Submitted:
24 December 2023
Posted:
27 December 2023
You are already at the latest version
Abstract
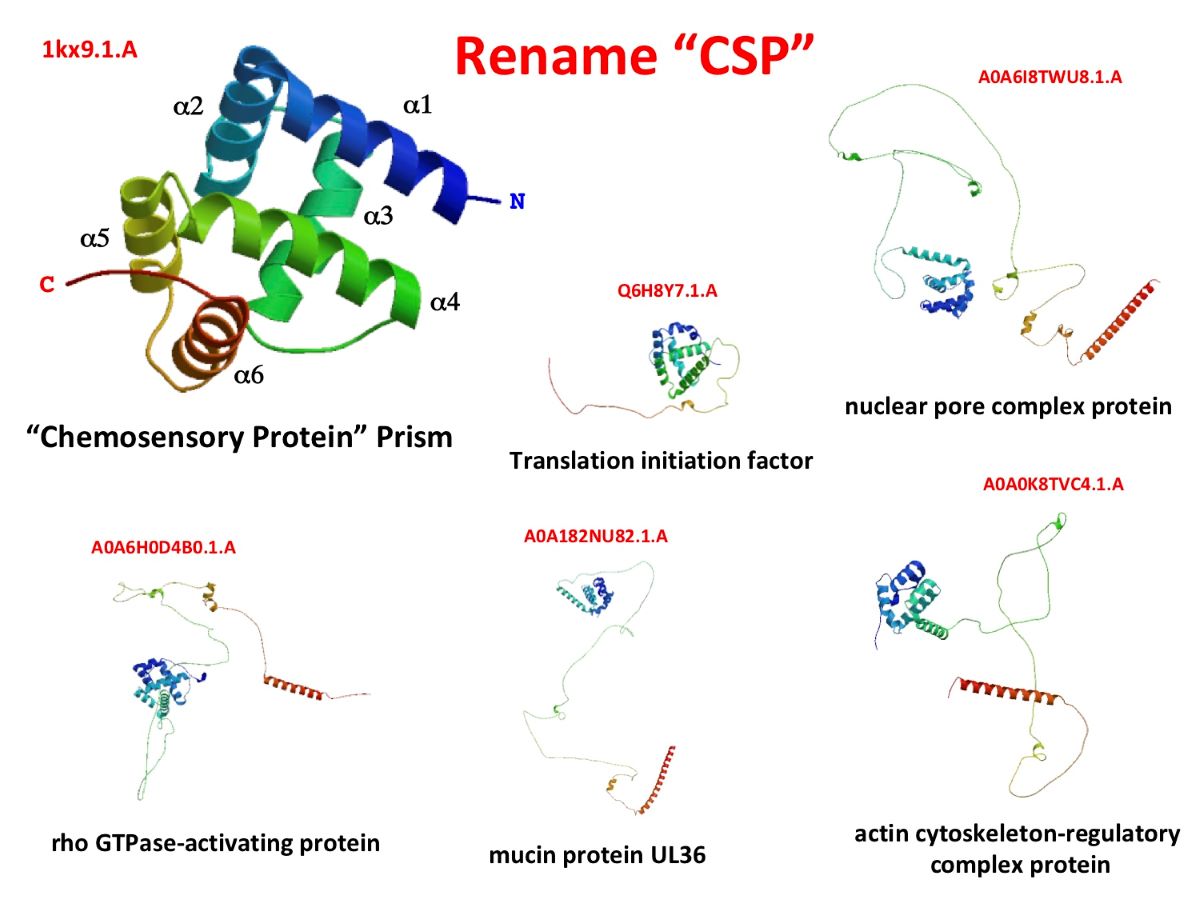
Keywords:
