Submitted:
20 December 2023
Posted:
21 December 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction of Shrews (Soricidae)
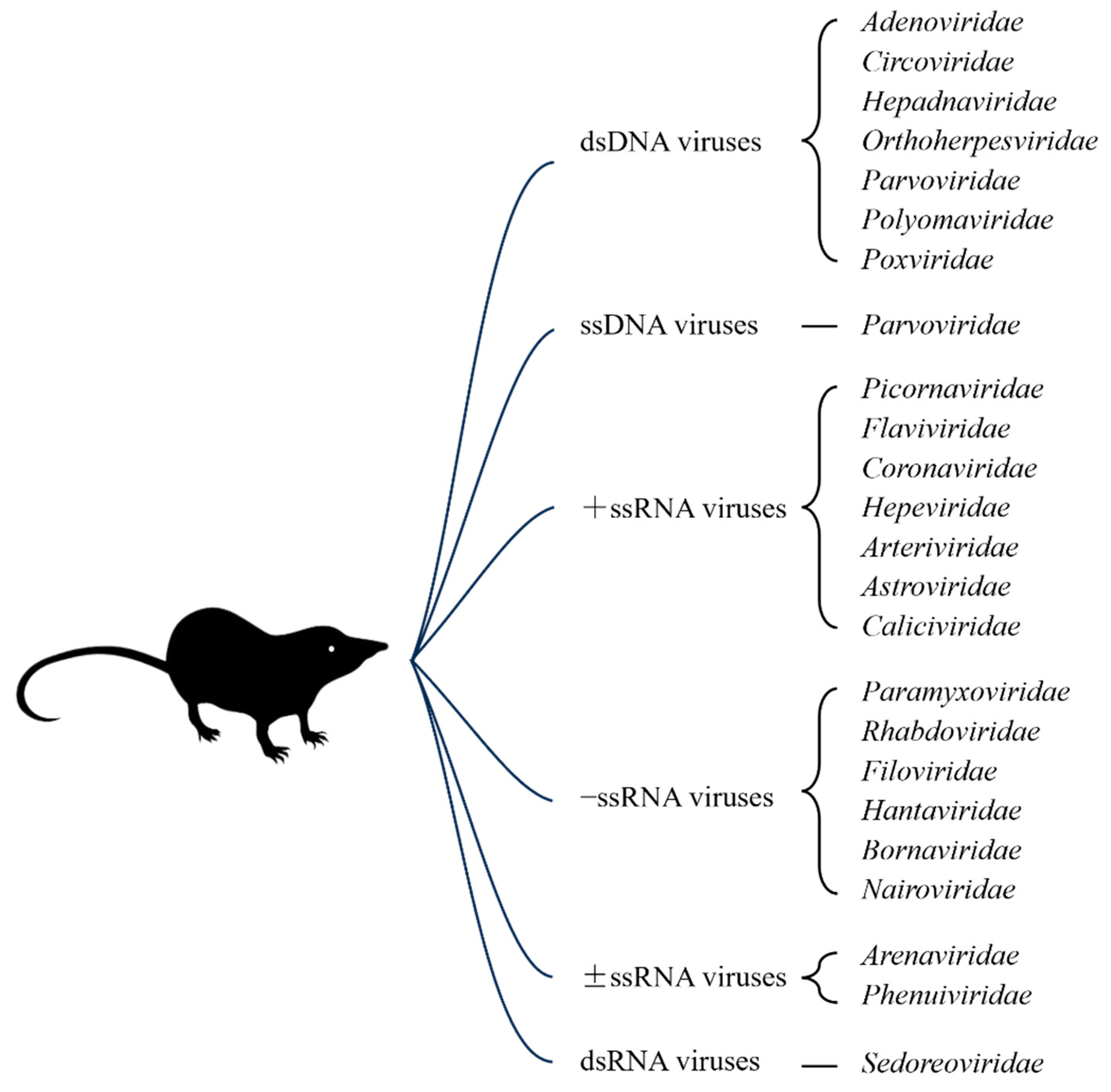
2. Paramyxoviruses in Shrews
3. Hantaviruses in Shrews
4. Sedoreoviruses in Shrews
5. Parvoviruses in Shrews
6. Nairoviruses in Shrews
7. Coronaviruses in Shrews
8. Flaviviruses in Shrews
9. Arteriviruses in Shrews
10. Other Viruses in Shrews
11. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jürgens, K. D. , Etruscan shrew muscle: The consequences of being small. J Exp Biol 2002, 205 Pt 15, 2161–2166. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z. , Animal biodiversity: An outline of higher-level classification and survey of taxonomic richness (Addenda 2013). Zootaxa 2013, 3703, 1–82. [Google Scholar] [CrossRef] [PubMed]
- Chung, D. J.; Madison, G. P.; Aponte, A. M.; Singh, K.; Li, Y.; Pirooznia, M.; Bleck, C. K. E.; Darmani, N. A.; Balaban, R. S. , Metabolic design in a mammalian model of extreme metabolism, the North American least shrew (Cryptotis parva). J Physiol 2022, 600, 547–567. [Google Scholar] [CrossRef] [PubMed]
- Bedford, J. M.; Mock, O. B.; Goodman, S. M. , Novelties of conception in insectivorous mammals (Lipotyphla), particularly shrews. Biol Rev Camb Philos Soc 2004, 79, 891–909. [Google Scholar] [CrossRef] [PubMed]
- Kowalski, K.; Rychlik, L. , Venom Use in Eulipotyphlans: An Evolutionary and Ecological Approach. Toxins (Basel) 2021, 13, (3). [Google Scholar] [CrossRef] [PubMed]
- Chai, S.; Tian, R.; Rong, X.; Li, G.; Chen, B.; Ren, W.; Xu, S.; Yang, G. , Evidence of Echolocation in the Common Shrew from Molecular Convergence with Other Echolocating Mammals. Zool Stud 2020, 59, e4. [Google Scholar] [PubMed]
- Siemers, B. M.; Schauermann, G.; Turni, H.; von Merten, S. , Why do shrews twitter? Communication or simple echo-based orientation. Biol Lett 2009, 5, 593–596. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y. M.; Hu, S. J.; Lin, X. D.; Tian, J. H.; Lv, J. X.; Wang, M. R.; Luo, X. Q.; Pei, Y. Y.; Hu, R. X.; Song, Z. G.; Holmes, E. C.; Zhang, Y. Z. , Host traits shape virome composition and virus transmission in wild small mammals. Cell 2023, 186, 4662–4675. [Google Scholar] [CrossRef] [PubMed]
- Thibault, P. A.; Watkinson, R. E.; Moreira-Soto, A.; Drexler, J. F.; Lee, B. , Zoonotic Potential of Emerging Paramyxoviruses: Knowns and Unknowns. Adv Virus Res 2017, 98, 1–55. [Google Scholar] [PubMed]
- Zhang, X. A.; Li, H.; Jiang, F. C.; Zhu, F.; Zhang, Y. F.; Chen, J. J.; Tan, C. W.; Anderson, D. E.; Fan, H.; Dong, L. Y.; Li, C.; Zhang, P. H.; Li, Y.; Ding, H.; Fang, L. Q.; Wang, L. F.; Liu, W. , A Zoonotic Henipavirus in Febrile Patients in China. N Engl J Med 2022, 387, 470–472. [Google Scholar] [CrossRef]
- Lee, S. H.; Kim, K.; Kim, J.; No, J. S.; Park, K.; Budhathoki, S.; Lee, S. H.; Lee, J.; Cho, S. H.; Cho, S.; Lee, G. Y.; Hwang, J.; Kim, H. C.; Klein, T. A.; Uhm, C. S.; Kim, W. K.; Song, J. W. , Discovery and Genetic Characterization of Novel Paramyxoviruses Related to the Genus Henipavirus in Crocidura Species in the Republic of Korea. Viruses 2021, 13, (10). [Google Scholar] [CrossRef] [PubMed]
- Vanmechelen, B.; Meurs, S.; Horemans, M.; Loosen, A.; Joly Maes, T.; Laenen, L.; Vergote, V.; Koundouno, F. R.; Magassouba, N.; Konde, M. K.; Condé, I. S.; Carroll, M. W.; Maes, P. , The characterization of multiple novel paramyxoviruses highlights the diverse nature of the subfamily Orthoparamyxovirinae. Virus Evol 2022, 8, veac061. [Google Scholar] [CrossRef] [PubMed]
- Horemans, M.; Van Bets, J.; Joly Maes, T.; Maes, P.; Vanmechelen, B. , Discovery and genome characterization of six new orthoparamyxoviruses in small Belgian mammals. Virus Evol 2023, 9, vead065. [Google Scholar] [CrossRef] [PubMed]
- Chen, J. J.; Zhang, X. A.; Fan, H.; Jiang, F. C.; Jin, M. Z.; Dai, K.; Wang, N.; Zhang, P. H.; Li, X. K.; Li, H.; Shi, W.; Yang, Z. C.; Fang, L. Q.; Zhou, H. S.; Wei, Y. H.; Liu, W. , Distribution and characteristics of Beilong virus among wild rodents and shrews in China. Infect Genet Evol 2020, 85, 104454. [Google Scholar] [CrossRef] [PubMed]
- Parvate, A.; Williams, E. P.; Taylor, M. K.; Chu, Y. K.; Lanman, J.; Saphire, E. O.; Jonsson, C. B. , Diverse Morphology and Structural Features of Old and New World Hantaviruses. Viruses 2019, 11, (9). [Google Scholar] [CrossRef] [PubMed]
- Muyangwa, M.; Martynova, E. V.; Khaiboullina, S. F.; Morzunov, S. P.; Rizvanov, A. A. , Hantaviral Proteins: Structure, Functions, and Role in Hantavirus Infection. Front Microbiol 2015, 6, 1326. [Google Scholar] [CrossRef] [PubMed]
- Yashina, L. N.; Abramov, S. A.; Zhigalin, A. V.; Smetannikova, N. A.; Dupal, T. A.; Krivopalov, A. V.; Kikuchi, F.; Senoo, K.; Arai, S.; Mizutani, T.; Suzuki, M.; Cook, J. A.; Yanagihara, R. , Geographic Distribution and Phylogeny of Soricine Shrew-Borne Seewis Virus and Altai Virus in Russia. Viruses 2021, 13, (7). [Google Scholar] [CrossRef] [PubMed]
- Radosa, L.; Schlegel, M.; Gebauer, P.; Ansorge, H.; Heroldová, M.; Jánová, E.; Stanko, M.; Mošanský, L.; Fričová, J.; Pejčoch, M.; Suchomel, J.; Purchart, L.; Groschup, M. H.; Krüger, D. H.; Ulrich, R. G.; Klempa, B. , Detection of shrew-borne hantavirus in Eurasian pygmy shrew (Sorex minutus) in Central Europe. Infect Genet Evol 2013, 19, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Gu, S. H.; Nicolas, V.; Lalis, A.; Sathirapongsasuti, N.; Yanagihara, R. , Complete genome sequence and molecular phylogeny of a newfound hantavirus harbored by the Doucet's musk shrew (Crocidura douceti) in Guinea. Infect Genet Evol 2013, 20, 118–123. [Google Scholar] [CrossRef]
