1. Introduction
Malnutrition is highly prevalent in patients with chronic kidney disease (CKD), particularly those on hemodialysis and is associated with increased mortality rates, decreased physical function and poorer outcomes. The etiology of malnutrition is multifactorial and includes decreased protein-calorie intake, inflammation, hypercatabolism, physical inactivity, nutrient loss into the dialysate, metabolic acidosis, uremic toxicity, nutrient malabsorption, among others [
1]. Recently, our group described that a novel oral nutritional supplement (ONS), designed for malnourished (or at-risk) hemodialysis patients with a "similar to the Mediterranean diet" model (containing functional nutrients such as extra virgin olive oil, n-3 fatty acids, whey protein, fiber and antioxidants), improved caloric and protein intake, nutritional status (especially fat-free mass), and inflammation and oxidation biomarkers. Furthermore, the addition of probiotics may act synergistically with the ONS components to improve these biomarkers [
2].
MicroRNAs (miRNAs) are a class of small, single-stranded, non-coding RNA molecules that play an important role in the regulation of gene expression. Most miRNAs are transcribed from DNA sequences as primary miRNAs and processed into precursor miRNAs and finally mature miRNAs [
3]. In general, miRNAs interact with the 3' untranslated region (3' UTR) of target mRNAs, causing mRNA degradation and translational repression, but under certain conditions miRNAs can activate translation or regulate transcription [
4]. miRNAs are very stable in human biological fluids, including blood and serum, as they are packaged in the blood vessel membrane (such as exosomes, microparticles, and apoptotic bodies) and associated with RNA-binding proteins. Many studies highlight the role of circulating microRNAs as biomarkers for several diseases [
5]. miRNAs are involved in the normal kidney function and development. They are also implicated in several renal diseases, including CKD, where patients show specific circulating miRNA expression profiles associated with the regulation of metabolic, muscle and inflammation function and specific miRNAs have been identified as key players in the fibrosis process [
6,
7]. The expression of miRNAs is potentially altered by external factors such as diet [
8,
9] or the microbiota [
10]. These miRNA profiles do not seem to vary before or after dialysis since most of them are not removed by hemodialysis membranes [
11,
12]. This makes miRNAs potential biomarker candidates even in this stage of kidney disease. Therefore, it would be interesting to evaluate the effect of a nutritional intervention on protein energy malnutrition associated with CKD. For example, miR-21 is overexpressed in CKD in animal models and humans. It also regulates metabolic pathways involved in fibrogenesis, inflammation, oxidation-reduction activity and cell proliferation [
13,
14]. Recent studies have described the role of miR-29 family as a strong regulator in several diseases such as obesity and diabetes [
15] and a regulator of lipids metabolism during renal fibrosis [
12]. Specifically, miR-29a and miR-29b have been associated with various protective effects in renal disease, particularly in the context of fibrosis, inflammation, apoptosis and kidney injury [
16,
17]. Furthermore, circulating levels of mir-155 (related to inflammatory pathways) and miR-126 (involved in vascular homeostasis) are reduced in patients with hemodialysis [
12,
13,
18]. Other miRNAs have been related with CKD, such as miR-128a intervenes in mechanisms of muscle atrophy [
19], meanwhile miR-223 intervenes in inflammation processes and is associated with vascular complications [
18]. Finally, miR-378 is involved in angiogenic processes and adipogenesis, and can be modulated by inflammatory cytokines such as TNFalpha, IL6 and leptin [
20].
MiRNAs have been suggested as potential diagnostic and therapeutic tools for CKD, however further research is needed to fully understand the role of miRNAs in CKD and its potential as a biomarker or therapeutic target [
21,
22]. Furthermore, it is known that miRNA levels change in response to different dietary interventions [
23]: miRNAs levels can be regulated by carbohydrates, protein, fat, vitamins, minerals, dietary fiber or even isolated nutrients or bioactive compounds [
9,
24]. Further studies are needed to fully understand the mechanisms underlying these interactions and to develop effective strategies for using oral nutrition supplements to modulate miRNA expression. Our aim in this study was to evaluate whether the new ONS, with the action of probiotics, may produce changes in miR-21, miR-29a, miR-29b, miR-126, miR-128, miR-155, miR-223 and miR-378 expression and its target genes in malnourished hemodialysis patients, in comparison to individualized diet recommendations.
2. Results
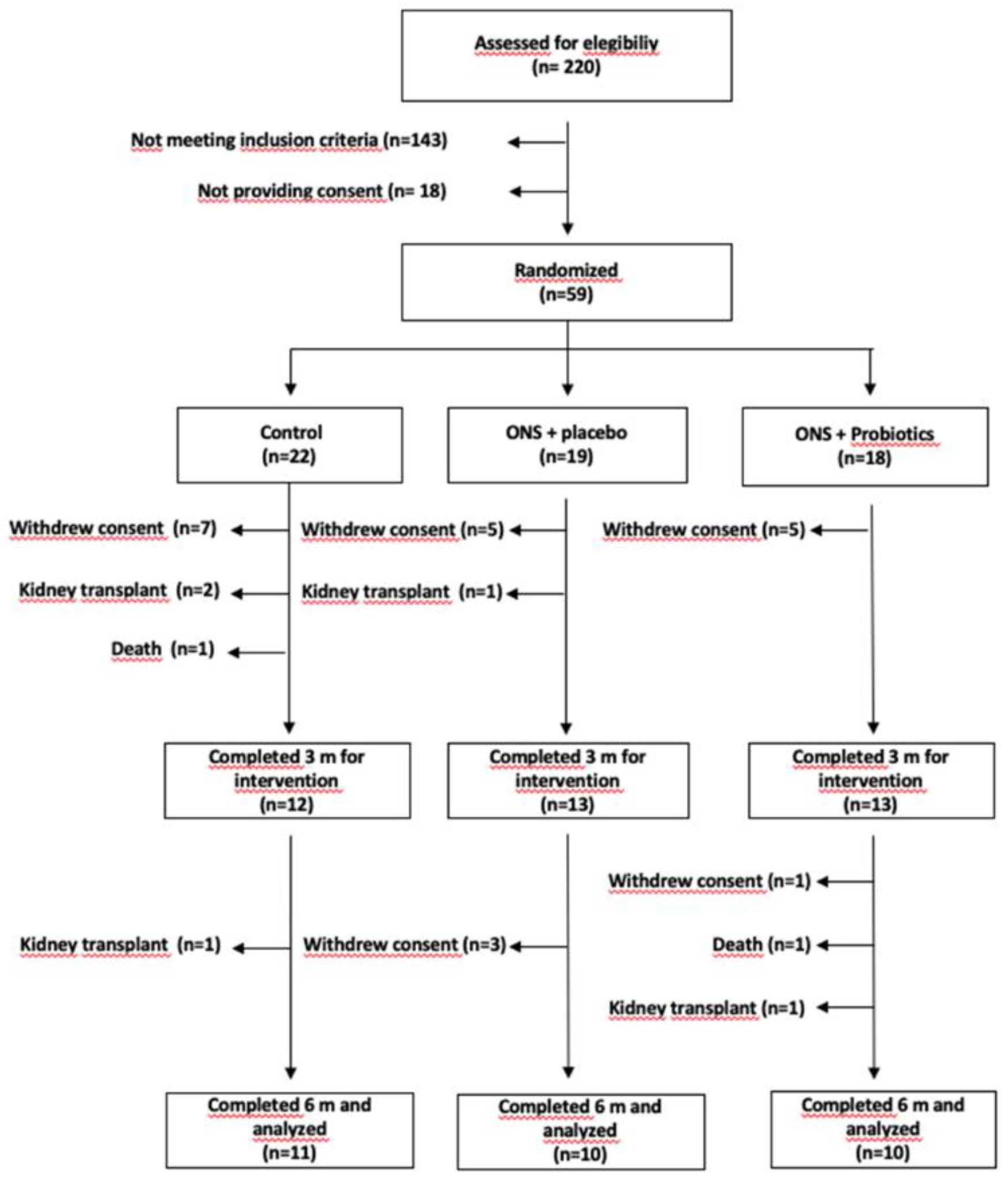
A total of 31 patients (11 corresponding to group C, 10 to ONS-PL, and 10 to ONS-PR) completed the 6-months trial (
Figure 1) [
2].