1. Definition
2. Protein structure and RNA sequence adaptability
3. Expression profiling in development, organisms, and tissues
4. The evolution and diversity of CSP genes
5. Multiple functions and binding properties
5. CSPs’ function in regulating gene transcription and nucleotide binding
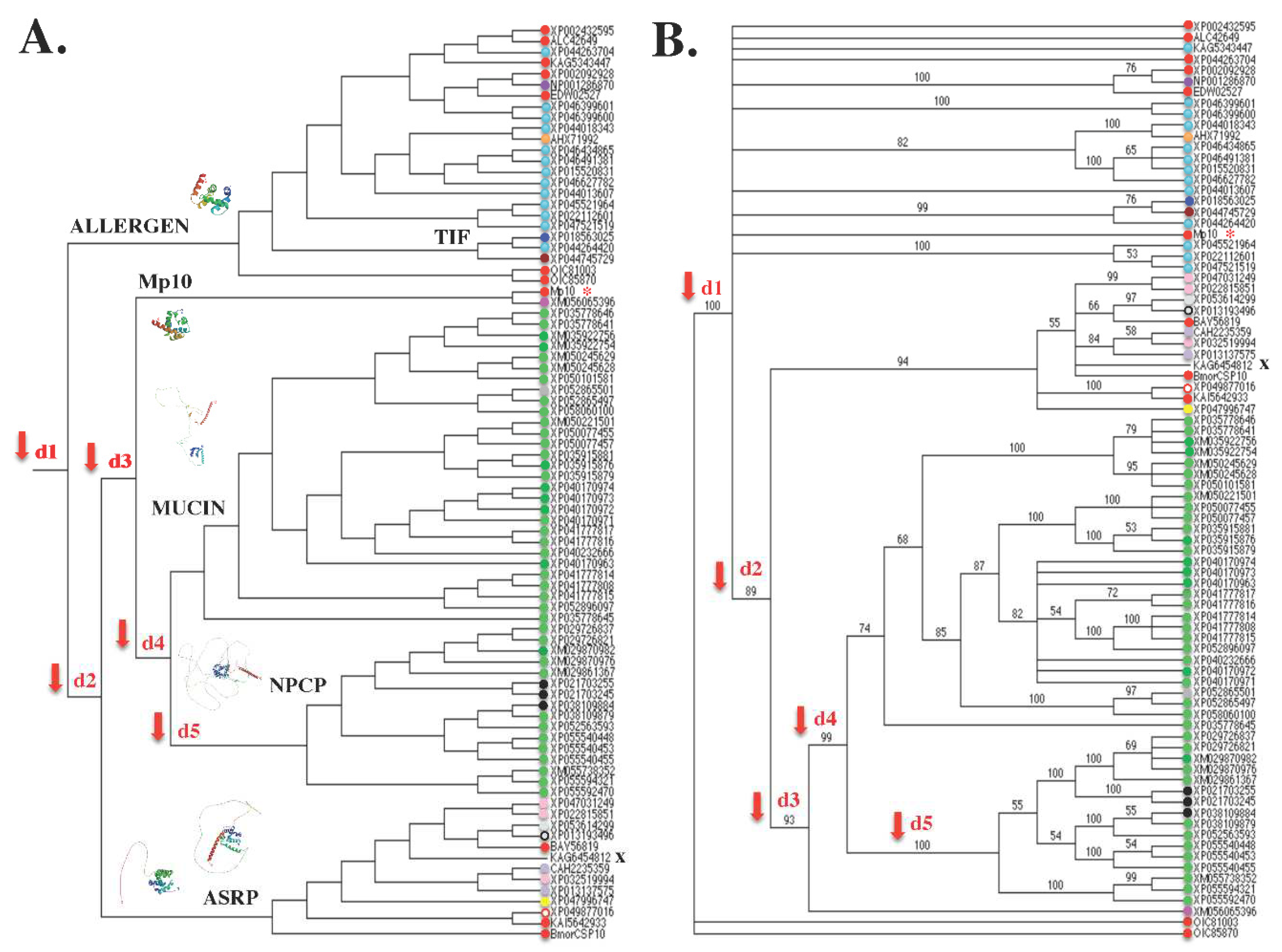
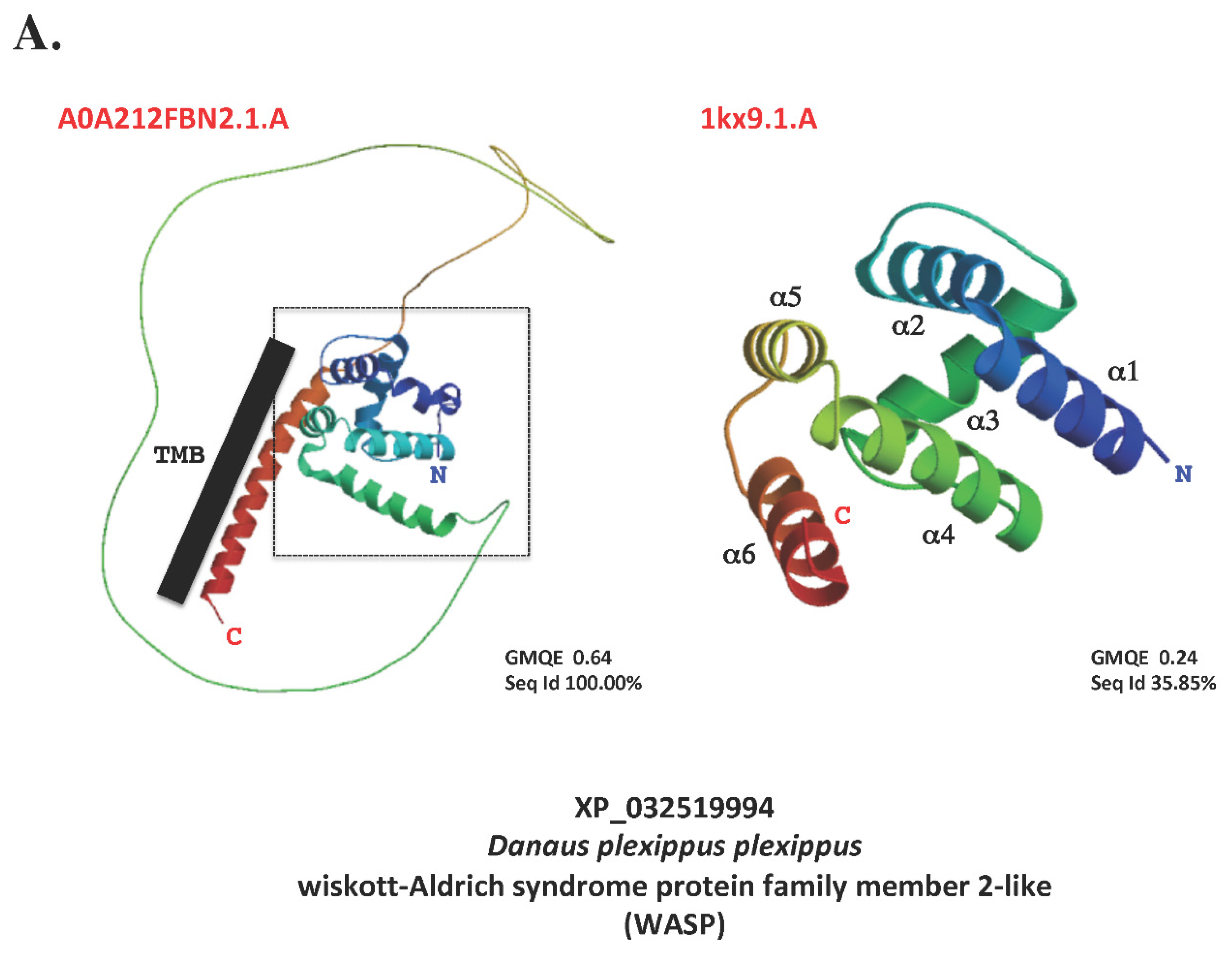
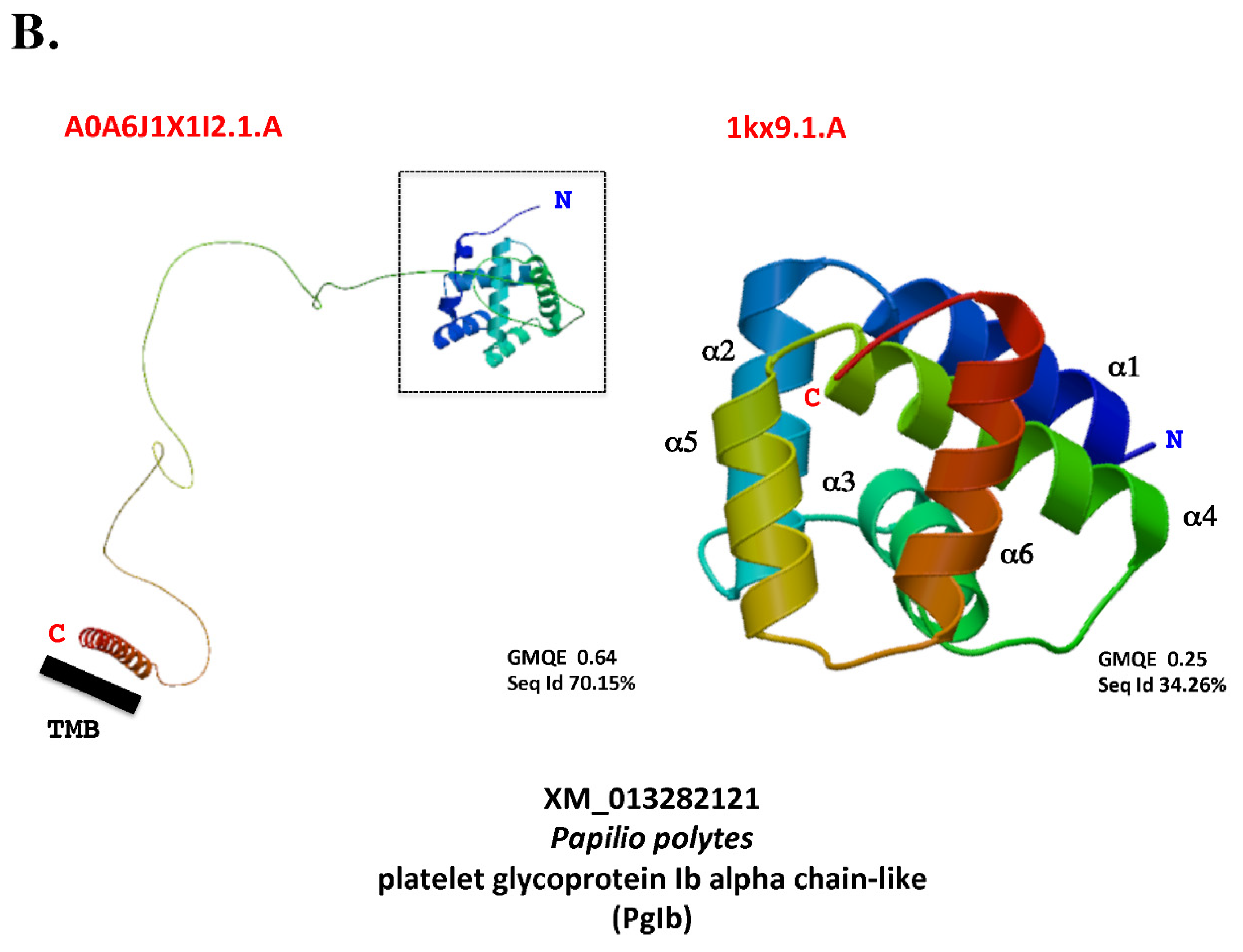
7. Terminology: function or structure?
8. Evolutionary Networks and Associated Diseases
9. Last reflections and conclusions
Supplementary Materials
Acknowledgements
References
- Abanes-De Mello, A.; Sun, Y.L.; Aung, S.; Pogliano, K. A cytoskeleton-like role for the bacterial cell wall during engulfment of the Bacillus subtilis forespore. Genes & Development 2002, 16, 3253–3264. [Google Scholar] [CrossRef]
- Abraham, D.; Löfstedt, C.; Picimbon, J.-F. Molecular characterization and evolution of pheromone binding protein genes in Agrotis moths. Insect Biochemistry and Molecular Biology 2005, 35, 1100–1111. [Google Scholar] [CrossRef]
- Adams, F.G.; Trappetti, C.; Waters, J.K.; Zang, M.; Brazel, E.B.; Paton, J.C.; Snel, M.F.; Eijkelkamp, B.A. To make or take: bacterial lipid homeostasis during infection. mBio 2021, 12, e00928-21. [Google Scholar] [CrossRef] [PubMed]
