Submitted:
19 December 2023
Posted:
20 December 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
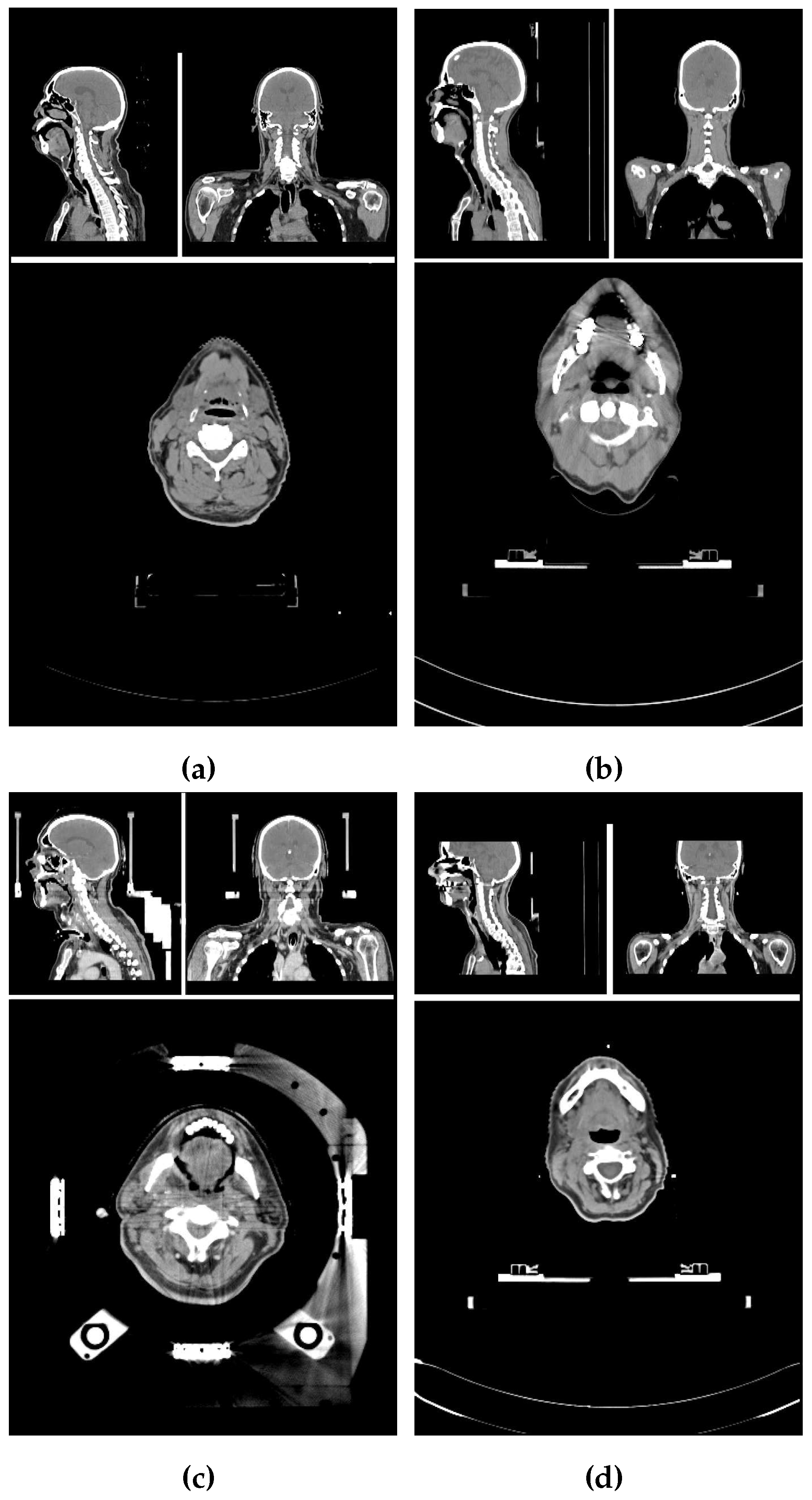
2.1. Image properties of the data set
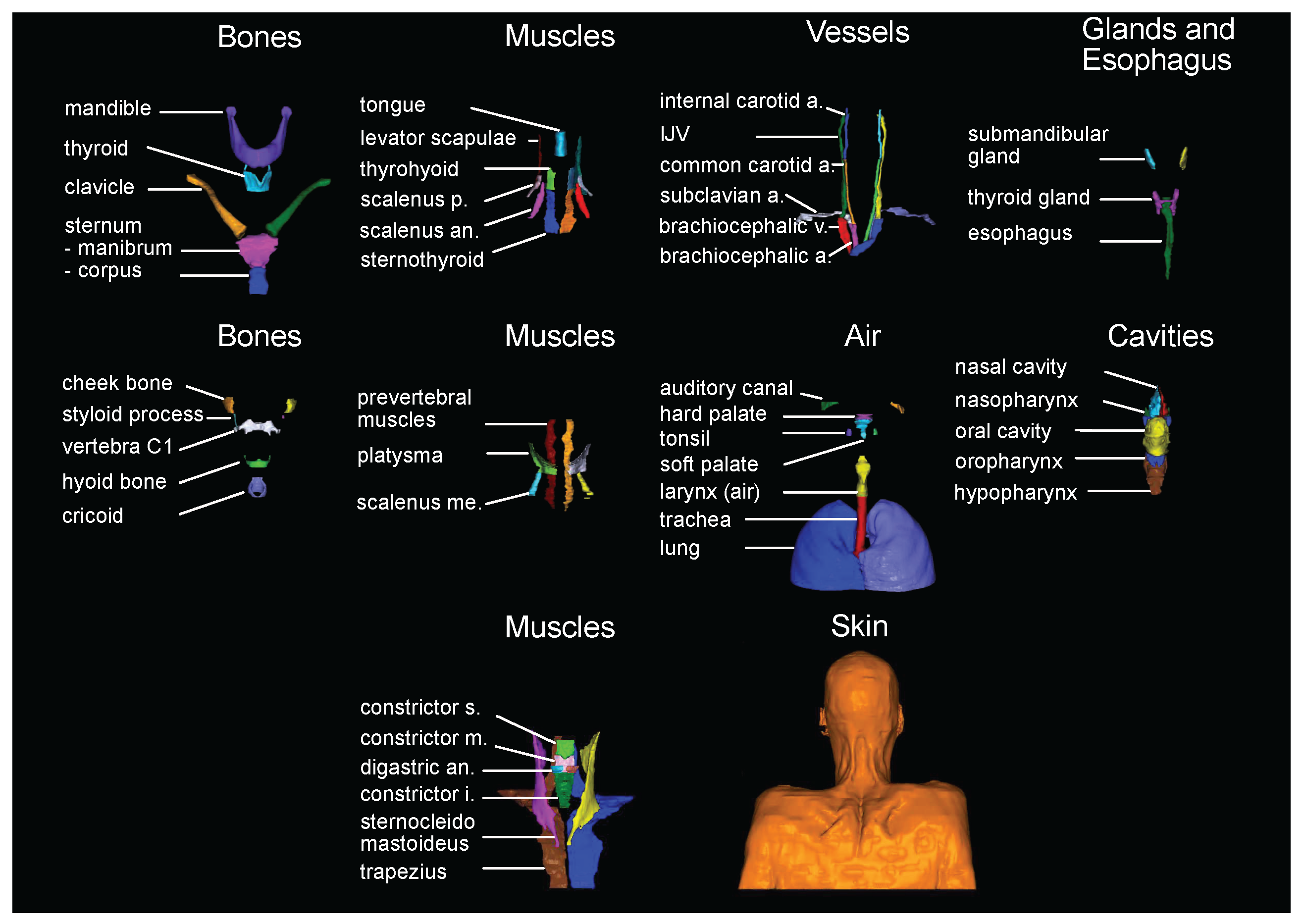
2.2. Label selection and generation of the manual labels
2.3. Network training and label prediction
2.4. Evaluation of predicted labels
3. Results
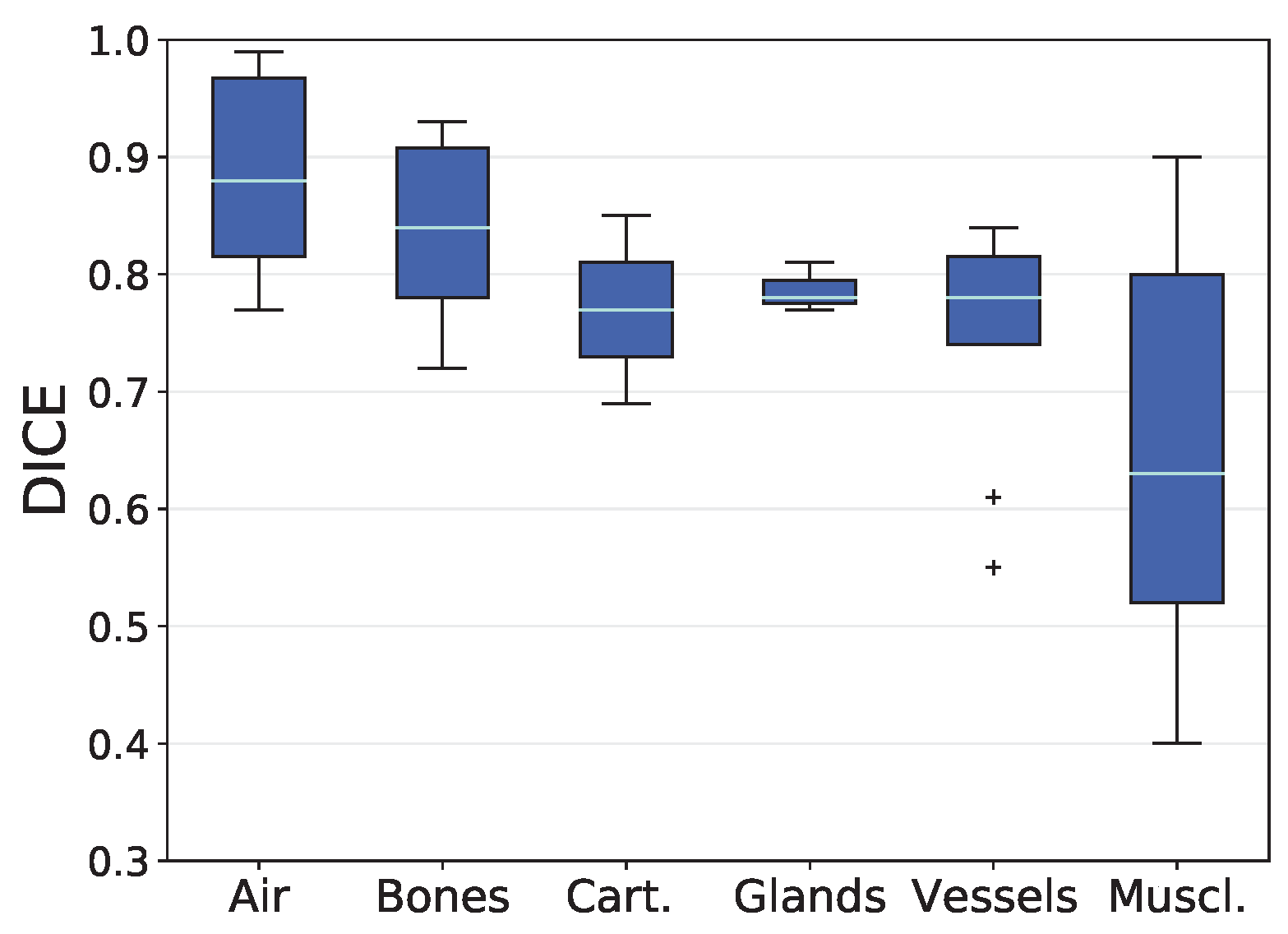
3.1. Analysis based on Volumetric Overlap
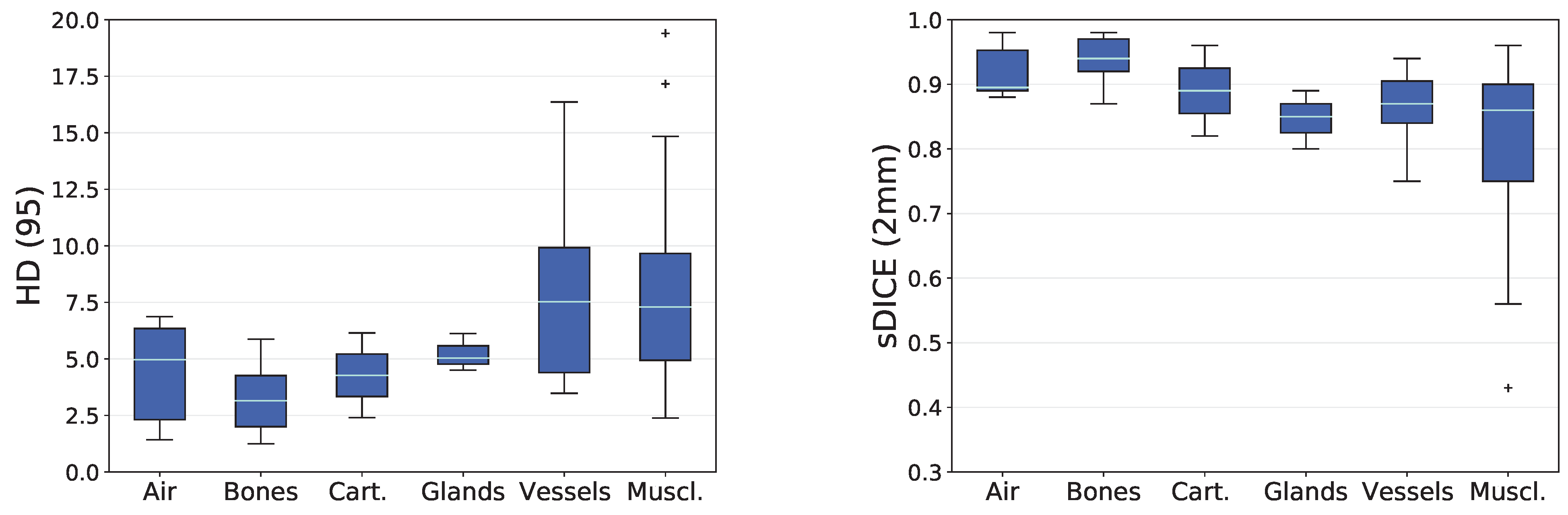
3.2. Analysis based on Distance-Based Metrics
3.3. Completeness of predicted label set
3.4. Analyzing only patients without tracheostoma
| Structure | DICE | HD (95) | sDICE (2 mm) |
|---|---|---|---|
| Trachea | 0.92 (0.13) | 5.64 (-7.40) | 0.93 (0.16) |
| Hyoid Bone | 0.83 (0.12) | 2.31 (-7.32) | 0.96 (0.09) |
| Thyroid Gland | 0.84 (0.14) | 5.90 (-1.32) | 0.92 (0.18) |
| Internal Carotid Artery (r) | 0.57 (0.10) | 11.77 (-12.50) | 0.77 (0.10) |
| Internal Jugular Vein (r) | 0.78 (0.15) | 8.09 (-0.98) | 0.89 (0.13) |
| Constrictors (s., m., i.) | 0.59 (0.19) | 7.14 (-0.32) | 0.90 (0.10) |
| Middle Constrictor | 0.48 (0.21) | 9.17 (-2.93) | 0.75 (0.15) |
| Superior Constrictor | 0.52 (0.23) | 11.32 (0.50) | 0.75 (0.14) |
| Digastric (r) | 0.51 (0.30) | 7.56 (-5.75) | 0.69 (0.33) |
| Platysma (r) | 0.54 (0.18) | 17.61 (-15.24) | 0.78 (0.20) |
| Sternothyroid (r) | 0.60 (0.21) | 4.66 (-3.01) | 0.91 (0.28) |
| Sternocleidomastoid (l) | 0.86 (0.12) | 3.63 (-7.86) | 0.93 (0.09) |
| Sternocleidomastoid (r) | 0.85 (0.26) | 5.17 (-42.80) | 0.92 (0.26) |
| Thyrohyoid (r) | 0.57 (0.09) | 2.85 (-1.79) | 0.91 (0.12) |
| Esophagus | 0.82 (0.12) | 5.41 (-4.44) | 0.90 (0.11) |
| Hypopharynx | 0.68 (0.23) | 5.95 (-4.73) | 0.86 (0.18) |
| Soft Palate | 0.63 (0.16) | 8.64 (-4.12) | 0.78 (0.14) |
3.5. Comparison to TotalSegmentator
4. Discussion
4.1. Reasons for impaired prediction accuracy
4.2. Inter-observer variability, and tracheostomy analysis
4.3. Comparison to TotalSegmentator
4.4. Impact on CTV delineation
4.5. Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| aDM | Anterior Belly of the Digastric Muscle |
| CCA | Common Carotid Artery |
| CM | Constrictor Muscle |
| CT | Computertomographie |
| CTV | Clinical Target Volume |
| DICE | Sørensen–Dice Coefficient |
| DL | Deep Learning |
| HD | Hausdorff Distance |
| ICA | Internal Carotid Artery |
| OAR | Organs At Risk |
| PM | Platysma Muscle |
| pSM | Posterior Scalene Muscle |
| Q1 | 25th Percentile |
| Q3 | 75th Percentile |
| sDICE | Surface DICE |
| SuA | Subclavian Artery |
| TS | TotalSegmentator |
Appendix A
Appendix A.1. Standard Operation Procedure
-
Nasopharynx
- Cranial boundary: up to the nasal septum
- Caudal boundary: from the hard palate
-
Oropharynx
- Cranial boundary: from the hard palate
- Caudal boundary: epiglottis
-
Hypopharynx
- Cranial boundary: epiglottis
- Caudal boundary: transition to the esophagus, along with the caudal end of the cricoid cartilage
- Segmentation note: No clear caudal boundary, orientation is based on the larynx-air structure
-
Tongue (muscle)
- Bounded by the oral cavity
- Caudal boundary: tongue base (ambiguous border)
-
Thyroid cartilage
- Segment in the larynx window
- Boundary: entire cartilage structure (typical shape was always recognizable in 3D view)
-
Sternocleidomastoid muscle
- Cranial boundary: mastoid cells, up to the skull
- Caudal boundary: clavicle and sternal manubrium, occasional branching near the origin may be visible
-
Thyroid gland
- Bright structure, merging caudally, variable cranial boundary
-
Hyoid bone
- Segment in the bone window
- Boundary: entire bone structure (typical shape was always recognizable in 3D view)
-
Cricoid cartilage
- Segment in the larynx window
- Boundary: entire cartilaginous structure (typical shape usually visible in 3D view)
- Special note: Caudal boundary simultaneously limits hypopharynx, larynx air, and inferior constrictor
-