At the baseline visit there were no basal significant differences between groups regarding age, sex, diabetes, Charlson comorbidity index, or intake of fermented milk or antibiotics during the month prior to inclusion. Furthermore, there were no baseline differences in any of the parameters for the nutritional assessment, dietary intake or biochemical data [
2].
2.1. Circulating miRNAs expression levels before and after intervention
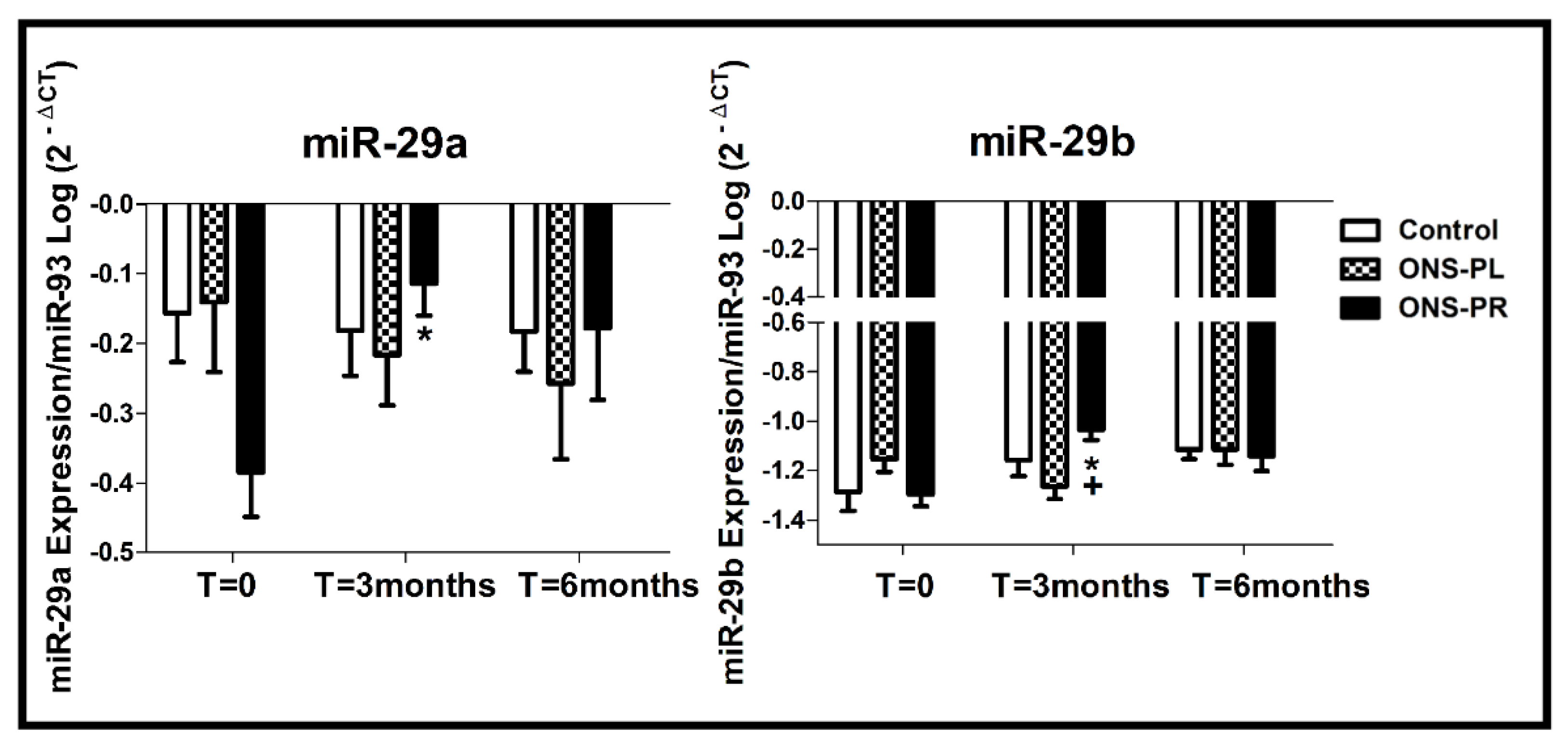
The changes in miRNAs expression levels are represented in
Figure 2.
We analyzed the miR29-a expression levels between the groups and we found a significant increase in the ONS-PR group in the 3-month intervention compared to baseline (p=0.03).
Regarding miR29-b expression levels, we found an increase in the ONS-PR group at 3-month intervention compared to baseline (p=0.023) and compared to the control group (p=0.027).
We did not find significant differences between the groups and between times of intervention for miR-21, miR-126, miR-128, miR-155, miR-233 and miR-378 expression levels.
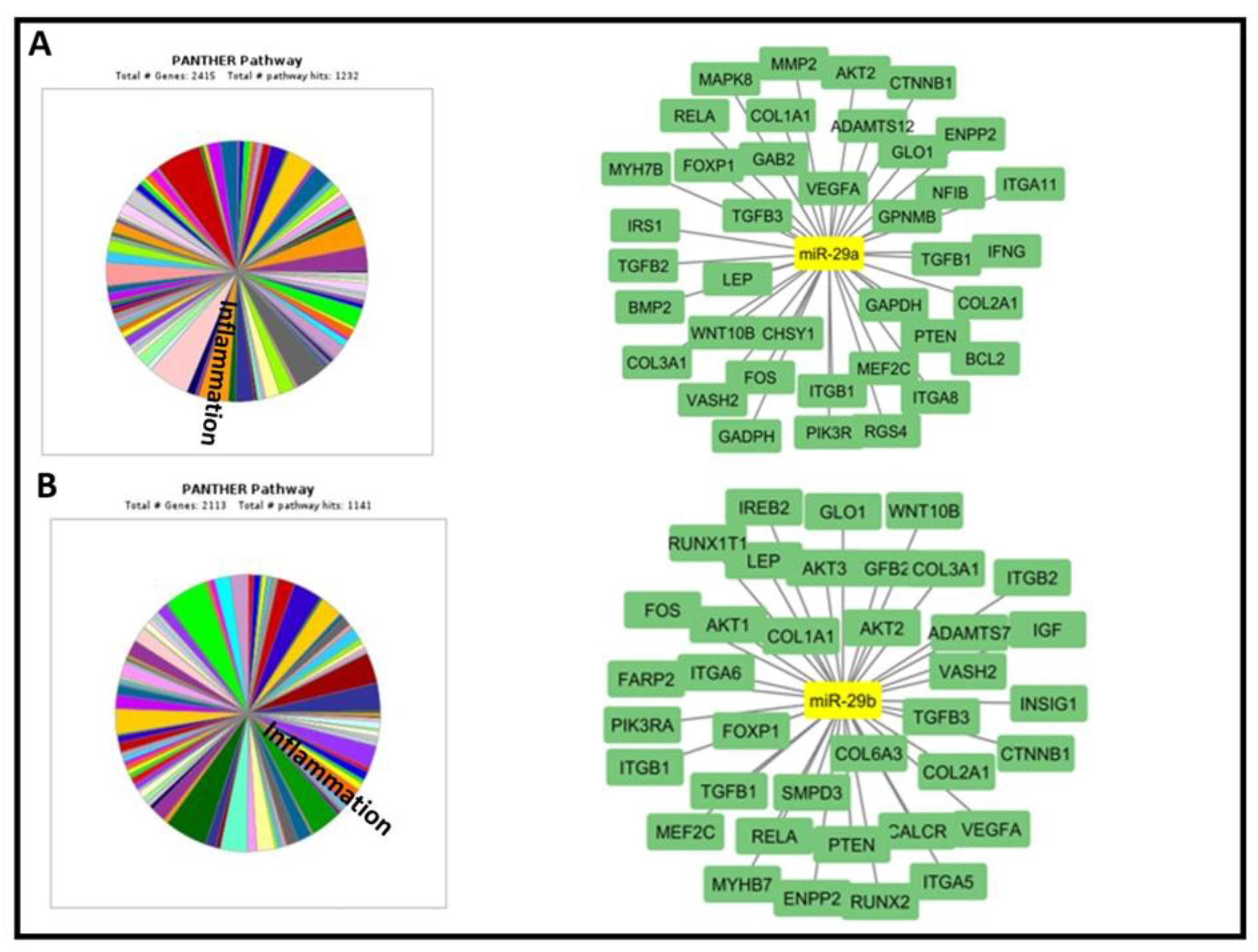
2.2. In silico identification of predicted and validated miR-29a and miR-29b target genes
The bioinformatic analysis has revealed 2900 potential TGs for miR-29a and 2600 miR-29b. Moreover, Panther and DAVID database have shown the implication of the examined miRNAs in various biological processes. We accorded priority to TGs and biological processes linked with our ONS and renal function including inflammation, lipid metabolism and bone formation (
Table 3,
Figure 3). Then, we chose 9 important TGs that regulate 2 or more of selected processes.
