Submitted:
20 December 2023
Posted:
20 December 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Monodentate ligands
2.1. N-donor
2.2. S-donor
3. Bidentate ligands
3.1. O, O-donor
3.2. N, N-donor
3.3. N, O-donor
3.4. N, N-donor
3.5. N, S-donor
4. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lakhdar, S.; Westermaier, M.; Terrier, F.; Goumont, R.; Boubaker, T.; Ofial, A. R.; Mayr, H. Nucleophilic reactivities of indoles. The Journal of organic chemistry 2006, 71, 9088–9095. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Zhang, X.; Qu, Y. Biodegradation and biotransformation of indole: advances and perspectives. Frontiers in microbiology 2018, 9, 2625. [Google Scholar] [CrossRef] [PubMed]
- Mondal, D.; Kalar, P. L.; Kori, S.; Gayen, S.; Das, K. Recent developments on synthesis of indole derivatives through green approaches and their pharmaceutical applications. Current Organic Chemistry 2020, 24, 2665–2693. [Google Scholar] [CrossRef]
- Heravi, M. M.; Amiri, Z.; Kafshdarzadeh, K.; Zadsirjan, V. Synthesis of indole derivatives as prevalent moieties present in selected alkaloids. RSC advances 2021, 11, 33540–33612. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, D.; Amin, A.; Qadir, T.; Sharma, P. K. Synthesis of medicinally important indole derivatives: A Review. The Open Medicinal Chemistry Journal 2021, 15. [Google Scholar] [CrossRef]
- Chen, L.; Zou, Y.-X. Recent progress in the synthesis of phosphorus-containing indole derivatives. Organic & Biomolecular Chemistry 2018, 16, 7544–7556. [Google Scholar] [CrossRef]
- Bischler, A. Ueber die entstehung einiger substituirter indole. Berichte der deutschen chemischen Gesellschaft 1892, 25, 2860–2879. [Google Scholar] [CrossRef]
- Fischer, E.; Jourdan, F. Ueber die hydrazine der brenztraubensäure. Berichte der deutschen chemischen Gesellschaft 1883, 16, 2241–2245. [Google Scholar] [CrossRef]
- Hemetsberger, H.; Knittel, D. Synthesis and thermolysis of α-azidoacrylates (ene-azides, IV) Enazide, 4. Mitt. Monatshefte für Chemie/Chemical Monthly 1972, 103, 194–204. [Google Scholar] [CrossRef]
- Baudin, J.-B.; Julia, S. A. Synthesis of indoles from N-aryl-1-alkenylsulphinamides. Tetrahedron letters 1986, 27, 837–840. [Google Scholar] [CrossRef]
- Dorababu, A. Indole–a promising pharmacophore in recent antiviral drug discovery. RSC Medicinal Chemistry 2020, 11, 1335–1353. [Google Scholar] [CrossRef]
- Al Awadh, A. A. Biomedical applications of selective metal complexes of Indole, Benzimidazole, Benzothiazole and Benzoxazole: A review (From 2015 to 2022). Saudi Pharmaceutical Journal 2023, 101698. [Google Scholar] [CrossRef]
- Zhang, M.-Z.; Chen, Q.; Yang, G.-F. A review on recent developments of indole-containing antiviral agents. European journal of medicinal chemistry 2015, 89, 421–441. [Google Scholar] [CrossRef]
- Chadha, N.; Silakari, O. Indoles as therapeutics of interest in medicinal chemistry: Bird's eye view. European Journal of Medicinal Chemistry 2017, 134, 159–184. [Google Scholar] [CrossRef] [PubMed]
- de Sa Alves, F. R.; Barreiro, E. J.; Manssour Fraga, C. A. From nature to drug discovery: the indole scaffold as a ‘privileged structure’. Mini reviews in medicinal chemistry 2009, 9, 782–793. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Kumar, N. M.; Tantak, M. P.; Ogura, M.; Kusaka, E.; Ito, T. Synthesis and identification of α-cyano bis (indolyl) chalcones as novel anticancer agents. Bioorganic & medicinal chemistry letters 2014, 24, 5170–5174. [Google Scholar] [CrossRef]
- Shimazaki, Y.; Yajima, T.; Takani, M.; Yamauchi, O. Metal complexes involving indole rings: Structures and effects of metal–indole interactions. Coordination Chemistry Reviews 2009, 253, 479–492. [Google Scholar] [CrossRef]
- Zhang, M.-Z.; Mulholland, N.; Beattie, D.; Irwin, D.; Gu, Y.-C.; Chen, Q.; Yang, G.-F.; Clough, J. Synthesis and antifungal activity of 3-(1, 3, 4-oxadiazol-5-yl)-indoles and 3-(1, 3, 4-oxadiazol-5-yl) methyl-indoles. European Journal of Medicinal Chemistry 2013, 63, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Lal, S.; J Snape, T. 2-Arylindoles: a privileged molecular scaffold with potent, broad-ranging pharmacological activity. Current medicinal chemistry 2012, 19, 4828–4837. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Rakesh, K.; Leng, J.; Fang, W.-Y.; Ravindar, L.; Gowda, D. C.; Qin, H.-L. Amino acids/peptides conjugated heterocycles: A tool for the recent development of novel therapeutic agents. Bioorganic chemistry 2018, 76, 113–129. [Google Scholar] [CrossRef] [PubMed]
- Bajad, N. G.; Singh, S. K.; Singh, S. K.; Singh, T. D.; Singh, M. Indole: A promising scaffold for the discovery and development of potential anti-tubercular agents. Current Research in Pharmacology and Drug Discovery 2022, 100119. [Google Scholar] [CrossRef] [PubMed]
- Fang, W.-Y.; Ravindar, L.; Rakesh, K.; Manukumar, H.; Shantharam, C.; Alharbi, N. S.; Qin, H.-L. Synthetic approaches and pharmaceutical applications of chloro-containing molecules for drug discovery: A critical review. European journal of medicinal chemistry 2019, 173, 117–153. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, P. A.; Santos, M. M. Recent progress in the development of indole-based compounds active against malaria, trypanosomiasis and leishmaniasis. Molecules 2022, 27, 319. [Google Scholar] [CrossRef] [PubMed]
- Moku, B.; Ravindar, L.; Rakesh, K.; Qin, H.-L. The significance of N-methylpicolinamides in the development of anticancer therapeutics: Synthesis and structure-activity relationship (SAR) studies. Bioorganic Chemistry 2019, 86, 513–537. [Google Scholar] [CrossRef]
- Bianucci, A. M.; Da Settimo, A.; Da Settimo, F.; Primofiore, G.; Martini, C.; Giannaccini, G.; Lucacchini, A. Benzodiazepine receptor affinity and interaction of some N-(indol-3-ylglyoxylyl) amine derivatives. Journal of medicinal chemistry 1992, 35, 2214–2220. [Google Scholar] [CrossRef]
- Singh, D.; Grover, V.; Kumar, K.; Jain, K. Metal ion prompted macrocyclic complexes derived from indole-2, 3-dione (isatin) and O-phenylenediamine with their spectroscopic and antibacterial studies. Acta Chimica Slovenica 2010, 57, 775–780. [Google Scholar] [PubMed]
- Vendeville, S.; Lin, T.-I.; Hu, L.; Tahri, A.; McGowan, D.; Cummings, M. D.; Amssoms, K.; Canard, M.; Last, S.; Van den Steen, I. Finger loop inhibitors of the HCV NS5b polymerase. Part II. Optimization of tetracyclic indole-based macrocycle leading to the discovery of TMC647055. Bioorganic & medicinal chemistry letters 2012, 22, 4437–4443. [Google Scholar] [CrossRef]
- Au, V. S.; Bremner, J. B.; Coates, J.; Keller, P. A.; Pyne, S. G. Synthesis of some cyclic indolic peptoids as potential antibacterials. Tetrahedron 2006, 62, 9373–9382. [Google Scholar] [CrossRef]
- Álvarez, R.; López, V.; Mateo, C.; Medarde, M.; Peláez, R. New para–para stilbenophanes: Synthesis by McMurry coupling, conformational analysis and inhibition of tubulin polymerisation. Chemistry–A European Journal 2011, 17, 3406–3419. [Google Scholar] [CrossRef]
- Neochoritis, C. G.; Miraki, M. K.; Abdelraheem, E. M.; Surmiak, E.; Zarganes-Tzitzikas, T.; Łabuzek, B.; Holak, T. A.; Dömling, A. Design of indole-and MCR-based macrocycles as p53-MDM2 antagonists. Beilstein Journal of Organic Chemistry 2019, 15, 513–520. [Google Scholar] [CrossRef] [PubMed]
- Chen, R. Synthesis of novel Indole-based Macrocycles. UNSW Sydney, 2012.