Pharyngeal constrictor muscles (superior, medius, inferior)
- S. from the level of upper jaw teeth caudally
- M. from hyoid cranially (both structures "meet" in the middle)
- I. from hyoid caudally to the caudal end of the cricoid cartilage
-
Esophagus
- Cranial boundary: caudal end of the cricoid cartilage
- Caudal boundary irrelevant for head-neck region, as the esophagus ends in the stomach
-
C1/vertebral bodies
- Segment in the bone window
-
Soft palate
- Cranial boundary: transition to hard palate
- Caudal boundary: uvula
-
Hard palate
- Segment in the larynx window
-
Larynx
- Cranial boundary: epiglottis
- Caudal boundary: caudal end of the cricoid cartilage
-
Mandible
- Segment in the bone window
- Teeth not segmented
-
Digastric muscle
- Cranial boundary: mandible
- Caudal boundary: until no longer visible
-
Nasal cavities
- Cranial boundary: until no longer visible
- Caudal boundary: together with nasopharynx
- Note: Exclude ethmoid cells
-
Oral cavities
- Includes tongue, uvula
- External Auditory Canal
-
Tonsils
- Bilateral at the level of uvula
-
Common Carotid Artery
- Cranial boundary: until artery bifurcation
- Caudal boundary: branching from brachiocephalic trunk
-
Sternal manubrium
- Segment in the bone window
- Note: Manubrium is posterior at transition with corpus sterni
-
Sternum body
- Segment in the bone window
- Note: Corpus is anterior at transition with manubrium
-
Clavicle
- Segment in the bone window
-
Zygomatic arch
- Segment in the bone window
- Ventral boundary: continuation from posterior edge of maxillary sinus
- Dorsal boundary: up to mastoid cells
-
Styloid process
- Segment in the bone window
- Cranial boundary: first slice where not connected to mastoid
- Caudal boundary: until no longer visible
-
Lung
- Segment in the lung window
- Often already exists
- ’Region growing’ with upper threshold = -300 and ’remove holes’, but avoid including trachea/air outside the patient (sometimes segmented, correct manually)
-
Trachea
- Cranial boundary: larynx air
- Caudal boundary: bifurcation
- Excludes bronchi
-
Internal Carotid Artery
- Cranial boundary: entry into the skull
- Caudal boundary: separation of common carotid
-
Internal Jugular Vein
- Cranial boundary: entry into the skull
- Caudal boundary: brachiocephalic vein
-
Trapezius muscles
- Cranial boundary: skull
- Caudal boundary: from the spine
- Note: At the clavicle, trapezius also extends anteriorly, creating a tight "hole" in segmentation where tendon lies
-
Platysma Muscle
- Boundaries not clear but segmented as long as visible course toward mandible
-
Brachiocephalic Artery
- Cranial boundary: up to division into common carotids
- Caudal boundary: from aortic arch
-
Brachiocephalic vein
- Cranial boundary: up to division into IJV
- Caudal boundary: from SVC division
-
Submandibular Gland
- Segment as long as visible within platysma
-
Levator Scapulae Muscle
- Cranial boundary: as far as possible
- Caudal boundary: from scapula
-
Scalenus muscles (anterior, medius, posterior)
- A. and M. around subclavian artery
- A. and M. originate from first rib
- P. often unclear, originates from second rib
- All three structures traced cranially as far as possible
-
Subclavian Artery
- Lateral boundary: up to cranial boundary of sternum
-
Skin
- Adopt from outline or external contour and correct significant errors from automatic contouring
-
Sterno-thyroid muscle
- Cranial boundary: first slice where thyroid cartilage is ventrally united
- Caudal boundary: first slice from manubrium
-
Thyro-hyoid muscle
- Cranial boundary: first slice where hyoid is visible
- Caudal boundary: first slice after sternothyroid
-
Pre-vertebral muscles (longus colli + longus capiti)
- Cranial boundary: up to visible dens axis
- Caudal boundary: as far as possible
Appendix A.2. Previously Reported DICE Values for Comparison
| Structure | Previously reported DICE (mean ± std) |
|---|---|
| Mandible | 0.86 ± 0.121[56], 0.90 ± 0.04 [54], 0.91 ± 0.02 [55], 0.94 ± 0.01 [52], 0.99 ± 0.01 [55] |