2.3. Gene expression levels from human blood before and after intervention
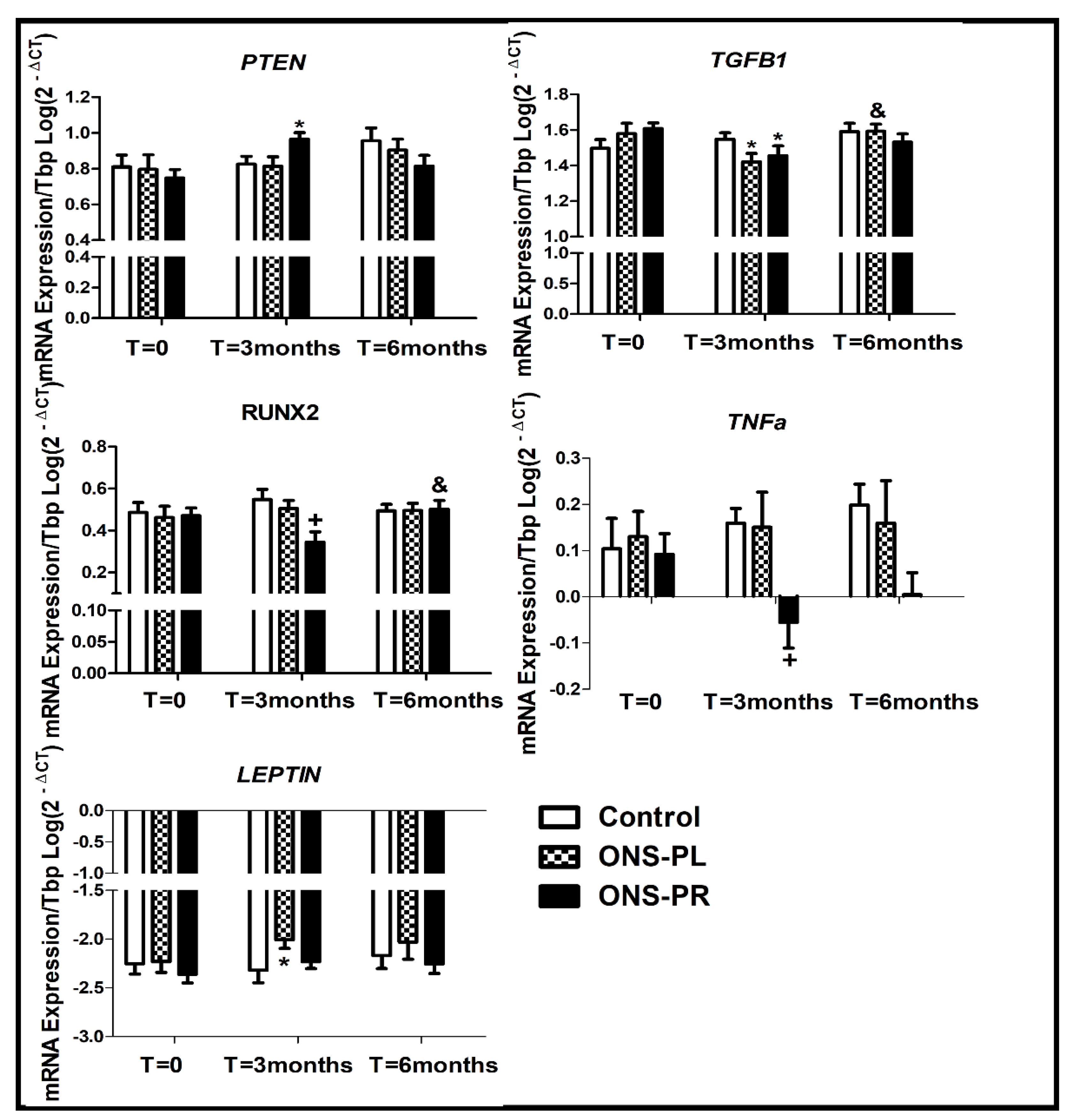
The changes in miR-29a and miR-29b target genes expression are represented in
Figure 4.
PTEN expression was increased at 3-month intervention compared to baseline in the ONS-PR group (p=0.023) and a slight but not significant increased expression compared to the control group (p=0.080). We found no differences between PTEN expression in 6-month intervention and baseline.
The expression of TGFB1 was decreased at 3-month intervention compared to baseline in the ONS-PR (p=0.019) and ONS-PL group (p=0.027). We observed an increase in TGFB1 expression at 6-month intervention in the ONS-PL group, when compared to 3-month intervention (p=0.016).
Regarding RUNX2 expression, we observed a decrease in the ONS-PR group compared to the control group at 3-month intervention (p=0.012). In the ONS-PR group, we observed a slight decrease in RUNX2 expression at 3-month intervention compared to baseline (p=0.079) and a later increase in 6-month intervention (p=0.023).
The LEPTIN expression was increased at 3-month intervention compared to baseline in the ONS-PL group (p=0.04). We found no significant differences between 6-month intervention and baseline.
Regarding TNFa expression, we observed a decrease in ONS-PR group at 3- month intervention compared to the control group (p=0.041). Furthermore, we observed a slight but not significant decrease in the ONS-PR group at 3-month intervention compared to baseline (p=0.098).
We also measured AKT1, RELA, IL1B, IL6, MYH7B, HEF2C, CTNNB1, GHRL, RASA1 and PPARG expression gene levels, for being target genes of the studied miRNAs, but we found no significant differences between the groups and the times of intervention.