- Song, J. W.; Kang, H. J.; Song, K. J.; Truong, T. T.; Bennett, S. N.; Arai, S.; Truong, N. U.; Yanagihara, R. , Newfound hantavirus in Chinese mole shrew, Vietnam. Emerg Infect Dis 2007, 13, 1784–1787. [Google Scholar] [CrossRef]
- Song, J. W.; Kang, H. J.; Gu, S. H.; Moon, S. S.; Bennett, S. N.; Song, K. J.; Baek, L. J.; Kim, H. C.; O'Guinn, M. L.; Chong, S. T.; Klein, T. A.; Yanagihara, R. , Characterization of Imjin virus, a newly isolated hantavirus from the Ussuri white-toothed shrew (Crocidura lasiura). J Virol 2009, 83, 6184–6191. [Google Scholar] [CrossRef] [PubMed]
- Arai, S.; Gu, S. H.; Baek, L. J.; Tabara, K.; Bennett, S. N.; Oh, H. S.; Takada, N.; Kang, H. J.; Tanaka-Taya, K.; Morikawa, S.; Okabe, N.; Yanagihara, R.; Song, J. W. , Divergent ancestral lineages of newfound hantaviruses harbored by phylogenetically related crocidurine shrew species in Korea. Virology 2012, 424, 99–105. [Google Scholar] [CrossRef]
- Kang, H. J.; Arai, S.; Hope, A. G.; Cook, J. A.; Yanagihara, R. , Novel hantavirus in the flat-skulled shrew (Sorex roboratus). Vector Borne Zoonotic Dis 2010, 10, 593–597. [Google Scholar] [CrossRef] [PubMed]
- Song, J. W.; Gu, S. H.; Bennett, S. N.; Arai, S.; Puorger, M.; Hilbe, M.; Yanagihara, R. , Seewis virus, a genetically distinct hantavirus in the Eurasian common shrew (Sorex araneus). Virol J 2007, 4, 114. [Google Scholar] [CrossRef]
- Guo, W. P.; Lin, X. D.; Wang, W.; Tian, J. H.; Cong, M. L.; Zhang, H. L.; Wang, M. R.; Zhou, R. H.; Wang, J. B.; Li, M. H.; Xu, J.; Holmes, E. C.; Zhang, Y. Z. , Phylogeny and origins of hantaviruses harbored by bats, insectivores, and rodents. PLoS Pathog 2013, 9, e1003159. [Google Scholar] [CrossRef]
- Sun, X. F.; Zhao, L.; Zhang, Z. T.; Liu, M. M.; Xue, Z. F.; Wen, H. L.; Ma, D. Q.; Huang, Y. T.; Sun, Y.; Zhou, C. M.; Luo, L. M.; Liu, J. W.; Li, W. Q.; Yu, H.; Yu, X. J. , Detection of Imjin Virus and Seoul Virus in Crocidurine Shrews in Shandong Province, China. Vector Borne Zoonotic Dis 2017, 17, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Kang, H. J.; Gu, S. H.; Yashina, L. N.; Cook, J. A.; Yanagihara, R. , Highly Divergent Genetic Variants of Soricid-Borne Altai Virus (Hantaviridae) in Eurasia Suggest Ancient Host-Switching Events. Viruses 2019, 11, (9). [Google Scholar] [CrossRef]
- Arai, S.; Kang, H. J.; Gu, S. H.; Ohdachi, S. D.; Cook, J. A.; Yashina, L. N.; Tanaka-Taya, K.; Abramov, S. A.; Morikawa, S.; Okabe, N.; Oishi, K.; Yanagihara, R. , Genetic Diversity of Artybash Virus in the Laxmann's Shrew (Sorex caecutiens). Vector Borne Zoonotic Dis 2016, 16, 468–475. [Google Scholar] [CrossRef] [PubMed]
- Gu, S. H.; Arai, S.; Yu, H. T.; Lim, B. K.; Kang, H. J.; Yanagihara, R. , Genetic variants of Cao Bang hantavirus in the Chinese mole shrew (Anourosorex squamipes) and Taiwanese mole shrew (Anourosorex yamashinai). Infect Genet Evol 2016, 40, 113–118. [Google Scholar] [CrossRef]
- Arai, S.; Bennett, S. N.; Sumibcay, L.; Cook, J. A.; Song, J. W.; Hope, A.; Parmenter, C.; Nerurkar, V. R.; Yates, T. L.; Yanagihara, R. , Phylogenetically distinct hantaviruses in the masked shrew (Sorex cinereus) and dusky shrew (Sorex monticolus) in the United States. Am J Trop Med Hyg 2008, 78, 348–351. [Google Scholar] [CrossRef]
- Arai, S.; Song, J. W.; Sumibcay, L.; Bennett, S. N.; Nerurkar, V. R.; Parmenter, C.; Cook, J. A.; Yates, T. L.; Yanagihara, R. , Hantavirus in northern short-tailed shrew, United States. Emerg Infect Dis 2007, 13, 1420–1423. [Google Scholar] [CrossRef] [PubMed]
- Gu, S. H.; Markowski, J.; Kang, H. J.; Hejduk, J.; Sikorska, B.; Liberski, P. P.; Yanagihara, R. , Boginia virus, a newfound hantavirus harbored by the Eurasian water shrew (Neomys fodiens) in Poland. Virol J 2013, 10, 160. [Google Scholar] [CrossRef] [PubMed]
- Kang, H. J.; Kadjo, B.; Dubey, S.; Jacquet, F.; Yanagihara, R. , Molecular evolution of Azagny virus, a newfound hantavirus harbored by the West African pygmy shrew (Crocidura obscurior) in Côte d'Ivoire. Virol J 2011, 8, 373. [Google Scholar] [CrossRef]
- Ling, J.; Sironen, T.; Voutilainen, L.; Hepojoki, S.; Niemimaa, J.; Isoviita, V. M.; Vaheri, A.; Henttonen, H.; Vapalahti, O. , Hantaviruses in Finnish soricomorphs: Evidence for two distinct hantaviruses carried by Sorex araneus suggesting ancient host-switch. Infect Genet Evol 2014, 27, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Kang, H. J.; Stanley, W. T.; Esselstyn, J. A.; Gu, S. H.; Yanagihara, R. , Expanded host diversity and geographic distribution of hantaviruses in sub-Saharan Africa. J Virol 2014, 88, 7663–7667. [Google Scholar] [CrossRef] [PubMed]
- Zuo, S. Q.; Gong, Z. D.; Fang, L. Q.; Jiang, J. F.; Zhang, J. S.; Zhao, Q. M.; Cao, W. C. , A new hantavirus from the stripe-backed shrew (Sorex cylindricauda) in the People's Republic of China. Virus Res 2014, 184, 82–86. [Google Scholar] [CrossRef] [PubMed]
- Carey, D. E.; Reuben, R.; Panicker, K. N.; Shope, R. E.; Myers, R. M. , Thottapalayam virus: A presumptive arbovirus isolated from a shrew in India. Indian J Med Res 1971, 59, 1758–1760. [Google Scholar] [PubMed]