| Submandibular Gland (r) | 0.73 ± 0.09 [54], 0.78 ± 0.10 [52], 0.79 [51], 0.95 ± 0.07 [55], 0.98 ± 0.03 [55] |
| Submandibular Gland (l) | 0.70 ± 0.13 [54], 0.77 ± 0.12 [52], 0.79 [51], 0.91 ± 0.08 [55], 0.97 ± 0.05 [55] |
| Internal Carotid Artery (r) | 0.81 [49], 0.86 ± 0.02 [50] |
| Internal Carotid Artery (l) | 0.81 [49], 0.86 ± 0.02 [50] |
| Superior Constrictor | 0.67 ± 0.11 [59], 0.76 ± 0.13 [55], 0.83 ± 0.15 [55] |
| Middle Constrictor | 0.60 ± 0.19 [59], 0.76 ± 0.10 [55], 0.84 ± 0.01 [55] |
| Inferior Constrictor | 0.65 ± 0.12 [59], 0.71 ± 0.21 [55], 0.80 ± 0.24 [55] |
| Constrictors (s., m., i.) | 0.52 [51], 0.68 ± 0.08 [52] |
| Esophagus | 0.85 ± 0.10 [55], 0.91 ± 0.03[52], 0.93 ± 0.07 [55] |
| Oral Cavity | 0.85 ± 0.10 [55], 0.91 ± 0.03 [52], 0.93 ± 0.07 [55] |
| 1 |
https://github.com/wasserth/TotalSegmentator [Accessed: 2023-10-31] |
| 2 |
https://metrics-reloaded.dkfz.de/ [Accessed: 2023-10-20] |
References
- van der Veen, J.; Gulyban, A.; Nuyts, S. Interobserver variability in delineation of target volumes in head and neck cancer. Radiotherapy and Oncology 2019, 137, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Jeanneret Sozzi, W. The reasons for discrepancies in target volume delineation: a SASRO study on head-and-neck and prostate cancer. PhD thesis, Université de Lausanne, Faculté de biologie et médecine, 2006.
- Isgum, I.; Staring, M.; Rutten, A.; Prokop, M.; Viergever, M.A.; Van Ginneken, B. Multi-atlas-based segmentation with local decision fusion—application to cardiac and aortic segmentation in CT scans. IEEE transactions on medical imaging 2009, 28, 1000–1010. [Google Scholar] [CrossRef]
- Wu, M.; Rosano, C.; Lopez-Garcia, P.; Carter, C.S.; Aizenstein, H.J. Optimum template selection for atlas-based segmentation. NeuroImage 2007, 34, 1612–1618. [Google Scholar] [CrossRef]
- Cabezas, M.; Oliver, A.; Lladó, X.; Freixenet, J.; Cuadra, M.B. A review of atlas-based segmentation for magnetic resonance brain images. Computer methods and programs in biomedicine 2011, 104, e158–e177. [Google Scholar] [CrossRef]
- Young, A.V.; Wortham, A.; Wernick, I.; Evans, A.; Ennis, R.D. Atlas-based segmentation improves consistency and decreases time required for contouring postoperative endometrial cancer nodal volumes. International Journal of Radiation Oncology* Biology* Physics 2011, 79, 943–947. [Google Scholar] [CrossRef] [PubMed]
- Daisne, J.F.; Blumhofer, A. Atlas-based automatic segmentation of head and neck organs at risk and nodal target volumes: a clinical validation. Radiation oncology 2013, 8, 1–11. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, 5-9 October 2015; Proceedings, Part III 18. Springer, 2015; pp. 234–241. [Google Scholar]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.; Petersen, J.; Maier-Hein, K.H. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nature methods 2021, 18, 203–211. [Google Scholar] [CrossRef]
- Antonelli, M.; Reinke, A.; Bakas, S.; Farahani, K.; Kopp-Schneider, A.; Landman, B.A.; Litjens, G.; Menze, B.; Ronneberger, O.; Summers, R.M.; others. The medical segmentation decathlon. Nature communications 2022, 13, 4128. [Google Scholar] [CrossRef]
- Wasserthal, J.; Meyer, M.; Breit, H.C.; Cyriac, J.; Yang, S.; Segeroth, M. TotalSegmentator: robust segmentation of 104 anatomical structures in CT images. arXiv 2022, arXiv:2208.05868 2022. [Google Scholar] [CrossRef]
- Kirillov, A.; Mintun, E.; Ravi, N.; Mao, H.; Rolland, C.; Gustafson, L.; Xiao, T.; Whitehead, S.; Berg, A.C.; Lo, W.Y. ; others. Segment anything. arXiv 2023, arXiv:2304.02643 2023. [Google Scholar]
- Gare, G.R.; Schoenling, A.; Philip, V.; Tran, H.V.; Bennett, P.d.; Rodriguez, R.L.; Galeotti, J.M. Dense pixel-labeling for reverse-transfer and diagnostic learning on lung ultrasound for COVID-19 and pneumonia detection. 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI). IEEE, 2021, pp. 1406–1410.