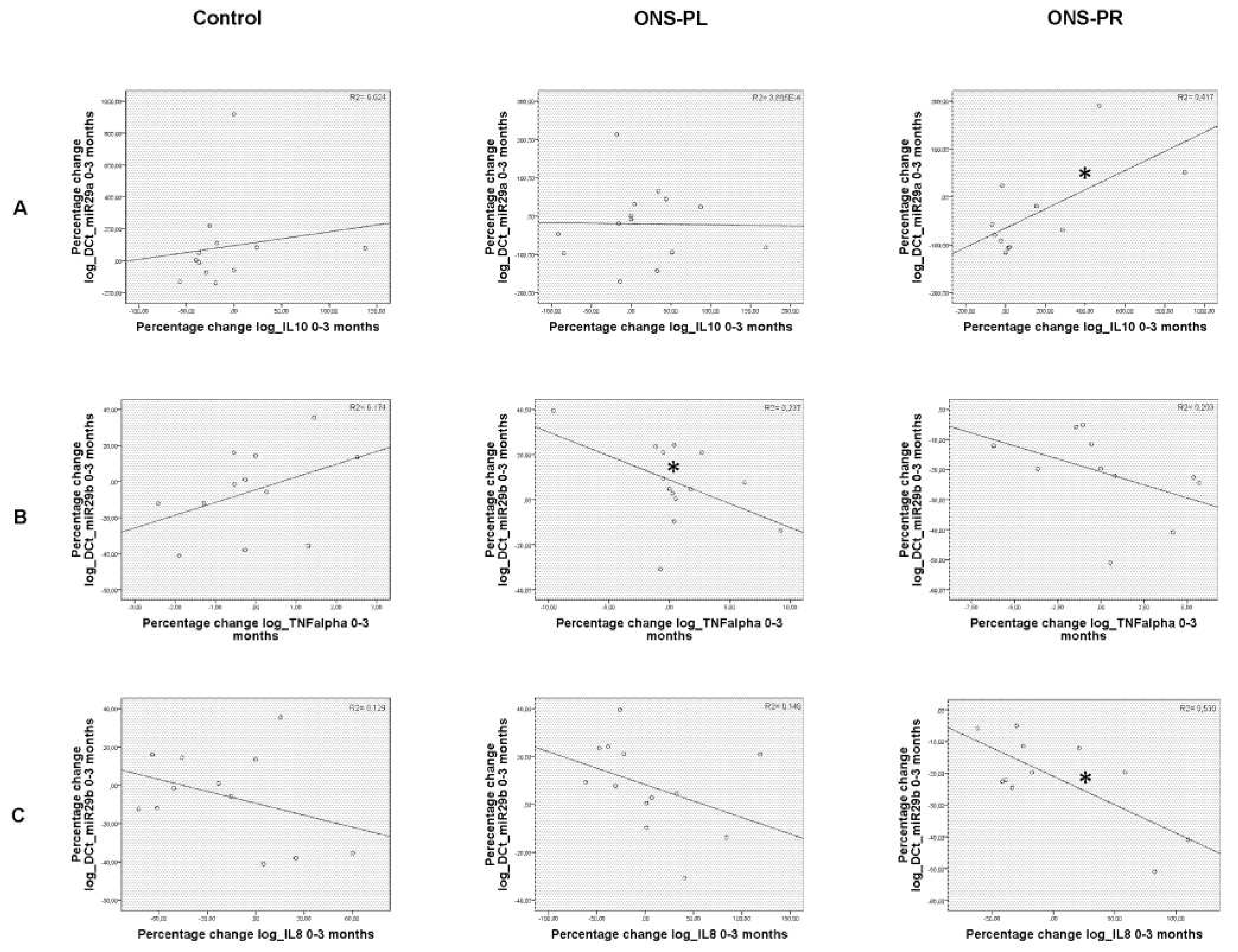
2.4. Correlations between miRNAs levels with serum biomarkers and expression levels of target genes
We have previously described significant changes in inflammation biomarkers in the groups receiving supplements along the intervention [
2]. Now we analyzed the correlations between the percentage change between these variables and miRNAs levels, represented in
Figure 5.
When comparing baseline and 3-month intervention, we found a significant negative correlation between the expression of miR-29b with TNFα (p=0.017) in the ONS-PL group. In the ONS-PR group, we found a significant positive correlation between miR-29a and IL-10 (p=0.025) and a significant negative correlation between miR-29b with IL-8 (p=0.020).
Regarding the correlations between miRNAs levels and expression levels of target genes we did not find significant results.
3. Discussion
In this study we analyzed for the first time the effects of a new oral supplement combined or not with probiotics on miRNA expression and its target genes in malnourished hemodialysis patients. The results provide valuable insights into the intricate relationship between nutritional interventions, miRNA regulation, and potential therapeutic outcomes. Notably, we observed that the expression of miR-29a and b increased significantly in patients with ONS-PR at 3 months in comparison with baseline. Moreover, miR-29b expression levels were elevated in patients receiving ONS-PR compared to control at month 3. Our previous study has shown that ONS improved body composition by inducing anti-inflammatory and antioxidant processes [
2]. It is well known that miR-29a and miR-29b are implicated in the pathophysiology of various diseases, including obesity, diabetes, cardiovascular diseases and cancers [
15,
25,
26,
27]. Also, recent studies highlighted the protective effects of miR-29a and miR-29b against chronic kidney disease (CKD) and renal fibrosis, which are characterized by excessive accumulation of collagen and fibronectin in extracellular matrix (ECM), inflammation and oxidative stress [
28,
29,
30,
31]. Moreover, in the present study, we revealed changes in
TNFα expression and a positive correlation between miR-29a and miR-29b with anti-inflammatory markers such as IL-10, and a negative correlation between miR-29b with pro-inflammatory markers TNFα and IL-8. All these findings may underscore the potential anti-inflammatory effects of the oral supplement and probiotics within miR-29a and miR-29b regulation. The decrease in
TNFα expression aligns with the notion that these supplements might contribute to decreasing the chronic inflammatory state associated with malnutrition and renal dysfunction. Specially, the decreased
TNFα expression observed in the ONS-PR group could be indicating a possible synergistic nutrition-probiotic effect. We observed that the expression of miR-29a and miR-29b increased significantly in patients with ONS-PR at 3 months in comparison with baseline, stabilizing at the sixth month.
Regarding the expression levels of
TGFB1, we observed a decrease of this transcription factor in both ONS groups at month 3 with respect to control. Consistent with these results, several studies, have emphasized the role of
TGFB1 in the induction of renal fibrosis and the progression of CKD by promoting inflammation and ECM modulation [
32,
33]. Moreover,
RUNX2, well-known for its crucial role in osteoblast differentiation and bone formation, has also been shown to be involved in fibrosis induction in various organs [
34,
35]. Furthermore, some studies have reported a tight correlation between
RUNX family and
TGFB1 with fibrosis and CKD development [
36,
37,
38,
39]. In our study, we observed a decrease in
RUNX2 expression in the ONS-PR group compared to the control group at 3-month, followed by a slight decrease at 3-month compared to baseline, and a subsequent increase at 6-month intervention. In addition, Wang and collaborators have revealed that miR-29a attenuates kidney fibrosis in Unilateral Ureteral Obstruction (UUO) mice by inhibiting fibrotic proteins and
TGFB1 [
17]. Moreover,
in vitro upregulation of miR-29b inhibited the expression of
TGFB1 [
30]. These observations are in accordance with our results, suggesting that these microRNAs could exert protective effects in the context of CKD by inhibiting a pro-fibrotic state mediated by
TGFB1 and
RUNX2. Concerning
PTEN expression, we found that it was significantly elevated in the ONS-PR group at 3-months in comparison with the control group. Higgins and collaborators have demonstrated that
PTEN prevents kidney fibrosis and renal injury by inhibiting PI3K/Akt and by counteracting the fibrotic effect of
TGFB1 in cultured renal cells and UUO model [
40]. In addition, another study has revealed that increased
TGFB expression during acute kidney injury (AKI) was associated with the loss of
PTEN expression [
41]. Also, the recovery of
PTEN was accompanied with normal tubule repair and less fibrosis. Furthermore, inhibition of
PTEN expression increased renal fibrosis in AKI mice model [
36]. Additionally, other researchers have found that
PTEN improves cellular fibrotic changes and renal fibrosis via inhibiting FAK/AKT signaling pathway [
42]. In line with these results, one study has shown that Homeobox transcript antisense RNA (HOTAIR) downregulates
PTEN via miR-29b inhibition during liver fibrosis [
43]. Moreover, other study has revealed that miR-29b contributed to the regulation of renal interstitial fibrosis via targeting PI3K/Akt signaling pathway and
PTEN expression [
44]. These findings suggest the role that could play miR-29a and b and its target genes in kidney fibrosis and renal injury within CKD.
