitelliform macular dystrophy-2 (VMD2), promoter |
Tetracycline/ Doxycycline | - Untargeted insertion of Cre. - “Leaky” Cre expression - Mosaic Cre expression - Cre recombinase undetectable by immunostaining - Weak ectopic Cre expression in the optic nerve |
[57,58,59] |
| Inducible MCT3-Cre | Monocarboxylate transporter 3 (Mct3) promoter | Tamoxifen | - Untargeted insertion of Cre - Mosaic Cre activity. Only 5-20% of RPE show Cre activity - Cre activity also in the choroid plexus epithelium of brain* |
[4] |
| Inducible Trp1-Cre | Tyrosinase-related protein 1 (Trp1) promoter | Tamoxifen | - Untargeted insertion of Cre. - Mosaic Cre activity - Ectopic Cre activity in some cells of neural retina, iris, ciliary body and optic nerve |
[60] |
| Inducible Tyr-Cre | Tyrosinase (Tyr) promoter | Tamoxifen | - Untargeted insertion of Cre - Mosaic Cre activity with better expression in embryonic RPE - Cre activity also observed in the ciliary body* - Weak ectopic Cre function observed in inner nuclear layer without any cell-type specificity |
[61] |
| Inducible Best1-Cre | Bestrophin-1 (Best1) promoter | Tamoxifen | - Targeted insertion of Cre gene into the Rosa26 locus - Cre function also in the testis* - Cre activity in >90% of RPE cells - Minimal/negligible (<1%) ectopic Cre function in Muller glia |
[45] |
| Inducible RPE65-Cre | Retinal pigment epithelium-specific 65 kDa protein (RPE65) promoter | Tamoxifen | - Targeted knock-in of sequence for P2A-CreERT2 fused in-frame with RPE65 gene - Cre activity in >90% of RPE cells - Levels of Cre recombinase undetectable by immunoblotting |
[52] |
| Inducible Pmel-Cre | Premelanosome protein (Pmel) promoter | Tetracycline/ Doxycycline | - Untargeted insertion of Cre. - No mosaic Cre expression - Cre expression also in most melanocytes* - Ectopic Cre expression in lung and heart mesothelial cells |
[62] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





