Submitted:
22 December 2023
Posted:
25 December 2023
You are already at the latest version
Abstract
Keywords:
Contents
- Introduction
- Major signaling pathways in PNETs
-
Epigenetic regulation of PNET-related signaling pathways
- 1.1
- DNA methylation
- 1.2
- Histone modifications
- 1.3
- Non-coding RNAs
- Future directions for epigenetic research and clinical applications in PNET patient care
- Conclusion
1. Introduction
2. Major Signaling Pathways and Molecules in PNETs
3. Epigenetic Regulation of PNET-Related Signaling Pathways
3.1. DNA Methylation
3.1.1. MEN1
3.1.3. Hypoxia-Induced Factor 1α (HIF1α)-VHL
3.1.4. RAS-MAPK-NF1
3.1.5. ATRX/DAXX
3.1.6. CDKN2A-RB1
3.1.8. Notch Signaling
3.1.9. Wnt/β-catenin
3.1.10. NF-κB
3.1.11. Somatostatin Receptor 2 (SSTR2)
3.1.12. Smad3
3.2. Histone Modifications
3.2.1. MEN1
3.2.3. HIF1α-VHL
3.2.4. RAS-MAPK-NF1
3.2.5. ATRX/DAXX
3.2.6. CDKN2A/RB1
3.2.7. P53
3.2.8. Notch Signaling
3.2.9. Wnt/β-catenin
3.2.10. NFκB
3.2.11. SSTR2
3.2.12. Smad3
3.3. Non-Coding RNAs
3.3.1. MEN1
3.3.3. HIF1α
3.3.4. Ras-MAPK
3.3.5. DAXX/ATRX
3.3.6. RB1
3.3.8. Notch
3.3.9. Wnt/β-catenin
3.3.10. NFκB
3.3.11. SSTR2
3.3.12. Smad3
4. Future Directions for Epigenetic Research and Clinical Applications in PNET Patient Care
5. Conclusion
Funding
Conflicts of Interest
References
- Pipinikas, C.P.; Berner, A.M.; Sposito, T.; Thirlwell, C. The evolving (epi)genetic landscape of pancreatic neuroendocrine tumours. Endocrine-Related Cancer 2019, 26, R519–R544. [Google Scholar] [CrossRef]
- Nigri, G.; Petrucciani, N.; Debs, T.; Mangogna, L.M.; Crovetto, A.; Moschetta, G.; Persechino, R.; Aurello, P.; Ramacciato, G. Treatment options for PNET liver metastases: a systematic review. World J. Surg. Oncol. 2018, 16, 142. [Google Scholar] [CrossRef]
- Shen, X.; Wang, X.; Lu, X.; Zhao, Y.; Guan, W. Molecular biology of pancreatic neuroendocrine tumors: From mechanism to translation. Front. Oncol. 2022, 12, 967071. [Google Scholar] [CrossRef]
- Scarpa, A.; Chang, D.K.; Nones, K.; Corbo, V.; Patch, A.-M.; Bailey, P.; Lawlor, R.T.; Johns, A.L.; Miller, D.K.; Mafficini, A.; et al. Whole-genome landscape of pancreatic neuroendocrine tumours. Nature 2017, 543, 65–71. [Google Scholar] [CrossRef]
- Noë, M.; Pea, A.; Luchini, C.; Felsenstein, M.; Barbi, S.; Bhaijee, F.; Yonescu, R.; Ning, Y.; Adsay, N.V.; Zamboni, G.; et al. Whole-exome sequencing of duodenal neuroendocrine tumors in patients with neurofibromatosis type 1. Mod. Pathol. 2018, 31, 1532–1538. [Google Scholar] [CrossRef]
- Mohindroo, C.; McAllister, F.; De Jesus-Acosta, A. Genetics of Pancreatic Neuroendocrine Tumors. Hematol. Oncol. Clin. North Am. 2022, 36, 1033–1051. [Google Scholar] [CrossRef]
- Ciobanu, O.A.; Martin, S.C.; Herlea, V.; Fica, S. Insights into Epigenetic Changes Related to Genetic Variants and Cells-of-Origin of Pancreatic Neuroendocrine Tumors: An Algorithm for Practical Workup. Cancers 2022, 14, 4444. [Google Scholar] [CrossRef]
- Sahafnejad, Z.; Ramazi, S.; Allahverdi, A. An Update of Epigenetic Drugs for the Treatment of Cancers and Brain Diseases: A Comprehensive Review. Genes 2023, 14, 873. [Google Scholar] [CrossRef]
- Di Domenico, A.; et al. Epigenetic landscape of pancreatic neuroendocrine tumours reveals distinct cells of origin and means of tumour progression. Commun Biol, 2020, 3, 740. [Google Scholar] [CrossRef]
- Maharjan, C.K.; Ear, P.H.; Tran, C.G.; Howe, J.R.; Chandrasekharan, C.; Quelle, D.E. Pancreatic Neuroendocrine Tumors: Molecular Mechanisms and Therapeutic Targets. Cancers 2021, 13, 5117. [Google Scholar] [CrossRef]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Mafficini, A.; Scarpa, A. Genetics and Epigenetics of Gastroenteropancreatic Neuroendocrine Neoplasms. Endocr. Rev. 2019, 40, 506–536. [Google Scholar] [CrossRef]
- Crabtree, J.S. Epigenetic Regulation in Gastroenteropancreatic Neuroendocrine Tumors. Front. Oncol. 2022, 12, 901435. [Google Scholar] [CrossRef]
- Maharjan, C.K.; Ear, P.H.; Tran, C.G.; Howe, J.R.; Chandrasekharan, C.; Quelle, D.E. Pancreatic Neuroendocrine Tumors: Molecular Mechanisms and Therapeutic Targets. Cancers 2021, 13, 5117. [Google Scholar] [CrossRef]
- Blair, L.P.; Yan, Q. Epigenetic Mechanisms in Commonly Occurring Cancers. DNA Cell Biol. 2012, 31 (Suppl. S1), S49–S61. [Google Scholar] [CrossRef]
- Tulsyan, S.; Aftab, M.; Sisodiya, S.; Khan, A.; Chikara, A.; Tanwar, P.; Hussain, S. Molecular basis of epigenetic regulation in cancer diagnosis and treatment. Front. Genet. 2022, 13, 885635. [Google Scholar] [CrossRef]
- Feehley, T.; O’donnell, C.W.; Mendlein, J.; Karande, M.; McCauley, T. Drugging the epigenome in the age of precision medicine. Clin. Epigenetics 2023, 15, 1–13. [Google Scholar] [CrossRef]
- Zhang, C.; Sun, C.; Zhao, Y.; Wang, Q.; Guo, J.; Ye, B.; Yu, G. Overview of MicroRNAs as Diagnostic and Prognostic Biomarkers for High-Incidence Cancers in 2021. Int. J. Mol. Sci. 2022, 23, 11389. [Google Scholar] [CrossRef]
- Lu, Y.; Chan, Y.-T.; Tan, H.-Y.; Li, S.; Wang, N.; Feng, Y. Epigenetic regulation in human cancer: the potential role of epi-drug in cancer therapy. Mol. Cancer 2020, 19, 79. [Google Scholar] [CrossRef]
- Soczomski, P.; Jurecka-Lubieniecka, B.; Krzywon, A.; Cortez, A.J.; Zgliczynski, S.; Rogozik, N.; Oczko-Wojciechowska, M.; Pawlaczek, A.; Bednarczuk, T.; Jarzab, B. A Direct Comparison of Patients With Hereditary and Sporadic Pancreatic Neuroendocrine Tumors: Evaluation of Clinical Course, Prognostic Factors and Genotype–Phenotype Correlations. Front. Endocrinol. 2021, 12, 681013. [Google Scholar] [CrossRef]
- Chatani, P.D.; Agarwal, S.K.; Sadowski, S.M. Molecular Signatures and Their Clinical Utility in Pancreatic Neuroendocrine Tumors. Front. Endocrinol. 2021, 11, 575620. [Google Scholar] [CrossRef]
- Ye, Z., et al., MEN1 promotes ferroptosis by inhibiting mTOR-SCD1 axis in pancreatic neuroendocrine tumors. Acta Biochim Biophys Sin (Shanghai), 2022, 54, 1599-1609.
- Corbo, V.; Dalai, I.; Scardoni, M.; Barbi, S.; Beghelli, S.; Bersani, S.; Albarello, L.; Doglioni, C.; Schott, C.; Capelli, P.; et al. MEN1 in pancreatic endocrine tumors: analysis of gene and protein status in 169 sporadic neoplasms reveals alterations in the vast majority of cases. Endocrine-Related Cancer 2010, 17, 771–783. [Google Scholar] [CrossRef]
- Ishida, N.; Miyazu, T.; Tamura, S.; Suzuki, S.; Tani, S.; Yamade, M.; Iwaizumi, M.; Osawa, S.; Hamaya, Y.; Shinmura, K.; et al. Tuberous sclerosis patient with neuroendocrine carcinoma of the esophagogastric junction: A case report. World J. Gastroenterol. 2020, 26, 7263–7271. [Google Scholar] [CrossRef]
- Tirosh, A.; Kebebew, E. Genetic and epigenetic alterations in pancreatic neuroendocrine tumors. J. Gastrointest. Oncol. 2020, 11, 567–577. [Google Scholar] [CrossRef]
- Chandrasekharappa, S.C.; Guru, S.C.; Manickam, P.; Olufemi, S.-E.; Collins, F.S.; Emmert-Buck, M.R.; Debelenko, L.V.; Zhuang, Z.; Lubensky, I.A.; Liotta, L.A.; et al. Positional Cloning of the Gene for Multiple Endocrine Neoplasia-Type 1. Science 1997, 276, 404–407. [Google Scholar] [CrossRef]
- Conemans, E.B.; Lodewijk, L.; Moelans, C.B.; A Offerhaus, G.J.; Pieterman, C.R.C.; Morsink, F.H.; Dekkers, O.M.; de Herder, W.W.; Hermus, A.R.; van der Horst-Schrivers, A.N.; et al. DNA methylation profiling in MEN1-related pancreatic neuroendocrine tumors reveals a potential epigenetic target for treatment. Eur. J. Endocrinol. 2018, 179, 153–160. [Google Scholar] [CrossRef]
- Massey, S.; Khan, M.A.; Rab, S.O.; Mustafa, S.; Khan, A.; Malik, Z.; Shaik, R.; Verma, M.K.; Deo, S.; Husain, S.A. Evaluating the role of MEN1 gene expression and its clinical significance in breast cancer patients. PLOS ONE 2023, 18, e0288482. [Google Scholar] [CrossRef]
- Brown, E.J.; Albers, M.W.; Shin, T.B.; Ichikawa, K.; Keith, C.T.; Lane, W.S.; Schreiber, S.L. A mammalian protein targeted by G1-arresting rapamycin–receptor complex. Nature 1994, 369, 756–758. [Google Scholar] [CrossRef]
- Zeng, J.-D.; Wu, W.K.; Wang, H.-Y.; Li, X.-X. Serine and one-carbon metabolism, a bridge that links mTOR signaling and DNA methylation in cancer. Pharmacol. Res. 2019, 149, 104352. [Google Scholar] [CrossRef]
- Fattahi, S.; Amjadi-Moheb, F.; Tabaripour, R.; Ashrafi, G.H.; Akhavan-Niaki, H. PI3K/AKT/mTOR signaling in gastric cancer: Epigenetics and beyond. Life Sci. 2020, 262, 118513. [Google Scholar] [CrossRef]
- Wang, C.; et al. A novel methylated cation channel TRPM4 inhibited colorectal cancer metastasis through Ca(2+)/Calpain-mediated proteolysis of FAK and suppression of PI3K/Akt/mTOR signaling pathway. Int. J. Biol. Sci. 2022, 18, 5575–5590. [Google Scholar] [CrossRef]
- Huang, J.; Li, J.; Tang, J.; Wu, Y.; Dai, F.; Yi, Z.; Wang, Y.; Li, Y.; Wu, Y.; Ren, G.; et al. ZDHHC22-mediated mTOR palmitoylation restrains breast cancer growth and endocrine therapy resistance. Int. J. Biol. Sci. 2022, 18, 2833–2850. [Google Scholar] [CrossRef]
- Zhuang, Z.; O Vortmeyer, A.; Pack, S.; Huang, S.; A Pham, T.; Wang, C.; Park, W.S.; Agarwal, S.K.; Debelenko, L.V.; Kester, M. Somatic mutations of the MEN1 tumor suppressor gene in sporadic gastrinomas and insulinomas. Cancer Res. 1997, 57, 4682–4686. [Google Scholar]
- Görtz, B.; Roth, J.; Krähenmann, A.; de Krijger, R.R.; Muletta-Feurer, S.; Rütimann, K.; Saremaslani, P.; Speel, E.J.; Heitz, P.U.; Komminoth, P. Mutations and Allelic Deletions of the MEN1 Gene Are Associated with a Subset of Sporadic Endocrine Pancreatic and Neuroendocrine Tumors and Not Restricted to Foregut Neoplasms. Am. J. Pathol. 1999, 154, 429–436. [Google Scholar] [CrossRef]
- Shan, L.; Nakamura, Y.; Nakamura, M.; Yokoi, T.; Tsujimoto, M.; Arima, R.; Kameya, T.; Kakudo, K. Somatic mutations of multiple endocrine neoplasia type 1 gene in the sporadic endocrine tumors. Lab Invest 1998, 78, 471–475. [Google Scholar]
- Lawrence, B.; Blenkiron, C.; Parker, K.; Tsai, P.; Fitzgerald, S.; Shields, P.; Robb, T.; Yeong, M.L.; Kramer, N.; James, S.; et al. Recurrent loss of heterozygosity correlates with clinical outcome in pancreatic neuroendocrine cancer. npj Genom. Med. 2018, 3, 18. [Google Scholar] [CrossRef]
- Ghayouri, M.; Boulware, D.; Nasir, A.; Strosberg, J.; Kvols, L.; Coppola, D. Activation of the serine/theronine protein kinase Akt in enteropancreatic neuroendocrine tumors. Anticancer Res. 2010, 30, 5063–5067. [Google Scholar]
- Shida, T.; Kishimoto, T.; Furuya, M.; Nikaido, T.; Koda, K.; Takano, S.; Kimura, F.; Shimizu, H.; Yoshidome, H.; Ohtsuka, M.; et al. Expression of an activated mammalian target of rapamycin (mTOR) in gastroenteropancreatic neuroendocrine tumors. Cancer Chemother. Pharmacol. 2009, 65, 889–893. [Google Scholar] [CrossRef]