In relation to
LEPTIN levels, the increase of its expression in the ONS-PL group raises interesting questions about the possible link between the oral supplement and appetite regulation in malnourished patients. Leptin is a peptide hormone primarily synthesized in adipose tissue and plays an important role in regulating appetite and energy balance [
45]. Moreover, leptin exerts a pivotal role in inducing angiogenesis and adipogenesis in adipose tissue [
46]. In addition, it has been described that low levels of leptin are associated with protein energy wasting in people with CKD and hemodialysis and with a worse prognosis [
47,
48,
49], even though a correlation with the amount of fat mass is maintained [
50]. In our previous study [
2] both groups that received the ONS increased weight at the end of the intervention, improving the nutritional status at the expense of, specially fat-free mass, but also with an increase in fat mass (around 1kg). Furthermore, dietary total fat increased significantly especially in the ONS-PL group [
2]. In this context, the significant increase in fat intake could have stimulated the
LEPTIN expression as an appetite-regulating response also associated with the increase in body fat mass. Furthermore, the probiotics in the ONS-PR group may have influenced the gut microbiota composition, mitigating a significant leptin response through various mechanisms [
51].
A major limitation of this study was the relatively high dropout rate, which was primarily due to the challenges posed by the SARS-CoV-2 pandemic and the difficulty of maintaining ongoing patient follow-up [
2], which has contributed to not finding significant changes in other circulating miRNAs. We highlight the study's strengths as being a randomized clinical trial (double-blind in the case of probiotics), long-term (6 months) follow-up, multicenter characteristics, and multiple parameter measurements, the measurement of multiple parameters (including biomarkers, miRNAs and its target genes), and the comparison to a control group following a personalized diet prescribed by certified dietitians.
4. Materials and Methods
4.1. Design
The trial design is set out in the previous publication [
2]. Briefly: it is a randomized, multicenter, parallel-group study divided in 3 groups, open for the intake of ONS or individualized dietary advice, but double-blind for probiotics consumption. Patients were assigned to one of the following three groups (using a computer-generated random number table):
- 1:
Control (C): received individualized dietary recommendations (IDR).
- 2:
ONS + placebo (ONS-PL): received ONS + IDR.
- 3:
ONS + probiotics (ONS-PR): received ONS + probiotics + IDR.
The Renacare® ONS was designed for malnourished hemodialysis patients. It is a high-calorie (2 kcal/mL) and high-protein supplement containing several functional nutrients (extra virgin olive oil, omega 3 fatty acids, whey protein, antioxidants, low-glycemic index carbohydrates, fiber and carnitine). The composition is shown on the website:
https://adventiapharma.com/nutricion-clinica/productos/enteral-oral/bi1-renacare-dial
Inclusion and exclusion criteria are detailed in our previous publication [
2]. Comprised adult subjects (>18 years) undergoing hemodialysis (standard therapy or on-line hemodiafiltration with high-reinfusion rate therapy not being modified within 3 months before inclusion for more than 6 months before inclusion) and who were malnourished. Written informed consent was obtained from all patients.
Nutritional requirements were assessed according to the International Society of Renal Nutrition and Metabolism guidelines. The target protein intake was at least 1.2 g/kg/day [
52]. All participants completed a personal interview with a nutritionist at baseline and 3 and 6 months after. Patients randomly assigned to the ONS group were advised to consume two bricks of 400 ml per day (minimum 1 day - 200 ml). Patients' daily ONS intakes were recorded on a data sheet. Probiotics and placebo were delivered in visually indistinguishable capsules (one 380g tablet). Each probiotic capsule contains the following live bacteria:
Bifidobacterium breve CNCM I-4035 (1.00E+09 Colony Forming Units -CFU-),
Bifidobacterium animalis lactis BPL1 CECT 8145 (3.50E+09 CFU), and
Lactobacillus paracasei CNCM I-4034 (5.00E+08 CFU).
The Research Ethics Committee of the Province of Malaga approved this study and the procedure follows the ethical standards of the Declaration of Helsinki. Study registration: NCT03924089.
4.2. Outcomes
Examinations were performed at baseline and after 3 and 6 months. Blood samples were taken before starting dialysis; plasma and serum were aliquoted and stored at −8 °C in the Hospital-IBIMA biobank, Andalusian Public Health System Biobank, which belongs to the National Biobank Platform (exp. PT20/00101).
4.3. miRNA and RNA extraction
miRNAs were extracted from plasma samples by means of automated method with Maxwell® 16 miRNA Tissue Kit (Promega Biotech Ibérica S.L., Madrid, Spain) according to the manufacturer’s protocol. For each sample, 300 μL of plasma were consequently mixed with the following reagents: 200 μL of homogenization solution, 200 μL of lysis buffer and 15 μL of Proteinase K. Samples were incubated for 20 min on a heat block at 37 °C, 300 rpm. After the incubation, samples were transferred to RSC cartridges followed by automated RNA extraction with the Maxwell instrument. Finally, total RNA was eluted in 30 μL nuclease-free water and stored at −80 °C until use.
Total RNA was extracted from PAXgene Blood RNA Tubes (Qiagen, USA). Briefly, the the content of the thawed tube was transferred to a 50 ml conical tube with 3 ml 1X phosphate buffered serum (PBS), then vortexed for 30 seconds, and later centrifuged at 4000xg in 4 °C for 20 min. The RNA pellet was further lysed with QIAzol Lysis reagent (Qiagen, USA), followed by incubation with 200 μL chloroform. After the phase separation, the upper aqueous phase was mixed with isopropanol and incubated for 10 minutes on ice. After centrifugation, the RNA pellet was washed with 75% ethanol. After centrifugation, the RNA pellet was air-dried at room temperature for 10 minutes. Total RNA was resuspended in 25 μL of nuclease-free water and stored at −80 °C until use.
The purity of miRNA and total RNA was determined by the absorbance 260/280 ratio on a Nanodrop ND-2000 spectrophotometer (Thermo Fisher Scientific Inc., USA).
4.4. RT-qPCR
cDNA was synthesized from miRNAs and mRNAs by reverse transcription with the Universal cDNA Synthesis Kit (Exiqon, Denmark) following the manufacturer’s recommendations for each kit.
Measurements of miRNA expression levels (miR-21-5p, miR-29a-3p, miR-29b-3p, miR-126-3p, miR-128-3p, miR-155-5p, miR-233-3p and miR-378-3p) were performed in duplicated by qPCR real time in 384 plates in a LightCycler 480 (Roche Diagnostics, S.L, Barcelona, Spain) at the Genomics platform of IBIMA-Plataforma Bionand. The master mix was prepared following the guidelines of Exiqon with GoTaq® qPCR Master Mix (Promega Biotech Ibérica S.L., Madrid, Spain) and the specific LNA™ miRNA PCR primer Assy (Exiqon A/S Vedbaek, Denmark) (
Table 1).
Gene expression levels (TBP,
TGFB, RUNX2, PTEN, AKT1, RELA, TNF, IL1B, IL6, MYH7B, HEF2C, CTNNB1, LEP, GHRL, RASA1 and
PPARG) were analyzed in duplicate by qPCR using QuantStudio 12K Flex Real Time PCR 384 system (Thermo Fisher Scientific Inc., USA) with TaqMan technology. Predesigned Thermo Fisher primers were used to detect the canonical isoform of the gene of interest in humans, built based on the GRCh38 reference genome (
Table 2). These primers have been validated in silico and in vitro by Thermo Fisher (
https://www.thermofisher.com/taqman/gene-expression/assay). To carry out the qPCR assays, the recommendations established by the manufacturer were followed, using TaqMan® Fast Advanced Master Mix.