- Albert, B.; Kos-Braun, I.C.; Henras, A.K.; Dez, C.; Rueda, M.P.; Zhang, X.; Gadal, O.; Kos, M.; Shore, D. A ribosome assembly stress response regulates transcription to maintain proteome homeostasis. eLife 2019, 8, e45002. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.; Hrithik, M.T.H.; Roy, M.C.; Bode, H.; Kim, Y. Phurelipids, produced by the entomopathogenic bacteria, Photorhabdus, mimic juvenile hormone to suppress insect immunity and immature development. Journal of Invertebrate Pathology 2022, 193, 107799. [Google Scholar] [CrossRef] [PubMed]
- Allen, P.E.; Martinez, J.J. Modulation of host lipid pathways by pathogenic intracellular bacteria. Pathogens 2020, 9, 614. [Google Scholar] [CrossRef] [PubMed]
- Angeli, S.; Ceron, F.; Scaloni, A.; Monti, M.; Monteforti, G.; Minnocci, A.; Petacchi, R.; Pelosi, P. Purification, structural characterization, cloning and immunocytochemical localization of chemoreception proteins from Schistocerca gregaria. European Journal of Biochemistry 1999, 262, 745–754. [Google Scholar] [CrossRef] [PubMed]
- Anton, S.; Rossler, W. Plasticity and modulation of olfactory circuits in insects. Cell and Tissue Research 2021, 383, 149–164. [Google Scholar] [CrossRef] [PubMed]
- Badiaga, S.; Brouqui, P. Human louse-transmitted infectious diseases. Clinical Microbiology and Infection 2012, 18, 332–337. [Google Scholar] [CrossRef] [PubMed]