- Jaafar, F. M.; Attoui, H.; Mertens, P. P. C.; de Micco, P.; de Lamballerie, X. , Structural organization of an encephalitic human isolate of Banna virus (genus Seadornavirus, family Reoviridae). J Gen Virol 2005, 86 Pt 4, 1147–1157. [Google Scholar] [CrossRef] [PubMed]
- Belhouchet, M.; Mohd Jaafar, F.; Firth, A. E.; Grimes, J. M.; Mertens, P. P.; Attoui, H. , Detection of a fourth orbivirus non-structural protein. PLoS ONE 2011, 6, e25697. [Google Scholar] [CrossRef]
- Li, K.; Lin, X. D.; Huang, K. Y.; Zhang, B.; Shi, M.; Guo, W. P.; Wang, M. R.; Wang, W.; Xing, J. G.; Li, M. H.; Hong, W. S.; Holmes, E. C.; Zhang, Y. Z. , Identification of novel and diverse rotaviruses in rodents and insectivores, and evidence of cross-species transmission into humans. Virology 2016, 494, 168–177. [Google Scholar] [CrossRef]
- Alexander, K. A.; MacLachlan, N. J.; Kat, P. W.; House, C.; O'Brien, S. J.; Lerche, N. W.; Sawyer, M.; Frank, L. G.; Holekamp, K.; Smale, L.; et al. , Evidence of natural bluetongue virus infection among African carnivores. Am J Trop Med Hyg 1994, 51, 568–576. [Google Scholar] [CrossRef] [PubMed]
- Cotmore, S. F.; Tattersall, P. , Parvoviruses: Small Does Not Mean Simple. Annu Rev Virol 2014, 1, 517–537. [Google Scholar] [CrossRef] [PubMed]
- Cotmore, S. F.; Tattersall, P. , Genome packaging sense is controlled by the efficiency of the nick site in the right-end replication origin of parvoviruses minute virus of mice and LuIII. J Virol 2005, 79, 2287–2300. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, M.; Orba, Y.; Anindita, P. D.; Ishii, A.; Ueno, K.; Hang'ombe, B. M.; Mweene, A. S.; Ito, K.; Sawa, H. , Distinct Lineages of Bufavirus in Wild Shrews and Nonhuman Primates. Emerg Infect Dis 2015, 21, 1230–1233. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y. Q.; You, F. F.; Chen, X. J.; Chen, Y. X.; Wen, Y. Q.; Chen, Q. , Detection and phylogenetic analysis of porcine bocaviruses carried by murine rodents and house shrews in China. Transbound Emerg Dis 2019, 66, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y. Q.; Zhang, M. Y.; Zhou, J. H.; Li, Y. Z.; You, F. F.; Wen, Y. Q.; He, W. Q.; Chen, Q. , A Molecular Epidemiological Investigation of Carriage of the Adeno-Associated Virus in Murine Rodents and House Shrews in China. Intervirology 2018, 61, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Liu, X.; Wang, X.; Dong, H.; Ma, C.; Wang, J.; Liu, B.; Mao, Y.; Wang, Y.; Li, T.; Yang, C.; Guo, Y. , Structural and Functional Diversity of Nairovirus-Encoded Nucleoproteins. J Virol 2015, 89, 11740–11749. [Google Scholar] [CrossRef]
- Garrison, A. R.; Alkhovsky Альхoвский Сергей Владимирoвич, S. V.; Avšič-Županc, T.; Bente, D. A.; Bergeron, É.; Burt, F.; Di Paola, N.; Ergünay, K.; Hewson, R.; Kuhn, J. H.; Mirazimi, A.; Papa, A.; Sall, A. A.; Spengler, J. R.; Palacios, G.; Consortium, I. R., ICTV Virus Taxonomy Profile: Nairoviridae. J Gen Virol 2020, 101, 798–799. [Google Scholar] [CrossRef]
- Ozeki, T.; Abe, H.; Ushijima, Y.; Nze-Nkogue, C.; Akomo-Okoue, E. F.; Ella, G. W. E.; Koumba, L. B. M.; Nso, B.; Mintsa-Nguema, R.; Makouloutou-Nzassi, P.; Makanga, B. K.; Nguelet, F. L. M.; Ondo, G. N.; Mbadinga, M.; Igasaki, Y.; Okada, S.; Hirano, M.; Yoshii, K.; Lell, B.; Bonney, L. C.; Hewson, R.; Kurosaki, Y.; Yasuda, J. , Identification of novel orthonairoviruses from rodents and shrews in Gabon, Central Africa. J Gen Virol 2022, 103, (10). [Google Scholar] [CrossRef]
- Chastel, C.; Main, A. J.; Richard, P.; Le Lay, G.; Legrand-Quillien, M. C.; Beaucournu, J. C. , Erve virus, a probable member of Bunyaviridae family isolated from shrews (Crocidura russula) in France. Acta Virol 1989, 33, 270–280. [Google Scholar]
- Walker, P. J.; Widen, S. G.; Firth, C.; Blasdell, K. R.; Wood, T. G.; Travassos da Rosa, A. P.; Guzman, H.; Tesh, R. B.; Vasilakis, N. , Genomic Characterization of Yogue, Kasokero, Issyk-Kul, Keterah, Gossas, and Thiafora Viruses: Nairoviruses Naturally Infecting Bats, Shrews, and Ticks. Am J Trop Med Hyg 2015, 93, 1041–1051. [Google Scholar] [CrossRef] [PubMed]
- Low, D. H. W.; Ch'ng, L.; Su, Y. C. F.; Linster, M.; Zhang, R.; Zhuang, Y.; Kwak, M. L.; Borthwick, S. A.; Hitch, A. T.; Smith, G. J. D.; Mendenhall, I. H. , Cencurut virus: A novel Orthonairovirus from Asian house shrews (Suncus murinus) in Singapore. One Health 2023, 16, 100529. [Google Scholar] [CrossRef] [PubMed]
- Bárcena, M.; Oostergetel, G. T.; Bartelink, W.; Faas, F. G.; Verkleij, A.; Rottier, P. J.; Koster, A. J.; Bosch, B. J. , Cryo-electron tomography of mouse hepatitis virus: Insights into the structure of the coronavirion. Proc Natl Acad Sci U S A 2009, 106, 582–587. [Google Scholar] [CrossRef] [PubMed]
- Bukhari, K.; Mulley, G.; Gulyaeva, A. A.; Zhao, L.; Shu, G.; Jiang, J.; Neuman, B. W. , Description and initial characterization of metatranscriptomic nidovirus-like genomes from the proposed new family Abyssoviridae, and from a sister group to the Coronavirinae, the proposed genus Alphaletovirus. Virology 2018, 524, 160–171. [Google Scholar] [CrossRef] [PubMed]