Quantitative measures were obtained using the 2
−ΔCt method and expression results were given as the expression ratio relative to miR-93 for miRNAs after an analysis with RefFinder (
https://blooge.cn/RefFinder/), found it to be the most stable miRNA. TBP was used as the normalizing gene for gene expression, selected after performing a screening (TaqMan
TM Array Human Endogenous Controls Plate, Fast 96-well), with 32 possible constituent genes in triplicate, in a study subpopulation made up of 3 subjects from each of the 3 groups, a total of 9 subjects. Also, in order to detect the most stable gene, the RefFinder program was used.
For data normalization, negative controls and a specific calibrator in all plates for interplate normalization were included. Those determinations with Ct >38 were considered under the limit of detection and were taken out of the analysis.
4.5. In silico identification of predicted and validated miRNA target genes
4.6. Biomarkers
Blood samples were taken before starting dialysis; plasma and serum were aliquoted and stored at −80 °C in the Hospital-IBIMA Biobank until analysis. IL-10, IL-8 and TNFα were measured in 25 ul of serum using the ProcartaPlex multiplex Immunoassay (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions [
2]. All measurements were performed in duplicate and a standard curve was used to obtain serum concentrations.
4.7. Statistical Analysis
Data analysis was performed with the IBM SPSS Statistics software version 26 (IBM Corp. Released 2019. IBM SPSS Statistics for Windows, Version 26.0. Armonk, NY: IBM Corp). Data are expressed as the mean ± SEM. Comparisons between multiple groups were performed using one-way ANOVA, and for comparisons between two groups Student’s t test was used. To compare variables by the different groups and the changes over time (baseline, 3, and 6 months), ANOVA for repeated variables was used. The level of significance considered was 5%. For multiple comparisons (post-hoc) Bonferroni’s correction was used. The percentage of increased or decreased expression of miRNAs or serum parameters with respect to the basal levels were calculated as follows:
Correlations were analyzed using Pearson’s correlation coefficient using the percentage change between times of intervention between the groups. Normality was assessed by Shapiro–Wilk test and all the variables that did not presented normal distribution (p > 0.05) were log transformed in order to normalize; thus, we performed parametric tests for our analysis. GraphPad Prism version 9.0.0 for Windows, GraphPad Software (Boston, Massachusetts USA,
www.graphpad.com) was used for miRNAs expression graphics.
5. Conclusions
The new Oral Nutritional Supplement specifically designed for malnourished (or at risk) hemodialysis patients with a "similar to the Mediterranean diet" pattern, associated with probiotics; increases the expression levels of miR-29a and miR-29b after 3 months of intervention, modifying the expression of target genes with anti-inflammatory and anti-fibrotic actions.
This study highlights the potential benefit of this Oral Nutritional Supplement, especially associated with probiotics, in malnourished patients with chronic renal disease on hemodialysis.
Author Contributions
Conceptualization, Gabriel Olveira; Data curation, Corina Sasso, Said Lhamyani, Ana Lago-Sampedro and Gabriel Olveira; Formal analysis, Corina Sasso, Said Lhamyani, Ana Lago-Sampedro and Gabriel Olveira; Funding acquisition, Gabriel Olveira; Investigation, Corina Sasso, Said Lhamyani, Francisco Hevilla, Marina Padial, María Blanca, Guillermina Barril, Tamara Jiménez-Salcedo, Enrique Sanz-Martínez, Ángel Nogueira, Ana Lago-Sampedro and Gabriel Olveira; Methodology, Gabriel Olveira; Project administration, Gabriel Olveira; Resources, Gabriel Olveira; Supervision, Gabriel Olveira; Validation, Gabriel Olveira; Writing – original draft, Corina Sasso, Said Lhamyani, Ana Lago-Sampedro and Gabriel Olveira; Writing – review & editing, Corina Sasso, Said Lhamyani, Ana Lago-Sampedro and Gabriel Olveira.
Funding
This research was funded by Ministerio de Ciencia, Innovación y Universidades Exp. RTC-2017-5959-1 and by ISCIII Exp. PI18/01041” and co-financed by “Fondo Europeo de Desarrollo Regional (FEDER) “Una manera de hacer Europa”. MP holds a contract (IFI20/00034) from the Carlos III National Health Institute, cofunded by European Social Fund 2014-2020 “The FSE invests in your future.” The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Institutional Review Board Statement
This study was approved by the Provincial Research Ethics Committee of Málaga. The ethical principles stated in the latest revision of the Declaration of Helsinki and good clinical practice standards were applied. The study was registered with the following code: NCT03924089.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Acknowledgments
The authors would like to thank the staff from the dialysis centres: Laura Fuentes-Sánchez, Elvira Esquivias, Alejandro Jiménez-Herrador; Diana López-Espinosa, Marta Sousah, Luis Cermeño, Alicia Martínez-Domínguez, Antonio Romero-Alcántara, Inmaculada Morales, Lourdes Blanca, Elena Vaquero, Enrique Sanz, María López-Picasso, Teresa Andrino-Llorente y Graciela Alvarez-García and all patients participating in the study.
Conflicts of Interest
FH received honoraria support for a presentation from Adventia Pharma. GO received occasional honoraria support for presentations, attending meetings and travel from Adventia Pharma. The funder had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Ikizler, T.A.; Burrowes, J.D.; Byham-Gray, L.D.; Campbell, K.L.; Carrero, J.J.; Chan, W.; Fouque, D.; Friedman, A.N.; Ghaddar, S.; Goldstein-Fuchs, D.J.; et al. KDOQI Clinical Practice Guideline for Nutrition in CKD: 2020 Update. American Journal of Kidney Diseases 2020, 76, S1–S107. [Google Scholar] [CrossRef] [PubMed]
- Hevilla, F.; Padial, M.; Blanca, M.; Barril, G.; Jiménez-Salcedo, T.; Ramirez-Ortiz, M.; Nogueira, Á.; Gentile, A.; García-Escobar, E.; Romero-Zerbo, S.Y.; et al. Effect on Nutritional Status and Biomarkers of Inflammation and Oxidation of an Oral Nutritional Supplement (with or without Probiotics) in Malnourished Hemodialysis Patients. A Multicenter Randomized Clinical Trial “Renacare Trial.” Front Nutr 2023, 10. [Google Scholar] [CrossRef]