- Bajguz, A.; Piotrowska-Niczyporuk, A. Biosynthetic pathways of hormones in plants. Metabolites 2023, 13, 884. [Google Scholar] [CrossRef] [PubMed]
- Barák, I.; Muchová, K. The role of lipid domains in bacterial cell processes. International Journal of Molecular Sciences 2013, 14, 4050–4065. [Google Scholar] [CrossRef] [PubMed]
- Bendary, M.M.; Abdel-Hamid, M.I.; Alshareef, W.A.; Alshareef, H.M.; Mosbah, R.A.; Omar, N.N.; Al-Sanea, M.M.; Alhomrani, M.; Alamri, A.S.; Moustafa, W.H. Comparative analysis of human and animal E. coli: serotyping, antimicrobial resistance, and virulence gene profiling. Antibiotics (Basel) 2022, 11, 552. [Google Scholar] [CrossRef] [PubMed]
- Beukes, N. Biogeochemistry: early options in photosynthesis. Nature 2004, 431, 522–523. [Google Scholar] [CrossRef] [PubMed]
- Bian, H.X.; Chen, D.B.; Zheng, X.X.; Ma, H.F.; Li, Y.P.; Li, Q.; Xia, R.X.; Wang, H.; Jiang, Y.R.; Liu, Y.Q.; Qin, L. Transcriptomic analysis of the prothoracic gland from two lepidopteran insects, domesticated silkmoth Bombyx mori and wild silkmoth Antheraea pernyi. Scientific Reports 2019, 9, 5313. [Google Scholar] [CrossRef] [PubMed]
- Blackman, R.L.; Takada, H.; Kawakami, K. Chromosomal rearrangement involved in insecticide resistance of Myzus persicae. Nature 1978, 271, 450–452. [Google Scholar] [CrossRef]