- Cheekatla, S. R.; Barik, D.; Anand, G.; Mol KM, R.; Porel, M. Indole-Based Macrocyclization by Metal-Catalyzed Approaches. Organics 2023, 4, 333–363. [Google Scholar] [CrossRef]
- Zhang, Y.-C.; Jiang, F.; Shi, F. Organocatalytic asymmetric synthesis of indole-based chiral heterocycles: strategies, reactions, and outreach. Accounts of chemical research 2019, 53, 425–446. [Google Scholar] [CrossRef]
- Lin, C.-H.; Wu, Y.-R.; Kung, P.-J.; Chen, W.-L.; Lee, L.-C.; Lin, T.-H.; Chao, C.-Y.; Chen, C.-M.; Chang, K.-H.; Janreddy, D. The potential of indole and a synthetic derivative for polyQ aggregation reduction by enhancement of the chaperone and autophagy systems. ACS Chemical Neuroscience 2014, 5, 1063–1074. [Google Scholar] [CrossRef] [PubMed]
- Kung, P.-J.; Tao, Y.-C.; Hsu, H.-C.; Chen, W.-L.; Lin, T.-H.; Janreddy, D.; Yao, C.-F.; Chang, K.-H.; Lin, J.-Y.; Su, M.-T. Indole and synthetic derivative activate chaperone expression to reduce polyQ aggregation in SCA17 neuronal cell and slice culture models. Drug design, development and therapy 2014, 1929–1939. [Google Scholar]
- Chen, C.-M.; Chen, W.-L.; Hung, C.-T.; Lin, T.-H.; Chao, C.-Y.; Lin, C.-H.; Wu, Y.-R.; Chang, K.-H.; Yao, C.-F.; Lee-Chen, G.-J. The indole compound NC009-1 inhibits aggregation and promotes neurite outgrowth through enhancement of HSPB1 in SCA17 cells and ameliorates the behavioral deficits in SCA17 mice. Neurotoxicology 2018, 67, 259–269. [Google Scholar] [CrossRef]
- Wei, P.-C.; Lee-Chen, G.-J.; Chen, C.-M.; Wu, Y.-R.; Chen, Y.-J.; Lin, J.-L.; Lo, Y.-S.; Yao, C.-F.; Chang, K.-H. Neuroprotection of indole-derivative compound NC001-8 by the regulation of the NRF2 pathway in Parkinson’s disease cell models. Oxidative medicine and cellular longevity 2019, 2019. [Google Scholar] [CrossRef] [PubMed]