- Ntumvi, N. F.; Mbala Kingebeni, P.; Tamoufe, U.; Kumakamba, C.; Ndze, V.; Ngay Lukusa, I.; LeBreton, M.; Atibu Losoma, J.; Le Doux Diffo, J.; N'Kawa, F.; Takuo, J. M.; Mulembakani, P.; Nwobegahay, J.; Makuwa, M.; Muyembe Tamfum, J. J.; Gillis, A.; Harris, S.; Rimoin, A. W.; Hoff, N. A.; Fair, J. M.; Monagin, C.; Ayukekbong, J.; Rubin, E. M.; Wolfe, N. D.; Lange, C. E. , High Herpesvirus Diversity in Wild Rodent and Shrew Species in Central Africa. Intervirology 2018, 61, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Tsoleridis, T.; Onianwa, O.; Horncastle, E.; Dayman, E.; Zhu, M.; Danjittrong, T.; Wachtl, M.; Behnke, J. M.; Chapman, S.; Strong, V.; Dobbs, P.; Ball, J. K.; Tarlinton, R. E.; McClure, C. P. , Discovery of Novel Alphacoronaviruses in European Rodents and Shrews. Viruses 2016, 8, 84. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Lu, L.; Du, J.; Yang, L.; Ren, X.; Liu, B.; Jiang, J.; Yang, J.; Dong, J.; Sun, L.; Zhu, Y.; Li, Y.; Zheng, D.; Zhang, C.; Su, H.; Zheng, Y.; Zhou, H.; Zhu, G.; Li, H.; Chmura, A.; Yang, F.; Daszak, P.; Wang, J.; Liu, Q.; Jin, Q. , Comparative analysis of rodent and small mammal viromes to better understand the wildlife origin of emerging infectious diseases. Microbiome 2018, 6, 178. [Google Scholar] [CrossRef] [PubMed]
- Barrows, N. J.; Campos, R. K.; Liao, K. C.; Prasanth, K. R.; Soto-Acosta, R.; Yeh, S. C.; Schott-Lerner, G.; Pompon, J.; Sessions, O. M.; Bradrick, S. S.; Garcia-Blanco, M. A. , Biochemistry and Molecular Biology of Flaviviruses. Chem Rev 2018, 118, 4448–4482. [Google Scholar] [CrossRef] [PubMed]
- Bartholomeusz, A. I.; Wright, P. J. , Synthesis of dengue virus RNA in vitro: Initiation and the involvement of proteins NS3 and NS5. Arch Virol 1993, 128, (1–2), 111. [Google Scholar] [CrossRef]
- Diagne, M. M.; Ndione, M. H. D.; Di Paola, N.; Fall, G.; Bedekelabou, A. P.; Sembène, P. M.; Faye, O.; Zanotto, P. M. A.; Sall, A. A. , Usutu Virus Isolated from Rodents in Senegal. Viruses 2019, 11, (2). [Google Scholar] [CrossRef]
- Goethert, H. K.; Mather, T. N.; Johnson, R. W.; Telford, S. R. , 3rd, Incrimination of shrews as a reservoir for Powassan virus. Commun Biol 2021, 4, 1319. [Google Scholar] [CrossRef] [PubMed]
- Nosek, J.; Grulich, I. , The relationship between the tick-borne encephalitis virus and the ticks and mammals of the Tribec mountain range. Bull World Health Organ 1967, (Suppl 1), 31–47. [Google Scholar]
- Banerjee, K.; Ilkal, M. A.; Deshmukh, P. K. , Susceptibility of Cynopterus sphinx (frugivorus bat) & Suncus murinus (house shrew) to Japanese encephalitis virus. Indian J Med Res 1984, 79, 8–12. [Google Scholar] [PubMed]
- Guo, H.; Cai, C.; Wang, B.; Zhuo, F.; Jiang, R.; Wang, N.; Li, B.; Zhang, W.; Zhu, Y.; Fan, Y.; Chen, W.; Chen, W.; Yang, X.; Shi, Z. , Novel hepacivirus in Asian house shrew, China. Sci China Life Sci 2019, 62, 701–704. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y. W.; Wan, Z. W.; Wu, Y.; Li, X. F.; Tang, S. X. , PCR-based screening and phylogenetic analysis of rat pegivirus (RPgV) carried by rodents in China. J Vet Med Sci 2020, 82, 1464–1471. [Google Scholar] [CrossRef] [PubMed]
- Spilman, M. S.; Welbon, C.; Nelson, E.; Dokland, T. , Cryo-electron tomography of porcine reproductive and respiratory syndrome virus: Organization of the nucleocapsid. J Gen Virol 2009, 90 Pt 3, 527–535. [Google Scholar] [CrossRef] [PubMed]
- Vanmechelen, B.; Vergote, V.; Laenen, L.; Koundouno, F. R.; Bore, J. A.; Wada, J.; Kuhn, J. H.; Carroll, M. W.; Maes, P. , Expanding the Arterivirus Host Spectrum: Olivier's Shrew Virus 1, A Novel Arterivirus Discovered in African Giant Shrews. Sci Rep 2018, 8, 11171. [Google Scholar] [CrossRef] [PubMed]
- Calvete, C.; Mendoza, M.; Sarto, M. P.; Bagüés, M. P. J.; Luján, L.; Molín, J.; Calvo, A. J.; Monroy, F.; Calvo, J. H. , Detection of Rabbit Hemorrhagic Disease Virus GI.2/RHDV2/b in the Mediterranean Pine Vole ( Microtus duodecimcostatus) and White-Toothed Shrew ( Crocidura russula). J Wildl Dis 2019, 55, 467–472. [Google Scholar] [PubMed]
- Guo, L.; Liu, S.; Song, J.; Han, L.; Zhang, H.; Wu, C.; Wang, C.; Zhou, H.; Wang, J. , Seroprevalence of Wenzhou virus in China. Biosaf Health 2020, 2, 152–156. [Google Scholar] [CrossRef]
- Li, K.; Lin, X. D.; Wang, W.; Shi, M.; Guo, W. P.; Zhang, X. H.; Xing, J. G.; He, J. R.; Wang, K.; Li, M. H.; Cao, J. H.; Jiang, M. L.; Holmes, E. C.; Zhang, Y. Z. , Isolation and characterization of a novel arenavirus harbored by Rodents and Shrews in Zhejiang province, China. Virology 2015, 476, 37–42. [Google Scholar] [CrossRef]
- Ushijima, Y.; Abe, H.; Ozeki, T.; Ondo, G. N.; Mbadinga, M.; Bikangui, R.; Nze-Nkogue, C.; Akomo-Okoue, E. F.; Ella, G. W. E.; Koumba, L. B. M.; Nso, B.; Mintsa-Nguema, R.; Makouloutou-Nzassi, P.; Makanga, B. K.; Nguelet, F. L. M.; Zadeh, V. R.; Urata, S.; Mbouna, A. V. N.; Massinga-Loembe, M.; Agnandji, S. T.; Lell, B.; Yasuda, J. , Identification of potential novel hosts and the risk of infection with lymphocytic choriomeningitis virus in humans in Gabon, Central Africa. Int J Infect Dis 2021, 105, 452–459. [Google Scholar] [CrossRef] [PubMed]
- Hilbe, M.; Herrsche, R.; Kolodziejek, J.; Nowotny, N.; Zlinszky, K.; Ehrensperger, F. , Shrews as reservoir hosts of borna disease virus. Emerg Infect Dis 2006, 12, 675–677. [Google Scholar] [CrossRef]