- Lv, W.; Fan, F.; Wang, Y.; Gonzalez-Fernandez, E.; Wang, C.; Yang, L.; Booz, G.W.; Roman, R.J. Therapeutic Potential of MicroRNAs for the Treatment of Renal Fibrosis and CKD. Physiol Genomics 2018, 50, 20–34. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front Endocrinol (Lausanne) 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Mompeón, A.; Ortega-Paz, L.; Vidal-Gómez, X.; Costa, T.J.; Pérez-Cremades, D.; Garcia-Blas, S.; Brugaletta, S.; Sanchis, J.; Sabate, M.; Novella, S.; et al. Disparate MiRNA Expression in Serum and Plasma of Patients with Acute Myocardial Infarction: A Systematic and Paired Comparative Analysis. Sci Rep 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Carmona, A.; Guerrero, F.; Jimenez, M.J.; Ariza, F.; Agüera, M.L.; Obrero, T.; Noci, V.; Muñoz-Castañeda, J.R.; Rodríguez, M.; Soriano, S.; et al. Inflammation, Senescence and MicroRNAs in Chronic Kidney Disease. Front Cell Dev Biol 2020, 8, 739. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Yu, C.; Feng, B.; Zhan, X.; Luo, N.; Yu, X.; Zhou, Q. Intrarenal MicroRNA Signature Related to the Fibrosis Process in Chronic Kidney Disease: Identification and Functional Validation of Key MiRNAs. BMC Nephrol 2019, 20, 1–13. [Google Scholar] [CrossRef]
- Izzotti, A.; Cartiglia, C.; Steele, V.E.; De Flora, S. MicroRNAs as Targets for Dietary and Pharmacological Inhibitors of Mutagenesis and Carcinogenesis. Mutation Research/Reviews in Mutation Research 2012, 751, 287–303. [Google Scholar] [CrossRef] [PubMed]
- Quintanilha, B.J.; Reis, B.Z.; Silva Duarte, G.B.; Cozzolino, S.M.F.; Rogero, M.M. Nutrimiromics: Role of MicroRNAs and Nutrition in Modulating Inflammation and Chronic Diseases. Nutrients 2017, Vol. 9, Page 1168 2017, 9, 1168. [Google Scholar] [CrossRef]
- Belcheva, A. MicroRNAs at the Epicenter of Intestinal Homeostasis. BioEssays 2017, 39, 1600200. [Google Scholar] [CrossRef]
- Martino, F.; Lorenzen, J.; Schmidt, J.; Schmidt, M.; Broll, M.; Görzig, Y.; Kielstein, J.T.; Thum, T. Circulating MicroRNAs Are Not Eliminated by Hemodialysis. PLoS One 2012, 7, e38269. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Peng, W.; Shen, X.; Huang, Y.; Ouyang, X.; Dai, Y. Circulating Levels of Inflammation-Associated MiR-155 and Endothelial-Enriched MiR-126 in Patients with End-Stage Renal Disease. Brazilian Journal of Medical and Biological Research 2012, 45, 1308. [Google Scholar] [CrossRef] [PubMed]
- Gomez, I.G.; Nakagawa, N.; Duffield, J.S. MicroRNAs as Novel Therapeutic Targets to Treat Kidney Injury and Fibrosis. Am J Physiol Renal Physiol 2016, 310, F931–F944. [Google Scholar] [CrossRef] [PubMed]
- Zununi Vahed, S.; Poursadegh Zonouzi, A.; Ghanbarian, H.; Ghojazadeh, M.; Samadi, N.; Omidi, Y.; Ardalan, M. Differential Expression of Circulating MiR-21, MiR-142-3p and MiR-155 in Renal Transplant Recipients with Impaired Graft Function. Int Urol Nephrol 2017, 49, 1681–1689. [Google Scholar] [CrossRef] [PubMed]
- Dooley, J.; Garcia-Perez, J.E.; Sreenivasan, J.; Schlenner, S.M.; Vangoitsenhoven, R.; Papadopoulou, A.S.; Tian, L.; Schonefeldt, S.; Serneels, L.; Deroose, C.; et al. The MicroRNA-29 Family Dictates the Balance Between Homeostatic and Pathological Glucose Handling in Diabetes and Obesity. Diabetes 2016, 65, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Komers, R.; Carew, R.; Winbanks, C.E.; Xu, B.; Herman-Edelstein, M.; Koh, P.; Thomas, M.; Jandeleit-Dahm, K.; Gregorevic, P.; et al. Suppression of MicroRNA-29 Expression by TGF-Β1 Promotes Collagen Expression and Renal Fibrosis. Journal of the American Society of Nephrology 2012, 23, 252–265. [Google Scholar] [CrossRef]
- Wang, H.; Wang, B.; Zhang, A.; Hassounah, F.; Seow, Y.; Wood, M.; Ma, F.; Klein, J.D.; Price, S.R.; Wang, X.H. Exosome-Mediated MiR-29 Transfer Reduces Muscle Atrophy and Kidney Fibrosis in Mice. Molecular Therapy 2019, 27, 571–583. [Google Scholar] [CrossRef]
- Brigant, B.; Metzinger-Le Meuth, V.; Massy, Z.A.; McKay, A.; Liabeuf, S.; Pelletier, M.; Sallée, M.; M’Baya-Moutoula, L.; Paul, P.; Drueke, T.B.; et al. Serum MicroRNAs Are Altered in Various Stages of Chronic Kidney Disease: A Preliminary Study. Clin Kidney J 2017, 10, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Pivarcsi, A.; Meisgen, F.; Xu, N.; Sonkoly, E. Changes in the Level of Serum MicroRNAs in Patients with Psoriasis after Antitumour Necrosis Factor-α Therapy. British Journal of Dermatology 2013, 169, 563–570. [Google Scholar] [CrossRef]
- Xu, L. lian; Shi, C. mei; Xu, G. feng; Chen, L.; Zhu, L. ling; Zhu, L.; Guo, X. rong; Xu, M. yu; Ji, C. bo TNF-α, IL-6, and Leptin Increase the Expression of MiR-378, an Adipogenesis-Related MicroRNA in Human Adipocytes. Cell Biochem Biophys 2014, 70, 771–776. [Google Scholar] [CrossRef]
- Peters, L.J.F.; Floege, J.; Biessen, E.A.L.; Jankowski, J.; van der Vorst, E.P.C. MicroRNAs in Chronic Kidney Disease: Four Candidates for Clinical Application. International Journal of Molecular Sciences 2020, Vol. 21, Page 6547 2020, 21, 6547. [Google Scholar] [CrossRef] [PubMed]
- Motshwari, D.D.; Matshazi, D.M.; Erasmus, R.T.; Kengne, A.P.; Matsha, T.E.; George, C. MicroRNAs Associated with Chronic Kidney Disease in the General Population and High-Risk Subgroups-A Systematic Review. Int J Mol Sci 2023, 24. [Google Scholar] [CrossRef] [PubMed]
- Giardina, S.; Hernández-Alonso, P.; Díaz-López, A.; Salas-Huetos, A.; Salas-Salvadó, J.; Bulló, M. Changes in Circulating MiRNAs in Healthy Overweight and Obese Subjects: Effect of Diet Composition and Weight Loss. Clinical Nutrition 2019, 38, 438–443. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Zhang, J.; Wang, J.; Sun, B. MicroRNAs: The Novel Mediators for Nutrient-Modulating Biological Functions. Trends Food Sci Technol 2021, 114, 167–175. [Google Scholar] [CrossRef]
- Gondaliya, P.; Jash, K.; Srivastava, A.; Kalia, K. MiR-29b Modulates DNA Methylation in Promoter Region of MiR-130b in Mouse Model of Diabetic Nephropathy. J Diabetes Metab Disord 2023, 1–11. [Google Scholar] [CrossRef]
- Deng, L.; Huang, Y.; Li, L.; Chen, H.; Su, J. Serum MiR-29a/b Expression in Gestational Diabetes Mellitus and Its Influence on Prognosis Evaluation. Journal of International Medical Research 2020, 48. [Google Scholar] [CrossRef] [PubMed]
- Muluhngwi, P.; Klinge, C.M. Identification and Roles of Mir-29b-1-3p and Mir29a-3p-Regulated and Non-Regulated Lncrnas in Endocrine-Sensitive and Resistant Breast Cancer Cells. Cancers (Basel) 2021, 13, 3530. [Google Scholar] [CrossRef] [PubMed]