- Afanas’eva, I. B.; Ostrakhovitch, E. A.; Mikhal’chik, E. V.; Ibragimova, G. A.; Korkina, L. G. Enhancement of antioxidant and anti-inflammatory activities of bioflavonoid rutin by complexation with transition metals. Biochemical pharmacology 2001, 61, 677–684. [Google Scholar] [CrossRef]
- Ejidike, I. P.; Ajibade, P. A. Synthesis, characterization and biological studies of metal (II) complexes of (3 E)-3-[(2-{(E)-[1-(2, 4-dihydroxyphenyl) ethylidene] amino} ethyl) imino]-1-phenylbutan-1-one Schiff base. Molecules 2015, 20, 9788–9802. [Google Scholar] [CrossRef]
- Zhu, X.; Li, Y.; Wei, Y.; Wang, S.; Zhou, S.; Zhang, L. Reactivity of 3-imino-functionalized indoles with rare-earth-metal amides: Unexpected substituent effects on C–H activation pathways and assembly of rare-earth-metal complexes. Organometallics 2016, 35, 1838–1846. [Google Scholar] [CrossRef]
- Abdulghani, A. J.; Hussain, R. K. Synthesis and Characterization of Schiff Base Metal Complexes Derived from Cefotaxime with 1H-indole-2, 3-dione (Isatin) and 4-N, N-dimethyl-aminobenzaldehyde. Open Journal of Inorganic Chemistry 2015, 5, 83. [Google Scholar] [CrossRef]
- EL-Gammal, O. A.; Alshater, H.; El-Boraey, H. A. Schiff base metal complexes of 4-methyl-1H-indol-3-carbaldehyde derivative as a series of potential antioxidants and antimicrobial: Synthesis, spectroscopic characterization and 3D molecular modeling. Journal of Molecular Structure 2019, 1195, 220–230. [Google Scholar] [CrossRef]
- Deng, X.-j.; Yu, Q.; Bian, H.-D.; Ju, H.-D.; Wang, B.-L. Syntheses, structures and properties of complexes of indole-3-propionic acid. Transition Metal Chemistry 2016, 41, 591–598. [Google Scholar] [CrossRef]