- Xiong, Y. Q.; Mo, Y.; Chen, M. J.; Cai, W.; He, W. Q.; Chen, Q. , Detection and phylogenetic analysis of torque teno virus (TTV) carried by murine rodents and house shrews in China. Virology 2018, 516, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Fevola, C.; Rossi, C.; Rosso, F.; Girardi, M.; Rosà, R.; Manica, M.; Delucchi, L.; Rocchini, D.; Garzon-Lopez, C. X.; Arnoldi, D.; Bianchi, A.; Buzan, E.; Charbonnel, N.; Collini, M.; Ďureje, L.; Ecke, F.; Ferrari, N.; Fischer, S.; Gillingham, E. L.; Hörnfeldt, B.; Kazimírová, M.; Konečný, A.; Maas, M.; Magnusson, M.; Miller, A.; Niemimaa, J.; Nordström, Å.; Obiegala, A.; Olsson, G.; Pedrini, P.; Piálek, J.; Reusken, C. B.; Rizzolli, F.; Romeo, C.; Silaghi, C.; Sironen, T.; Stanko, M.; Tagliapietra, V.; Ulrich, R. G.; Vapalahti, O.; Voutilainen, L.; Wauters, L.; Rizzoli, A.; Vaheri, A.; Jääskeläinen, A. J.; Henttonen, H.; Hauffe, H. C. , Geographical Distribution of Ljungan Virus in Small Mammals in Europe. Vector Borne Zoonotic Dis 2020, 20, 692–702. [Google Scholar] [CrossRef] [PubMed]
- Drexler, J. F.; Corman, V. M.; Lukashev, A. N.; van den Brand, J. M.; Gmyl, A. P.; Brünink, S.; Rasche, A.; Seggewiβ, N.; Feng, H.; Leijten, L. M.; Vallo, P.; Kuiken, T.; Dotzauer, A.; Ulrich, R. G.; Lemon, S. M.; Drosten, C. , Evolutionary origins of hepatitis A virus in small mammals. Proc Natl Acad Sci U S A 2015, 112, 15190–15195. [Google Scholar] [CrossRef] [PubMed]
- Gedvilaite, A.; Tryland, M.; Ulrich, R. G.; Schneider, J.; Kurmauskaite, V.; Moens, U.; Preugschas, H.; Calvignac-Spencer, S.; Ehlers, B. , Novel polyomaviruses in shrews (Soricidae) with close similarity to human polyomavirus 12. J Gen Virol 2017, 98, 3060–3067. [Google Scholar] [CrossRef] [PubMed]
- Orba, Y.; Sasaki, M.; Yamaguchi, H.; Ishii, A.; Thomas, Y.; Ogawa, H.; Hang'ombe, B. M.; Mweene, A. S.; Morikawa, S.; Saijo, M.; Sawa, H. , Orthopoxvirus infection among wildlife in Zambia. J Gen Virol 2015, 96 Pt 2, 390–394. [Google Scholar] [CrossRef] [PubMed]
- Sandvik, T.; Tryland, M.; Hansen, H.; Mehl, R.; Moens, U.; Olsvik, O.; Traavik, T. , Naturally occurring orthopoxviruses: Potential for recombination with vaccine vectors. J Clin Microbiol 1998, 36, 2542–2547. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Chmura, A. A.; Li, J.; Zhu, G.; Desmond, J. S.; Zhang, Y.; Zhang, W.; Epstein, J. H.; Daszak, P.; Shi, Z. , Detection of diverse novel astroviruses from small mammals in China. J Gen Virol 2014, 95 Pt 11, 2442–2449. [Google Scholar] [CrossRef] [PubMed]
- Morvan, J. M.; Deubel, V.; Gounon, P.; Nakouné, E.; Barrière, P.; Murri, S.; Perpète, O.; Selekon, B.; Coudrier, D.; Gautier-Hion, A.; Colyn, M.; Volehkov, V. , Identification of Ebola virus sequences present as RNA or DNA in organs of terrestrial small mammals of the Central African Republic. Microbes Infect 1999, 1, 1193–1201. [Google Scholar] [CrossRef]
- Liu, J. W.; Wen, H. L.; Fang, L. Z.; Zhang, Z. T.; He, S. T.; Xue, Z. F.; Ma, D. Q.; Zhang, X. S.; Wang, T.; Yu, H.; Zhang, Y.; Zhao, L.; Yu, X. J. , Prevalence of SFTSV among Asian house shrews and rodents, China, January-August 2013. Emerg Infect Dis 2014, 20, 2126–2128. [Google Scholar] [CrossRef] [PubMed]
- Kemp, G. E.; Causey, O. R.; Moore, D. L.; Odelola, A.; Fabiyi, A. , Mokola virus. Further studies on IbAn 27377, a new rabies-related etiologic agent of zoonosis in nigeria. Am J Trop Med Hyg 1972, 21, 356–359. [Google Scholar] [CrossRef] [PubMed]
- Guan, D.; Li, W.; Su, J.; Fang, L.; Takeda, N.; Wakita, T.; Li, T. C.; Ke, C. , Asian musk shrew as a reservoir of rat hepatitis E virus, China. Emerg Infect Dis 2013, 19, 1341–1343. [Google Scholar] [CrossRef] [PubMed]
- Nie, F. Y.; Tian, J. H.; Lin, X. D.; Yu, B.; Xing, J. G.; Cao, J. H.; Holmes, E. C.; Ma, R. Z.; Zhang, Y. Z. , Discovery of a highly divergent hepadnavirus in shrews from China. Virology 2019, 531, 162–170. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X. Y.; Qiu, M.; Ke, X. M.; Guan, W. J.; Li, J. M.; Huo, S. T.; Chen, S. W.; Zhong, X. S.; Zhou, W.; Xiong, Y. Q.; Ge, J.; Chen, Q. , Detection of novel adenoviruses in fecal specimens from rodents and shrews in southern China. Virus Genes 2016, 52, 417–421. [Google Scholar] [CrossRef] [PubMed]
- Diffo, J.; Ndze, V. N.; Ntumvi, N. F.; Takuo, J. M.; Mouiche, M. M. M.; Tamoufe, U.; Nwobegahay, J.; LeBreton, M.; Gillis, A.; Schneider, B. S.; Fair, J. M.; Monagin, C.; McIver, D. J.; Joly, D. O.; Wolfe, N. D.; Rubin, E. M.; Lange, C. E. , DNA of diverse adenoviruses detected in Cameroonian rodent and shrew species. Arch Virol 2019, 164, 2359–2366. [Google Scholar] [CrossRef]
- Harima, H.; Sasaki, M.; Kajihara, M.; Mori-Kajihara, A.; Hang'ombe, B. M.; Changula, K.; Orba, Y.; Ogawa, H.; Simuunza, M.; Yoshida, R.; Mweene, A.; Takada, A.; Sawa, H. , Detection of novel orthoreovirus genomes in shrew (Crocidura hirta) and fruit bat (Rousettus aegyptiacus). J Vet Med Sci 2020, 82, 162–167. [Google Scholar] [CrossRef]

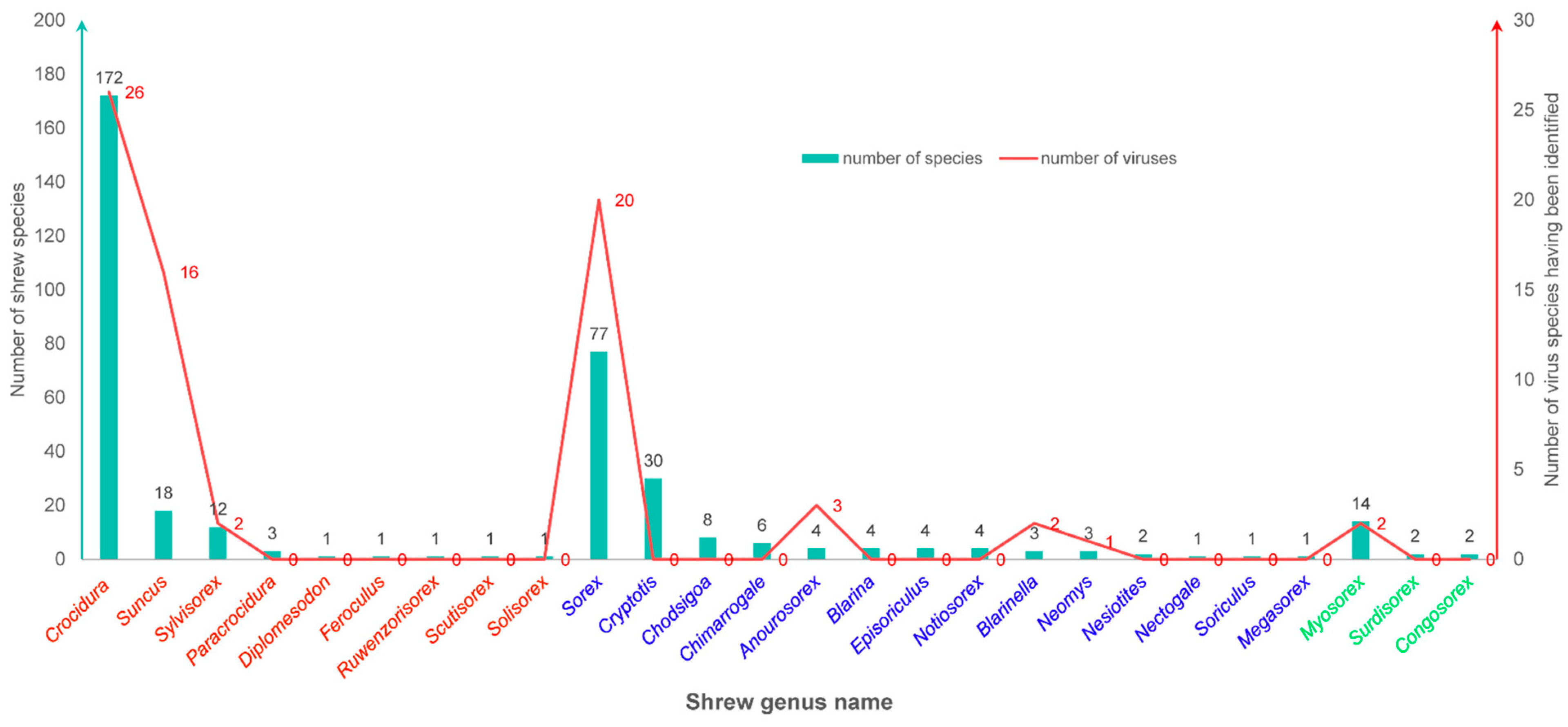
| Shrew Genus | Geographical Distribution |
Viral Distribution (Shrew Host Species Were Given in Parentheses) |