- Blank, H.M.; Maitra, N.; Polymenis, M. Lipid biosynthesis: When the cell cycle meets protein synthesis? Cell Cycle 2017a, 16, 905–906. [Google Scholar] [CrossRef]
- Blank, H.M.; Perez, R.; He, C.; Maitra, N.; Metz, R.; Hill, J.; Lin, Y.; Johnson, C.D.; Bankaitis, V.A.; Kennedy, B.K.; Aramayo, R.; Polymenis, M. Translational control of lipogenic enzymes in the cell cycle of synchronous, growing yeast cells. EMBO Journal 2017b, 36, 487–502. [Google Scholar] [CrossRef]
- Blobel, G. Protein targeting (Nobel lecture). ChemBioChem 2000, 1, 86–102. [Google Scholar] [CrossRef] [PubMed]
- Blomquist, G.J., Tittiger, C. and Jurenka, R. (2018) Cuticular hydrocarbons and pheromones of arthropods. Oils and Lipids: Diversity, Origin, Chemistry and Fate. (ed. H. Wilkes) Hydrocarbons, Handbook of Hydrocarbon and Lipid Microbiology, pp. 1–32, Springer Cham, Swizerland. [CrossRef]
- Bos, J.I.; Prince, D.; Pitino, M.; Maffei, M.E.; Win, J.; Hogenhout, S.A. A functional genomics approach identifies candidate effectors from the aphid species Myzus persicae (green peach aphid). PLoS Genetics 2010, 6, e1001210. [Google Scholar] [CrossRef] [PubMed]
- Browning, D.; Busby, S. The regulation of bacterial transcription initiation. Nature Reviews Microbiology 2004, 2, 57–65. [Google Scholar] [CrossRef]
- Cahoon, E.B.; Lindqvist, Y.; Schneider, G.; Shanklin, J. Redesign of soluble fatty acid desaturases from plants for altered substrate specificity and double bond position. Proceedings of the National Academy of Sciences 1997, 94, 4872–4877. [Google Scholar] [CrossRef] [PubMed]