- Scheikl, P.M.; Laschewski, S.; Kisilenko, A.; Davitashvili, T.; Müller, B.; Capek, M.; Müller-Stich, B.P.; Wagner, M.; Mathis-Ullrich, F. Deep learning for semantic segmentation of organs and tissues in laparoscopic surgery. Current Directions in Biomedical Engineering. De Gruyter, 2020, Vol. 6, p. 20200016.
- Bauer, C.J.; Teske, H.; Walter, A.; Hoegen, P.; Adeberg, S.; Debus, J.; Jäkel, O.; Giske, K. Biofidelic image registration for head and neck region utilizing an in-silico articulated skeleton as a transformation model. Physics in Medicine & Biology 2023, 68, 095006. [Google Scholar]
- Billot, B.; Greve, D.N.; Puonti, O.; Thielscher, A.; Van Leemput, K.; Fischl, B.; Dalca, A.V.; Iglesias, J.E.; others. SynthSeg: Segmentation of brain MRI scans of any contrast and resolution without retraining. Medical image analysis 2023, 86, 102789. [Google Scholar] [CrossRef] [PubMed]
- Siddique, N.; Paheding, S.; Elkin, C.P.; Devabhaktuni, V. U-net and its variants for medical image segmentation: A review of theory and applications. Ieee Access 2021, 9, 82031–82057. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, X.; Xiao, B.; Wang, S.; Miao, Z.; Sun, Y.; Zhang, F. Segmentation of organs-at-risk in cervical cancer CT images with a convolutional neural network. Physica Medica 2020, 69, 184–191. [Google Scholar] [CrossRef]
- Nikolov, S.; Blackwell, S.; Zverovitch, A.; Mendes, R.; Livne, M.; De Fauw, J.; Patel, Y.; Meyer, C.; Askham, H.; Romera-Paredes, B.; others. Clinically applicable segmentation of head and neck anatomy for radiotherapy: deep learning algorithm development and validation study. Journal of medical Internet research 2021, 23, e26151. [Google Scholar] [CrossRef]
- Kosmin, M.; Ledsam, J.; Romera-Paredes, B.; Mendes, R.; Moinuddin, S.; de Souza, D.; Gunn, L.; Kelly, C.; Hughes, C.; Karthikesalingam, A.; others. Rapid advances in auto-segmentation of organs at risk and target volumes in head and neck cancer. Radiotherapy and Oncology 2019, 135, 130–140. [Google Scholar] [CrossRef] [PubMed]
- Bi, N.; Wang, J.; Zhang, T.; Chen, X.; Xia, W.; Miao, J.; Xu, K.; Wu, L.; Fan, Q.; Wang, L.; others. Deep learning improved clinical target volume contouring quality and efficiency for postoperative radiation therapy in non-small cell lung cancer. Frontiers in oncology 2019, 9, 1192. [Google Scholar] [CrossRef] [PubMed]
- Cardenas, C.E.; Beadle, B.M.; Garden, A.S.; Skinner, H.D.; Yang, J.; Rhee, D.J.; McCarroll, R.E.; Netherton, T.J.; Gay, S.S.; Zhang, L.; others. Generating High-Quality Lymph Node Clinical Target Volumes for Head and Neck Cancer Radiation Therapy Using a Fully Automated Deep Learning-Based Approach. International Journal of Radiation Oncology* Biology* Physics 2021, 109, 801–812. [Google Scholar] [CrossRef]
- Weissmann, T.; Huang, Y.; Fischer, S.; Roesch, J.; Mansoorian, S.; Ayala Gaona, H.; Gostian, A.O.; Hecht, M.; Lettmaier, S.; Deloch, L.; others. Deep learning for automatic head and neck lymph node level delineation provides expert-level accuracy. Frontiers in Oncology 2023, 13, 1115258. [Google Scholar] [CrossRef]
- Grøvik, E.; Yi, D.; Iv, M.; Tong, E.; Rubin, D.; Zaharchuk, G. Deep learning enables automatic detection and segmentation of brain metastases on multisequence MRI. Journal of Magnetic Resonance Imaging 2020, 51, 175–182. [Google Scholar] [CrossRef]
- on Radiation Units, I.C. Prescribing, recording, and reporting photon beam therapy; Vol. 50, International Commission on Radiation Units & Measurements, 1993.
- Vorwerk, H.; Hess, C.F. Guidelines for delineation of lymphatic clinical target volumes for high conformal radiotherapy: head and neck region. Radiation Oncology 2011, 6, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Grégoire, V.; Ang, K.; Budach, W.; Grau, C.; Hamoir, M.; Langendijk, J.A.; Lee, A.; Le, Q.T.; Maingon, P.; Nutting, C.; others. Delineation of the neck node levels for head and neck tumors: a 2013 update. DAHANCA, EORTC, HKNPCSG, NCIC CTG, NCRI, RTOG, TROG consensus guidelines. Radiotherapy and Oncology 2014, 110, 172–181. [Google Scholar] [CrossRef] [PubMed]
- Grégoire, V.; Evans, M.; Le, Q.T.; Bourhis, J.; Budach, V.; Chen, A.; Eisbruch, A.; Feng, M.; Giralt, J.; Gupta, T.; others. Delineation of the primary tumour clinical target volumes (ctv-p) in laryngeal, hypopharyngeal, oropharyngeal and oral cavity squamous cell carcinoma: Airo, caca, dahanca, eortc, georcc, gortec, hknpcsg, hncig, iag-kht, lprhht, ncic ctg, ncri, nrg oncology, phns, sbrt, somera, sro, sshno, trog consensus guidelines. Radiotherapy and Oncology 2018, 126, 3–24. [Google Scholar]
- Haas-Kogan, D.A.; Barani, I.J.; Hayden, M.G.; Edwards, M.S.; Fisher, P.G. 53 - Pediatric Central Nervous System Tumors. In Leibel and Phillips Textbook of Radiation Oncology (Third Edition), Third Edition ed.; Hoppe, R.T., Phillips, T.L., Roach, M., Eds.; W.B. Saunders: Philadelphia, 2010; pp. 1111–1129. [Google Scholar] [CrossRef]