| Crocidura in the subfamily Crocidurinae covers 172 species | Distributed across all tropical and temperate regions of Africa, Europe, and Asia, from northern South Africa to Europe, and across the entire eastern part of Asia, extending to the Malay Archipelago | Langya virus (Crocidura shantungensis, Crocidura lasiura); Gamak virus and Daeryong virus (Crocidura shantungensis, Crocidura lasiura); Melian virus (Crocidura grandiceps); Denwin virus (Crocidura russula); Beilong virus (Crocidura shantungensis); Bowé virus (Crocidura douceti); Imjin virus (Crocidura lasiura); Jeju virus (Crocidura lasiura); Seoul virus (Crocidura lasiura); Azagny virus (Crocidura obscurior); Bufavirus (Crocidura hirta); Lamusara virus (Crocidura sp.); Lamgora virus (Crocidura sp.); Erve virus (Crocidura russula); Thiafora virus (Crocidura spp.); Usutu virus (Crocidura sp.); Olivier's shrew virus 1 (Crocidura olivieri guineensis); lymphocytic choriomeningitis virus (Crocidura goliath); Borna disease virus (Crocidura leucodon); Ljungan virus (Crocidura leucodon); Monkeypox virus (Crocidura spp.); Shrew herpesviruses (Crocidura spp.); Mokola virus (Crocidura spp.); rabbit hemorrhagic disease virus 2b (Crocidura russula); Shrew orthoreovirus (Crocidura hirta) |
| Suncus in the subfamily Crocidurinae covers 18 species | Distributed in all continents except Oceania | Thottapalayam virus (Suncus murinus); Rotavirus A (Suncus murinus); Porcine Bocavirus G4 (Suncus murinus); adeno-associated virus (Suncus murinus); Japanese encephalitis virus (Suncus murinus); Suncus murinus hepacivirus (Suncus murinus); rat pegivirus (Suncus murinus); Wenzhou virus (Suncus murinus); torque teno virus (Suncus murinus); astroviruses (Suncus murinus); severe fever with thrombocytopenia syndrome virus (Suncus murinus); hepatitis E virus (Suncus murinus); shrew hepatitis B virus (Suncus murinus); Asian house shrew adenovirus (Suncus murinus); Cencurut virus (Suncus murinus); Wénchéng shrew virus (Suncus murinus) |
| Sylvisorex in the subfamily Crocidurinae covers 12 species | Distributed in Africa | Ebola virus (Sylvisorex ollula); Unnamed shrew adenovirus (Sylvisorex sp.) |
|
Diplomesodon in the subfamily Crocidurinae covers 1 species |
Distributed in Caspian region | No viruses have been identified in this shrew genus |
|
Feroculus in the subfamily Crocidurinae covers 1 species |
Distributed in southern Sri Lanka and southern India | No viruses have been identified in this shrew genus |
| Paracrocidura in the subfamily Crocidurinae covers 3 species | Distributed in Burundi, the Democratic Republic of the Congo, Rwanda, Uganda, Gabon, the Central African Republic, and Equatorial Guinea in Africa | No viruses have been identified in this shrew genus |
|
Ruwenzorisorex in the subfamily Crocidurinae covers 1 species |
Distributed in Found in Burundi, the Democratic Republic of the Congo, Rwanda, and Uganda | No viruses have been identified in this shrew genus |
|
Scutisorex in the subfamily Crocidurinae covers 1 species |
Distributed in the Republic of the Congo | No viruses have been identified in this shrew genus |
|
Solisorex in the subfamily Crocidurinae covers 1 species |
Distributed in Sri Lanka and southern India | No viruses have been identified in this shrew genus |
|
Sorex in the subfamily Soricinae covers 77 species |
Distributed Eurasia and North America | Ninorex virus (Sorex minutus); Lena mobatvirus (Sorex caecutiens, Sorex roboratus); Asikkala virus (Sorex minutus); Asikkala virus (Sorex minutus); Kenkeme virus (Sorex roboratus); Seewis virus (Sorex araneus); Yakeshi virus (Sorex unguiculatus); Altai virus (Sorex araneus); Artybash virus (Sorex caecutiens); Artybash virus (Sorex caecutiens); Ash River virus (Sorex cinereus) Jemez Springs virus (Sorex monticolus); Puumala virus (Sorex araneus); Qian hu shan virus (Sorex cylindricauda); common shrew coronavirus Tibet-2014 (Sorex araneus); Ljungan Virus (Sorex antinorii); Hepatitis A virus (sorex araneus); human polyomavirus 12 (sorex araneus, sorex coronatus, sorex minutus); cowpox virus (Sorex araneus); tick-borne encephalitis virus (Sorex araneus) |
|
Anourosorex in the subfamily Soricinae covers 4 species |
Distributed in China, Taiwan, India, and Indochina | Cao Bang virus (Anourosorex squamipes); Lianghe virus (Anourosorex squamipes); Xinyi virus (Anourosorex yamashinai) |
|
Blarinella in the subfamily Soricinae covers 3 species |
Distributed in India, China, and Myanmar | Camp Ripley virus (Blarina brevicauda); Powassan virus type 2 (Blarina brevicauda) |
|
Neomys in the subfamily Soricinae covers 3 species |
Distributed in Europe and northern Asia | Boginia virus (Neomys fodiens) |
|
Blarina in the subfamily Soricinae covers 4 species |
Distributed in North America | No viruses have been identified in this shrew genus |
|
Cryptotis in the subfamily Soricinae covers 30 species |
Distributed in North America and Central America | No viruses have been identified in this shrew genus |
|
Chimarrogale in the subfamily Soricinae covers 6 species |
Distributed in Asia | No viruses have been identified in this shrew genus |
|
Chodsigoa in the subfamily Soricinae covers 8 species |
Distributed in Asia | No viruses have been identified in this shrew genus |
|
Episoriculus in the subfamily Soricinae covers 4 species |
Distributed in Asia | No viruses have been identified in this shrew genus |
|
Nectogale in the subfamily Soricinae covers 1 species |
Distributed in India and China | No viruses have been identified in this shrew genus |
|
Nesiotites in the subfamily Soricinae covers 2 species |
Distributed in Europe and North Africa | No viruses have been identified in this shrew genus |