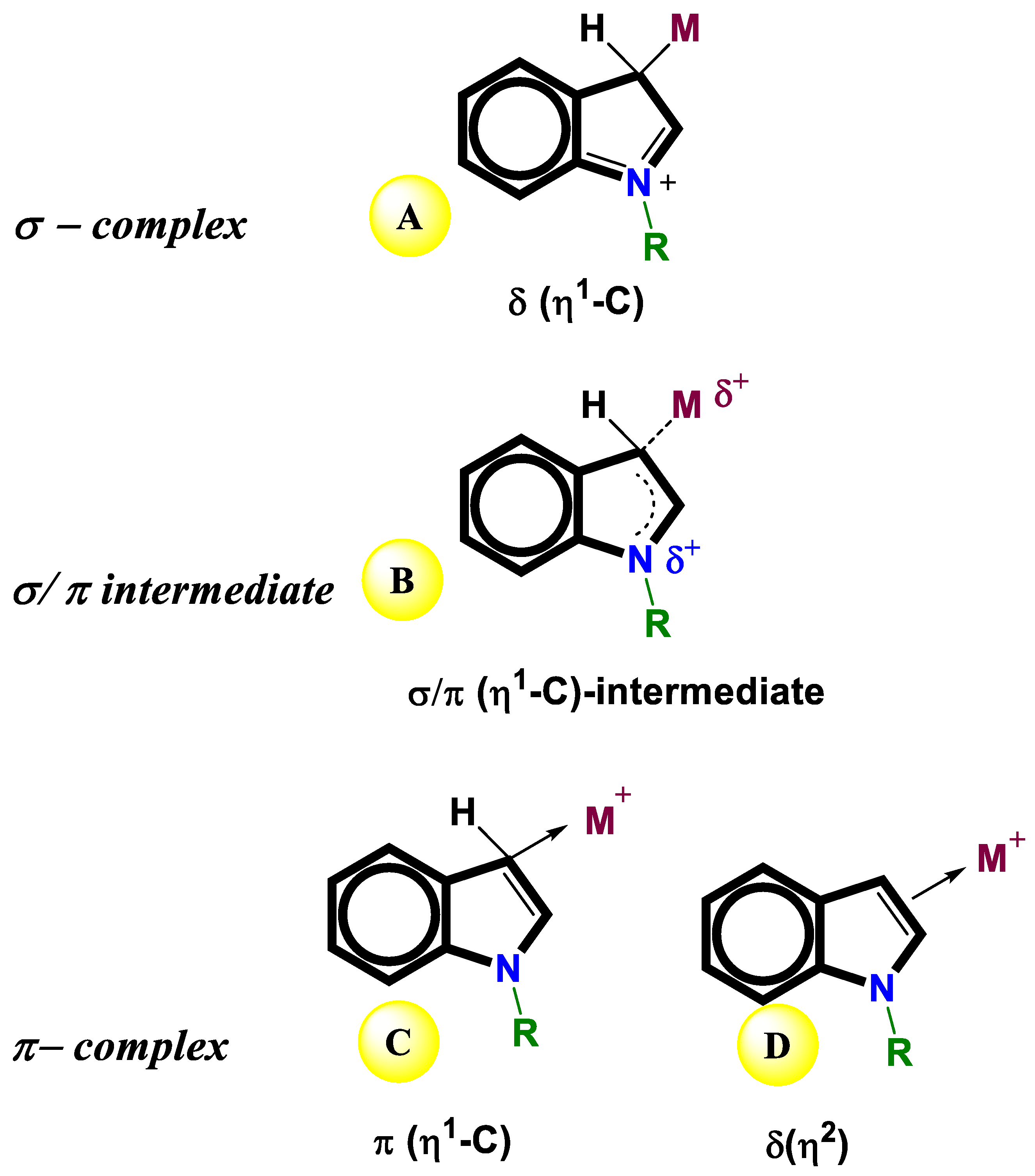
- Yamamoto, K.; Kimura, S.; Murahashi, T. σ–π Continuum in indole–Palladium (II) complexes. Angewandte Chemie International Edition 2016, 55, 5322–5326. [Google Scholar] [CrossRef]
- Shimazaki, Y.; Yajima, T.; Yamauchi, O. Properties of the indole ring in metal complexes. A comparison with the phenol ring. Journal of Inorganic Biochemistry 2015, 148, 105–115. [Google Scholar] [CrossRef]
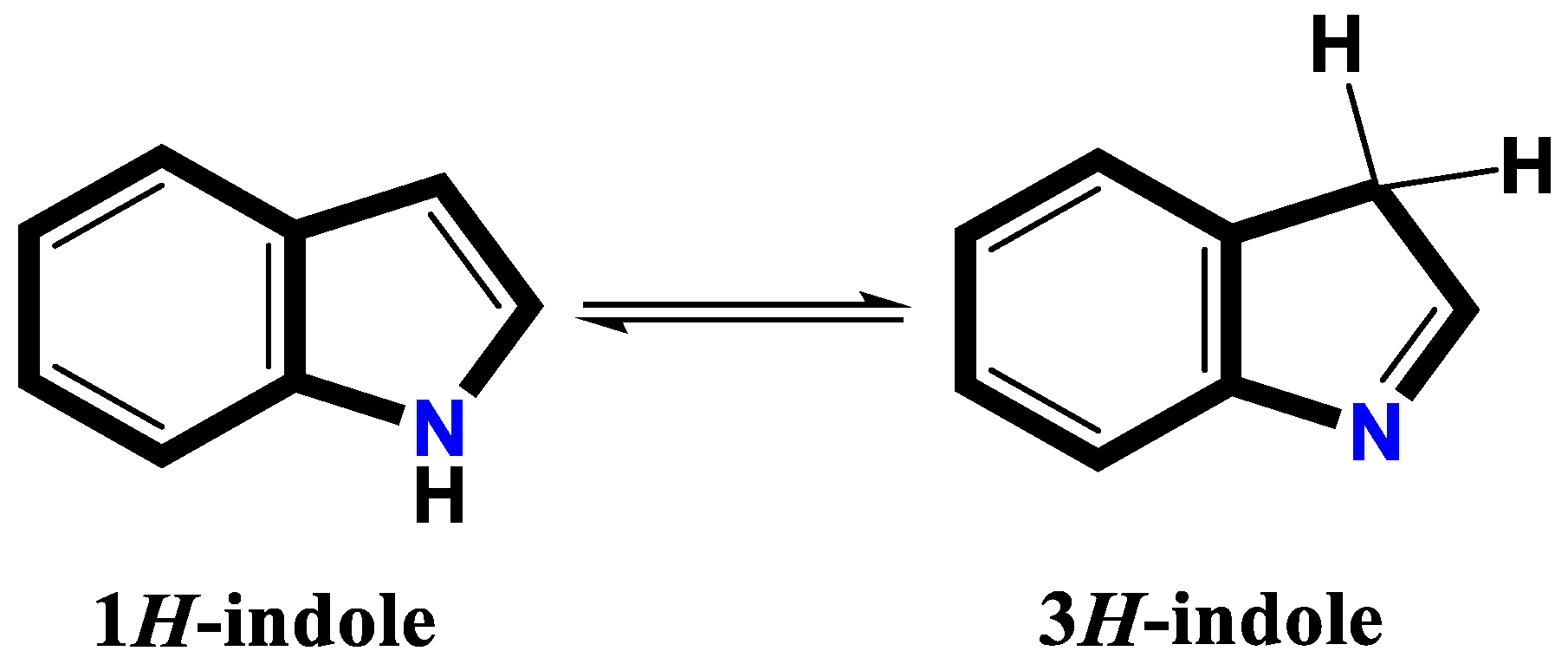
- Yamauchi, O.; Takani, M.; Toyoda, K.; Masuda, H. Indole nitrogen-palladium (II) bonding. Chemical and structural characterization of palladium (II) complexes of alkylindoles and intermediacy of the 3H-indole ring. Inorganic Chemistry 1990, 29, 1856–1860. [Google Scholar] [CrossRef]
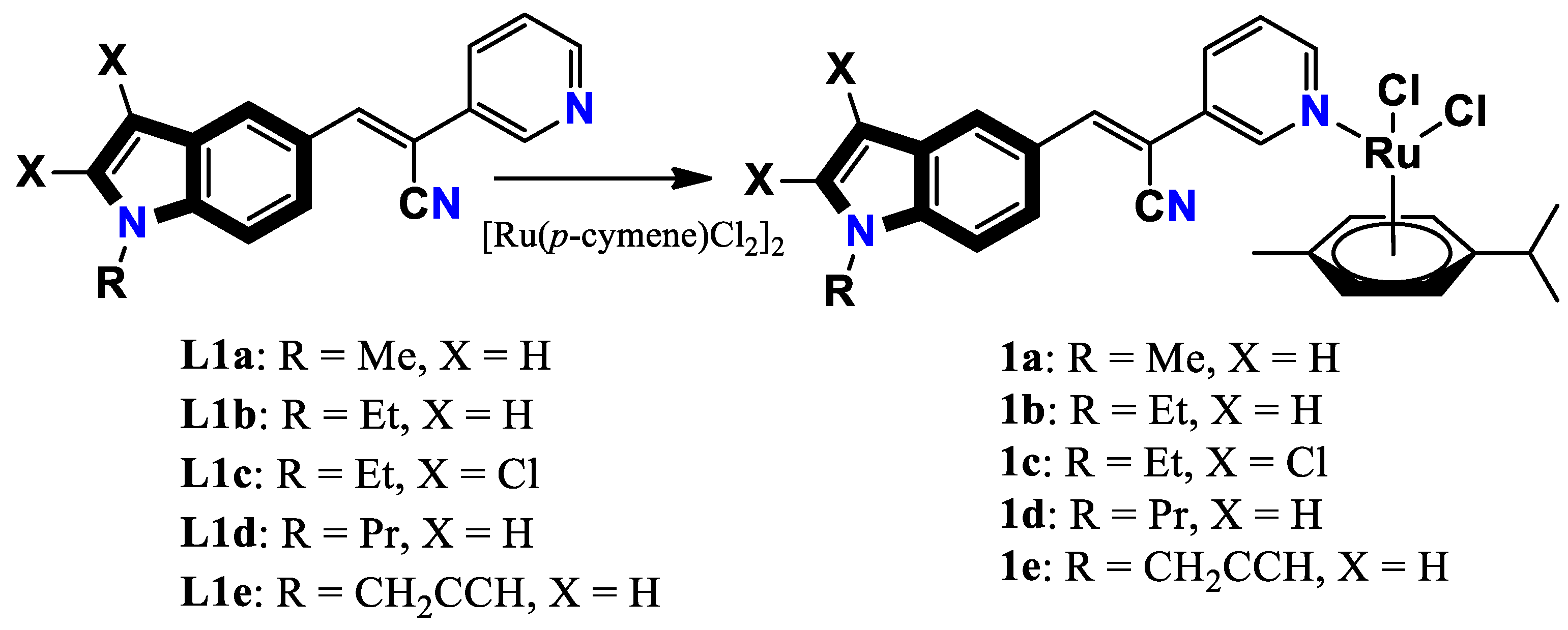
- Oberhuber, N.; Ghosh, H.; Nitzsche, B.; Dandawate, P.; Höpfner, M.; Schobert, R.; Biersack, B. Synthesis and Anticancer Evaluation of New Indole-Based Tyrphostin Derivatives and Their (p-Cymene) dichloridoruthenium (II) Complexes. International Journal of Molecular Sciences 2023, 24, 854. [Google Scholar] [CrossRef]
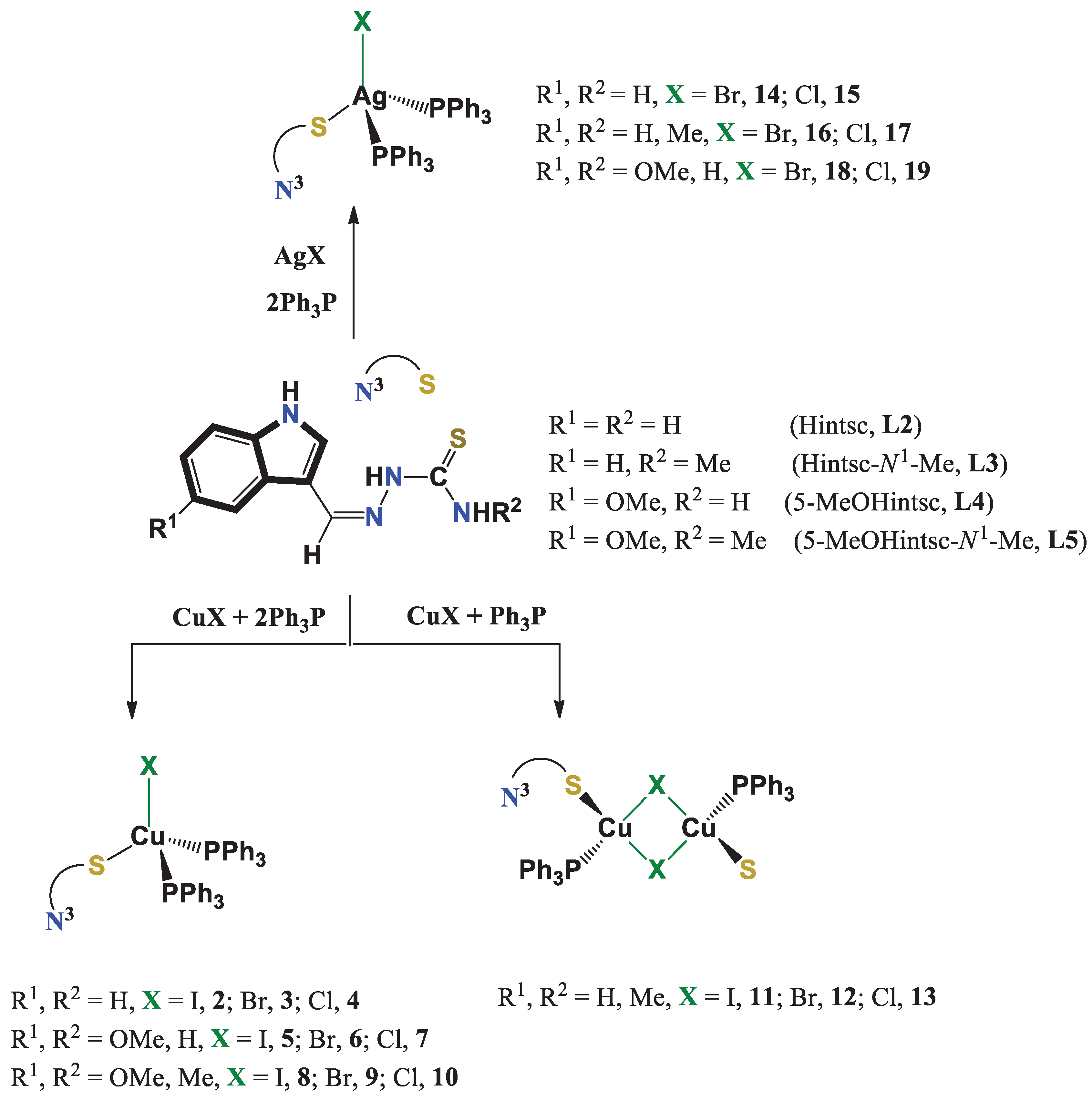
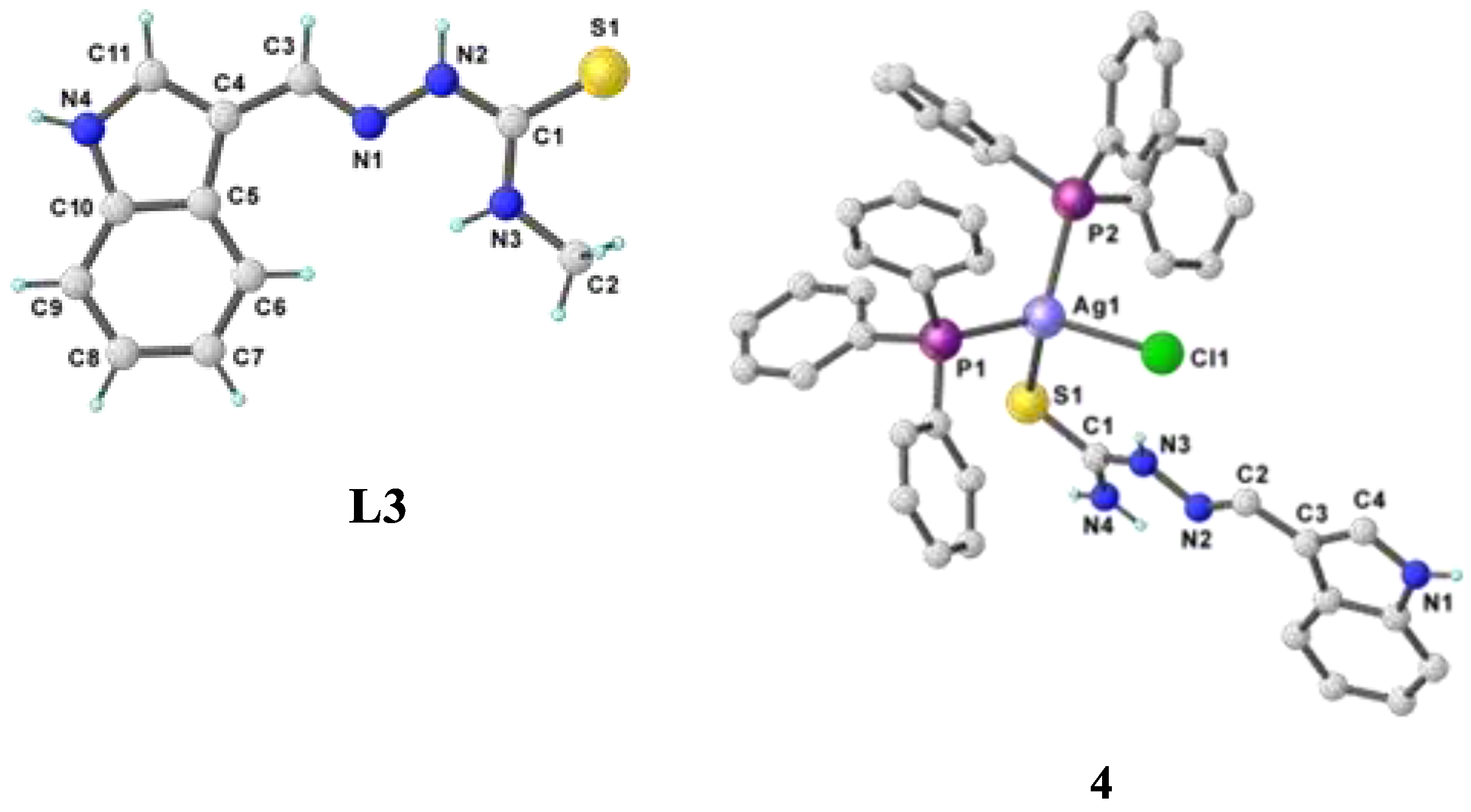
- Khan, A.; Jasinski, J. P.; Smolenski, V. A.; Hotchkiss, E. P.; Kelley, P. T.; Shalit, Z. A.; Kaur, M.; Paul, K.; Sharma, R. Enhancement in anti-tubercular activity of indole based thiosemicarbazones on complexation with copper (I) and silver (I) halides: structure elucidation, evaluation and molecular modelling. Bioorganic Chemistry 2018, 80, 303–318. [Google Scholar] [CrossRef] [PubMed]
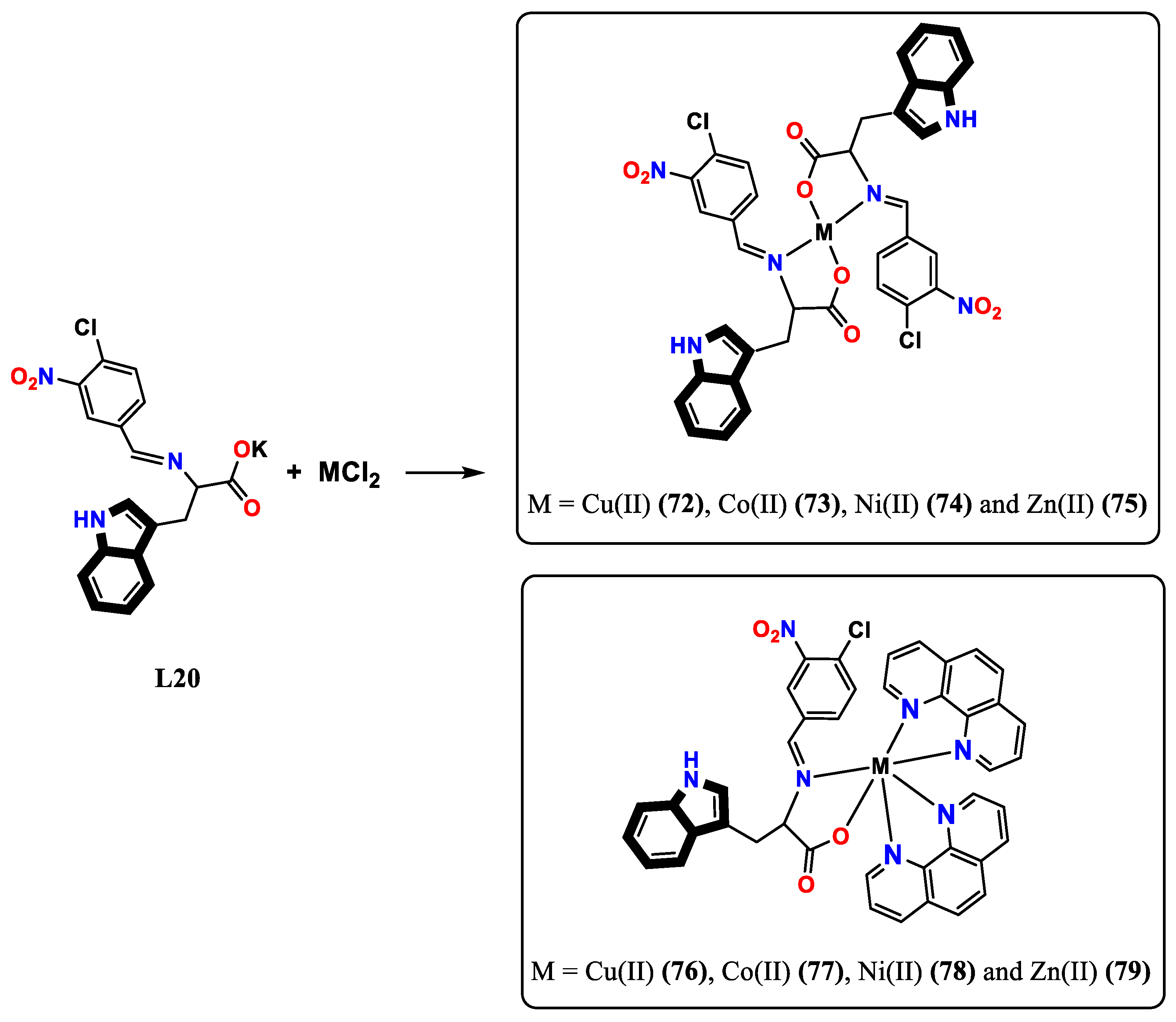