- Chang, T.M.; et al. The regulatory role of aberrant Phosphatase and Tensin Homologue and Liver Kinase B1 on AKT/mTOR/c-Myc axis in pancreatic neuroendocrine tumors. Oncotarget 2017, 8, 98068–98083. [Google Scholar] [CrossRef]
- Stefanoli, M.; La Rosa, S.; Sahnane, N.; Romualdi, C.; Pastorino, R.; Marando, A.; Capella, C.; Sessa, F.; Furlan, D. Prognostic Relevance of Aberrant DNA Methylation in G1 and G2 Pancreatic Neuroendocrine Tumors. Neuroendocrinology 2014, 100, 26–34. [Google Scholar] [CrossRef]
- Peng, X.; Gao, H.; Xu, R.; Wang, H.; Mei, J.; Liu, C. The interplay between HIF-1α and noncoding RNAs in cancer. J. Exp. Clin. Cancer Res. 2020, 39, 1–19. [Google Scholar] [CrossRef]
- Maxwell, P.H.; Wiesener, M.S.; Chang, G.-W.; Clifford, S.C.; Vaux, E.C.; Cockman, M.E.; Wykoff, C.C.; Pugh, C.W.; Maher, E.R.; Ratcliffe, P.J. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 1999, 399, 271–275. [Google Scholar] [CrossRef]
- Carmeliet, P.; et al. Role of HIF-1α or in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature 1998, 394, 485–490. [Google Scholar] [CrossRef]
- Nielsen, S.M.; Rhodes, L.; Blanco, I.; Chung, W.K.; Eng, C.; Maher, E.R.; Richard, S.; Giles, R.H. Von Hippel-Lindau Disease: Genetics and Role of Genetic Counseling in a Multiple Neoplasia Syndrome. J. Clin. Oncol. 2016, 34, 2172–2181. [Google Scholar] [CrossRef]
- Mamo, M.; Ye, I.C.; DiGiacomo, J.W.; Park, J.Y.; Downs, B.; Gilkes, D.M. Hypoxia Alters the Response to Anti-EGFR Therapy by Regulating EGFR Expression and Downstream Signaling in a DNA Methylation–Specific and HIF-Dependent Manner. Cancer Res 2020, 80, 4998–5010. [Google Scholar] [CrossRef]
- Cimmino, F.; Avitabile, M.; Lasorsa, V.A.; Montella, A.; Pezone, L.; Cantalupo, S.; Visconte, F.; Corrias, M.V.; Iolascon, A.; Capasso, M. HIF-1 transcription activity: HIF1A driven response in normoxia and in hypoxia. BMC Med Genet. 2019, 20, 37. [Google Scholar] [CrossRef]
- Perigny, M.; et al. Pancreatic endocrine microadenomatosis in patients with von Hippel-Lindau disease: characterization by VHL/HIF pathway proteins expression. Am. J. Surg. Pathol. 2009, 33, 739–748. [Google Scholar]
- Speisky, D.; Duces, A.; Bièche, I.; Rebours, V.; Hammel, P.; Sauvanet, A.; Richard, S.; Bedossa, P.; Vidaud, M.; Murat, A.; et al. Molecular Profiling of Pancreatic Neuroendocrine Tumors in Sporadic and Von Hippel-Lindau Patients. Clin. Cancer Res. 2012, 18, 2838–2849. [Google Scholar] [CrossRef]
- Schmitt, A.M.; Schmid, S.; Rudolph, T.; Anlauf, M.; Prinz, C.; Klöppel, G.; Moch, H.; Heitz, P.U.; Komminoth, P.; Perren, A. VHL inactivation is an important pathway for the development of malignant sporadic pancreatic endocrine tumors. Endocrine-Related Cancer 2009, 16, 1219–1227. [Google Scholar] [CrossRef]
- Chakraborty, J.; Chakraborty, S.; Chakraborty, S.; Narayan, M.N. Entanglement of MAPK pathways with gene expression and its omnipresence in the etiology for cancer and neurodegenerative disorders. Biochim. et Biophys. Acta (BBA) - Gene Regul. Mech. 2023, 1866, 194988. [Google Scholar] [CrossRef]
- Pudewell, S.; Wittich, C.; Jasemi, N.S.K.; Bazgir, F.; Ahmadian, M.R. Accessory proteins of the RAS-MAPK pathway: moving from the side line to the front line. Commun. Biol. 2021, 4, 1–10. [Google Scholar] [CrossRef]
- Chan, D.W.; et al. Genome-wide DNA methylome analysis identifies methylation signatures associated with survival and drug resistance of ovarian cancers. Clin. Epigenetics 2021, 13, 142. [Google Scholar] [CrossRef]
- Gutierrez, A.; Demond, H.; Brebi, P.; Ili, C.G. Novel Methylation Biomarkers for Colorectal Cancer Prognosis. Biomolecules 2021, 11, 1722. [Google Scholar] [CrossRef]
- Yun, D.; Yang, Z.; Zhang, S.; Yang, H.; Liu, D.; Grützmann, R.; Pilarsky, C.; Britzen-Laurent, N. An m5C methylation regulator-associated signature predicts prognosis and therapy response in pancreatic cancer. Front. Cell Dev. Biol. 2022, 10, 975684. [Google Scholar] [CrossRef]
- Shi, J.; Lu, D.; Gu, R.; Xie, J.; Yu, L.; Sun, X.; Zhang, Y. Integrated Analysis of Transcriptome and Differential Methylation of Neurofibromatosis Type 2 Vestibular Schwannomas. World Neurosurg. 2021, 157, e66–e76. [Google Scholar] [CrossRef]
- Zhang, Q.; et al. Activation of RAS/MAPK pathway confers MCL-1 mediated acquired resistance to BCL-2 inhibitor venetoclax in acute myeloid leukemia. Signal. Transduct. Target Ther. 2022, 7, 51. [Google Scholar] [CrossRef]
- Xiao, Y.; Dong, J. The Hippo Signaling Pathway in Cancer: A Cell Cycle Perspective. Cancers 2021, 13, 6214. [Google Scholar] [CrossRef]
- Dubois, F.; Bergot, E.; Zalcman, G.; Levallet, G. RASSF1A, puppeteer of cellular homeostasis, fights tumorigenesis, and metastasis—an updated review. Cell Death Dis. 2019, 10, 928. [Google Scholar] [CrossRef]
- Khandelwal, M.; Anand, V.; Appunni, S.; Seth, A.; Singh, P.; Mathur, S.; Sharma, A. RASSF1A–Hippo pathway link in patients with urothelial carcinoma of bladder: plausible therapeutic target. Mol. Cell. Biochem. 2019, 464, 51–63. [Google Scholar] [CrossRef]
- Bakavicius, A.; Daniunaite, K.; Zukauskaite, K.; Barisiene, M.; Jarmalaite, S.; Jankevicius, F. Urinary DNA methylation biomarkers for prediction of prostate cancer upgrading and upstaging. Clin. Epigenetics 2019, 11, 115. [Google Scholar] [CrossRef]
- Sangtani, A.; Wang, C.; Weaver, A.; Hoppman, N.L.; Kerr, S.E.; Abyzov, A.; Shridhar, V.; Staub, J.; Kocher, J.-P.A.; Voss, J.S.; et al. Combining copy number, methylation markers, and mutations as a panel for endometrial cancer detection via intravaginal tampon collection. Gynecol. Oncol. 2019, 156, 387–392. [Google Scholar] [CrossRef]
- Aibel, C.; De Peña, A.C.; Tripathi, A. An Optimized CoBRA Method for the Microfluidic Electrophoresis Detection of Breast Cancer Associated RASSF1 Methylation. BioTech 2023, 12, 7. [Google Scholar] [CrossRef]
- Raos, D.; Ulamec, M.; Bojanac, A.K.; Bulic-Jakus, F.; Jezek, D.; Sincic, N. Epigenetically inactivated RASSF1A as a tumor biomarker. Bosn. J. Basic Med Sci. 2020, 21, 386–397. [Google Scholar] [CrossRef]
- Mashayekhi, M.; Asadi, M.; Hashemzadeh, S.; Vahedi, A.; Shanehbandi, D.; Al-Omar, A.F.; Akbari, M.; Raeisi, M. Promoter methylation levels of RASSF1 and ATIC genes are associated with lung cancer in Iranian patients. Horm. Mol. Biol. Clin. Investig. 2023, 44, 145–152. [Google Scholar] [CrossRef]
- Daniunaite, K.; Sestokaite, A.; Kubiliute, R.; Stuopelyte, K.; Kettunen, E.; Husgafvel-Pursiainen, K.; Jarmalaite, S. Frequent DNA methylation changes in cancerous and noncancerous lung tissues from smokers with non-small cell lung cancer. Mutagenesis 2020, 35, 373–379. [Google Scholar] [CrossRef]
- Khatami, F.; Larijani, B.; Heshmat, R.; Nasiri, S.; Saffar, H.; Tavangar, S.M. Promoter Methylation of Four Tumor Suppressor Genes in Human Papillary Thyroid Carcinoma. Iran. J. Pathol. 2019, 14, 290–298. [Google Scholar] [CrossRef]
- House, M.G.; Herman, J.G.; Guo, M.Z.; Hooker, C.M.; Schulick, R.D.; Lillemoe, K.D.; Cameron, J.L.; Hruban, R.H.; Maitra, A.; Yeo, C.J. Aberrant Hypermethylation of Tumor Suppressor Genes in Pancreatic Endocrine Neoplasms. Ann. Surg. 2003, 238, 423–432, discussion 431–432. [Google Scholar] [CrossRef]
- Pyronnet, S.; Bousquet, C.; Najib, S.; Azar, R.; Laklai, H.; Susini, C. Antitumor effects of somatostatin. Mol. Cell. Endocrinol. 2008, 286, 230–237. [Google Scholar] [CrossRef]
- Malpeli, G.; et al. Methylation-associated down-regulation of RASSF1A and up-regulation of RASSF1C in pancreatic endocrine tumors. BMC Cancer 2011, 11, 351. [Google Scholar] [CrossRef]
- Ratner, N.; Miller, S.J. A RASopathy gene commonly mutated in cancer: the neurofibromatosis type 1 tumour suppressor. Nat. Rev. Cancer 2015, 15, 290–301. [Google Scholar] [CrossRef]
- Gauci, J.; et al. Neurofibromatosis Type 1 A Rare Predisposition for Gastrinomas and Other Neuroendocrine Tumors. Pancreas 2022, 51, 559–562. [Google Scholar] [CrossRef]
- Dyer, M.A.; Qadeer, Z.A.; Valle-Garcia, D.; Bernstein, E. ATRX and DAXX: Mechanisms and Mutations. Cold Spring Harb. Perspect. Med. 2017, 7, a026567. [Google Scholar] [CrossRef] [PubMed]
- Han, B.; Cai, J.; Gao, W.; Meng, X.; Gao, F.; Wu, P.; Duan, C.; Wang, R.; Dinislam, M.; Lin, L.; et al. Loss of ATRX suppresses ATM dependent DNA damage repair by modulating H3K9me3 to enhance temozolomide sensitivity in glioma. Cancer Lett. 2018, 419, 280–290. [Google Scholar] [CrossRef] [PubMed]
- Cavalcante, S.G.; Pereira, B.J.; Lerario, A.M.; Sola, P.R.; Oba-Shinjo, S.M.; Marie, S.K. The chromatin remodeler complex ATRX-DAXX-H3.3 and telomere length in meningiomas. Clin. Neurol. Neurosurg. 2021, 210, 106962. [Google Scholar] [CrossRef]
- Heaphy, C.M.; Bi, W.L.; Coy, S.; Davis, C.; Gallia, G.L.; Santagata, S.; Rodriguez, F.J. Telomere length alterations and ATRX/DAXX loss in pituitary adenomas. Mod. Pathol. 2020, 33, 1475–1481. [Google Scholar] [CrossRef] [PubMed]
- Yachida, S.; Vakiani, E.; White, C.M.B.; Zhong, Y.; Saunders, T.H.; Morgan, R.; de Wilde, R.F.; Maitra, A.M.; Hicks, J.B.; DeMarzo, A.M.; et al. Small Cell and Large Cell Neuroendocrine Carcinomas of the Pancreas are Genetically Similar and Distinct From Well-differentiated Pancreatic Neuroendocrine Tumors. Am. J. Surg. Pathol. 2012, 36, 173–184. [Google Scholar] [CrossRef] [PubMed]
- Yachida, S.; Totoki, Y.; Noë, M.; Nakatani, Y.; Horie, M.; Kawasaki, K.; Nakamura, H.; Saito-Adachi, M.; Suzuki, M.; Takai, E.; et al. Comprehensive Genomic Profiling of Neuroendocrine Carcinomas of the Gastrointestinal System. Cancer Discov. 2022, 12, 692–711. [Google Scholar] [CrossRef] [PubMed]
- Ji, Z.; Huo, C.; Yang, P. Genistein inhibited the proliferation of kidney cancer cells via CDKN2a hypomethylation: role of abnormal apoptosis. Int. Urol. Nephrol. 2020, 52, 1049–1055. [Google Scholar] [CrossRef] [PubMed]
- Xia, L., W. Zhang, and L. Gao, Clinical and prognostic effects of CDKN2A, CDKN2B and CDH13 promoter methylation in ovarian cancer: a study using meta-analysis and TCGA data. Biomarkers 2019, 24, 700–711. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.-C.; Kuo, Y.-C.; Hu, J.-M.; Chang, P.-K.; Sun, C.-A.; Yang, T.; Li, C.-W.; Chen, C.-Y.; Lin, F.-H.; Hsu, C.-H.; et al. MTNR1B polymorphisms with CDKN2A and MGMT methylation status are associated with poor prognosis of colorectal cancer in Taiwan. World J. Gastroenterol. 2021, 27, 5737–5752. [Google Scholar] [CrossRef]
- Kong, R.; Wang, N.; Han, W.; Bao, W.; Lu, J. Fenofibrate Exerts Antitumor Effects in Colon Cancer via Regulation of DNMT1 and CDKN2A. PPAR Res. 2021, 2021, 6663782. [Google Scholar] [CrossRef]
- Truong, P.K., T. D. Lao, and T.A.H. Le, Methylation - an Epigenetic Biomarker for Cervical Cancer Risk: A Meta-Analysis. Pharmacophore 2020, 11, 21–29. [Google Scholar]