- Dawson, L.A.; Anzai, Y.; Marsh, L.; Martel, M.K.; Paulino, A.; Ship, J.A.; Eisbruch, A. Patterns of local-regional recurrence following parotid-sparing conformal and segmental intensity-modulated radiotherapy for head and neck cancer. International Journal of Radiation Oncology* Biology* Physics 2000, 46, 1117–1126. [Google Scholar] [CrossRef]
- Chao, K.C.; Ozyigit, G.; Tran, B.N.; Cengiz, M.; Dempsey, J.F.; Low, D.A. Patterns of failure in patients receiving definitive and postoperative IMRT for head-and-neck cancer. International Journal of Radiation Oncology* Biology* Physics 2003, 55, 312–321. [Google Scholar] [CrossRef]
- Evans, E.; Radhakrishna, G.; Gilson, D.; Hoskin, P.; Miles, E.; Yuille, F.; Dickson, J.; Gwynne, S. Target volume delineation training for clinical oncology trainees: the Role of ARENA and COPP. Clinical Oncology 2019, 31, 341–343. [Google Scholar] [CrossRef]
- Cardenas, C.E.; Anderson, B.M.; Aristophanous, M.; Yang, J.; Rhee, D.J.; McCarroll, R.E.; Mohamed, A.S.; Kamal, M.; Elgohari, B.A.; Elhalawani, H.M.; others. Auto-delineation of oropharyngeal clinical target volumes using 3D convolutional neural networks. Physics in Medicine & Biology 2018, 63, 215026. [Google Scholar]
- Strijbis, V.I.; Dahele, M.; Gurney-Champion, O.J.; Blom, G.J.; Vergeer, M.R.; Slotman, B.J.; Verbakel, W.F. Deep Learning for Automated Elective Lymph Node Level Segmentation for Head and Neck Cancer Radiotherapy. Cancers 2022, 14, 5501. [Google Scholar] [CrossRef]
- Kazemimoghadam, M.; Yang, Z.; Chen, M.; Rahimi, A.; Kim, N.; Alluri, P.; Nwachukwu, C.; Lu, W.; Gu, X. A deep learning approach for automatic delineation of clinical target volume in stereotactic partial breast irradiation (S-PBI). Physics in Medicine & Biology 2023, 68, 105011. [Google Scholar]
- Shi, J.; Ding, X.; Liu, X.; Li, Y.; Liang, W.; Wu, J. Automatic clinical target volume delineation for cervical cancer in CT images using deep learning. Medical Physics 2021, 48, 3968–3981. [Google Scholar] [CrossRef]
- Balagopal, A.; Nguyen, D.; Morgan, H.; Weng, Y.; Dohopolski, M.; Lin, M.H.; Barkousaraie, A.S.; Gonzalez, Y.; Garant, A.; Desai, N.; others. A deep learning-based framework for segmenting invisible clinical target volumes with estimated uncertainties for post-operative prostate cancer radiotherapy. Medical image analysis 2021, 72, 102101. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Liu, X.; Guan, H.; Zhen, H.; Sun, Y.; Chen, Q.; Chen, Y.; Wang, S.; Qiu, J. Development and validation of a deep learning algorithm for auto-delineation of clinical target volume and organs at risk in cervical cancer radiotherapy. Radiotherapy and Oncology 2020, 153, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Reinke, A.; Tizabi, M.D.; Baumgartner, M.; Eisenmann, M.; Heckmann-Nötzel, D.; Kavur, A.E.; Rädsch, T.; Sudre, C.H.; Acion, L.; Antonelli, M.; others. Understanding metric-related pitfalls in image analysis validation. ArXiv 2023. [Google Scholar] [CrossRef] [PubMed]
- Bejarano, T.; De Ornelas Couto, M.; Mihaylov, I.B. Head-and-neck squamous cell carcinoma patients with CT taken during pre-treatment, mid-treatment, and post-treatment Dataset. The Cancer Imaging Archive 2018, 10, K9. [Google Scholar]
- Bejarano, T.; De Ornelas-Couto, M.; Mihaylov, I.B. Longitudinal fan-beam computed tomography dataset for head-and-neck squamous cell carcinoma patients. Medical physics 2019, 46, 2526–2537. [Google Scholar] [CrossRef] [PubMed]
- Clark, K.; Vendt, B.; Smith, K.; Freymann, J.; Kirby, J.; Koppel, P.; Moore, S.; Phillips, S.; Maffitt, D.; Pringle, M.; others. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. Journal of digital imaging 2013, 26, 1045–1057. [Google Scholar] [CrossRef]
- Giske, K.; Stoiber, E.M.; Schwarz, M.; Stoll, A.; Muenter, M.W.; Timke, C.; Roeder, F.; Debus, J.; Huber, P.E.; Thieke, C.; others. Local setup errors in image-guided radiotherapy for head and neck cancer patients immobilized with a custom-made device. International Journal of Radiation Oncology* Biology* Physics 2011, 80, 582–589. [Google Scholar] [CrossRef]
- Stoiber, E.M.; Bougatf, N.; Teske, H.; Bierstedt, C.; Oetzel, D.; Debus, J.; Bendl, R.; Giske, K. Analyzing human decisions in IGRT of head-and-neck cancer patients to teach image registration algorithms what experts know. Radiation Oncology 2017, 12, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Isensee, F. nnU-Net V2. 2023. https://github.com/MIC-DKFZ/nnUNet/releases/tag/v2.0 (accessed on 31 October 2023).
- Dice, L.R. Measures of the amount of ecologic association between species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]