- Chilwal, A.; Malhotra, P.; Narula, A. Synthesis, characterization, thermal, and antibacterial studies of organotin (IV) complexes of indole-3-butyric acid and indole-3-propionic acid. Phosphorus, Sulfur, and Silicon and the Related Elements 2014, 189, 410–421. [Google Scholar] [CrossRef]
- Li, J.; Gao, T.; Zhang, W.; Sun, W.-H. Synthesis and characterization of 2-imino-indole nickel complexes and their ethylene oligomerization study. Inorganic Chemistry Communications 2003, 6, 1372–1374. [Google Scholar] [CrossRef]
- Soldevila-Barreda, J. J.; Fawibe, K. B.; Azmanova, M.; Rafols, L.; Pitto-Barry, A.; Eke, U. B.; Barry, N. P. Synthesis, characterisation and in vitro anticancer activity of catalytically active indole-based half-sandwich complexes. Molecules 2020, 25, 4540. [Google Scholar] [CrossRef]
- Babijczuk, K.; Warżajtis, B.; Starzyk, J.; Mrówczyńska, L.; Jasiewicz, B.; Rychlewska, U. Synthesis, Structure and Biological Activity of Indole–Imidazole Complexes with ZnCl2: Can Coordination Enhance the Functionality of Bioactive Ligands? Molecules 2023, 28, 4132. [Google Scholar] [CrossRef] [PubMed]
- Lo, K. K.-W.; Tsang, K. H.-K.; Hui, W.-K.; Zhu, N. Synthesis, characterization, crystal structure, and electrochemical, photophysical, and protein-binding properties of luminescent rhenium (I) diimine indole complexes. Inorganic chemistry 2005, 44, 6100–6110. [Google Scholar] [CrossRef] [PubMed]
- Wittmann, C.; Sivchenko, A. S.; Bacher, F.; Tong, K. K.; Guru, N.; Wilson, T.; Gonzales, J.; Rauch, H.; Kossatz, S.; Reiner, T. Inhibition of microtubule dynamics in cancer cells by indole-modified latonduine derivatives and their metal complexes. Inorganic Chemistry 2022, 61, 1456–1470. [Google Scholar] [CrossRef] [PubMed]
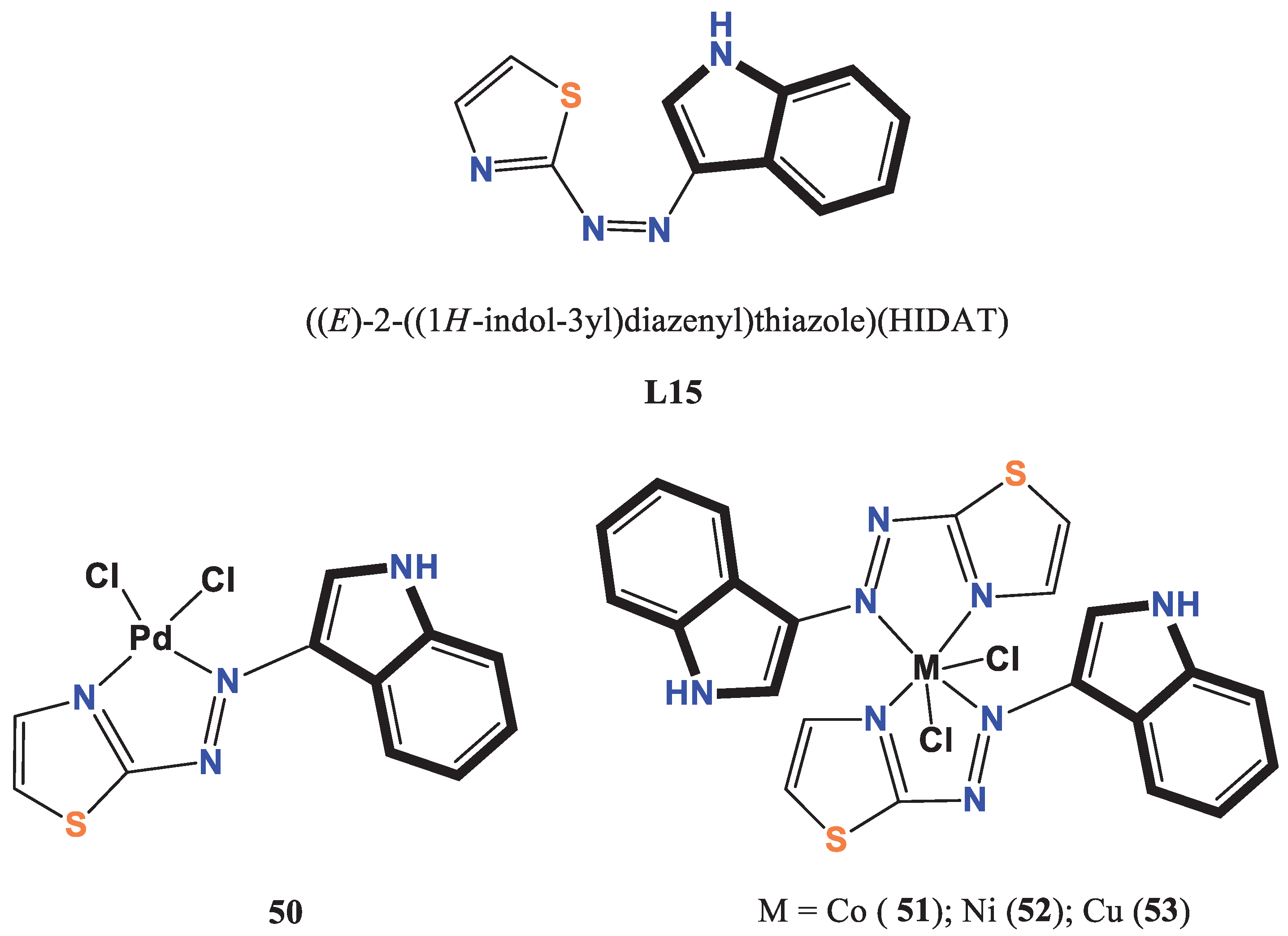
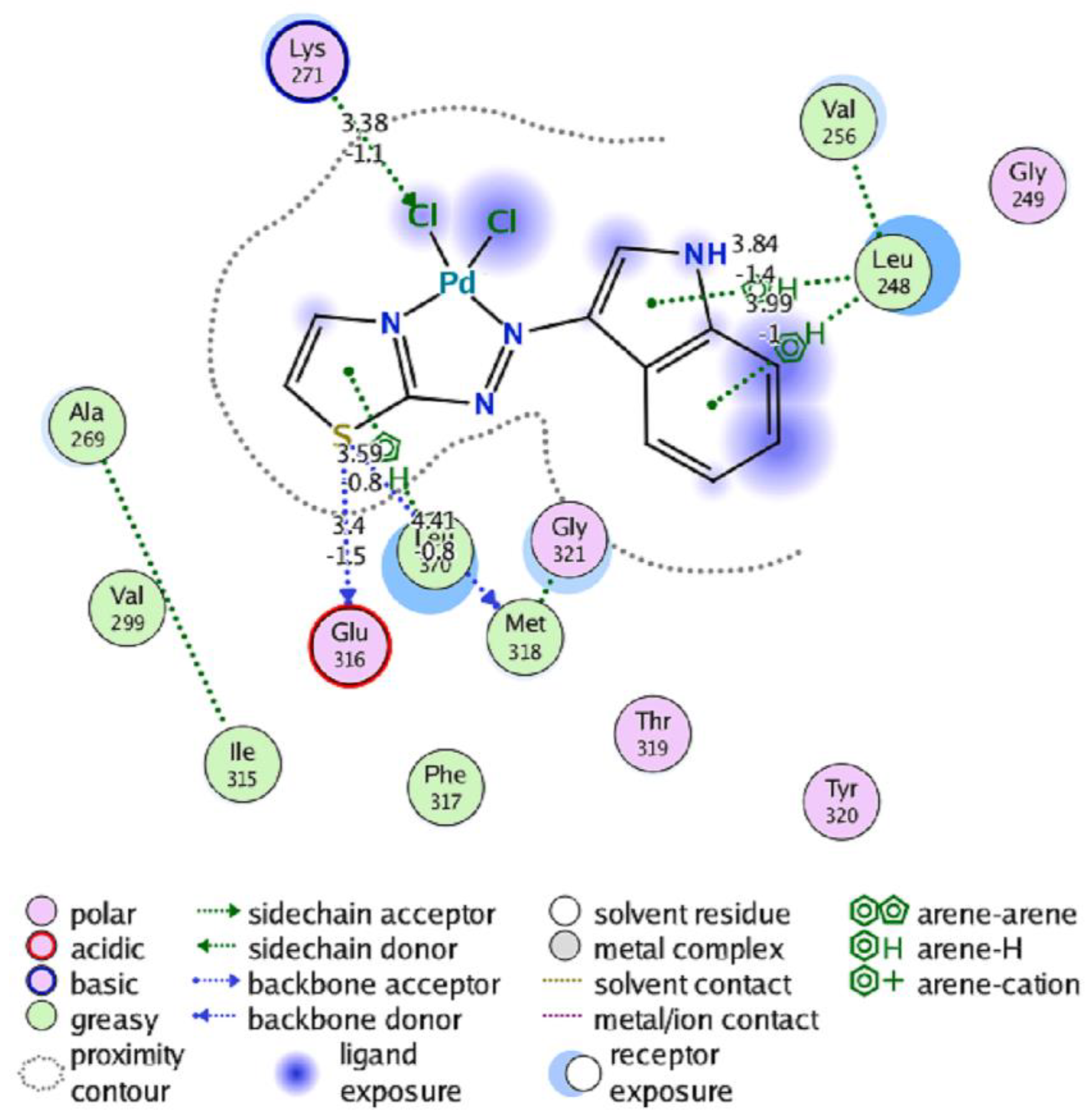