- Xu, J.; Li, N.; Deng, W.; Luo, S. Discovering the mechanism and involvement of the methylation of cyclin-dependent kinase inhibitor 2A (CDKN2A) gene and its special locus region in gastric cancer. Bioengineered 2021, 12, 1286–1298. [Google Scholar] [CrossRef]
- Li, P.; et al. P16 methylation increases the sensitivity of cancer cells to the CDK4/6 inhibitor palbociclib. PLoS One 2019, 14, e0223084. [Google Scholar] [CrossRef]
- Hsu, J.Y.; et al. Clinical Utility of CDK4/6 Inhibitors in Sarcoma: Successes and Future Challenges. JCO Precis Oncol. 2022, 6, e2100211. [Google Scholar] [CrossRef]
- Anwar, S.L. and U. Lehmann, Detection of Aberrant DNA Methylation Patterns in the RB1 Gene. Methods Mol. Biol. 2018, 1726, 35–47. [Google Scholar]
- Bartsch, D.K.; Kersting, M.; Wild, A.; Ramaswamy, A.; Gerdes, B.; Schuermann, M.; Simon, B.; Rothmund, M. Low Frequency of p16INK4a Alterations in Insulinomas. Digestion 2000, 62, 171–177. [Google Scholar] [CrossRef]
- Cao, L.Q.; et al. Exosomal miR-21 regulates the TETs/PTENp1/PTEN pathway to promote hepatocellular carcinoma growth. Mol. Cancer, 2019, 18, 148. [Google Scholar] [CrossRef]
- Lindberg, D., G. Akerstrom, and G. Westin, Evaluation of CDKN2C/p18, CDKN1B/p27 and CDKN2B/p15 mRNA expression, and CpG methylation status in sporadic and MEN1-associated pancreatic endocrine tumours. Clin. Endocrinol. (Oxf), 2008, 68, 271–277. [Google Scholar] [CrossRef]
- Khan, M.A.; Tiwari, D.; Dongre, A.; Sadaf, *!!! REPLACE !!!*; Mustafa, S.; Das, C.R.; Massey, S.; Bose, P.D.; Bose, S.; Husain, S.A. Exploring the p53 connection of cervical cancer pathogenesis involving north-east Indian patients. PLoS ONE 2020, 15, e0238500. [Google Scholar] [CrossRef]
- Chatterjee, B.; Ghosh, K.; Kanade, S.R. Resveratrol modulates epigenetic regulators of promoter histone methylation and acetylation that restores BRCA1, p53, p21CIP1 in human breast cancer cell lines. BioFactors 2019, 45, 818–829. [Google Scholar] [CrossRef]
- Liu, Z.L.; et al. Resveratrol induces p53 in colorectal cancer through SET7/9. Oncol. Lett. 2019, 17, 3783–3789. [Google Scholar] [CrossRef]
- Kang, K.A.; Piao, M.J.; Hyun, Y.J.; Zhen, A.X.; Cho, S.J.; Ahn, M.J.; Yi, J.M.; Hyun, J.W. Luteolin promotes apoptotic cell death via upregulation of Nrf2 expression by DNA demethylase and the interaction of Nrf2 with p53 in human colon cancer cells. Exp. Mol. Med. 2019, 51, 1–14. [Google Scholar] [CrossRef]
- Wu, C.; Guo, E.; Ming, J.; Sun, W.; Nie, X.; Sun, L.; Peng, S.; Luo, M.; Liu, D.; Zhang, L.; et al. Radiation-Induced DNMT3B Promotes Radioresistance in Nasopharyngeal Carcinoma through Methylation of p53 and p21. Mol. Ther. - Oncolytics 2020, 17, 306–319. [Google Scholar] [CrossRef]
- Zhang, L.; Li, W.; Cao, L.; Xu, J.; Qian, Y.; Chen, H.; Zhang, Y.; Kang, W.; Gou, H.; Wong, C.C.; et al. PKNOX2 suppresses gastric cancer through the transcriptional activation of IGFBP5 and p53. Oncogene 2019, 38, 4590–4604. [Google Scholar] [CrossRef]
- Vandamme, T.; Peeters, M.; Dogan, F.; Pauwels, P.; Van Assche, E.; Beyens, M.; Mortier, G.; Vandeweyer, G.; de Herder, W.; Van Camp, G.; et al. Whole-exome characterization of pancreatic neuroendocrine tumor cell lines BON-1 and QGP-1. J. Mol. Endocrinol. 2015, 54, 137–147. [Google Scholar] [CrossRef]
- Ohki, R.; Saito, K.; Chen, Y.; Kawase, T.; Hiraoka, N.; Saigawa, R.; Minegishi, M.; Aita, Y.; Yanai, G.; Shimizu, H.; et al. PHLDA3 is a novel tumor suppressor of pancreatic neuroendocrine tumors. Proc. Natl. Acad. Sci. 2014, 111, E2404–E2413. [Google Scholar] [CrossRef]
- Wharton, K.A.; Johansen, K.M.; Xu, T.; Artavanis-Tsakonas, S. Nucleotide sequence from the neurogenic locus Notch implies a gene product that shares homology with proteins containing EGF-like repeats. Cell 1985, 43, 567–581. [Google Scholar] [CrossRef]
- Ferrante, F.; Giaimo, B.D.; Bartkuhn, M.; Zimmermann, T.; Close, V.; Mertens, D.; Nist, A.; Stiewe, T.; Meier-Soelch, J.; Kracht, M.; et al. HDAC3 functions as a positive regulator in Notch signal transduction. Nucleic Acids Res. 2020, 48, 3496–3512. [Google Scholar] [CrossRef]
- Jeong, G.-Y.; Park, M.K.; Choi, H.-J.; An, H.W.; Park, Y.-U.; Choi, H.-J.; Park, J.; Kim, H.-Y.; Son, T.; Lee, H.; et al. NSD3-Induced Methylation of H3K36 Activates NOTCH Signaling to Drive Breast Tumor Initiation and Metastatic Progression. Cancer Res 2021, 81, 77–90. [Google Scholar] [CrossRef]
- Yousefi, H.; Bahramy, A.; Zafari, N.; Delavar, M.R.; Nguyen, K.; Haghi, A.; Kandelouei, T.; Vittori, C.; Jazireian, P.; Maleki, S.; et al. Notch signaling pathway: a comprehensive prognostic and gene expression profile analysis in breast cancer. BMC Cancer 2022, 22, 1282. [Google Scholar] [CrossRef]
- Qiu, L.; Yang, X.; Wu, J.; Huang, C.; Miao, Y.; Fu, Z. HIST2H2BF Potentiates the Propagation of Cancer Stem Cells via Notch Signaling to Promote Malignancy and Liver Metastasis in Colorectal Carcinoma. Front. Oncol. 2021, 11, 677646. [Google Scholar] [CrossRef]
- Chai, H.; Pan, C.; Zhang, M.; Huo, H.; Shan, H.; Wu, J. Histone methyltransferase SETD1A interacts with notch and promotes notch transactivation to augment ovarian cancer development. BMC Cancer 2023, 23, 96. [Google Scholar] [CrossRef]
- Kadian, L.K.; Gulshan, G.; Ahuja, P.; Singhal, G.; Sharma, S.; Nanda, S.; Yadav, R. Aberrant promoter methylation of NOTCH1 and NOTCH3 and its association with cervical cancer risk factors in North Indian population. 2020, 12, 2814–2826.
- Biran, A.; et al. Activation of Notch and Myc Signaling via B-cell-Restricted Depletion of Dnmt3a Generates a Consistent Murine Model of Chronic Lymphocytic Leukemia. Cancer Res. 2021, 81, 6117–6130. [Google Scholar] [CrossRef]
- Wang, H.; Chen, Y.; Castillo, C.F.-D.; Yilmaz, O.; Deshpande, V. Heterogeneity in signaling pathways of gastroenteropancreatic neuroendocrine tumors: a critical look at notch signaling pathway. Mod. Pathol. 2013, 26, 139–147. [Google Scholar] [CrossRef]
- Greenblatt, D.Y.; Vaccaro, A.M.; Jaskula-Sztul, R.; Ning, L.; Haymart, M.; Kunnimalaiyaan, M.; Chen, H. Valproic Acid Activates Notch-1 Signaling and Regulates the Neuroendocrine Phenotype in Carcinoid Cancer Cells. Oncol. 2007, 12, 942–951. [Google Scholar] [CrossRef]
- Kunnimalaiyaan, M.; Traeger, K.; Chen, H. Conservation of the Notch1 signaling pathway in gastrointestinal carcinoid cells. Am. J. Physiol. Liver Physiol. 2005, 289, G636–G642. [Google Scholar] [CrossRef]
- Adler, J.T.; Hottinger, D.G.; Kunnimalaiyaan, M.; Chen, H. Histone deacetylase inhibitors upregulate Notch-1 and inhibit growth in pheochromocytoma cells. Surgery 2008, 144, 956–962, discussion 961–962. [Google Scholar] [CrossRef]
- Mohammed, T.A.; Holen, K.D.; Jaskula-Sztul, R.; Mulkerin, D.; Lubner, S.J.; Schelman, W.R.; Eickhoff, J.; Chen, H.; LoConte, N.K. A Pilot Phase II Study of Valproic Acid for Treatment of Low-Grade Neuroendocrine Carcinoma. Oncol. 2011, 16, 835–843. [Google Scholar] [CrossRef]
- Liu, J.; et al. Wnt/beta-catenin signalling: function, biological mechanisms, and therapeutic opportunities. Signal. Transduct. Target Ther. 2022, 7, 3. [Google Scholar] [CrossRef]
- You, H.; Li, Q.; Kong, D.; Liu, X.; Kong, F.; Zheng, K.; Tang, R. The interaction of canonical Wnt/β-catenin signaling with protein lysine acetylation. Cell. Mol. Biol. Lett. 2022, 27, 1–14. [Google Scholar] [CrossRef]
- Mehdi, S.; et al. LY75 Ablation Mediates Mesenchymal-Epithelial Transition (MET) in Epithelial Ovarian Cancer (EOC) Cells Associated with DNA Methylation Alterations and Suppression of the Wnt/beta-Catenin Pathway. Int. J. Mol. Sci. 2020, 21. [Google Scholar] [CrossRef]
- Yen, H.Y.; et al. Regulation of carcinogenesis and modulation through Wnt/beta-catenin signaling by curcumin in an ovarian cancer cell line. Sci. Rep. 2019, 9, 17267. [Google Scholar] [CrossRef]
- Tao, C.; et al. The tumor suppressor Zinc finger protein 471 suppresses breast cancer growth and metastasis through inhibiting AKT and Wnt/beta-catenin signaling. Clin. Epigenetics 2020, 12, 173. [Google Scholar] [CrossRef]
- Meng, F.; et al. SMYD2 suppresses APC2 expression to activate the Wnt/beta-catenin pathway and promotes epithelial-mesenchymal transition in colorectal cancer. Am. J. Cancer Res. 2020, 10, 997–1011. [Google Scholar]
- Zhang, W.; et al. KDM1A promotes thyroid cancer progression and maintains stemness through the Wnt/beta-catenin signaling pathway. Theranostics 2022, 12, 1500–1517. [Google Scholar] [CrossRef]
- Papadopoulou, S.; Edlund, H. Attenuated Wnt Signaling Perturbs Pancreatic Growth but Not Pancreatic Function. Diabetes 2005, 54, 2844–2851. [Google Scholar] [CrossRef]
- Weiss, V.; et al. Immunohistochemical analysis of the Wnt/beta-catenin signaling pathway in pancreatic neuroendocrine neoplasms. World J. Gastrointest Oncol. 2016, 8, 615–622. [Google Scholar] [CrossRef]
- Kim, J.T.; Li, J.; Jang, E.R.; Gulhati, P.; Rychahou, P.G.; Napier, D.L.; Wang, C.; Weiss, H.L.; Lee, E.Y.; Anthony, L.; et al. Deregulation of Wnt/β-catenin signaling through genetic or epigenetic alterations in human neuroendocrine tumors. Carcinog. 2013, 34, 953–961. [Google Scholar] [CrossRef]
- de Mestier, L.; et al. Critical appraisal of MGMT in digestive NET treated with alkylating agents. Endocr Relat. Cancer 2020, 27, R391–R405. [Google Scholar] [CrossRef]
- Brighi, N.; Lamberti, G.; Andrini, E.; Mosconi, C.; Manuzzi, L.; Donati, G.; Lisotti, A.; Campana, D. Prospective Evaluation of MGMT-Promoter Methylation Status and Correlations with Outcomes to Temozolomide-Based Chemotherapy in Well-Differentiated Neuroendocrine Tumors. Curr. Oncol. 2023, 30, 1381–1394. [Google Scholar] [CrossRef]
- Chauhan, A.; et al. Phytochemicals targeting NF-kappaB signaling: Potential anti-cancer interventions. J. Pharm. Anal. 2022, 12, 394–405. [Google Scholar] [CrossRef]
- Hartley, A.V.; et al. PRMT5-mediated methylation of YBX1 regulates NF-κB activity in colorectal cancer. Sci. Rep. 2020, 10. [Google Scholar]
- Kawaguchi, K.; et al. Aberrant DNA methylation-mediated NF-icB/fatty acid-binding protein 5 (FABP5) feed-forward loop promotes malignancy of colorectal cancer cells. Biochim. Et Biophys. Acta-Mol. Cell Biol. Lipids, 2023; 1868. [Google Scholar]
- Tan, Y.Q.; et al. Tumor suppressor DRD2 facilitates M1 macrophages and restricts NF-κB signaling to trigger pyroptosis in breast cancer. Theranostics 2021, 11, 5214–5231. [Google Scholar] [CrossRef]
- Chen, J.; Liu, A.; Lin, Z.; Wang, B.; Chai, X.; Chen, S.; Lu, W.; Zheng, M.; Cao, T.; Zhong, M.; et al. Downregulation of the circadian rhythm regulator HLF promotes multiple-organ distant metastases in non-small cell lung cancer through PPAR/NF-κb signaling. Cancer Lett. 2020, 482, 56–71. [Google Scholar] [CrossRef]
- Vitali, E.; et al. Pancreatic neuroendocrine tumor progression and resistance to everolimus: the crucial role of NF-kB and STAT3 interplay. J Endocrinol Invest, 2023.