- Sorensen, T. A method of establishing groups of equal amplitude in plant sociology based on similarity of species content and its application to analyses of the vegetation on Danish commons. Biologiske skrifter 1948, 5, 1–34. [Google Scholar]
- Rogers, C.A. Hausdorff measures; Cambridge University Press, 1998.
- Nikan, S.; Van Osch, K.; Bartling, M.; Allen, D.G.; Rohani, S.A.; Connors, B.; Agrawal, S.K.; Ladak, H.M. PWD-3DNet: a deep learning-based fully-automated segmentation of multiple structures on temporal bone CT scans. IEEE Transactions on Image Processing 2020, 30, 739–753. [Google Scholar] [CrossRef] [PubMed]
- Ke, J.; Lv, Y.; Ma, F.; Du, Y.; Xiong, S.; Wang, J.; Wang, J. Deep learning-based approach for the automatic segmentation of adult and pediatric temporal bone computed tomography images. Quantitative Imaging in Medicine and Surgery 2023, 13, 1577. [Google Scholar] [CrossRef]
- Thomson, D.; Boylan, C.; Liptrot, T.; Aitkenhead, A.; Lee, L.; Yap, B.; Sykes, A.; Rowbottom, C.; Slevin, N. Evaluation of an automatic segmentation algorithm for definition of head and neck organs at risk. Radiation Oncology 2014, 9, 1–12. [Google Scholar] [CrossRef]
- Van Dijk, L.V.; Van den Bosch, L.; Aljabar, P.; Peressutti, D.; Both, S.; Steenbakkers, R.J.; Langendijk, J.A.; Gooding, M.J.; Brouwer, C.L. Improving automatic delineation for head and neck organs at risk by Deep Learning Contouring. Radiotherapy and Oncology 2020, 142, 115–123. [Google Scholar] [CrossRef]
- Gite, S.; Mishra, A.; Kotecha, K. Enhanced lung image segmentation using deep learning. Neural Computing and Applications 2022, 1–15. [Google Scholar] [CrossRef]
- Ibragimov, B.; Xing, L. Segmentation of organs-at-risks in head and neck CT images using convolutional neural networks. Medical physics 2017, 44, 547–557. [Google Scholar] [CrossRef]
- Van der Veen, J.; Willems, S.; Deschuymer, S.; Robben, D.; Crijns, W.; Maes, F.; Nuyts, S. Benefits of deep learning for delineation of organs at risk in head and neck cancer. Radiotherapy and Oncology 2019, 138, 68–74. [Google Scholar] [CrossRef]
- Watkins, W.T.; Qing, K.; Han, C.; Hui, S.; Liu, A. Auto-segmentation for total marrow irradiation. Frontiers in Oncology 2022, 12, 970425. [Google Scholar] [CrossRef] [PubMed]
- Belal, S.L.; Sadik, M.; Kaboteh, R.; Enqvist, O.; Ulén, J.; Poulsen, M.H.; Simonsen, J.; Høilund-Carlsen, P.F.; Edenbrandt, L.; Trägårdh, E. Deep learning for segmentation of 49 selected bones in CT scans: first step in automated PET/CT-based 3D quantification of skeletal metastases. European journal of radiology 2019, 113, 89–95. [Google Scholar] [CrossRef]
- Liu, S.; Xie, Y.; Reeves, A.P. Segmentation of the sternum from low-dose chest CT images. Medical Imaging 2015: Computer-Aided Diagnosis. SPIE, 2015, Vol. 9414, pp. 8–17.
- Li, Y.; Rao, S.; Chen, W.; Azghadi, S.F.; Nguyen, K.N.B.; Moran, A.; Usera, B.M.; Dyer, B.A.; Shang, L.; Chen, Q.; others. Evaluating automatic segmentation for swallowing-related organs for head and neck cancer. Technology in Cancer Research & Treatment 2022, 21, 15330338221105724. [Google Scholar]
- Weber, K.A.; Abbott, R.; Bojilov, V.; Smith, A.C.; Wasielewski, M.; Hastie, T.J.; Parrish, T.B.; Mackey, S.; Elliott, J.M. Multi-muscle deep learning segmentation to automate the quantification of muscle fat infiltration in cervical spine conditions. Scientific reports 2021, 11, 16567. [Google Scholar] [CrossRef] [PubMed]
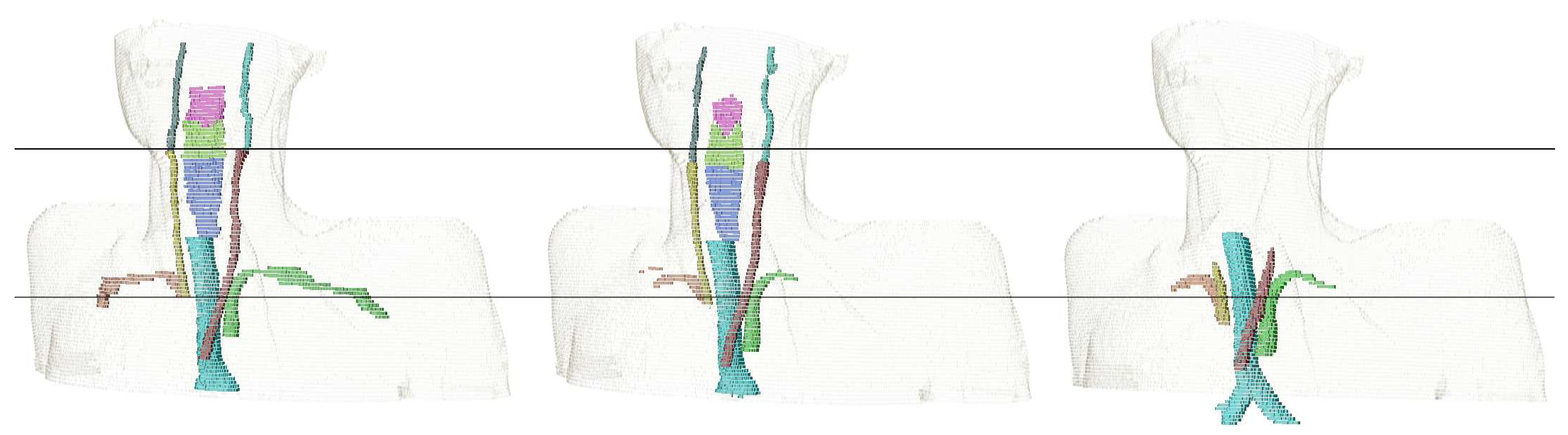
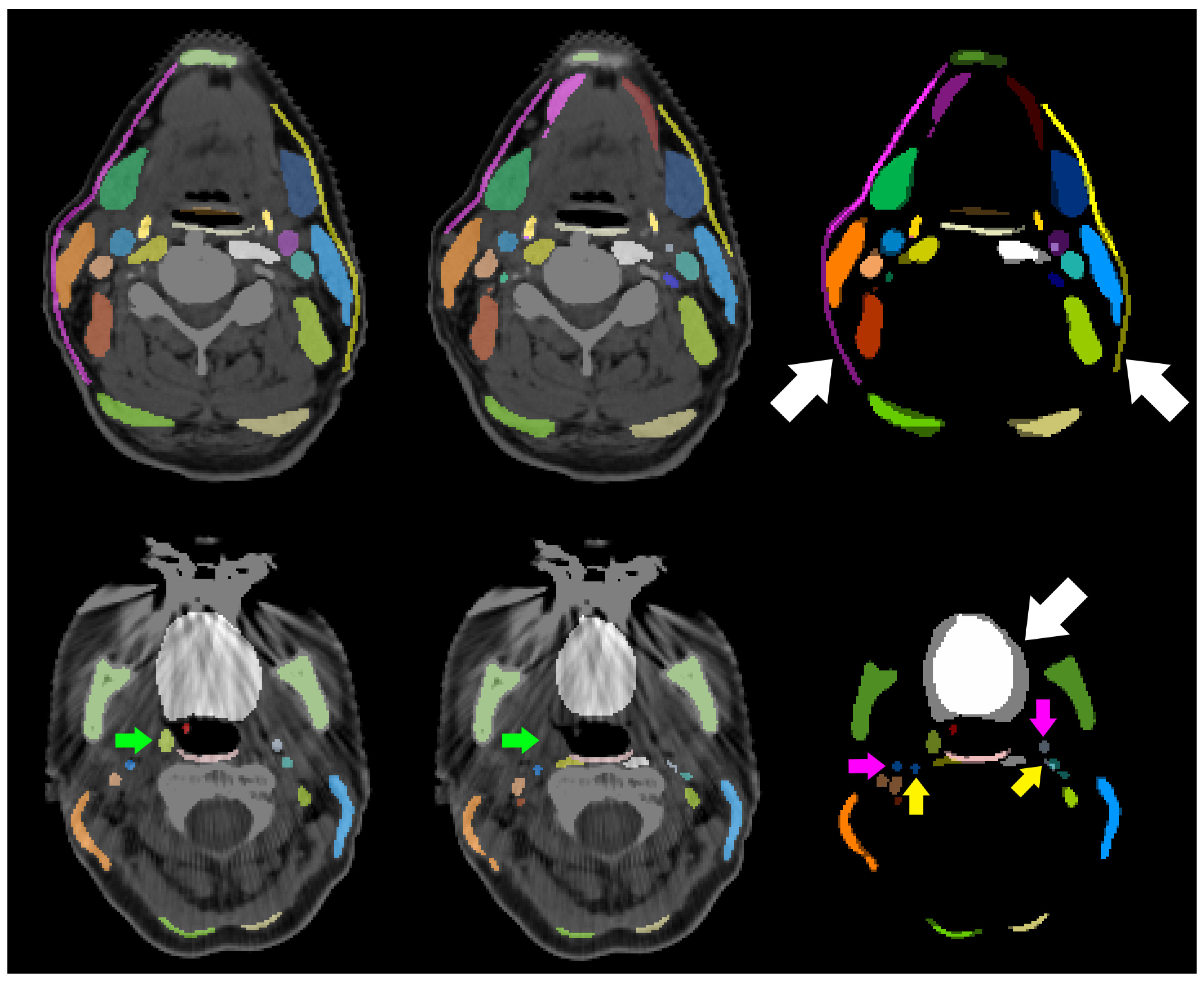
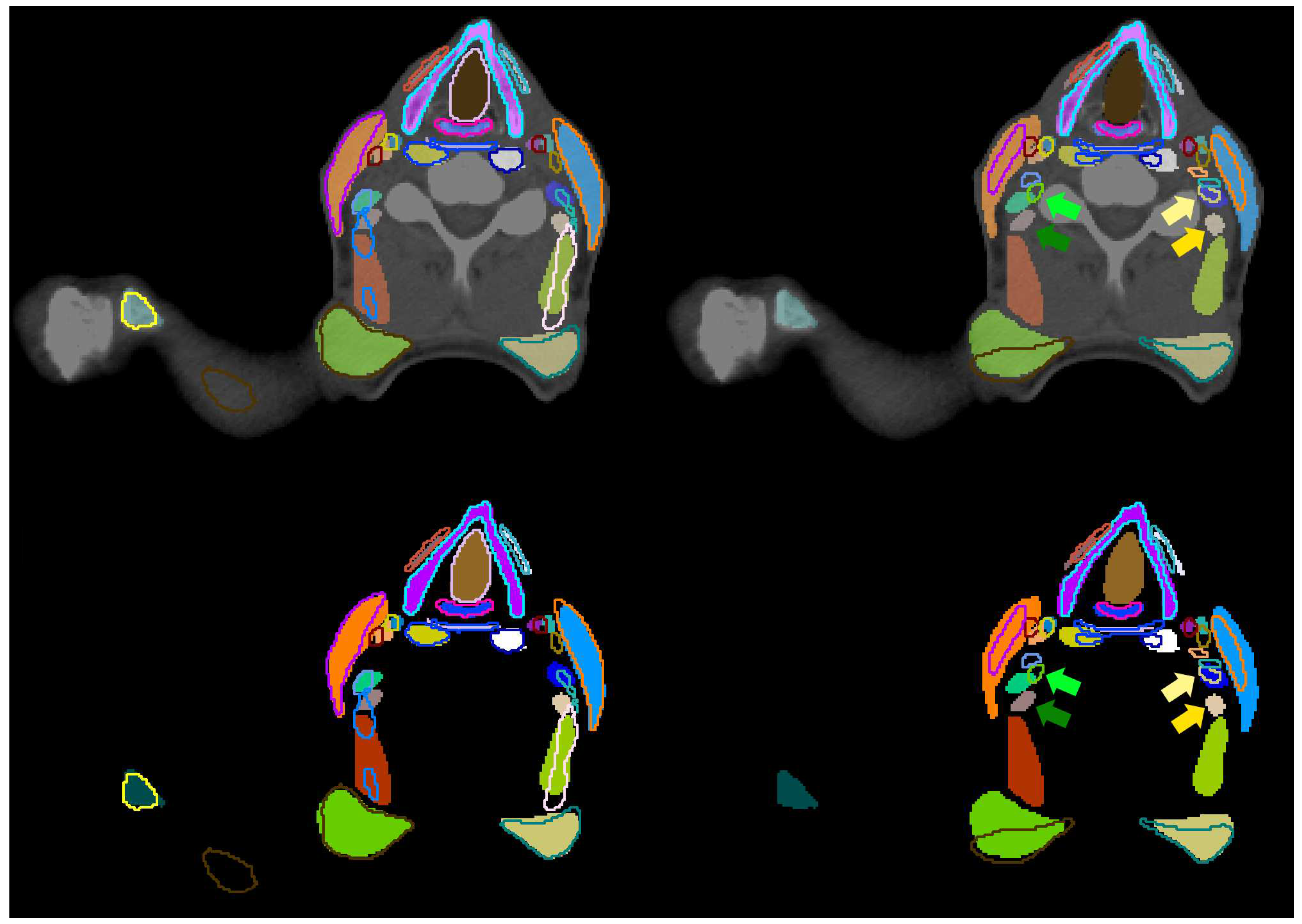
| Structure | pred. vs. manual | interobserver | literature | |
|---|---|---|---|---|
| Air | Auditory Canal (l) | 0.77 ± 0.09 | 0.83 ± 0.02 [50]2 | |
| Auditory Canal (r) | 0.80 ± 0.10 | 0.83 ± 0.02 [50]2 | ||
| Larynx (air) | 0.86 ± 0.06 | |||
| Lung (l) | 0.99 ± 0.01 | 0.98 [53]1, 2 | ||
| Lung (r) | 0.99 ± 0.01 | 0.98 [53]1, 2 | ||
| Trachea | 0.90 ± 0.07 | |||
| Bones | Cheek Bone (l) | 0.78 ± 0.04 | ||
| Cheek Bone (r) | 0.78 ± 0.06 | |||
| Clavicle (l) | 0.93 ± 0.02 | |||
| Clavicle (r) | 0.93 ± 0.01 | |||
| Hyoid Bone | 0.82 ± 0.07 | 0.76 | ||
| Mandible | 0.88 ± 0.06 | 0.78 | [0.86 - 0.99] [52,54,55,56] | |
| Sternum (M., C.) | 0.93 ± 0.04 | 0.83 [57]1 | ||
| Sternum Corpus | 0.82 ± 0.22 | 0.90 ± 0.03 [58]1 | ||
| Sternum Manubrium | 0.90 ± 0.06 | 0.88 | ||
| Styloid Process (l) | 0.72 ± 0.14 | |||
| Styloid Process (r) | 0.77 ± 0.08 | |||
| Vertebra C1 | 0.86 ± 0.04 | 0.84 | ||
| Ca. | Cricoid Cartilage | 0.69 ± 0.15 | 0.78 | 0.66 ± 0.12 [52] |
| Thyroid Cartilage | 0.85 ± 0.06 | 0.85 | ||
| Gland | Submandibular Gland (l) | 0.77 ± 0.17 | [0.70 - 0.97] [51,52,54,55] | |
| Submandibular Gland (r) | 0.78 ± 0.13 | [0.73 - 0.98] [51,52,54,55] | ||
| Thyroid Gland | 0.81 ± 0.13 | 0.83 ± 0.08 [52] | ||