- Sahar, Y. J.; Mohammed, H.; Al-Abady, Z. N. Synthesis and characterization of new metal complexes containing azo-indole moiety and anti-leukemia human (HL-60) study of its palladium (II) complex. Results in Chemistry 2023, 5, 100847. [Google Scholar] [CrossRef]
- Bhanumathy, K.K.; Balagopal, A.; Vizeacoumar, F. S.; Vizeacoumar, F. J.; Freywald, A.; Giambra, V. Protein tyrosine kinases: their roles and their targeting in leukemia. Cancers 2021, 13, 184. [Google Scholar] [CrossRef]
- Babahan, I.; Özmen, A.; Aksel, M.; Bilgin, M. D.; Gumusada, R.; Gunay, M. E.; Eyduran, F. A novel bidentate ligand containing oxime, hydrazone and indole moieties and its BF2+ bridged transition metal complexes and their efficiency against prostate and breast cancer cells. Applied Organometallic Chemistry 2020, 34, e5632. [Google Scholar] [CrossRef]
- Alanazi, R. L.; Zaki, M.; Bawazir, W. A. Synthesis and characterization of new metal complexes containing Triazino [5, 6–b] indole moiety: In vitro DNA and HSA binding studies. Journal of Molecular Structure 2021, 1246, 131203. [Google Scholar] [CrossRef]
- Shakir, Y. G.; Shamran, M. H. Synthesis and characterization of indole azo metal complexes and study of their biological activity. Res. J. Chem. Environ. 2022, 2022, 12. [Google Scholar]
- Tümer, M. Synthesis and spectral characterization of metal complexes containing tetra-and pentadentate Schiff base ligands. Sythesis and Reactivity in Inorganic and Matel-Organic Chemistry 2000, 30, 1139–1158. [Google Scholar]
- Arunadevi, A.; Raman, N. Indole-derived water-soluble N, O bi-dentate ligand-based mononuclear transition metal complexes: in silico and in vitro biological screening, molecular docking and macromolecule interaction studies. Journal of Biomolecular Structure and Dynamics 2019. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Scientific reports 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed]