- Lehman, J.M.; Hoeksema, M.D.; Staub, J.; Qian, J.; Harris, B.; Callison, J.C.; Miao, J.; Shi, C.; Eisenberg, R.; Chen, H.; et al. Somatostatin receptor 2 signaling promotes growth and tumor survival in small-cell lung cancer. Int. J. Cancer 2018, 144, 1104–1114. [Google Scholar] [CrossRef]
- Si, Y.; Kim, S.; Ou, J.; Lu, Y.; Ernst, P.; Chen, K.; Whitt, J.; Carter, A.M.; Markert, J.M.; Bibb, J.A.; et al. Anti-SSTR2 antibody-drug conjugate for neuroendocrine tumor therapy. Cancer Gene Ther. 2020, 28, 799–812. [Google Scholar] [CrossRef]
- Torrisani, J.; Hanoun, N.; Laurell, H.; Lopez, F.; Maoret, J.-J.; Souque, A.; Susini, C.; Cordelier, P.; Buscail, L. Identification of an Upstream Promoter of the Human Somatostatin Receptor, hSSTR2, Which Is Controlled by Epigenetic Modifications. Endocrinology 2008, 149, 3137–3147. [Google Scholar] [CrossRef]
- Evans, J.S.; Beaumont, J.; Braga, M.; Masrour, N.; Mauri, F.; Beckley, A.; Butt, S.; Karali, C.S.; Cawthorne, C.; Archibald, S.; et al. Epigenetic potentiation of somatostatin-2 by guadecitabine in neuroendocrine neoplasias as a novel method to allow delivery of peptide receptor radiotherapy. Eur. J. Cancer 2022, 176, 110–120. [Google Scholar] [CrossRef]
- Zhao, M., L. Mishra, and C.X. Deng, The role of TGF-beta/SMAD4 signaling in cancer. Int. J. Biol. Sci. 2018, 14, 111–123. [Google Scholar] [CrossRef]
- Kidd, M.; et al. EGFR/TGFalpha and TGFbeta/CTGF Signaling in Neuroendocrine Neoplasia: Theoretical Therapeutic Targets. Neuroendocrinology 2013, 97, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Wimmel, A., B. Wiedenmann, and S. Rosewicz, Autocrine growth inhibition by transforming growth factor beta-1 (TGFbeta-1) in human neuroendocrine tumour cells. Gut 2003, 52, 1308–1316. [Google Scholar] [CrossRef] [PubMed]
- Shattuck, T.M.; et al. Mutational analysis of Smad3, a candidate tumor suppressor implicated in TGF-beta and menin pathways, in parathyroid adenomas and enteropancreatic endocrine tumors. J. Clin. Endocrinol. Metab. 2002, 87, 3911–3914. [Google Scholar] [PubMed]
- Sehrawat, A.; et al. SMAD7 enhances adult beta-cell proliferation without significantly affecting beta-cell function in mice. J. Biol. Chem. 2020, 295, 4858–4869. [Google Scholar] [CrossRef] [PubMed]
- Norollahi, S.E.; et al. DNA Methylation Profiling of MYC, SMAD2/3 and DNMT3A in Colorectal Cancer. Oman Med. J. 2021, 36, e315. [Google Scholar] [CrossRef] [PubMed]
- Ansar, M.; Wang, C.-J.; Wang, Y.-H.; Shen, T.-H.; Hung, C.-S.; Chang, S.-C.; Lin, R.-K. SMAD3 Hypomethylation as a Biomarker for Early Prediction of Colorectal Cancer. Int. J. Mol. Sci. 2020, 21, 7395. [Google Scholar] [CrossRef] [PubMed]
- Arnold, C.N.; Sosnowski, A.; Schmitt-Gräff, A.; Arnold, R.; Blum, H.E. Analysis of molecular pathways in sporadic neuroendocrine tumors of the gastro-entero-pancreatic system. Int. J. Cancer 2007, 120, 2157–2164. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Gao, Z.; Li, L.; Jiang, X.; Shan, A.; Cai, J.; Peng, Y.; Li, Y.; Jiang, X.; Huang, X.; et al. Whole exome sequencing of insulinoma reveals recurrent T372R mutations in YY1. Nat. Commun. 2013, 4, 2810. [Google Scholar] [CrossRef] [PubMed]
- Chan, A.O.-O.; Kim, S.G.; Bedeir, A.; Issa, J.-P.; Hamilton, S.R.; Rashid, A. CpG island methylation in carcinoid and pancreatic endocrine tumors. Oncogene 2003, 22, 924–934. [Google Scholar] [CrossRef]
- Li, G.; Tian, Y.; Zhu, W.-G. The Roles of Histone Deacetylases and Their Inhibitors in Cancer Therapy. Front. Cell Dev. Biol. 2020, 8, 576946. [Google Scholar] [CrossRef]
- Klomp, M.; Dalm, S.; de Jong, M.; Feelders, R.; Hofland, J.; Hofland, L. Epigenetic regulation of somatostatin and somatostatin receptors in neuroendocrine tumors and other types of cancer. Rev. Endocr. Metab. Disord. 2020, 22, 495–510. [Google Scholar] [CrossRef] [PubMed]
- Hess, L.; Moos, V.; Lauber, A.A.; Reiter, W.; Schuster, M.; Hartl, N.; Lackner, D.; Boenke, T.; Koren, A.; Guzzardo, P.M.; et al. A toolbox for class I HDACs reveals isoform specific roles in gene regulation and protein acetylation. PLOS Genet. 2022, 18, e1010376. [Google Scholar] [CrossRef] [PubMed]
- Audia, J.E.; Campbell, R.M. Histone Modifications and Cancer. Cold Spring Harb. Perspect. Biol. 2016, 8, a019521. [Google Scholar] [CrossRef] [PubMed]
- Weichert, W.; Denkert, C.; Noske, A.; Darb-Esfahani, S.; Dietel, M.; Kalloger, S.E.; Huntsman, D.G.; Köbel, M. Expression of Class I Histone Deacetylases Indicates Poor Prognosis in Endometrioid Subtypes of Ovarian and Endometrial Carcinomas. Neoplasia 2008, 10, 1021–1027. [Google Scholar] [CrossRef]
- Mori, M.; Sudo, T.; Mimori, K.; Nishida, N.; Kogo, R.; Iwaya, T.; Tanaka, F.; Shibata, K.; Fujita, H.; Shirouzu, K. Histone deacetylase 1 expression in gastric cancer. Oncol. Rep. 2011, 26, 777–782. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; et al. Regulation of cyclin B2 expression and cell cycle G2/m transition by menin. J. Biol. Chem. 2010, 285, 18291–18300. [Google Scholar] [CrossRef]
- Baradari, V.; Huether, A.; Höpfner, M.; Schuppan, D.; Scherübl, H. Antiproliferative and proapoptotic effects of histone deacetylase inhibitors on gastrointestinal neuroendocrine tumor cells. Endocrine-Related Cancer 2006, 13, 1237–1250. [Google Scholar] [CrossRef]
- Adler, J.T.; Hottinger, D.G.; Kunnimalaiyaan, M.; Chen, H. Combination Therapy with Histone Deacetylase Inhibitors and Lithium Chloride: A Novel Treatment for Carcinoid Tumors. Ann. Surg. Oncol. 2008, 16, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.; Laddha, S.V.; Tang, L.; Vosburgh, E.; Levine, A.J.; Normant, E.; Sandy, P.; Harris, C.R.; Chan, C.S.; Xu, E.Y. The bromodomain and extra-terminal inhibitor CPI203 enhances the antiproliferative effects of rapamycin on human neuroendocrine tumors. Cell Death Dis. 2014, 5, e1450. [Google Scholar] [CrossRef]
- Arvidsson, Y.; Johanson, V.; Pfragner, R.; Wängberg, B.; Nilsson, O. Cytotoxic Effects of Valproic Acid on Neuroendocrine Tumour Cells. Neuroendocrinology 2015, 103, 578–591. [Google Scholar] [CrossRef]
- Lines, K.E.; Vas Nunes, R.P.; Frost, M.; Yates, C.J.; Stevenson, M.; Thakker, R.V. A MEN1 pancreatic neuroendocrine tumour mouse model under temporal control. Endocr. Connect. 2017, 6, 232–242. [Google Scholar] [CrossRef]
- Yokoyama, A.; Wang, Z.; Wysocka, J.; Sanyal, M.; Aufiero, D.J.; Kitabayashi, I.; Herr, W.; Cleary, M.L. Leukemia Proto-Oncoprotein MLL Forms a SET1-Like Histone Methyltransferase Complex with Menin To Regulate Hox Gene Expression. Mol. Cell. Biol. 2004, 24, 5639–5649. [Google Scholar] [CrossRef]
- Chen, X.; Li, Y.; Zhu, F.; Xu, X.; Estrella, B.; Pazos, M.A.; McGuire, J.T.; Karagiannis, D.; Sahu, V.; Mustafokulov, M.; et al. Context-defined cancer co-dependency mapping identifies a functional interplay between PRC2 and MLL-MEN1 complex in lymphoma. Nat. Commun. 2023, 14, 4259. [Google Scholar] [CrossRef] [PubMed]
- Silva-Figueroa, A.M.; Perrier, N.D. Epigenetic processes in sporadic parathyroid neoplasms. Mol. Cell. Endocrinol. 2018, 469, 54–59. [Google Scholar] [CrossRef]
- Fennell, L.J.; Kane, A.; Liu, C.; McKeone, D.; Fernando, W.; Su, C.; Bond, C.; Jamieson, S.; Dumenil, T.; Patch, A.-M.; et al. APC Mutation Marks an Aggressive Subtype of BRAF Mutant Colorectal Cancers. Cancers 2020, 12, 1171. [Google Scholar] [CrossRef] [PubMed]
- Masui, K.; Tanaka, K.; Akhavan, D.; Babic, I.; Gini, B.; Matsutani, T.; Iwanami, A.; Liu, F.; Villa, G.R.; Gu, Y.; et al. mTOR Complex 2 Controls Glycolytic Metabolism in Glioblastoma through FoxO Acetylation and Upregulation of c-Myc. Cell Metab. 2013, 18, 726–739. [Google Scholar] [CrossRef]
- Harachi, M.; Masui, K.; Honda, H.; Muragaki, Y.; Kawamata, T.; Cavenee, W.K.; Mischel, P.S.; Shibata, N. Dual Regulation of Histone Methylation by mTOR Complexes Controls Glioblastoma Tumor Cell Growth via EZH2 and SAM. Mol. Cancer Res. 2020, 18, 1142–1152. [Google Scholar] [CrossRef] [PubMed]
- Leslie, N.R.; et al. The regulation of cell migration by PTEN. Biochem Soc. Trans. 2005, 33, 1507–1508. [Google Scholar] [CrossRef] [PubMed]
- Fusco, N.; et al. Alterations and Their Role in Cancer Management: Are We Making Headway on Precision Medicine? Genes 2020, 11. [Google Scholar]
- Okumura, K.; Mendoza, M.; Bachoo, R.M.; DePinho, R.A.; Cavenee, W.K.; Furnari, F.B. PCAF Modulates PTEN Activity. J. Biol. Chem. 2006, 281, 26562–26568. [Google Scholar] [CrossRef]
- Meng, Z.; Jia, L.-F.; Gan, Y.-H. PTEN activation through K163 acetylation by inhibiting HDAC6 contributes to tumour inhibition. Oncogene 2015, 35, 2333–2344. [Google Scholar] [CrossRef]
- Zhu, Q.-Y.; He, Z.-M.; Cao, W.-M.; Li, B. The role of TSC2 in breast cancer: a literature review. Front. Oncol. 2023, 13. [Google Scholar] [CrossRef]
- Uysal, S.P.; Şahi̇n, M. Tuberous sclerosis: a review of the past, present, and future. Turk. J. Med Sci. 2020, 50, 1665–1676. [Google Scholar] [CrossRef]
- Sauter, M.; Belousova, E.; Benedik, M.P.; Carter, T.; Cottin, V.; Curatolo, P.; Dahlin, M.; D’amato, L.; D’augères, G.B.; de Vries, P.J.; et al. Rare manifestations and malignancies in tuberous sclerosis complex: findings from the TuberOus SClerosis registry to increAse disease awareness (TOSCA). Orphanet J. Rare Dis. 2021, 16, 301. [Google Scholar] [CrossRef]