| Vessels | Brachiocephalic Artery | 0.84 ± 0.06 | 0.85 | |
| Brachiocephalic Vein (l) | 0.82 ± 0.10 | 0.77 | ||
| Brachiocephalic Vein (r) | 0.82 ± 0.07 | 0.76 | ||
| Common Carotid Artery (l) | 0.81 ± 0.08 | 0.72 | ||
| Common Carotid Artery (r) | 0.78 ± 0.10 | 0.50 | ||
| Internal Carotid Artery (l) | 0.61 ± 0.15 | 0.25 | 0.81, 0.86 [49,50]2 | |
| Internal Carotid Artery (r) | 0.55 ± 0.22 | 0.49 | 0.81, 0.86 [49,50]2 | |
| Internal Jugular Vein (l) | 0.78 ± 0.13 | 0.45 | ||
| Internal Jugular Vein (r) | 0.75 ± 0.18 | 0.53 | ||
| Subclavian Artery (l) | 0.74 ± 0.09 | 0.54 | ||
| Subclavian Artery (r) | 0.74 ± 0.13 | 0.34 | ||
| Muscles | Constrictors (s., m., i.) | 0.56 ± 0.12 | 0.74 | 0.52, 0.68 [51,52] |
| Inferior Constrictor | 0.44 ± 0.16 | 0.54 | [0.65 - 0.80] [55,59] | |
| Middle Constrictor | 0.45 ± 0.18 | 0.66 | [0.60 - 0.84] [55,59] | |
| Superior Constrictor | 0.48 ± 0.19 | 0.42 | [0.67 - 0.83] [55,59] | |
| Digastric (l) | 0.52 ± 0.24 | 0.39 | ||
| Digastric (r) | 0.46 ± 0.28 | 0.33 | ||
| Levator Scapulae (l) | 0.87 ± 0.05 | 0.76 ± 0.01 [60] | ||
| Levator Scapulae (r) | 0.83 ± 0.07 | 0.76 ± 0.01 [60] | ||
| Platysma (l) | 0.59 ± 0.12 | |||
| Platysma (r) | 0.52 ± 0.16 | |||
| Prevertebral (l) | 0.74 ± 0.07 | 0.53 | 0.70 ± 0.01 [60] | |
| Prevertebral (r) | 0.76 ± 0.06 | 0.50 | 0.71 ± 0.01 [60] | |
| Scalene (an., me., p.) (l) | 0.74 ± 0.09 | 0.44 | ||
| Scalene (an., me., p.) (r) | 0.71 ± 0.11 | 0.03 | ||
| Anterior Scalene (l) | 0.82 ± 0.06 | 0.60 | ||
| Anterior Scalene (r) | 0.80 ± 0.06 | 0.00 | ||
| Muscles | Medius Scalene (l) | 0.68 ± 0.10 | 0.14 | |
| Medius Scalene (r) | 0.66 ± 0.16 | 0.03 | ||
| Posterior Scalene (l) | 0.40 ± 0.20 | 0.01 | ||
| Posterior Scalene (r) | 0.42 ± 0.28 | 0.00 | ||
| Sternothyroid (l) | 0.58 ± 0.08 | |||
| Sternothyroid (r) | 0.59 ± 0.09 | |||
| Sternocleidomastoid (l) | 0.84 ± 0.07 | 0.51 | 0.73 ± 0.02 [60] | |
| Sternocleidomastoid (r) | 0.81 ± 0.15 | 0.52 | 0.74 ± 0.02 [60] | |
| Thyrohyoid (l) | 0.50 ± 0.17 | 0.48 | ||
| Thyrohyoid (r) | 0.56 ± 0.12 | 0.56 | ||
| Trapezius (l) | 0.90 ± 0.03 | 0.65* | 0.41 ± 0.04 [60] | |
| Trapezius (r) | 0.89 ± 0.04 | 0.72* | 0.45 ± 0.04 [60] | |
| Tongue | 0.63 ± 0.17 | |||
| Esophagus | 0.80 ± 0.10 | [0.55 - 0.83] [52,55]3 | ||
| Hard Palate | 0.63 ± 0.13 | |||
| Hypopharynx | 0.64 ± 0.15 | 0.71 | ||
| Nasal Cavity (l) | 0.86 ± 0.03 | |||
| Nasal Cavity (r) | 0.86 ± 0.03 | |||
| Nasopharynx | 0.83 ± 0.09 | 0.74 | ||
| Oral Cavity | 0.85 ± 0.07 | [0.85 - 0.93] [52,55] | ||
| Oropharynx | 0.84 ± 0.09 | 0.83 | ||
| Pharynx (nasop., orop., hyp.) | 0.82 ± 0.07 | 0.83 | 0.69 ± 0.06 [54] | |
| Skin | 0.99 ± 0.00 | |||
| Soft Palate | 0.61 ± 0.19 | |||
| Tonsil (l) | 0.08 ± 0.13 | 0.12 | ||
| Tonsil (r) | 0.12 ± 0.15 | 0.15 |
| HD (95) | sDICE (2 mm) | ||||
| Structure | pred. vs. manual | interobserver | pred. vs. manual | interobserver | |
| Air | Auditory Canal (l) | 5.16 ± 2.94 | 0.88 ± 0.08 | ||
| Auditory Canal (r) | 4.76 ± 3.16 | 0.89 ± 0.09 | |||
| Larynx (air) | 6.74 ± 4.13 | 0.89 ± 0.06 | |||
| Lung (l) | 1.42 ± 1.00 | 0.97 ± 0.03 | |||
| Lung (r) | 1.50 ± 0.86 | 0.98 ± 0.02 | |||
| Trachea | 6.87 ± 5.49 | 0.90 ± 0.08 | |||
| Bones | Cheek Bone (l) | 4.23 ± 2.89 | 0.92 ± 0.05 | ||
| Cheek Bone (r) | 4.36 ± 3.37 | 0.92 ± 0.07 | |||
| Clavicle (l) | 1.33 ± 0.67 | 0.98 ± 0.02 | |||
| Clavicle (r) | 1.25 ± 0.49 | 0.98 ± 0.01 | |||
| Hyoid Bone | 3.23 ± 3.77 | 1.96 | 0.95 ± 0.06 | 0.97 | |
| Mandible | 2.31 ± 1.67 | 2.77 | 0.96 ± 0.04 | 0.88 | |
| Sternum (M., C.) | 1.98 ± 1.63 | 0.97 ± 0.04 | |||
| Sternum Corpus | 5.87 ± 6.69 | 0.87 ± 0.20 | |||
| Sternum Manubrium | 3.99 ± 4.18 | 3.00 | 0.93 ± 0.08 | 0.93 | |
| Styloid Process (l) | 5.72 ± 9.58 | 0.92 ± 0.13 | |||
| Styloid Process (r) | 2.01 ± 0.97 | 0.97 ± 0.03 | |||
| Vertebra C1 | 3.07 ± 1.24 | 3.16 | 0.93 ± 0.04 | 0.90 | |
| Ca. | Cricoid Cartilage | 6.15 ± 3.30 | 3.16 | 0.82 ± 0.14 | 0.92 |
| Thyroid Cartilage | 2.40 ± 2.10 | 0.98 | 0.96 ± 0.04 | 0.98 | |
| Gland | Submandibular Gland (l) | 5.04 ± 4.28 | 0.85 ± 0.15 | ||
| Submandibular Gland (r) | 4.50 ± 2.69 | 0.80 ± 0.23 | |||
| Thyroid Gland | 6.12 ± 9.45 | 0.89 ± 0.13 | |||
| Vessels | Brachiocephalic Artery | 3.90 ± 2.66 | 3.00 | 0.89 ± 0.09 | 0.96 |
| Brachiocephalic Vein (l) | 3.53 ± 1.58 | 6.00 | 0.90 ± 0.08 | 0.88 | |
| Brachiocephalic Vein (r) | 4.88 ± 2.09 | 4.08 | 0.86 ± 0.07 | 0.85 | |
| Common Carotid Artery (l) | 5.01 ± 7.04 | 2.94 | 0.94 ± 0.06 | 0.94 | |
| Common Carotid Artery (r) | 3.48 ± 2.69 | 4.38 | 0.92 ± 0.07 | 0.81 | |
| Internal Carotid Artery (l) | 7.53 ± 8.95 | 11.17 | 0.84 ± 0.12 | 0.38* | |
| Internal Carotid Artery (r) | 13.85 ± 15.86 | 4.38 | 0.75 ± 0.20 | 0.80 | |
| Internal Jugular Vein (l) | 9.57 ± 23.20 | 9.00 | 0.91 ± 0.10 | 0.64 | |