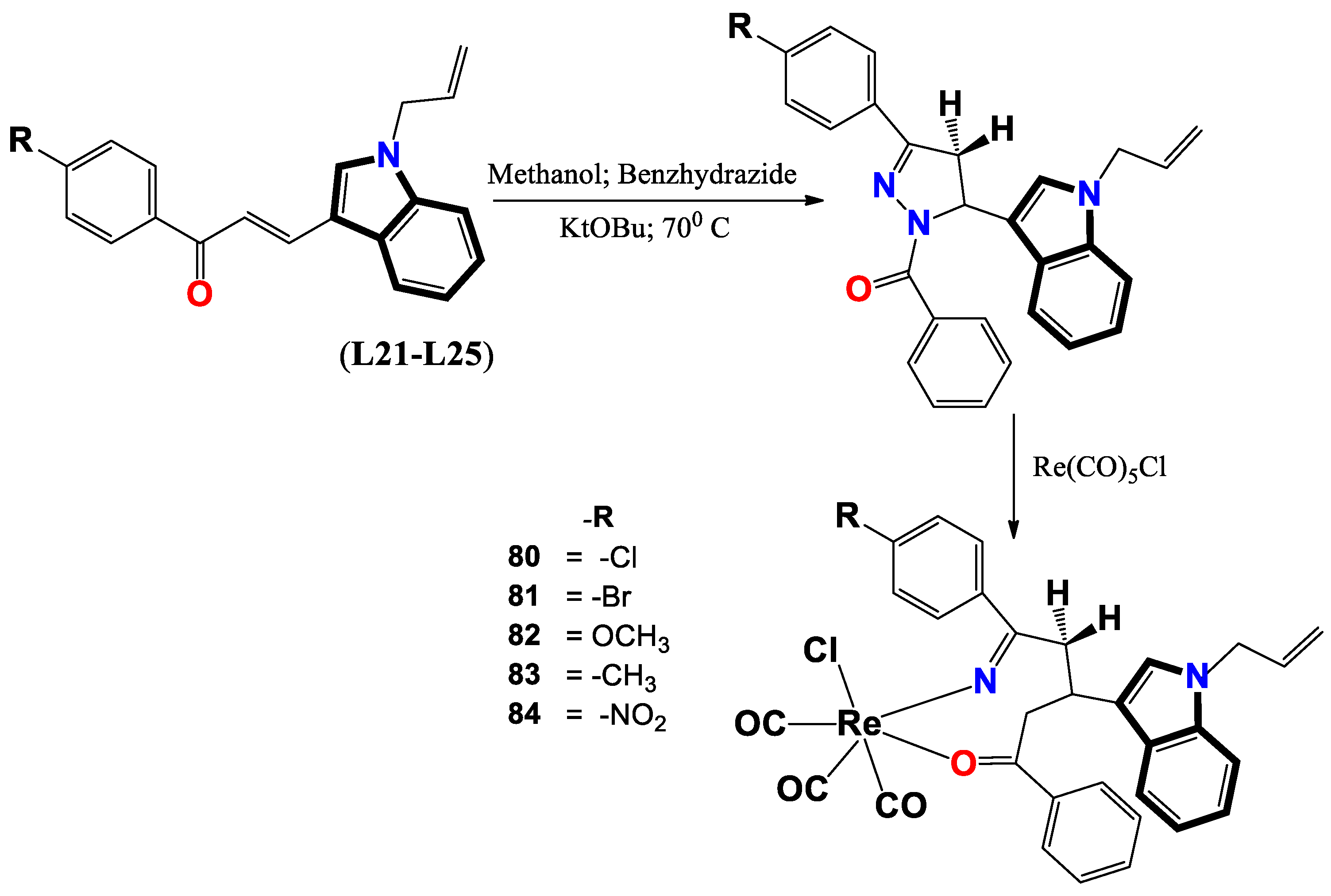
- Varma, R. R.; Pandya, J. G.; Vaidya, F. U.; Pathak, C.; Bhatt, B. S.; Patel, M. N. Biological activities of pyrazoline-indole based Re (I) carbonyls: DNA interaction, antibacterial, anticancer, ROS production, lipid peroxidation, in vivo and in vitro cytotoxicity studies. Chemico-Biological Interactions 2020, 330, 109231. [Google Scholar] [CrossRef]
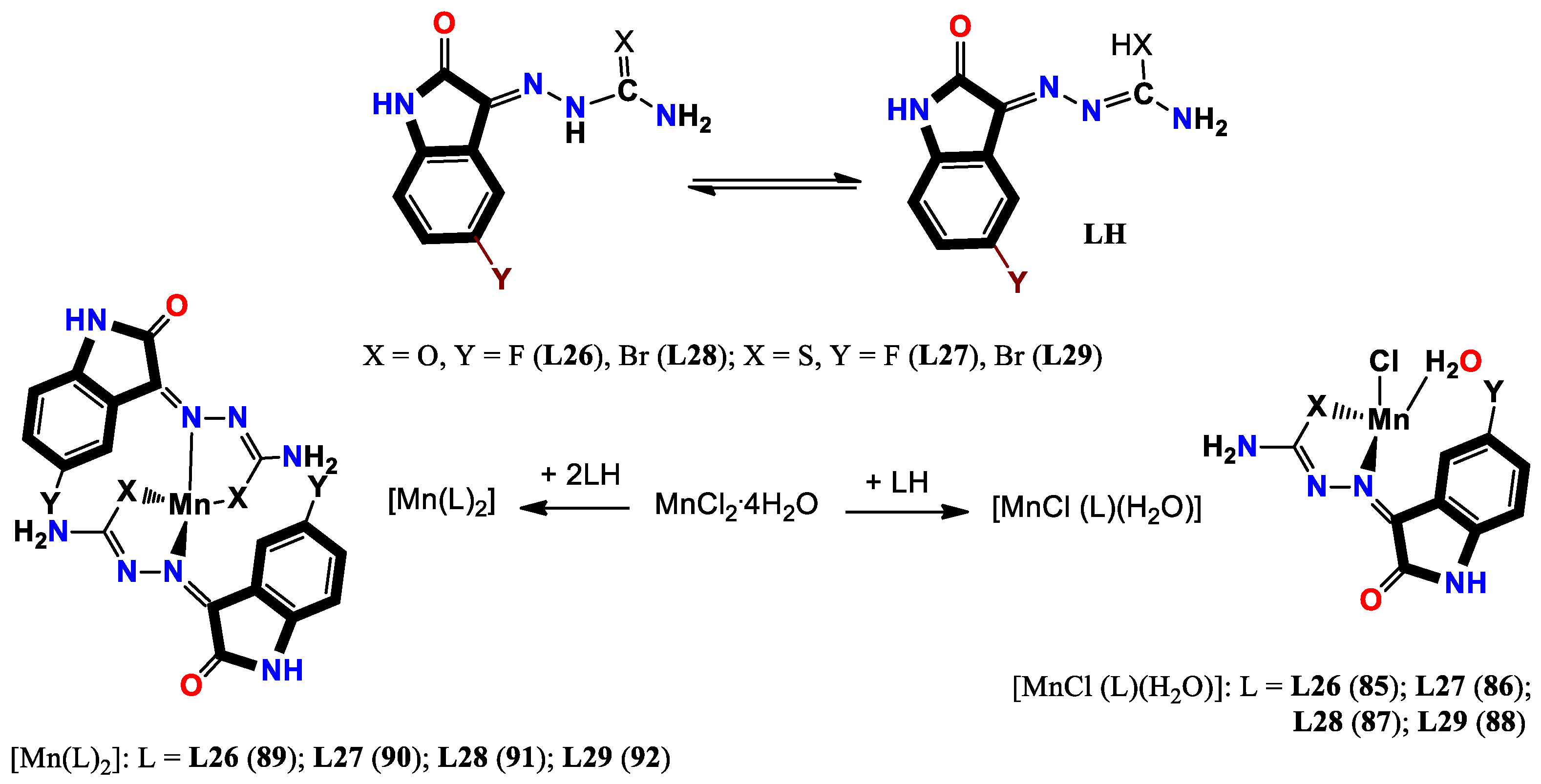
- Sharma, S.; Meena, R.; Satyawana, Y.; Fahmi, N. Manganese (II) complexes of biological relevance: Synthesis and spectroscopic characterization of novel manganese (II) complexes with monobasic bidentate ligands derived from halo-substituted 1 H-indole-2, 3-diones. Russian Journal of General Chemistry 2016, 86, 2807–2816. [Google Scholar] [CrossRef]
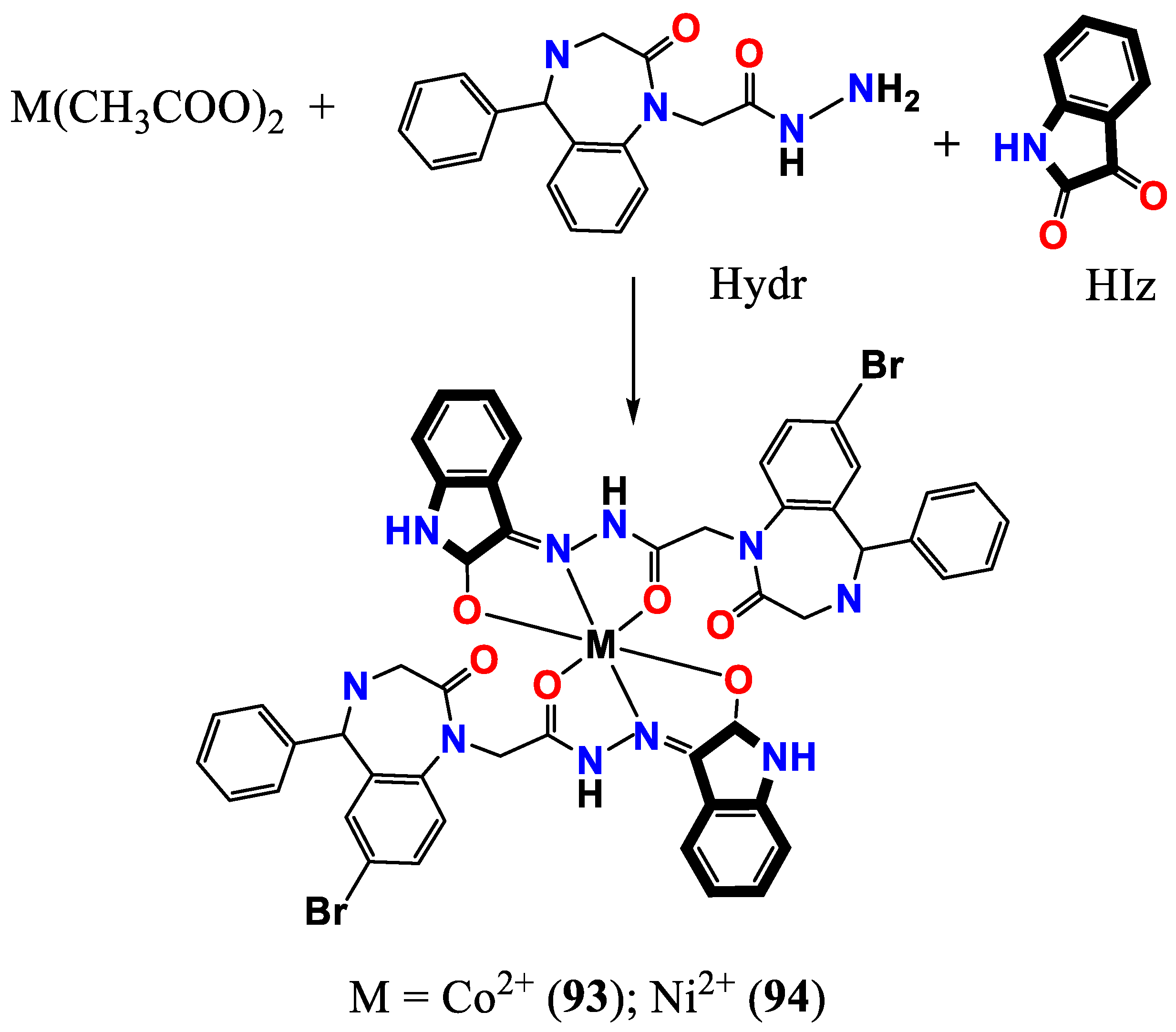
- Seifullina, I.; Skorokhod, L.; Pulya, A.; Vlasenko, V.; Trigub, A.; Rakipow, I. Synthesis, Structure, and Properties of Co 2+ and Ni 2+ Complexes with the Product of Condensation of 2-(7-Bromo-2-oxo-5-phenyl-3 H-1, 4-benzodiazepin-1-yl) acetohydrazide and 1 H-Indole-2, 3-dione. Russian Journal of General Chemistry 2020, 90, 1298–1303. [Google Scholar] [CrossRef]
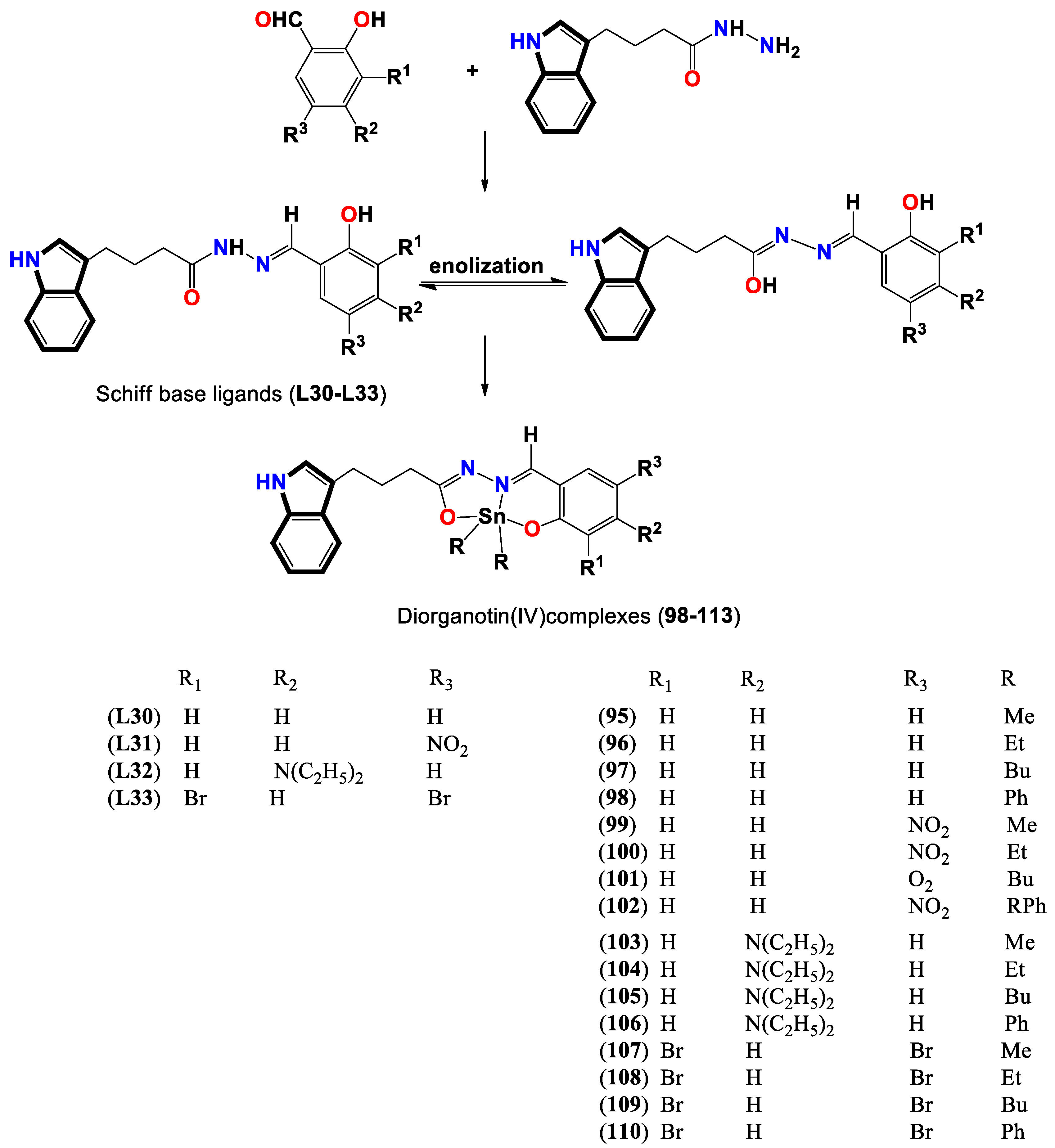
- Devi, J.; Yadav, J.; Kumar, D.; Jindal, D. K.; Basu, B. Synthesis, spectral analysis and in vitro cytotoxicity of diorganotin (IV) complexes derived from indole-3-butyric hydrazide. Applied Organometallic Chemistry 2020, 34, e5815. [Google Scholar] [CrossRef]
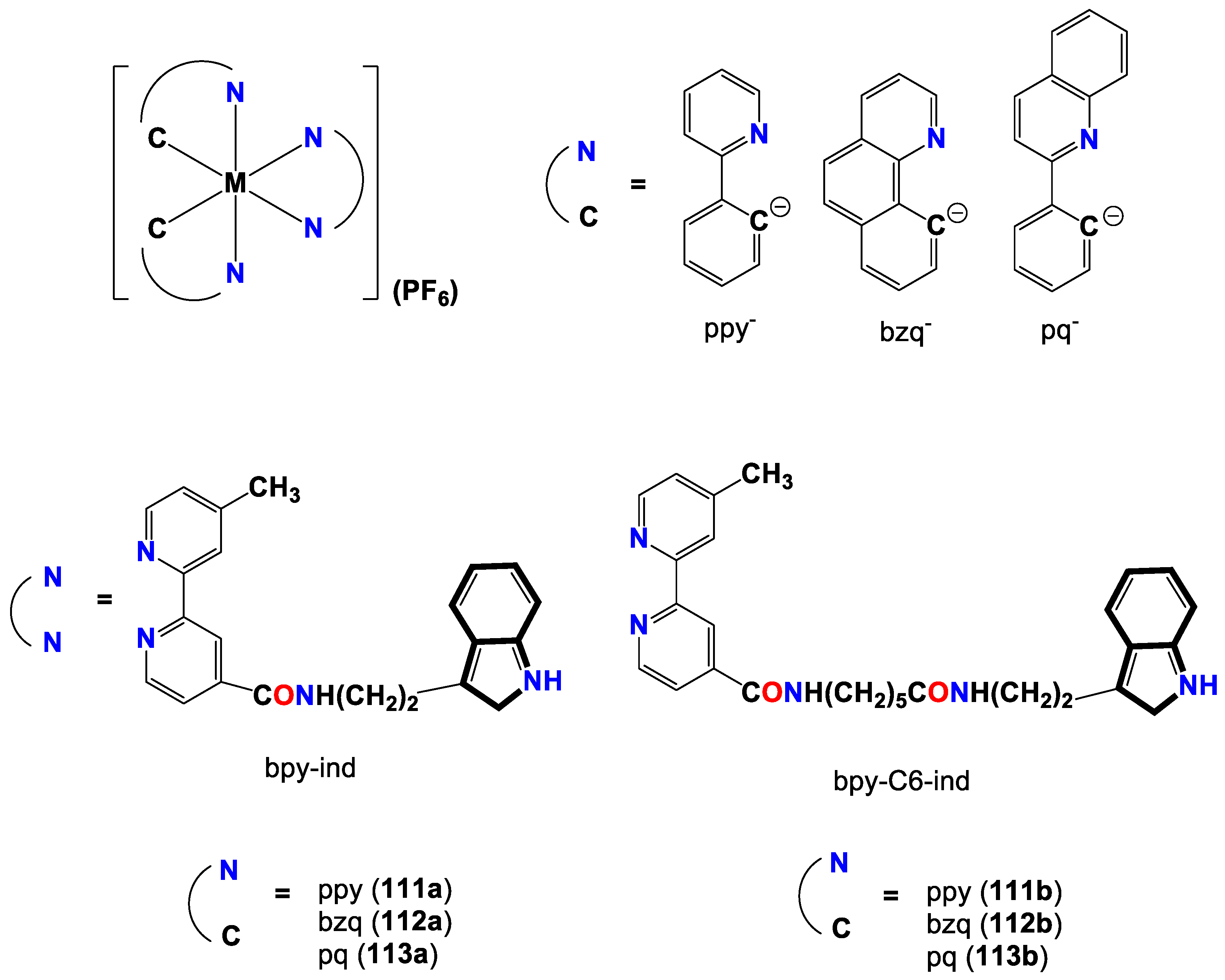