- Geng, H.; Liu, Q.; Xue, C.; David, L.L.; Beer, T.M.; Thomas, G.V.; Dai, M.-S.; Qian, D.Z. HIF1α Protein Stability Is Increased by Acetylation at Lysine 709. J. Biol. Chem. 2012, 287, 35496–35505. [Google Scholar] [CrossRef]
- Zhang, C.; Yang, C.; Feldman, M.J.; Wang, H.; Pang, Y.; Maggio, D.M.; Zhu, D.; Nesvick, C.L.; Dmitriev, P.; Bullova, P.; et al. Vorinostat suppresses hypoxia signaling by modulating nuclear translocation of hypoxia inducible factor 1 alpha. Oncotarget 2017, 8, 56110–56125. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, K.; Feng, X.; Wang, J.; Chu, Y.; Jia, C.; He, Q.; Chen, C. PRMT3 promotes tumorigenesis by methylating and stabilizing HIF1α in colorectal cancer. Cell Death Dis. 2021, 12, 1066. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.H.; et al. Regulation of RAS oncogenicity by acetylation. Proc. Natl. Acad. Sci. USA 2012, 109, 10843–10848. [Google Scholar] [CrossRef]
- Hałasa, M.; Wawruszak, A.; Przybyszewska, A.; Jaruga, A.; Guz, M.; Kałafut, J.; Stepulak, A.; Cybulski, M. H3K18Ac as a Marker of Cancer Progression and Potential Target of Anti-Cancer Therapy. Cells 2019, 8, 485. [Google Scholar] [CrossRef] [PubMed]
- Donninger, H.; Clark, J.; Rinaldo, F.; Nelson, N.; Barnoud, T.; Schmidt, M.L.; Hobbing, K.R.; Vos, M.D.; Sils, B.; Clark, G.J. The RASSF1A Tumor Suppressor Regulates XPA-Mediated DNA Repair. Mol. Cell. Biol. 2015, 35, 277–287. [Google Scholar] [CrossRef]
- Dubois, F.; et al. RASSF1A Suppresses the Invasion and Metastatic Potential of Human Non-Small Cell Lung Cancer Cells by Inhibiting YAP Activation through the GEF-H1/RhoB Pathway. Cancer Res. 2016, 76, 1627–1640. [Google Scholar] [CrossRef]
- Patel, A.J.; Liao, C.-P.; Chen, Z.; Liu, C.; Wang, Y.; Le, L.Q. BET Bromodomain Inhibition Triggers Apoptosis of NF1-Associated Malignant Peripheral Nerve Sheath Tumors through Bim Induction. Cell Rep. 2013, 6, 81–92. [Google Scholar] [CrossRef]
- Brosseau, J.-P.; Liao, C.-P.; Le, L.Q. Translating current basic research into future therapies for neurofibromatosis type 1. Br. J. Cancer 2020, 123, 178–186. [Google Scholar] [CrossRef]
- Agarwal, S.K.; Jothi, R. Genome-Wide Characterization of Menin-Dependent H3K4me3 Reveals a Specific Role for Menin in the Regulation of Genes Implicated in MEN1-Like Tumors. PLOS ONE 2012, 7, e37952. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Qi, Y.; Fowlkes, N.; Lazic, N.; Su, X.; Lozano, G.; Wasylishen, A.R. The histone chaperone function of Daxx is dispensable for embryonic development. Cell Death Dis. 2023, 14, 1–9. [Google Scholar] [CrossRef]
- Hoelper, D.; Huang, H.; Jain, A.Y.; Patel, D.J.; Lewis, P.W. Structural and mechanistic insights into ATRX-dependent and -independent functions of the histone chaperone DAXX. Nat. Commun. 2017, 8, 1–13. [Google Scholar] [CrossRef]
- Mahmud, I.; Liao, D. DAXX in cancer: phenomena, processes, mechanisms and regulation. Nucleic Acids Res. 2019, 47, 7734–7752. [Google Scholar] [CrossRef] [PubMed]
- Brehm, A.; Miska, E.A.; McCance, D.J.; Reid, J.L.; Bannister, A.J.; Kouzarides, T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature 1998, 391, 597–601. [Google Scholar] [CrossRef]
- Puri, P.L.; Iezzi, S.; Stiegler, P.; Chen, T.-T.; Schiltz, R.; Muscat, G.E.; Giordano, A.; Kedes, L.; Wang, J.Y.; Sartorelli, V. Class I Histone Deacetylases Sequentially Interact with MyoD and pRb during Skeletal Myogenesis. Mol. Cell 2001, 8, 885–897. [Google Scholar] [CrossRef]
- Huang, H.; Fu, Y.; Zhang, Y.; Peng, F.; Lu, M.; Feng, Y.; Chen, L.; Chen, Z.; Li, M.; Chen, Y. Dissection of Anti-tumor Activity of Histone Deacetylase Inhibitor SAHA in Nasopharyngeal Carcinoma Cells via Quantitative Phosphoproteomics. Front. Cell Dev. Biol. 2020, 8, 577784. [Google Scholar] [CrossRef]
- Zhou, Y.K.; et al. HDAC5 Loss Impairs RB Repression of Pro-Oncogenic Genes and Confers CDK4/6 Inhibitor Resistance in Cancer. Cancer Res. 2021, 81, 1486–1499. [Google Scholar] [CrossRef] [PubMed]
- Chun, P. Histone deacetylase inhibitors in hematological malignancies and solid tumors. Arch. Pharmacal Res. 2015, 38, 933–949. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; et al. Trichostatin A, a histone deacetylase inhibitor, suppresses proliferation and epithelial-mesenchymal transition in retinal pigment epithelium cells. J. Cell Mol. Med. 2014, 18, 646–655. [Google Scholar] [CrossRef] [PubMed]
- Mrakovcic, M.; Kleinheinz, J.; Fröhlich, L.F. p53 at the Crossroads between Different Types of HDAC Inhibitor-Mediated Cancer Cell Death. Int. J. Mol. Sci. 2019, 20, 2415. [Google Scholar] [CrossRef] [PubMed]
- Juengel, E.; Nowaz, S.; Makarevi, J.; Natsheh, I.; Werner, I.; Nelson, K.; Reiter, M.; Tsaur, I.; Mani, J.; Harder, S.; et al. HDAC-inhibition counteracts everolimus resistance in renal cell carcinoma in vitro by diminishing cdk2 and cyclin A. Mol. Cancer 2014, 13, 152. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Kao, G.D.; Garcia, B.A.; Shabanowitz, J.; Hunt, D.F.; Qin, J.; Phelan, C.; Lazar, M.A. A novel histone deacetylase pathway regulates mitosis by modulating Aurora B kinase activity. Genes Dev. 2006, 20, 2566–2579. [Google Scholar] [CrossRef] [PubMed]
- Maggi, E.C.; Trillo-Tinoco, J.; Struckhoff, A.P.; Vijayaraghavan, J.; Del Valle, L.; Crabtree, J.S. Retinoblastoma-binding protein 2 (RBP2) is frequently expressed in neuroendocrine tumors and promotes the neoplastic phenotype. Oncogenesis 2016, 5, e257. [Google Scholar] [CrossRef] [PubMed]
- Gu, W.; Roeder, R.G. Activation of p53 Sequence-Specific DNA Binding by Acetylation of the p53 C-Terminal Domain. Cell 1997, 90, 595–606. [Google Scholar] [CrossRef] [PubMed]
- Ito, A.; et al. p300/CBP-mediated p53 acetylation is commonly induced by p53-activating agents and inhibited by MDM2. Embo J. 2001, 20, 1331–1340. [Google Scholar] [CrossRef]
- Tang, Y.; Zhao, W.; Chen, Y.; Zhao, Y.; Gu, W. Acetylation Is Indispensable for p53 Activation. Cell 2008, 133, 612–626. [Google Scholar] [CrossRef]
- Kloster, M.M.; Naderi, E.H.; Haaland, I.; Gjertsen, B.T.; Blomhoff, H.K.; Naderi, S. cAMP signalling inhibits p53 acetylation and apoptosis via HDAC and SIRT deacetylases. Int. J. Oncol. 2013, 42, 1815–1821. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Pan, M.; Sun, J.; Lu, H.; Shen, Q.; Zhang, S.; Jiang, T.; Liu, L.; Jin, W.; Chen, Y.; et al. Histone deacetylase 3 inhibits expression of PUMA in gastric cancer cells. J. Mol. Med. 2012, 91, 49–58. [Google Scholar] [CrossRef]
- Chi, X.Z.; et al. RUNX3 suppresses gastric epithelial cell growth by inducing p21(WAF1/Cip1) expression in cooperation with transforming growth factor beta-activated SMAD. Mol. Cell Biol. 2005, 25, 8097–8107. [Google Scholar] [CrossRef]
- Fröhlich, L.F.; Mrakovcic, M.; Smole, C.; Zatloukal, K. Molecular mechanism leading to SAHA-induced autophagy in tumor cells: evidence for a p53-dependent pathway. Cancer Cell Int. 2016, 16, 68. [Google Scholar] [CrossRef]
- Montani, M.S.G.; Granato, M.; Santoni, C.; Del Porto, P.; Merendino, N.; D’orazi, G.; Faggioni, A.; Cirone, M. Histone deacetylase inhibitors VPA and TSA induce apoptosis and autophagy in pancreatic cancer cells. Cell. Oncol. 2017, 40, 167–180. [Google Scholar] [CrossRef]
- Jang, S.; Jin, H.; Roy, M.; Ma, A.L.; Gong, S.; Jaskula-Sztul, R.; Chen, H. Antineoplastic effects of histone deacetylase inhibitors in neuroendocrine cancer cells are mediated through transcriptional regulation of Notch1 by activator protein 1. Cancer Med. 2017, 6, 2142–2152. [Google Scholar] [CrossRef] [PubMed]
- Zhan, T., N. Rindtorff, and M. Boutros, Wnt signaling in cancer. Oncogene 2017, 36, 1461–1473. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.-Y.; Huang, G.-D.; Chen, L.; Zhang, C.; Chen, B.-D.; Li, Q.-Z.; Wang, X.; Zhang, X.-J.; Li, W.-P. Tanshinone IIA induces apoptosis via inhibition of Wnt/β-catenin/MGMT signaling in AtT-20 cells. Mol. Med. Rep. 2017, 16, 5908–5914. [Google Scholar] [CrossRef] [PubMed]
- Götze, S.; Coersmeyer, M.; Müller, O.; Sievers, S. Histone deacetylase inhibitors induce attenuation of Wnt signaling and TCF7L2 depletion in colorectal carcinoma cells. Int. J. Oncol. 2014, 45, 1715–1723. [Google Scholar] [CrossRef]
- Vallée, A.; Lecarpentier, Y.; Vallée, J.-N. Curcumin: a therapeutic strategy in cancers by inhibiting the canonical WNT/β-catenin pathway. J. Exp. Clin. Cancer Res. 2019, 38, 1–16. [Google Scholar] [CrossRef]
- Chen, J. and L.F. Chen, Methods to detect NF-kappaB acetylation and methylation. Methods Mol. Biol. 2015, 1280, 395–409. [Google Scholar]
- Chen, L.F., Y. Mu, and W.C. Greene, Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-kappaB. EMBO J. 2002, 21, 6539–6548. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; et al. Persistently activated Stat3 maintains constitutive NF-kappaB activity in tumors. Cancer Cell 2009, 15, 283–293. [Google Scholar] [CrossRef] [PubMed]
- Wanek, J.; Gaisberger, M.; Beyreis, M.; Mayr, C.; Helm, K.; Primavesi, F.; Jäger, T.; Di Fazio, P.; Jakab, M.; Wagner, A.; et al. Pharmacological Inhibition of Class IIA HDACs by LMK-235 in Pancreatic Neuroendocrine Tumor Cells. Int. J. Mol. Sci. 2018, 19, 3128. [Google Scholar] [CrossRef] [PubMed]