- Horita, M.; Farquharson, C.; Stephen, L.A. The Role of MiR-29 Family in Disease. J Cell Biochem 2021, 122, 696–715. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.C.; Chang, P.J.; Ho, C.; Huang, Y.T.; Shih, Y.H.; Wang, C.J.; Lin, C.L. Protective Effects of MiR-29a on Diabetic Glomerular Dysfunction by Modulation of DKK1/Wnt/β-Catenin Signaling. Scientific Reports 2016 6:1 2016, 6, 1–12. [Google Scholar] [CrossRef]
- Qin, W.; Chung, A.C.K.; Huang, X.R.; Meng, X.M.; Hui, D.S.C.; Yu, C.M.; Sung, J.J.Y.; Lan, H.Y. TGF-β/Smad3 Signaling Promotes Renal Fibrosis by Inhibiting MiR-29. J Am Soc Nephrol 2011, 22, 1462. [Google Scholar] [CrossRef]
- Drummond, C.A.; Fan, X.; Haller, S.T.; Kennedy, D.J.; Liu, J.; Tian, J. Na/K-ATPase Signaling Mediates MiR-29b-3p Regulation and Cardiac Fibrosis Formation in Mice with Chronic Kidney Disease. PLoS One 2018, 13, e0197688. [Google Scholar] [CrossRef] [PubMed]
- Huynh, P.; Chai, Z. Transforming Growth Factor β (TGFβ) and Related Molecules in Chronic Kidney Disease (CKD). Clin Sci 2019, 133, 287–313. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Ortega, M.; Rayego-Mateos, S.; Lamas, S.; Ortiz, A.; Rodrigues-Diez, R.R. Targeting the Progression of Chronic Kidney Disease. Nature Reviews Nephrology 2020 16:5 2020, 16, 269–288. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.; Zhao, J.; Huang, L.; Liu, Y.; Pang, X.; Zhan, K.; Li, S.; Xue, Q.; Pan, X.; Deng, L. Runx2 Activates Hepatic Stellate Cells to Promote Liver Fibrosis via Transcriptionally Regulating Itgav Expression. Clin Transl Med 2023, 13, e1316. [Google Scholar] [CrossRef] [PubMed]
- Mümmler, C.; Burgy, O.; Hermann, S.; Mutze, K.; Günther, A.; Königshoff, M. Cell-Specific Expression of Runt-Related Transcription Factor 2 Contributes to Pulmonary Fibrosis. The FASEB Journal 2018, 32, 703–716. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Luo, M.; Cai, W.; Zhou, S.; Feng, D.; Xu, C.; Wang, H. Runt-Related Transcription Factor 1 (RUNX1) Promotes TGF-β-Induced Renal Tubular Epithelial-to-Mesenchymal Transition (EMT) and Renal Fibrosis through the PI3K Subunit P110δ. EBioMedicine 2018, 31, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Wan, X.; Zou, Y.; Chen, Z.; Zhong, A. Transforming Growth Factor Beta (TGF-β) Is Activated by the CtBP2-P300-AP1 Transcriptional Complex in Chronic Renal Failure. Int J Biol Sci 2020, 16, 204. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.I.; Jang, H.S.; Jeong, J.H.; Noh, M.R.; Choi, J.Y.; Park, K.M. Defect in Runx2 Gene Accelerates Ureteral Obstruction-Induced Kidney Fibrosis via Increased TGF-β Signaling Pathway. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease 2013, 1832, 1520–1527. [Google Scholar] [CrossRef]
- Lee, K.-S.; Kim, H.-J.; Li, Q.-L.; Chi, X.-Z.; Ueta, C.; Komori, T.; Wozney, J.M.; Kim, E.-G.; Choi, J.-Y.; Ryoo, H.-M.; et al. Runx2 Is a Common Target of Transforming Growth Factor Β1 and Bone Morphogenetic Protein 2, and Cooperation between Runx2 and Smad5 Induces Osteoblast-Specific Gene Expression in the Pluripotent Mesenchymal Precursor Cell Line C2C12. Mol Cell Biol 2000, 20, 8783–8792. [Google Scholar] [CrossRef]
- Higgins, D.F.; Ewart, L.M.; Masterson, E.; Tennant, S.; Grebnev, G.; Prunotto, M.; Pomposiello, S.; Conde-Knape, K.; Martin, F.M.; Godson, C. BMP7-Induced-Pten Inhibits Akt and Prevents Renal Fibrosis. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease 2017, 1863, 3095–3104. [Google Scholar] [CrossRef]
- Lan, R.; Geng, H.; Polichnowski, A.J.; Singha, P.K.; Saikumar, P.; Mcewen, D.G.; Griffin, K.A.; Koesters, R.; Weinberg, J.M.; Bidani, A.K.; et al. PTEN Loss Defines a TGF-(β-Induced Tubule Phenotype of Failed Differentiation and JNK Signaling during Renal Fibrosis. Am J Physiol Renal Physiol 2012, 302. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Liu, P.; Chen, Z.; He, Y.; Zhang, B.; Dai, G.; Xia, W.; Liu, Y.; Chen, X. PTEN Improve Renal Fibrosis in Vitro and in Vivo through Inhibiting FAK/AKT Signaling Pathway. J Cell Biochem 2019, 120, 17887–17897. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Chen, B.; Dong, P.; Zheng, J. HOTAIR Epigenetically Modulates PTEN Expression via MicroRNA-29b: A Novel Mechanism in Regulation of Liver Fibrosis. Molecular Therapy 2017, 25, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Hu, S.; Xu, S.; Gao, Y.; Zeng, F.; Shui, H. MiR-29b Regulates Ang II-Induced EMT of Rat Renal Tubular Epithelial Cells via Targeting PI3K/AKT Signaling Pathway. Int J Mol Med 2018, 42, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Barrios-Correa, A.A.; Estrada, J.A.; Contreras, I. Leptin Signaling in the Control of Metabolism and Appetite: Lessons from Animal Models. Journal of Molecular Neuroscience 2018, 66, 390–402. [Google Scholar] [CrossRef] [PubMed]
- Hausman, G.J.; Richardson, R.L. Adipose Tissue Angiogenesis. J Anim Sci 2004, 82, 925–934. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Scherer, P.E. Immunologic and Endocrine Functions of Adipose Tissue: Implications for Kidney Disease. Nature Reviews Nephrology 2017 14:2 2017, 14, 105–120. [Google Scholar] [CrossRef] [PubMed]
- Canpolat, N.; Sever, L.; Agbas, A.; Tasdemir, M.; Oruc, C.; Ekmekci, O.B.; Caliskan, S. Leptin and Ghrelin in Chronic Kidney Disease: Their Associations with Protein-Energy Wasting. Pediatr Nephrol 2018, 33, 2113–2122. [Google Scholar] [CrossRef]
- Markaki, A.; Grammatikopoulou, M.G.; Venihaki, M.; Kyriazis, J.; Perakis, K.; Stylianou, K. Associations of Adiponectin and Leptin Levels with Protein-Energy Wasting, in End Stage Renal Disease Patients. Endocrinología y Nutrición 2016, 63, 449–457. [Google Scholar] [CrossRef]
- Yamamoto, T.; Carrero, J.J.; Lindholm, B.; Stenvinkel, P.; Axelsson, J. PROGRESS IN UREMIC TAXIN RESEARCH: Leptin and Uremic Protein-Energy Wasting—The Axis of Eating. Semin Dial 2009, 22, 387–390. [Google Scholar] [CrossRef]
- Hamamah, S.; Amin, A.; Al-Kassir, A.L.; Chuang, J.; Covasa, M. Dietary Fat Modulation of Gut Microbiota and Impact on Regulatory Pathways Controlling Food Intake. Nutrients 2023, 15. [Google Scholar] [CrossRef] [PubMed]
- Ikizler, T.A.; Cano, N.J.; Franch, H.; Fouque, D.; Himmelfarb, J.; Kalantar-Zadeh, K.; Kuhlmann, M.K.; Stenvinkel, P.; Terwee, P.; Teta, D.; et al. Prevention and Treatment of Protein Energy Wasting in Chronic Kidney Disease Patients: A Consensus Statement by the International Society of Renal Nutrition and Metabolism. Kidney Int 2013, 84, 1096–1107. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).