|
Soriculus in the subfamily Soricinae covers 1 species |
Distributed in Asia | No viruses have been identified in this shrew genus |
|
Megasorex in the subfamily Soricinae covers 1 species |
Distributed in the United States and Mexico | No viruses have been identified in this shrew genus |
|
Notiosorex in the subfamily Soricinae covers 4 species |
Distributed in the United States and Mexico | No viruses have been identified in this shrew genus |
|
Myosorex in the subfamily Myosoricinae covers 14 species |
Distributed in Africa | Kilimanjaro virus (Myosorex geata); Uluguru virus (Myosorex zinki) |
|
Surdisorex in the subfamily Myosoricinae covers 2 species |
Distributed in Kenya | No viruses have been identified in this shrew genus |
|
Congosorex in the subfamily Myosoricinae covers 2 species |
Distributed in Gabon, Central African Republic, Democratic Congo, Cameroon, Tanzania | No viruses have been identified in this shrew genus |
| Common Name of Shrews | Species Name of Shrews |
| American water shrew | Sorex palustris |
| Asian house shrews | Suncus murinus |
| Asian lesser white-toothed shrews | Crocidura shantungensis |
| bicolored shrews | Crocidura leucodon |
| Chinese mole shrews | Anourosorex squamipes |
| Doucet's musk shrews | Crocidura douceti |
| dusky shrews | Sorex monticolus |
| Eurasian pygmy shrews | Sorex minutus |
| Eurasian common shrews | Sorex Araneus |
| Eurasian water shrews | Neomys fodiens |
| flat-skulled shrews | Sorex roboratus |
| Geata mouse shrews | Myosorex geata |
| Goliath shrews | Crocidura goliath |
| greater forest shrews | Sylvisorex ollula |
| greater white-toothed shrews | Crocidura russula |
| Kilimanjaro mouse shrews | Myosorex zinki |
| large-headed shrews | Crocidura grandiceps |
| Laxmann's shrews | Sorex caecutiens |
| lesser red musk shrews | Crocidura hirta |
| long-clawed shrews | Sorex unguiculatus |
| masked shrews | Sorex cinereus |
| Millet's shrews | Sorex coronatus |
| northern short-tailed shrews | Blarina brevicauda |
| Olivier's shrews | Crocidura olivieri guineensis |
| stripe-backed shrews | Sorex cylindricauda |
| Taiwanese mole shrews | Anourosorex yamashinai |
| Ussuri white-toothed shrews | Crocidura lasiura |
| Valais shrews | Sorex antinorii |
| West African pygmy shrews | Crocidura obscurior |
| Family | Genus | Virus | Zoonotic Potential | Year / Country / Host Species |
| Paramyxoviridae | Henipavirus | Langya virus | Infects humans and some other mammals | 2022 / China / Crocidura lasiura, Crocidura shantungensis [10] |
| Paramyxoviridae | Henipavirus | Gamak virus | Unknown | 2021 / China / Crocidura lasiura, Crocidura shantungensis [11] |
| Paramyxoviridae | Henipavirus | Daeryong virus | Unknown | 2021 / China / Crocidura lasiura, Crocidura shantungensis [11] |
| Paramyxoviridae | Henipavirus | Melian virus | Unknown | 2022 / Guinea / Crocidura grandiceps [12] |
| Paramyxoviridae | Henipavirus | Denwin virus | Unknown | 2022 / Belgium / Crocidura russula [12] |
| Paramyxoviridae | Henipavirus | Ninorex virus | Unknown | 2023 / Belgian / Sorex minutus [13] |
| Paramyxoviridae | Jeilongvirus | Beilong virus | Unknown | 2019 / China / Crocidura shantungensis, Suncus murinus [14] |
| Hantaviridae | Mobatvirus | Lena mobatvirus | Unknown | 2021 / Siberian and Russia / Sorex caecutiens, Sorex roboratus [17] |
| Hantaviridae | Orthohantavirus | Asikkala virus | Unknown | 2010 / Finland / Sorex minutus [18] |
| Hantaviridae | Orthohantavirus | Bowé virus | Unknown | 2012 / Guinea / Crocidura douceti [19] |
| Hantaviridae | Orthohantavirus | Cao Bang virus | Unknown | 2006 / China / Anourosorex squamipes [20] |
| Hantaviridae | Orthohantavirus | Imjin virus | Unknown | 2009 / South Korea / Crocidura lasiura [21] |
| Hantaviridae | Orthohantavirus | Jeju virus | Unknown | 2012 / South Korea / Crocidura lasiura [22] |
| Hantaviridae | Orthohantavirus | Kenkeme virus | Unknown | 2010 / Sakha Republic, Siberia / Sorex roboratus [23] |
| Hantaviridae | Orthohantavirus | Seewis virus | Unknown | 2006 / Switzerland / Sorex araneus [24] |
| Hantaviridae | Orthohantavirus | Yakeshi virus | Unknown | 2013 / China / Sorex unguiculatus [25] |
| Hantaviridae | Orthohantavirus | Lianghe virus | Unknown | 2013 / China / Anourosorex squamipes [25] |
| Hantaviridae | Orthohantavirus | Seoul virus | Infects humans | 2014 / China / Crocidura lasiura [26] |
| Hantaviridae | Orthohantavirus | Altai virus | Unknown | 2007 / Altai Republic / Sorex araneus [27] |
| Hantaviridae | Orthohantavirus | Artybash virus | Unknown | 2006 / Altai Republic / Sorex caecutiens [28] |
| Hantaviridae | Orthohantavirus | Xinyi virus | Unknown | 2016 / Chian / Anourosorex yamashinai [29] |
| Hantaviridae | Orthohantavirus | Ash River virus | Unknown | 2008 / United States / Sorex cinereus, Sorex monticolus [30] |
| Hantaviridae | Orthohantavirus | Camp Ripley virus | Unknown | 2007 / United States / Blarina brevicauda [31] |
| Hantaviridae | Orthohantavirus | Boginia virus | Unknown | 2013 / Poland / Neomys fodiens [32] |
| Hantaviridae | Orthohantavirus | Azagny virus | Unknown | 2011 / Côte d'Ivoire / Crocidura obscurior [33] |
| Hantaviridae | Orthohantavirus | Puumala virus | Unknown | 2014 / Finland / Sorex araneus [34] |