- Veenstra, M.J.; van Koetsveld, P.M.; Dogan, F.; Farrell, W.E.; Feelders, R.A.; Lamberts, S.W.; de Herder, W.W.; Vitale, G.; Hofland, L.J. Epidrug-induced upregulation of functional somatostatin type 2 receptors in human pancreatic neuroendocrine tumor cells. Oncotarget 2016, 9, 14791–14802. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; et al. Valproic acid induces NET cell growth arrest and enhances tumor suppression of the receptor-targeted peptide-drug conjugate via activating somatostatin receptor type II. J. Drug Target. 2016, 24, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Ryu, T.Y.; Jung, E.; Han, T.-S.; Lee, J.; Kim, S.-K.; Na Roh, Y.; Lee, M.-S.; Jung, C.-R.; Lim, J.H.; et al. Epigenetic regulation of SMAD3 by histone methyltransferase SMYD2 promotes lung cancer metastasis. Exp. Mol. Med. 2023, 55, 952–964. [Google Scholar] [CrossRef]
- Tang, Y.-N.; Ding, W.-Q.; Guo, X.-J.; Yuan, X.-W.; Wang, D.-M.; Song, J.-G. Epigenetic regulation of Smad2 and Smad3 by profilin-2 promotes lung cancer growth and metastasis. Nat. Commun. 2015, 6, 8230. [Google Scholar] [CrossRef]
- Du, D.; et al. Smad3-mediated recruitment of the methyltransferase SETDB1/ESET controls Snail1 expression and epithelial-mesenchymal transition. EMBO Rep. 2018, 19, 135–155. [Google Scholar] [CrossRef]
- Lin, W.; Watanabe, H.; Peng, S.; Francis, J.M.; Kaplan, N.; Pedamallu, C.S.; Ramachandran, A.; Agoston, A.; Bass, A.J.; Meyerson, M. Dynamic Epigenetic Regulation by Menin During Pancreatic Islet Tumor Formation. Mol. Cancer Res. 2015, 13, 689–698. [Google Scholar] [CrossRef]
- Wang, F.; Xu, X.; Ye, Z.; Qin, Y.; Yu, X.; Ji, S. Prognostic Significance of Altered ATRX/DAXX Gene in Pancreatic Neuroendocrine Tumors: A Meta-Analysis. Front. Endocrinol. 2021, 12, 691557. [Google Scholar] [CrossRef] [PubMed]
- Lewis, P.W.; Elsaesser, S.J.; Noh, K.-M.; Stadler, S.C.; Allis, C.D. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc. Natl. Acad. Sci. 2010, 107, 14075–14080. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.J.; Hostetter, G.; Macfarlane, A.W.; Madaj, Z.; Ross, E.A.; Hinoue, T.; Kulchycki, J.R.; Burgos, R.S.; Tafseer, M.; Alpaugh, R.K.; et al. A Phase II Trial of Guadecitabine plus Atezolizumab in Metastatic Urothelial Carcinoma Progressing after Initial Immune Checkpoint Inhibitor Therapy. Clin. Cancer Res. 2023, 29, 2052–2065. [Google Scholar] [CrossRef] [PubMed]
- Goodall, G.J. and V.O. Wickramasinghe, RNA in cancer. Nat. Rev. Cancer 2021, 21, 22–36. [Google Scholar] [CrossRef] [PubMed]
- Moschovis, D. " Long non-coding RNA in pancreatic adenocarcinoma and pancreatic neuroendocrine tumors". Ann. Gastroenterol. 2017, 30, 622–628. [Google Scholar] [CrossRef] [PubMed]
- Moschovis, D.; Vasilaki, E.; Tzouvala, M.; Karamanolis, G.; Katifelis, H.; Legaki, E.; Vezakis, A.; Aravantinos, G.; Gazouli, M. Association between genetic polymorphisms in long non-coding RNAs and pancreatic cancer risk. Cancer Biomarkers 2019, 24, 117–123. [Google Scholar] [CrossRef]
- Ji, M.; et al. Long noncoding RNA-mRNA expression profiles and validation in pancreatic neuroendocrine neoplasms. Clin. Endocrinol. 2020, 92, 312–322. [Google Scholar] [CrossRef]
- Hwang, S.; Jeong, J.J.; Kim, S.H.; Chung, Y.J.; Song, S.Y.; Lee, Y.J.; Rhee, Y. Differential expression of miRNA199b-5p as a novel biomarker for sporadic and hereditary parathyroid tumors. Sci. Rep. 2018, 8, 12016. [Google Scholar] [CrossRef]
- Prinz, F.; Kapeller, A.; Pichler, M.; Klec, C. The Implications of the Long Non-Coding RNA NEAT1 in Non-Cancerous Diseases. Int. J. Mol. Sci. 2019, 20, 627. [Google Scholar] [CrossRef]
- Ding, N.; Wu, H.; Tao, T.; Peng, E. NEAT1 regulates cell proliferation and apoptosis of ovarian cancer by miR-34a-5p/BCL2. OncoTargets Ther. 2017, ume 10, 4905–4915. [Google Scholar] [CrossRef]
- Ouyang, M.; Su, W.; Xiao, L.; Rao, J.N.; Jiang, L.; Li, Y.; Turner, D.J.; Gorospe, M.; Wang, J.-Y. Modulation by miR-29b of intestinal epithelium homoeostasis through the repression of menin translation. Biochem. J. 2015, 465, 315–323. [Google Scholar] [CrossRef]
- Zhang, X.; Zhou, Y.; Mehta, K.R.; Danila, D.C.; Scolavino, S.; Johnson, S.R.; Klibanski, A. A Pituitary-Derived MEG3 Isoform Functions as a Growth Suppressor in Tumor Cells. J. Clin. Endocrinol. Metab. 2003, 88, 5119–5126. [Google Scholar] [CrossRef]
- Zhang, Y.; et al. Long noncoding RNA MEG3 inhibits breast cancer growth via upregulating endoplasmic reticulum stress and activating NF-kappaB and p53. J. Cell Biochem. 2019, 120, 6789–6797. [Google Scholar] [CrossRef]
- Zhang, Y.-Y.; Feng, H.-M. MEG3 Suppresses Human Pancreatic Neuroendocrine Tumor Cells Growth and Metastasis by Down-Regulation of Mir-183. Cell. Physiol. Biochem. 2017, 44, 345–356. [Google Scholar] [CrossRef]
- Modali, S.D.; Parekh, V.I.; Kebebew, E.; Agarwal, S.K. Epigenetic Regulation of the lncRNA MEG3 and Its Target c-MET in Pancreatic Neuroendocrine Tumors. Mol. Endocrinol. 2015, 29, 224–237. [Google Scholar] [CrossRef]
- Luzi, E.; Marini, F.; Giusti, F.; Galli, G.; Cavalli, L.; Brandi, M.L. The Negative Feedback-Loop between the Oncomir Mir-24-1 and Menin Modulates the Men1 Tumorigenesis by Mimicking the “Knudson’s Second Hit”. PLOS ONE 2012, 7, e39767. [Google Scholar] [CrossRef] [PubMed]
- Luzi, E.; Marini, F.; Ciuffi, S.; Galli, G.; Brandi, M.L. An autoregulatory network between menin and pri-miR-24-1 is required for the processing of its specific modulator miR-24-1 in BON1 cells. Mol. Biosyst. 2016, 12, 1922–1928. [Google Scholar] [CrossRef]
- Marini, F.; Brandi, M.L. Role of miR-24 in Multiple Endocrine Neoplasia Type 1: A Potential Target for Molecular Therapy. Int. J. Mol. Sci. 2021, 22, 7352. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Xue, J.; Yang, S.; Chen, Y.; Wang, Y.; Zhu, Y.; Wang, X.; Kuang, D.; Ruan, Q.; Duan, Y.; et al. Co-targeting of IGF1R/mTOR pathway by miR-497 and miR-99a impairs hepatocellular carcinoma development. Oncotarget 2017, 8, 47984–47997. [Google Scholar] [CrossRef]
- Li, Z.; et al. Knockdown of lncRNA-HOTAIR downregulates the drug-resistance of breast cancer cells to doxorubicin via the PI3K/AKT/mTOR signaling pathway. Exp. Ther. Med. 2019, 18, 435–442. [Google Scholar]
- Yang, Y.; Sun, D.; Yu, J.; Zhang, M.; Yi, C.; Yang, R.; Dan, B.; Li, A. Long noncoding RNA TUG1 promotes renal cell carcinoma cell proliferation, migration and invasion by downregulating microRNA-196a. Mol. Med. Rep. 2018, 18, 5791–5798. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; et al. miR-144/451 Promote Cell Proliferation via Targeting PTEN/AKT Pathway in Insulinomas. Endocrinology 2015, 156, 2429–2439. [Google Scholar] [CrossRef] [PubMed]
- Ji, M.; Yao, Y.; Liu, A.; Shi, L.; Chen, D.; Tang, L.; Yang, G.; Liang, X.; Peng, J.; Shao, C. lncRNA H19 binds VGF and promotes pNEN progression via PI3K/AKT/CREB signaling. Endocrine-Related Cancer 2019, 26, 643–658. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Wei, N.; Ma, R.; Jiang, S.; Song, D. A miR-210-3p regulon that controls the Warburg effect by modulating HIF-1α and p53 activity in triple-negative breast cancer. Cell Death Dis. 2020, 11, 731. [Google Scholar] [CrossRef] [PubMed]
- Yu, A.; et al. Transcription factor HIF1alpha promotes proliferation, migration, and invasion of cholangiocarcinoma via long noncoding RNA H19/microRNA-612/Bcl-2 axis. Transl. Res. 2020, 224, 26–39. [Google Scholar] [CrossRef]
- Derry, M.M.; Raina, K.; Balaiya, V.; Jain, A.K.; Shrotriya, S.; Huber, K.M.; Serkova, N.J.; Agarwal, R.; Agarwal, C. Grape Seed Extract Efficacy against Azoxymethane-Induced Colon Tumorigenesis in A/J Mice: Interlinking miRNA with Cytokine Signaling and Inflammation. Cancer Prev. Res. 2013, 6, 625–633. [Google Scholar] [CrossRef] [PubMed]
- Shih, C.-H.; Chuang, L.-L.; Tsai, M.-H.; Chen, L.-H.; Chuang, E.Y.; Lu, T.-P.; Lai, L.-C. Hypoxia-Induced MALAT1 Promotes the Proliferation and Migration of Breast Cancer Cells by Sponging MiR-3064-5p. Front. Oncol. 2021, 11, 658151. [Google Scholar] [CrossRef] [PubMed]
- Thorns, C.; Schurmann, C.; Gebauer, N.; Wallaschofski, H.; Kümpers, C.; Bernard, V.; Feller, A.C.; Keck, T.; Habermann, J.K.; Begum, N.; et al. Global microRNA profiling of pancreatic neuroendocrine neoplasias. . 2014, 34, 2249–2254. [Google Scholar] [PubMed]
- Huang, X.; Ding, L.; Bennewith, K.L.; Tong, R.T.; Welford, S.M.; Ang, K.K.; Story, M.; Le, Q.-T.; Giaccia, A.J. Hypoxia-Inducible mir-210 Regulates Normoxic Gene Expression Involved in Tumor Initiation. Mol. Cell 2009, 35, 856–867. [Google Scholar] [CrossRef]
- Pizzini, S.; Bisognin, A.; Mandruzzato, S.; Biasiolo, M.; Facciolli, A.; Perilli, L.; Rossi, E.; Esposito, G.; Rugge, M.; Pilati, P.; et al. Impact of microRNAs on regulatory networks and pathways in human colorectal carcinogenesis and development of metastasis. BMC Genom. 2013, 14, 589. [Google Scholar] [CrossRef]
- Zhao, Q.; Chen, S.; Zhu, Z.; Yu, L.; Ren, Y.; Jiang, M.; Weng, J.; Li, B. miR-21 promotes EGF-induced pancreatic cancer cell proliferation by targeting Spry2. Cell Death Dis. 2018, 9, 1157. [Google Scholar] [CrossRef] [PubMed]
- Sugito, N.; Heishima, K.; Akao, Y. Chemically modified MIR143-3p exhibited anti-cancer effects by impairing the KRAS network in colorectal cancer cells. Mol. Ther. - Nucleic Acids 2022, 30, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Tang, X.; Duan, S. Interference from LncRNA SPRY4-IT1 restrains the proliferation, migration, and invasion of melanoma cells through inactivating MAPK pathway by up-regulating miR-22-3p. Int. J. Clin. Exp. Pathol. 2019, 12, 477–487.