| Internal Jugular Vein (r) | 8.25 ± 14.72 | 6.20 | 0.87 ± 0.14 | 0.73 | |
| Subclavian Artery (l) | 16.36 ± 19.40 | 81.22* | 0.84 ± 0.11 | 0.54 | |
| Subclavian Artery (r) | 10.27 ± 12.35 | 75.01* | 0.83 ± 0.12 | 0.42* | |
| Muscles | Constrictors (s., m., i.) | 7.19 ± 6.40 | 3.00 | 0.89 ± 0.08 | 0.95 |
| Inferior Constrictor | 7.10 ± 6.16 | 2.77 | 0.82 ± 0.16 | 0.95 | |
| Middle Constrictor | 9.66 ± 6.41 | 9.00 | 0.72 ± 0.18 | 0.88 | |
| Superior Constrictor | 11.23 ± 8.38 | 9.00 | 0.73 ± 0.22 | 0.75 | |
| Digastric (l) | 6.08 ± 3.90 | 6.30 | 0.73 ± 0.22 | 0.58 | |
| Digastric (r) | 8.52 ± 5.28 | 6.96 | 0.64 ± 0.30 | 0.52 | |
| Levator Scapulae (l) | 3.86 ± 2.05 | 0.92 ± 0.05 | |||
| Levator Scapulae (r) | 5.26 ± 2.87 | 0.88 ± 0.07 | |||
| Platysma (l) | 13.02 ± 9.59 | 0.82 ± 0.12 | |||
| Platysma (r) | 19.40 ± 11.75 | 0.75 ± 0.17 | |||
| Prevertebral (l) | 7.35 ± 8.25 | 6.86 | 0.90 ± 0.05 | 0.75 | |
| Prevertebral (r) | 7.29 ± 8.51 | 6.28 | 0.91 ± 0.05 | 0.73* | |
| Scalene (an., me., p.) (l) | 5.74 ± 3.20 | 13.09 | 0.86 ± 0.08 | 0.64 | |
| Scalene (an., me., p.) (r) | 7.59 ± 5.19 | 15.80 | 0.82 ± 0.10 | 0.21* | |
| Anterior Scalene (l) | 7.36 ± 9.67 | 15.00 | 0.92 ± 0.07 | 0.85 | |
| Anterior Scalene (r) | 8.19 ± 9.73 | 16.69 | 0.89 ± 0.07 | 0.17* | |
| Medius Scalene (l) | 6.06 ± 2.84 | 9.82 | 0.81 ± 0.10 | 0.42* | |
| Medius Scalene (r) | 7.63 ± 4.11 | 19.16 | 0.78 ± 0.11 | 0.21 | |
| Posterior Scalene (l) | 14.84 ± 8.84 | 17.71 | 0.56 ± 0.23 | 0.14 | |
| Posterior Scalene (r) | 17.16 ± 16.53 | 19.45 | 0.57 ± 0.30 | 0.10 | |
| Sternothyroid (l) | 4.48 ± 2.36 | 0.89 ± 0.08 | |||
| Sternothyroid (r) | 4.87 ± 2.03 | 0.89 ± 0.08 | |||
| Sternocleidomastoid (l) | 4.94 ± 5.34 | 22.57 | 0.92 ± 0.08 | 0.50 | |
| Sternocleidomastoid (r) | 12.31 ± 24.65 | 20.98 | 0.88 ± 0.15 | 0.54 | |
| Thyrohyoid (l) | 4.16 ± 2.68 | 3.10 | 0.86 ± 0.12 | 0.91 | |
| Thyrohyoid (r) | 3.08 ± 1.18 | 4.04 | 0.90 ± 0.07 | 0.87 | |
| Trapezius (l) | 2.38 ± 0.76 | 12.96 | 0.96 ± 0.03 | 0.69 | |
| Trapezius (r) | 2.43 ± 0.59 | 9.42 | 0.95 ± 0.04 | 0.71 | |
| Tongue | 13.29 ± 5.51 | 0.43 ± 0.17 | |||
| Esophagus | 6.15 ± 5.92 | 0.88 ± 0.10 | |||
| Hard Palate | 7.60 ± 4.08 | 0.73 ± 0.12 | |||
| Hypopharynx | 6.74 ± 3.85 | 2.94 | 0.83 ± 0.12 | 0.93 | |
| Nasal Cavity (l) | 2.30 ± 0.79 | 0.96 ± 0.02 | |||
| Nasal Cavity (r) | 2.26 ± 0.74 | 0.96 ± 0.02 | |||
| Nasopharynx | 4.84 ± 3.35 | 4.94 | 0.84 ± 0.12 | 0.72 | |
| Oral Cavity | 7.56 ± 3.80 | 0.67 ± 0.12 | |||
| Oropharynx | 6.40 ± 4.89 | 6.00 | 0.88 ± 0.09 | 0.83 | |
| Pharynx (nasop., orop., hyp.) | 5.15 ± 2.78 | 3.30 | 0.89 ± 0.06 | 0.91 | |
| Skin | 1.88 ± 1.08 | 0.96 ± 0.05 | |||
| Soft Palate | 9.33 ± 7.89 | 0.75 ± 0.18 | |||
| Tonsil (l) | 10.57 ± 8.90 | 15.00 | 0.20 ± 0.23 | 0.26 | |
| Tonsil (r) | 11.15 ± 8.19 | 15.13 | 0.28 ± 0.27 | 0.31 |
| Structure | pred. vs. manual | diff. |
|---|---|---|
| Lung (l) | 0.98 ± 0.01 | -0.01 |
| Lung (r) | 0.98 ± 0.01 | -0.01 |
| Trachea | 0.80 ± 0.06 | -0.10 |
| Clavicle (l) | 0.89 ± 0.03 | -0.04 |
| Clavicle (r) | 0.88 ± 0.02 | -0.06 |
| Sternum (M., C.) | 0.90 ± 0.02 | -0.02 |
| Vertebra C1 | 0.81 ± 0.04 | -0.05 |
| Thyroid Gland | 0.71 ± 0.14 | -0.10 |
| Brachiocephalic Artery | 0.75 ± 0.07 | -0.09 |
| Brachiocephalic Vein (l) | 0.76 ± 0.10 | -0.05 |
| Brachiocephalic Vein (r) | 0.72 ± 0.08 | -0.10 |
| Common Carotid Artery (l) | 0.64 ± 0.13 | -0.17 |
| Common Carotid Artery (r) | 0.55 ± 0.18 | -0.23 |
| Subclavian Artery (l) | 0.67 ± 0.10 | -0.07 |
| Subclavian Artery (r) | 0.65 ± 0.14 | -0.09 |
| Esophagus | 0.77 ± 0.09 | -0.04 |
| HD (95) | sDICE (2 mm) | |||
|---|---|---|---|---|
| Structure | pred. vs. manual | diff. | pred. vs. manual | diff. |
| Lung (l) | 2.18 ± 1.31 | 0.76 | 0.97 ± 0.03 | -0.01 |
| Lung (r) | 1.91 ± 1.31 | 0.41 | 0.97 ± 0.01 | 0.00 |
| Trachea | 16.04 ± 6.73 | 9.17 | 0.80 ± 0.09 | -0.10 |
| Clavicle (l) | 2.54 ± 1.82 | 1.21 | 0.96 ± 0.03 | -0.02 |
| Clavicle (r) | 2.83 ± 1.69 | 1.57 | 0.94 ± 0.03 | -0.04 |
| Sternum (M., C.) | 2.98 ± 1.45 | 1.00 | 0.94 ± 0.03 | -0.03 |
| Vertebra C1 | 3.70 ± 1.52 | 0.63 | 0.90 ± 0.06 | -0.03 |
| Thyroid Gland | 8.89 ± 8.70 | 2.77 | 0.79 ± 0.15 | -0.11 |
| Brachiocephalic Artery | 9.29 ± 5.16 | 5.39 | 0.80 ± 0.08 | -0.09 |
| Brachiocephalic Vein (l) | 5.82 ± 2.07 | 2.28 | 0.86 ± 0.08 | -0.04 |
| Brachiocephalic Vein (r) | 7.68 ± 2.96 | 2.80 | 0.79 ± 0.08 | -0.07 |
| Common Carotid Artery (l) | 25.15 ± 17.16 | 20.14 | 0.80 ± 0.13 | -0.13 |
| Common Carotid Artery (r) | 28.41 ± 20.01 | 24.94 | 0.71 ± 0.17 | -0.22 |
| Subclavian Artery (l) | 23.94 ± 16.66 | 7.58 | 0.79 ± 0.10 | -0.05 |
| Subclavian Artery (r) | 20.88 ± 17.13 | 10.61 | 0.75 ± 0.14 | -0.08 |
| Esophagus | 9.80 ± 9.62 | 3.65 | 0.85 ± 0.10 | -0.03 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