| Hantaviridae | Orthohantavirus | Kilimanjaro virus | Unknown | 2014 / West Africa / Myosorex geata [35] |
| Hantaviridae | Orthohantavirus | Uluguru virus | Unknown | 2014 / West Africa / Myosorex zinki [35] |
| Hantaviridae | Orthohantavirus | Qian hu shan virus | Unknown | 2014 / China / Sorex cylindricauda [36] |
| Hantaviridae | Thottimvirus | Thottapalayam virus | Unknown | 1965 / India / Suncus murinus [37] |
| Sedoreoviridae | Rotavirus | Rotavirus A | Infects humans | 2016 / China / Suncus murinus [40] |
| Sedoreoviridae | Orbivirus | Bluetongue virus | Unknown | 1994 / Africa / unknown shrew species [41] |
| Parvoviridae | Protoparvovirus | Bufavirus | Infects humans | 2015 / Zambia / Crocidura hirta [44] |
| Parvoviridae | Bocaparvovirus | porcine bocavirus G4 | Infects pigs | 2019 / China / Suncus murinus [45] |
| Parvoviridae | Dependoparvovirus | adeno-associated virus | Unknown | 2020 / China / Suncus murinus [46] |
| Nairoviridae | Orthonairovirus | Lamusara virus | Unknown | 2022 / Gabon / Crocidura sp. [49] |
| Nairoviridae | Orthonairovirus | Lamgora virus | Unknown | 2022 / Gabon / Crocidura sp. [49] |
| Nairoviridae | Orthonairovirus | Erve virus | Unknown | 1989 / France / Crocidura russula [50] |
| Nairoviridae | Orthonairovirus | Thiafora virus | Unknown | 2015 / Senegal / Crocidura sp. [51] |
| Nairoviridae | Orthonairovirus | Cencurut virus | Unknown | 2023 / Singapore / Suncus murinus [52] |
| Coronaviridae | Alphacoronavirus | Wencheng shrew virus | Unknown | 2017 / China / Suncus murinus [55] |
| Coronaviridae | Alphacoronavirus | unnamed shrew coronavirus | Unknown | 2016 / United Kingdom / Sorex araneus [56] |
| Coronaviridae | Alphacoronavirus | common shrew coronavirus Tibet-2014 | Unknown | 2014 / China / Sorex araneus [57] |
| Flaviviridae | Orthoflavivirus | Usutu virus | Infects birds, arthropods, humans, bats, and horses | 2019 / Senegal / Crocidura sp. [60] |
| Flaviviridae | Orthoflavivirus | Powassan virus type 2 | Infects humans and causes encephalitis | 2021 / United States / Blarina brevicauda [61] |
| Flaviviridae | Orthoflavivirus | tick-borne encephalitis virus | Infects humans and causes encephalitis | 1967 / Slovakia / Sorex araneus [62] |
| Flaviviridae | Orthoflavivirus | Japanese encephalitis virus | Infects humans and various domestic animals and causes human encephalitis | 1984 / Japan / Suncus murinus [63] |
| Flaviviridae | Hepacivirus | Suncus murinus hepacivirus | Unknown | 2016 / China / Suncus murinus [64] |
| Flaviviridae | Pegivirus | rat pegivirus | Infects humans and other animals without symptoms | 2020 / China / Suncus murinus [65] |
| Arteriviridae | Muarterivirus | Olivier's shrew virus 1 | Unknown | 2018 / Guinea / Crocidura olivieri guineensis [67] |
| Caliciviridae | Lagovirus | Rabbit hemorrhagic disease virus | Infects rabbits | 2019 / Spain / Crocidura russula [68] |
| Arenaviridae | Mammarenavirus | Wenzhou virus | Infects humans | 2015 / China / Suncus murinus [69,70] |
| Arenaviridae | Mammarenavirus | lymphocytic choriomeningitis virus | Infects humans and various animals and dangerous to pregnant women and infants | 2021 / Gabon / Crocidura goliath [71] |
| Bornaviridae | Orthobornavirus | Borna disease virus | Infects humans, horses, sheep, and various other mammals and causes encephalitis | 2006 / Switzerland / Crocidura leucodon [72] |
| Circoviridae | Anellovirus | torque teno virus | Infects humans and causes various symptoms | 2018 / China / Suncus murinus [73] |
| Picornaviridae | Parechovirus | Ljungan Virus | Infects humans and causes various symptoms | 2020 / Italy / Crocidura leucodon, Sorex antinorii [74] |
| Picornaviridae | Hepatovirus | Hepatitis A virus | Unknown | 2015 / Germany / sorex araneus [75] |
| Polyomaviridae | Betapolyomavirus | human polyomavirus 12 | Infects humans | 2017 / Germany, Norway / sorex Araneus, sorex coronatus, sorex minutus [76] |
| Poxviridae | Orthopoxvirus | monkeypox virus | Infects humans and other primates | 2015 / Zambia, Congo / Crocidura spp. [77] |
| Poxviridae | Orthopoxvirus | shrew cowpox virus | Unknown | 1998 / Norway / Sorex araneus [78] |
| Astroviridae | Mamastrovirus | shrew astrovirus | Unknown | 2014 / China / Suncus murinus [79] |
| Filoviridae | Ebolavirus | Ebola virus | Infects humans with a high case fatality rate | 1999 / Central Africa / Sylvisorex ollula [80] |
| Phenuiviridae | Phlebovirus | severe fever with thrombocytopenia syndrome virus | Infects humans with a high case fatality rate | 2014 / China / Suncus murinus [81] |
| Herpesviridae | Unknown | shrew herpesvirus | Unknown | 2018 / Central Africa / Crocidura spp. [55] |
| Rhabdoviridae | Lyssavirus | Mokola virus | Unknown | 1968 / Nigeria / Crocidura spp. [82] |
| Hepeviridae | Paslahepevirus | shrew hepatitis E virus | Unknown | 2013 / China / Suncus murinus [83] |
| Hepadnaviridae | Orthohepadnavirus | shrew hepatitis B virus | Unknown | 2019 / China / Suncus murinus [84] |
| Adenoviridae | Atadenovirus | Asian house shrew adenovirus | Unknown | 2016 / China / Suncus murinus [85] |
| Adenoviridae | Atadenovirus | shrew adenovirus | Unknown | 2019 / Cameroon / Sylvisorex sp. [86] |
| Spinareoviridae | Orthoreovirus | shrew orthoreovirus | Unknown | 2019 / Zambia / Crocidura hirta [87] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