- Feichtenschlager, V.; Zheng, Y.J.; Ho, W.; Chen, L.; Callanan, C.; Chen, C.; Lee, A.; Ortiz, J.; Rappersberger, K.; Ortiz-Urda, S. Deconstructing the role of MALAT1 in MAPK-signaling in melanoma: insights from antisense oligonucleotide treatment. Oncotarget 2023, 14, 543–560. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; et al. miR-431 Promotes Metastasis of Pancreatic Neuroendocrine Tumors by Targeting DAB2 Interacting Protein, a Ras GTPase Activating Protein Tumor Suppressor. Am. J. Pathol. 2020, 190, 689–701. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, Q.; Luo, N.; Liu, J.; Ren, H.; Shao, X.; Zhang, L.; Yu, Y. MicroRNA-1269a Promotes Proliferation and Arrest of Apoptosis of Glioma Cells by Directly Targeting ATRX. Front. Oncol. 2020, 10, 563901. [Google Scholar] [CrossRef]
- Liu, X.; Abraham, J.M.; Cheng, Y.; Wang, Z.; Wang, Z.; Zhang, G.; Ashktorab, H.; Smoot, D.T.; Cole, R.N.; Boronina, T.N.; et al. Synthetic Circular RNA Functions as a miR-21 Sponge to Suppress Gastric Carcinoma Cell Proliferation. Mol. Ther. - Nucleic Acids 2018, 13, 312–321. [Google Scholar] [CrossRef] [PubMed]
- Ren, W.; Medeiros, N.; Warneford-Thomson, R.; Wulfridge, P.; Yan, Q.; Bian, J.; Sidoli, S.; Garcia, B.A.; Skordalakes, E.; Joyce, E.; et al. Disruption of ATRX-RNA interactions uncovers roles in ATRX localization and PRC2 function. Nat. Commun. 2020, 11, 2219. [Google Scholar] [CrossRef]
- Gill, P.; Kim, E.; Chua, T.C.; Clifton-Bligh, R.J.; Nahm, C.B.; Mittal, A.; Gill, A.J.; Samra, J.S. MiRNA-3653 Is a Potential Tissue Biomarker for Increased Metastatic Risk in Pancreatic Neuroendocrine Tumours. Endocr. Pathol. 2019, 30, 128–133. [Google Scholar] [CrossRef]
- Cai, K.; Wang, Y.; Bao, X. MiR-106b promotes cell proliferation via targeting RB in laryngeal carcinoma. J. Exp. Clin. Cancer Res. 2011, 30, 73. [Google Scholar] [CrossRef]
- Bandi, N.; Vassella, E. miR-34a and miR-15a/16 are co-regulated in non-small cell lung cancer and control cell cycle progression in a synergistic and Rb-dependent manner. Mol. Cancer 2011, 10, 55–55. [Google Scholar] [CrossRef]
- Ohtsuka, M.; et al. H19 Noncoding RNA, an Independent Prognostic Factor, Regulates Essential Rb-E2F and CDK8-beta-Catenin Signaling in Colorectal Cancer. EBioMedicine 2016, 13, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.-Z.; Cui, H.-P.; Lv, H.-J.; Feng, L. Knockdown of lncRNA PVT1 inhibits retinoblastoma progression by sponging miR-488-3p. Biomed. Pharmacother. 2019, 112, 108627. [Google Scholar] [CrossRef] [PubMed]
- Fang, D.; Ou, X.; Sun, K.; Zhou, X.; Li, Y.; Shi, P.; Zhao, Z.; He, Y.; Peng, J.; Xu, J. m6A modification–mediated lncRNA TP53TG1 inhibits gastric cancer progression by regulating CIP2A stability. Cancer Sci. 2022, 113, 4135–4150. [Google Scholar] [CrossRef] [PubMed]
- Idogawa, M.; et al. Prognostic Effect of Long Noncoding RNA NEAT1 Expression Depends on p53 Mutation Status in Cancer. J. Oncol. 2019, 2019, 4368068. [Google Scholar] [PubMed]
- Zhou, Y.; Zhong, Y.; Wang, Y.; Zhang, X.; Batista, D.L.; Gejman, R.; Ansell, P.J.; Zhao, J.; Weng, C.; Klibanski, A. Activation of p53 by MEG3 Non-coding RNA. J. Biol. Chem. 2007, 282, 24731–24742. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Zhang, Y.-Y.; Wu, Y.-Y.; Wang, X.-D.; Wan, L.-H.; Li, L.; Zhou, L.-M. Correlation of over-expressions of miR-21 and Notch-1 in human colorectal cancer with clinical stages. Life Sci. 2014, 106, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ai, F.; Li, X.; Tian, L.; Wang, X.; Shen, S.; Liu, F. MicroRNA-34a suppresses colorectal cancer metastasis by regulating Notch signaling. Oncol. Lett. 2017, 14, 2325–2333. [Google Scholar] [CrossRef] [PubMed]
- Guessous, F.; Zhang, Y.; Kofman, A.; Catania, A.; Li, Y.; Schiff, D.; Purow, B.; Abounader, R. microRNA-34a is tumor suppressive in brain tumors and glioma stem cells. Cell Cycle 2010, 9, 1031–1036. [Google Scholar] [CrossRef]
- Cai, H.; Yao, J.; An, Y.; Chen, X.; Chen, W.; Wu, D.; Luo, B.; Yang, Y.; Jiang, Y.; Sun, D.; et al. LncRNA HOTAIR acts as competing endogenous RNA to control the expression of Notch3 via sponging miR-613 in pancreatic cancer. Oncotarget 2017, 8, 32905–32917. [Google Scholar] [CrossRef]
- He, R.; Zhang, W.; Chen, S.; Liu, Y.; Yang, W.; Li, J. Transcriptional Profiling Reveals the Regulatory Role of DNER in Promoting Pancreatic Neuroendocrine Neoplasms. Front. Genet. 2020, 11, 587402. [Google Scholar] [CrossRef]
- Yu, Y.; et al. MicroRNA-21 induces stemness by downregulating transforming growth factor beta receptor 2 (TGFbetaR2) in colon cancer cells. Carcinogenesis 2012, 33, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; et al. miR-34a screened by miRNA profiling negatively regulates Wnt/beta-catenin signaling pathway in Aflatoxin B1 induced hepatotoxicity. Sci. Rep. 2015, 5, 16732. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; et al. Overexpression of long non-coding RNA HOTAIR leads to chemoresistance by activating the Wnt/beta-catenin pathway in human ovarian cancer. Tumour Biol. 2016, 37, 2057–2065. [Google Scholar] [CrossRef]
- Ling, H.; Spizzo, R.; Atlasi, Y.; Nicoloso, M.; Shimizu, M.; Redis, R.S.; Nishida, N.; Gafà, R.; Song, J.; Guo, Z.; et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 2013, 23, 1446–1461. [Google Scholar] [CrossRef]
- Wei, Y.L.; et al. LncNEN885 inhibits epithelial-mesenchymal transition by partially regulation of Wnt/beta-catenin signalling in gastroenteropancreatic neuroendocrine neoplasms. Cancer Sci. 2018, 109, 3139–3148. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; et al. miR-892b Silencing Activates NF-kappaB and Promotes Aggressiveness in Breast Cancer. Cancer Res. 2016, 76, 1101–1111. [Google Scholar] [CrossRef] [PubMed]
- Iliopoulos, D.; Jaeger, S.A.; Hirsch, H.A.; Bulyk, M.L.; Struhl, K. STAT3 Activation of miR-21 and miR-181b-1 via PTEN and CYLD Are Part of the Epigenetic Switch Linking Inflammation to Cancer. Mol. Cell 2010, 39, 493–506. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Liu, S.; Cai, G.; Kong, L.; Zhang, T.; Ren, Y.; Wu, Y.; Mei, M.; Zhang, L.; Wang, X. Long Non Coding RNA MALAT1 Promotes Tumor Growth and Metastasis by inducing Epithelial-Mesenchymal Transition in Oral Squamous Cell Carcinoma. Sci. Rep. 2015, 5, 15972. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Wang, X.; Zheng, L.; Li, D.; Liu, Z.; Teng, L. The lncRNA NEAT1 Inhibits miRNA-216b and Promotes Colorectal Cancer Progression by Indirectly Activating YY1. J. Oncol. 2022, 2022, 8130132. [Google Scholar] [CrossRef]
- Lee, Y.S.; Kim, H.; Kim, H.W.; Lee, J.-C.; Paik, K.-H.; Kang, J.; Kim, J.; Yoon, Y.-S.; Han, H.-S.; Sohn, I.; et al. High Expression of MicroRNA-196a Indicates Poor Prognosis in Resected Pancreatic Neuroendocrine Tumor. Medicine 2015, 94, e2224. [Google Scholar] [CrossRef]
- Døssing, K.B.V.; Kjær, C.; Vikeså, J.; Binderup, T.; Knigge, U.; Culler, M.D.; Kjær, A.; Federspiel, B.; Friis-Hansen, L. Somatostatin Analogue Treatment Primarily Induce miRNA Expression Changes and Up-Regulates Growth Inhibitory miR-7 and miR-148a in Neuroendocrine Cells. Genes 2018, 9, 337. [Google Scholar] [CrossRef] [PubMed]
- Jo, H.; Park, Y.; Kim, J.; Kwon, H.; Kim, T.; Lee, J.; Pyun, J.-C.; Lee, M.; Yun, M. Elevated miR-16-5p induces somatostatin receptor 2 expression in neuroendocrine tumor cells. PLOS ONE 2020, 15, e0240107. [Google Scholar] [CrossRef] [PubMed]
- Lai, X.N.; et al. MiRNAs and LncRNAs: Dual Roles in TGF-beta Signaling-Regulated Metastasis in Lung Cancer. Int. J. Mol. Sci. 2020, 21. [Google Scholar]
- Li, Q.; et al. MiR-21/Smad 7 signaling determines TGF-beta1-induced CAF formation. Sci. Rep. 2013, 3, 2038. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Li, M.; Li, J.; Lv, Y. Inhibition mechanism of lung cancer cell metastasis through targeted regulation of Smad3 by miR-15a. Oncol. Lett. 2019, 19, 1516–1522. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; et al. A Transforming Growth Factor-beta and H19 Signaling Axis in Tumor-Initiating Hepatocytes That Regulates Hepatic Carcinogenesis. Hepatology 2019, 69, 1549–1563. [Google Scholar] [CrossRef] [PubMed]
- Vijayaraghavan, J.; Maggi, E.C.; Crabtree, J.S. miR-24 regulates menin in the endocrine pancreas. Am. J. Physiol. Metab. 2014, 307, E84–E92. [Google Scholar] [CrossRef]
- Yan, X., H. Jia, and J. Zhao, LncRNA MEG3 attenuates the malignancy of retinoblastoma cells through inactivating PI3K /Akt/mTOR signaling pathway. Exp. Eye Res. 2023, 226, 109340. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; et al. miR-144/451 Promote Cell Proliferation via Targeting PTEN/AKT Pathway in Insulinomas. Endocrinology 2015, 156, 2429–2439. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Ding, L.; Bennewith, K.L.; Tong, R.T.; Welford, S.M.; Ang, K.K.; Story, M.; Le, Q.-T.; Giaccia, A.J. Hypoxia-Inducible mir-210 Regulates Normoxic Gene Expression Involved in Tumor Initiation. Mol. Cell 2009, 35, 856–867. [Google Scholar] [CrossRef]
- Blázquez-Encinas, R.; Moreno-Montilla, M.T.; García-Vioque, V.; Gracia-Navarro, F.; Alors-Pérez, E.; Pedraza-Arevalo, S.; Ibáñez-Costa, A.; Castaño, J.P. The uprise of RNA biology in neuroendocrine neoplasms: altered splicing and RNA species unveil translational opportunities. Rev. Endocr. Metab. Disord. 2022, 24, 267–282. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Ma, J.; Hua, X. Epigenetic regulation by the menin pathway. Endocrine-Related Cancer 2017, 24, T147–T159. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Hua, X.; Jin, G. Menin regulates endocrine diseases by controlling histone modification and gene transcription. Ann. d’Endocrinologie 2008, 69, 426–432. [Google Scholar] [CrossRef]
- Ehsanullah, S.; A Trikalinos, N. Synchronous AML and pancreatic neuroendocrine neoplasm, both successfully treated with somatostatin analogs and decitabine. Endocr. Oncol. 2022, 2, K1–K4. [Google Scholar] [CrossRef] [PubMed]
- Pollard, J.H.; Menda, Y.; Zamba, K.; Madsen, M.; O’Dorisio, M.S.; O’Dorisio, T.; Bushnell, D. Potential for Increasing Uptake of Radiolabeled 68Ga-DOTATOC and 123I-MIBG in Patients with Midgut Neuroendocrine Tumors Using a Histone Deacetylase Inhibitor Vorinostat. Cancer Biotherapy Radiopharm. 2021, 36, 632–641. [Google Scholar] [CrossRef]
- Doss, H.H.; Jones, S.F.; Infante, J.R.; Spigel, D.R.; Willcutt, N.; Lamar, R.; Barton, J.; Keegan, M.; Burris, H.A. A phase I trial of romidepsin in combination with gemcitabine in patients with pancreatic and other advanced solid tumors. J. Clin. Oncol. 2008, 26, 2567–2567. [Google Scholar] [CrossRef]
- Jin, N.; Lubner, S.J.; Mulkerin, D.L.; Rajguru, S.; Carmichael, L.; Chen, H.; Holen, K.D.; LoConte, N.K. A Phase II Trial of a Histone Deacetylase Inhibitor Panobinostat in Patients With Low-Grade Neuroendocrine Tumors. Oncol. 2016, 21, 785–786g. [Google Scholar] [CrossRef]
- Balasubramaniam, S.; Redon, C.E.; Peer, C.J.; Bryla, C.; Lee, M.-J.; Trepel, J.B.; Tomita, Y.; Rajan, A.; Giaccone, G.; Bonner, W.M.; et al. Phase I trial of belinostat with cisplatin and etoposide in advanced solid tumors, with a focus on neuroendocrine and small cell cancers of the lung. Anti-Cancer Drugs 2018, 29, 457–465. [Google Scholar] [CrossRef]
- Katti, A.; Diaz, B.J.; Caragine, C.M.; Sanjana, N.E.; Dow, L.E. CRISPR in cancer biology and therapy. Nat. Rev. Cancer 2022, 22, 259–279. [Google Scholar] [CrossRef]
- A English, K.; Thakker, R.V.; E Lines, K. The role of DNA methylation in human pancreatic neuroendocrine tumours. Endocr. Oncol. 2023, 3, e230003. [Google Scholar] [CrossRef]
- O’neill, H.; Lee, H.; Gupta, I.; Rodger, E.J.; Chatterjee, A. Single-Cell DNA Methylation Analysis in Cancer. Cancers 2022, 14, 6171. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, T.; et al. STC-1 Cells, in The Impact of Food Bioactives on Health: in vitro and ex vivo models, K. Verhoeckx, et al., Editors. 2015: Cham (CH). p. 211-220.