- Lau, J. S.-Y.; Lee, P.-K.; Tsang, K. H.-K.; Ng, C. H.-C.; Lam, Y.-W.; Cheng, S.-H.; Lo, K. K.-W. Luminescent cyclometalated iridium (III) polypyridine indole complexes synthesis, photophysics, electrochemistry, protein-binding properties, cytotoxicity, and cellular uptake. Inorganic Chemistry 2009, 48, 708–718. [Google Scholar] [CrossRef] [PubMed]
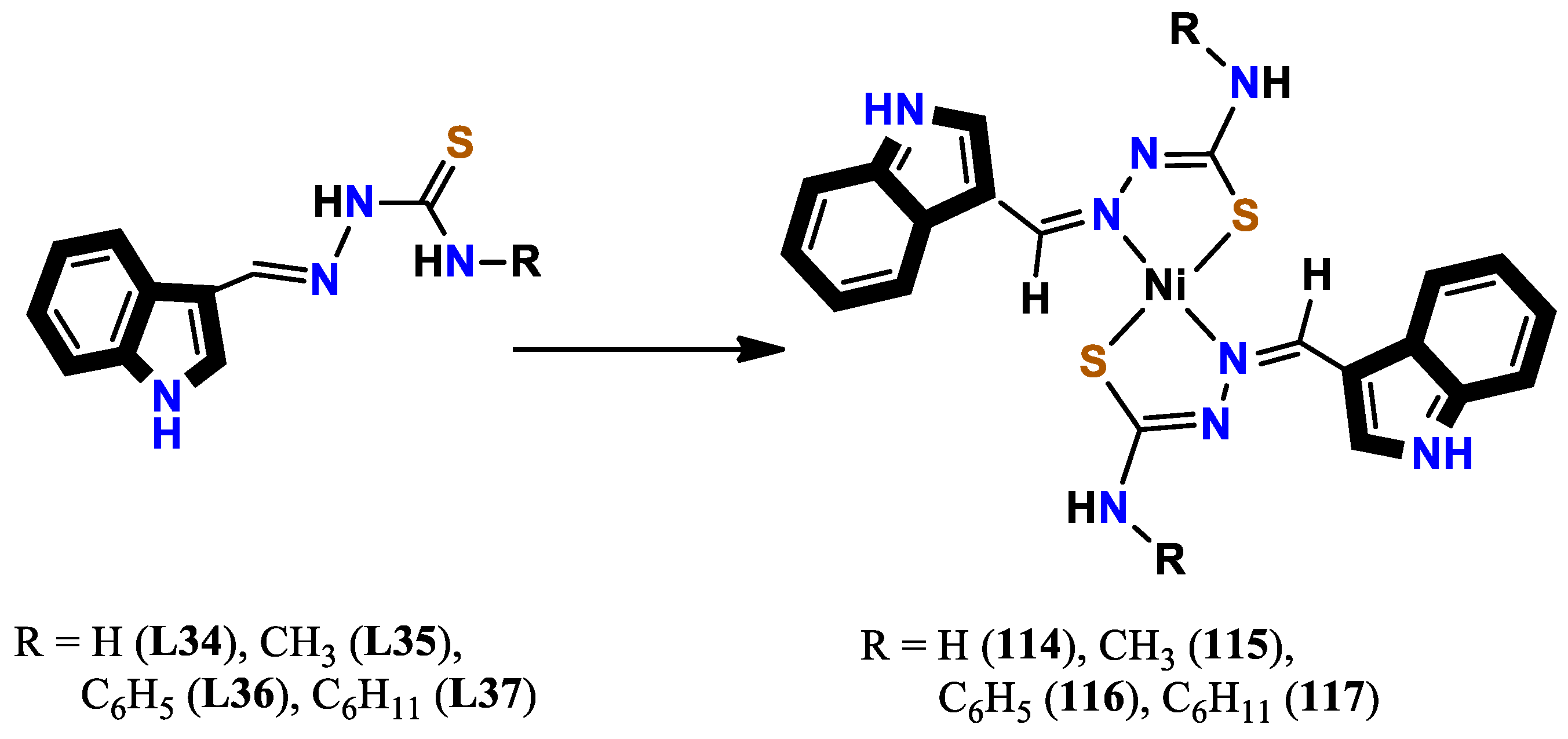
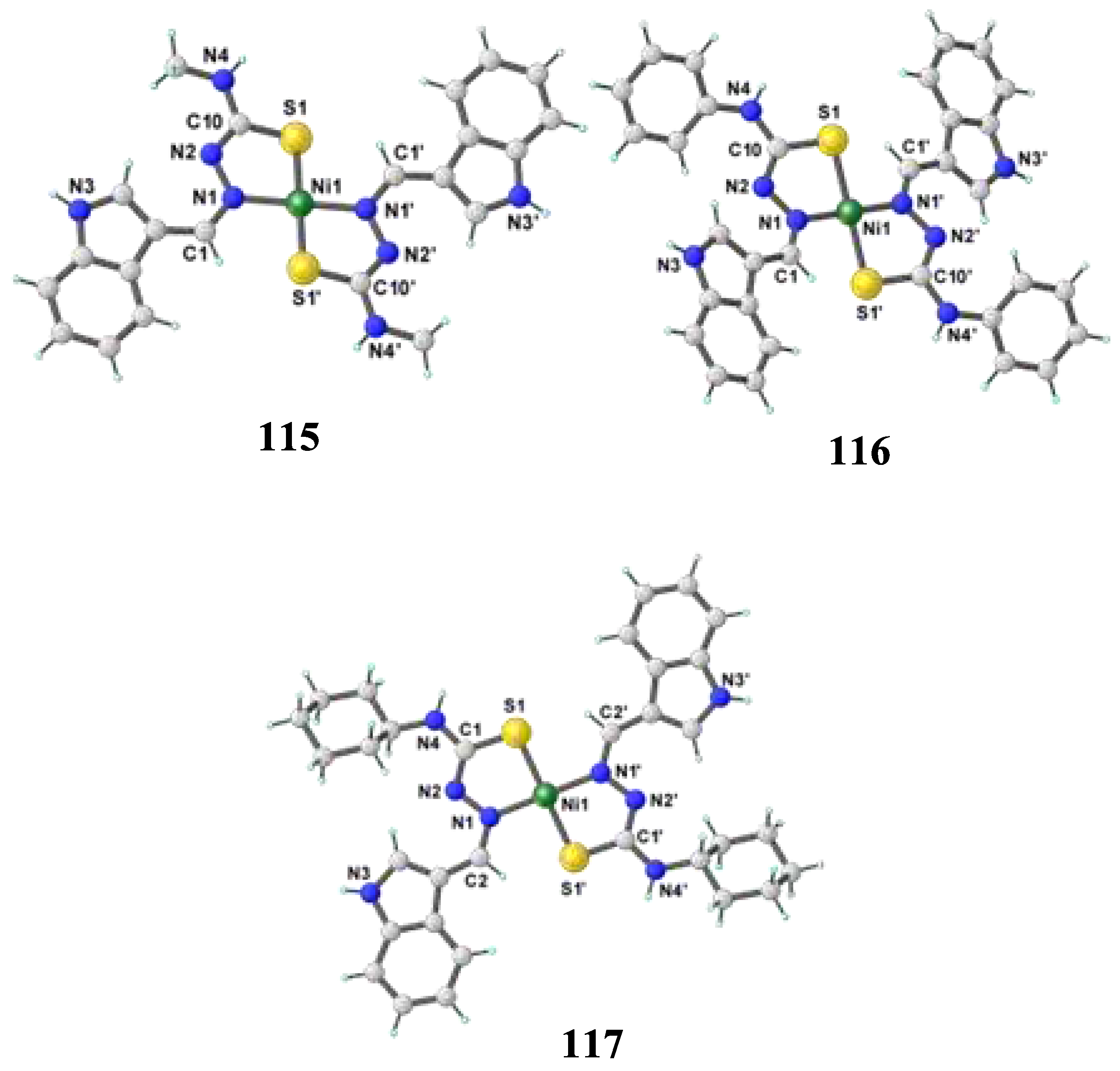
- Haribabu, J.; Jeyalakshmi, K.; Arun, Y.; Bhuvanesh, N. S.; Perumal, P. T.; Karvembu, R. Synthesis of Ni (II) complexes bearing indole-based thiosemicarbazone ligands for interaction with biomolecules and some biological applications. JBIC Journal of Biological Inorganic Chemistry 2017, 22, 461–480. [Google Scholar] [CrossRef]
- Haribabu, J.; Tamizh, M. M.; Balachandran, C.; Arun, Y.; Bhuvanesh, N. S.; Endo, A.; Karvembu, R. Synthesis, structures and mechanistic pathways of anticancer activity of palladium (II) complexes with indole-3-carbaldehyde thiosemicarbazones. New Journal of Chemistry 2018, 42, 10818–10832. [Google Scholar] [CrossRef]
- Nolan, V. C.; Rafols, L.; Harrison, J.; Soldevila-Barreda, J. J.; Crosatti, M.; Garton, N. J.; Wegrzyn, M.; Timms, D. L.; Seaton, C. C.; Sendron, H. Indole-containing arene-ruthenium complexes with broad spectrum activity against antibiotic-resistant bacteria. Current research in microbial sciences 2022, 3, 100099. [Google Scholar] [CrossRef]
- Balakrishnan, N.; Haribabu, J.; Krishnan, D. A.; Swaminathan, S.; Mahendiran, D.; Bhuvanesh, N. S.; Karvembu, R. Zinc (II) complexes of indole thiosemicarbazones: DNA/protein binding, molecular docking and in vitro cytotoxicity studies. Polyhedron 2019, 170, 188–201. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