- Zeng, Z.; Wong, C.J.; Yang, L.; Ouardaoui, N.; Li, D.; Zhang, W.; Gu, S.; Zhang, Y.; Liu, Y.; Wang, X.; et al. TISMO: syngeneic mouse tumor database to model tumor immunity and immunotherapy response. Nucleic Acids Res. 2021, 50, D1391–D1397. [Google Scholar] [CrossRef]
- Ney, A.; Canciani, G.; Hsuan, J.J.; Pereira, S.P. Modelling Pancreatic Neuroendocrine Cancer: From Bench Side to Clinic. Cancers 2020, 12, 3170. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Sandilos, G.; Williamson, J.; Emery, R.; Platoff, R.; Joneja, U.; Acharya, N.K.; Lin, A.; Badach, J.; Zilberman, B.; et al. Novel treatment strategy of targeting epigenetic dysregulation in pancreatic neuroendocrine tumors. Surgery 2023, 173, 1045–1051. [Google Scholar] [CrossRef]
- Wang, H.; Peng, R.; Wang, J.; Qin, Z.; Xue, L. Circulating microRNAs as potential cancer biomarkers: the advantage and disadvantage. Clin. Epigenetics 2018, 10, 59. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; et al. A novel circulating miRNA-based signature for the early diagnosis and prognosis prediction of non-small-cell lung cancer. J. Clin. Lab. Anal. 2020, 34, e23505. [Google Scholar] [CrossRef]
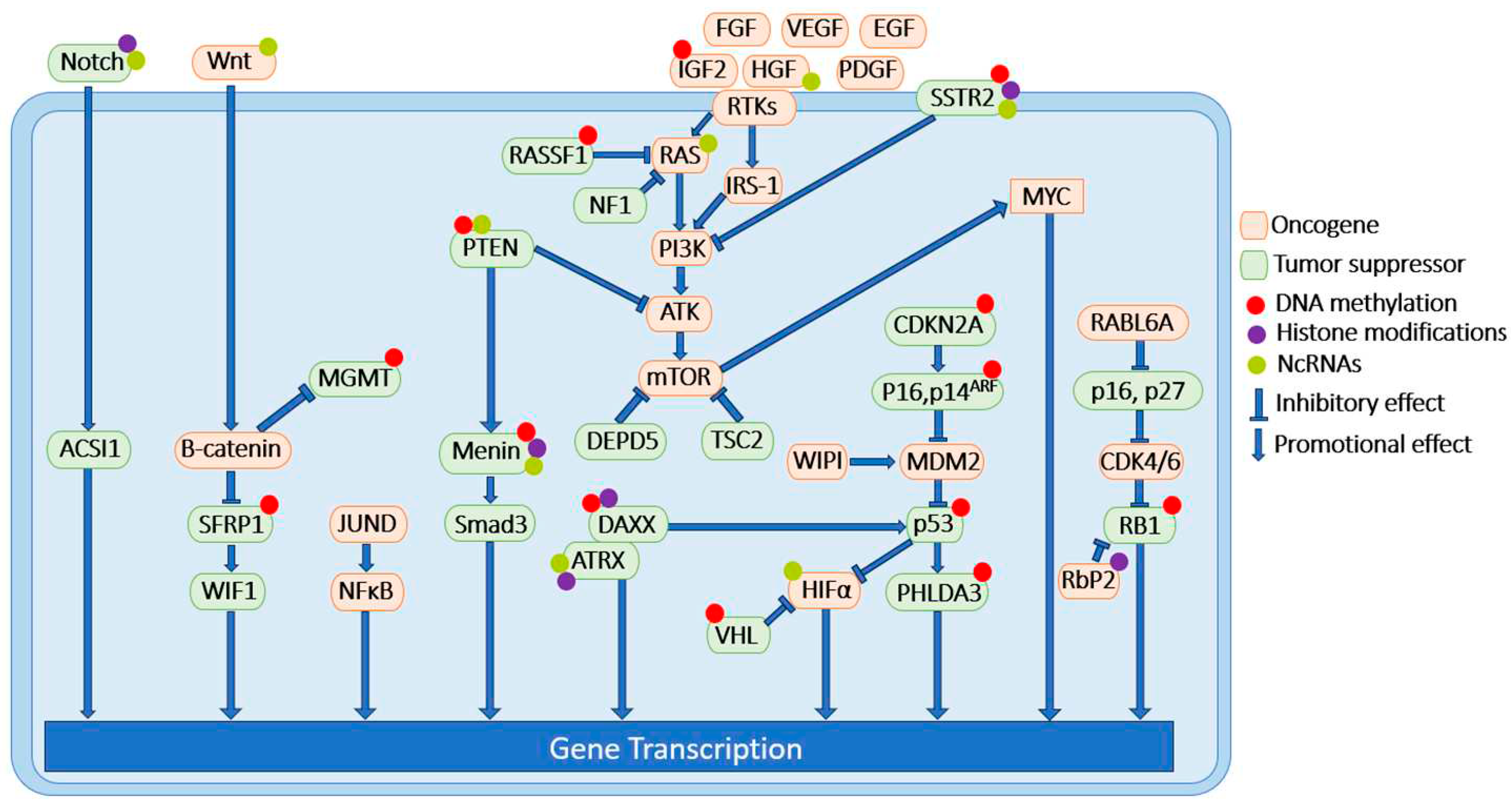
| Signaling pathway | Signaling molecules | DNA methylation status of promoter region | Experimental systems |
|---|---|---|---|
| MEN1 | MEN1 | Hypermethylated | human PNET samples [141] |
| PTEN/PI3K/AKT/mTOR/c-Myc/TSC/RTK1 | PTEN | Hypermethylated | human PNET samples [41] |
| TSC | no change | human PNET samples [41] | |
| IGF2 | hypermethylated | human PNET samples [142] | |
| HIF-1α/VHL | VHL | Hypermethylated | human PNET samples [50] |
| RAS/MAPK/NF1 | RASSF1 | Hypermethylated | human PNET samples [70] |
| ALT/DAXX/ATRX | DAXX | Hypermethylated | human PNET samples [78] |
| CDKN2A/RB1 | CDKN2A | Hypermethylated | human PNET samples [88] |
| P16, P14ARF | Hypermethylated | human PNET samples [143] | |
| P27 | no change | human PNET samples [90] | |
| RB1 | Hypermethylated | human PNET samples [41] | |
| P53 | P53 | Hypermethylated | human PNET samples [41] |
| PHLDA3 | Hypermethylated | human PNET samples [98] | |
| Wnt/β Catenin/MGMT | SFRP1 | Hypermethylated | BON-1, and QGP-1 cell lines [121] |
| WIF1 | no change | BON-1, and QGP-1 cell lines [121] | |
| MGMT | MGMT-promoter methylation status correlates with chemoresistance in well-differentiated PNET. | PNET patient samples[122,123] | |
| SSTR | SSTR2 | SSTR2 promoter is hypermethylated in PNETs comparing to non-NET tissue and is inversely correlated with SSTR2 protein expression. | human PNET samples BON-1, and QGP-1 cell lines xenograft mouse model [133] |
| Signaling pathway | Signaling molecules | Histone modification status | Experimental systems |
|---|---|---|---|
| MEN1 | MEN1 | Loss of menin causes H3K4me3 loss and sporadic PNET tumors. | PNET patient samples [178] |
| PTEN/PI3K/AKT/mTOR/ c-Myc/TSC/RTK1 |
IGF2 | Genome-wide studies of H3K4 methylation in pancreatic islets indicate that loss of MEN1 alters the epigenetic landscape of its target genes such as insulin like growth factor binding protein 2 (Igf2bp2), p18ink4c (CDKN2C) and p27kip1 (CDKN1B). | Pancreatic islets from MEN1-deficient mice [214] |
| DAXX/ATRX/ALT |
DAXX ATRX |
DAXX and TRX form a histone chaperone complex to deposit histone variant H3.3 at the telomeres and pericentric heterochromatin regions of the genome. They are frequently mutated in PNET samples. | Human PNET samples, Hela cells [215,216] |
| CDKN2A/RB1 | RB1 | Histone demethylase retinoblastoma binding protein 2 (Rbp2) was found overexpressed in PNET tumors. Aberrant expression of Rbp2 altered histone demethylation and contributed to PNET pathogenesis. | PNET patient samples, βlox5 cell, H727 cell, QGP-1 cell [191] |
| Notch | Notch1 | HDAC inhibitor causes increased Notch 1 expression in tumor cells and mouse tumor xenograft[108,217] | BON-1 cells [217], carcinoid cancer cells and mouse tumor xenograft [108] |
| SSTR2 | SSTR2 | Histone acetylation present on SSTR2. In addition, the combination treatment of HDACi (VPA) and camptothecin-somatostatin conjugate significantly reduced tumor growth comparing to monotherapies. | BON-1 and QGP-1 cells [208] [209], BON-1 xenograft mouse model [210] |
| Signaling pathway | Signaling molecules | Non-coding RNA status | Experimental systems |
|---|---|---|---|
| MEN1 | MEN1 | Menin negatively regulates miR-24-1 in a negative feedback loop manner. | BON-1 cells [230,231] |
| MiR-24 negatively regulates menin in endocrine pancreas. | MIN6 cells, βlox5 cells; floxed MEN1 mouse model [282] | ||
| Menin upregulates expression of MEG3. | Mouse insulinoma cells [229] | ||
| PTEN/PI3K/AKT/mTOR/c-Myc/ TSC/PRK1 | PTEN | MEG3 causes decreased p-PI3K, p-AKT, p-mTOR, and smaller tumor size. | human retinoblastoma cells [283] |
| PI3K | miR-144 causes decreased PTEN. | and xenograft mouse model [284] | |
| AKT | MiR-144 correlated with increased p-AKT. | MIN6 cells [236] | |
| mTOR | IncRNA H19 causes increased PI3K-AKT and PNET progression. | Human insulinoma samples, QGP-1, PNET primary Cells.QGP-1 xenograft model [237] | |
| HGF/MET | MEG3 downregulates c-MET in PNET. | MIN6 cells, mouse, PNET patient samples [229]. | |
| HIF-1α/VHL | HIF-1α | MiR-210 expression is positively correlated with PNET progression and was shown to regulate colorectal adenocarcinoma progression through HIF1α. | PNET patient samples [242] |
| FaDu head & neck cancer cell line, SU86.76 pancreatic cancer cell line, Xenograft mouse model [244,285]. | |||
| RAS/MAPK/NF1 | RAS | MiR-431 promotes PNET progression by silencing DAB21P, resulting in the activation of RAS pathway. | QGP-1 cell line, and xenograft mouse model [249]. |
| ALT/DAXX/ATRX | ATRX | ATRX negatively regulates miR-3653, which might serve as a risk factor of metastatic disease in PNET. | Microarray differential expression of human PNET tissue samples [253]. |
| Notch | Notch1,2,3, ASCL1 | LncRNA XLOC_221242 is positively correlated with Notch/Wnt signaling. | PNET patient samples [265,286]. |
| Wnt/β Catenin |
Wnt, β-Catenin, SFRP1, WIF1 |
LncNEN885 negatively regulates Wnt/β-catenin signaling, leading to reduction of EMT in PNET. LncNEN885 is negatively correlated with PNET progression. | BON-1 cells, and PNET patient samples [270]. |
| SSTR | SSTR2 | Upregulation of miR-16-5p induces SSTR2 expression. | INS-1 cell line [277] |
| Drug name | Drug target | Targeted disease | Trial in PNET | Status of trial |
|---|---|---|---|---|
| Azacitidine | DNMTi | AML, CML and MDS | NR | NA |
| 5-Aza-2’-deoxycytidine | DNMTi | AML, CML and MDS | Trial on synchronous AML and PNET | Treatment was successful with combination of somatostatin analogs and decitabine, but with severe side effects [289]. |
| Tazemetostat | HDMi | Advance epithelioid sarcoma | NR | NA |
| Enasidenib | HDMi | AML | NR | NA |
| Vorinostat | HDACi | CTCL | Pilot-imaging study to test efficacy of vorinostat on radionuclide uptake. | Statistically significant increase of radionuclide uptake was observed [290]. |
| Romidepsin | HDACi | CTCL and PTCL | Phase I trial of romidepsin in patients with pancreatic and other advanced solid tumors. | Stable disease status was observed when combined with treatment of gemcitabine [291]. |
| Panobinostat | HDACi | MM | Phase II-trial against low-Grade PNET | 15 patients were in the trial. No response was observed [292]. |
| Belinostat | HDACi | PTCL | Phase I-trial against NET and small cell lung cancer | Partial response was observed when patients were treated with belinostat combined with cisplatin and etoposide [293]. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