- Campanacci, V.; Lartigue, A.; Hällberg, B.M.; Jones, T.A.; Giuici-Orticoni, M.T.; Tegoni, M.; Cambillau, C. Moth chemosensory protein exhibits drastic conformational changes and cooperativity on ligand binding. Proceedings of the National Academy of Sciences of the United States of America 2003, 100, 5069–5074. [Google Scholar] [CrossRef] [PubMed]
- Cayre, M.; Strambi, C.; Strambi, A. Neurogenesis in an adult insect brain and its hormonal control. Nature 1994, 368, 57–59. [Google Scholar] [CrossRef]
- Celorio-Mancera, M.L.; Sundmalm, S.M.; Vogel, H.; Rutishauser, D.; Ytterberg, A.J.; Zubarv, R.A.; Janz, N. Chemosensory proteins, major salivary factors in caterpillar mandibular glands. Insect Biochemistry and Molecular Biology 2012, 42, 796–805. [Google Scholar] [CrossRef] [PubMed]
- Cerone, M.; Smith, T.K. Desaturases: Structural and mechanistic insights into the biosynthesis of unsaturated fatty acids. IUBMB Life 2022, 74, 1036–1051. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, A.; Ashraf, M.Z.; Modlinger, R.; Synek, J.; Schlyter, F.; Roy, A. Unravelling the gut bacteriome of Ips (Coleoptera: Curculionidae: Scolytinae): identifying core bacterial assemblage and their ecological relevance. Scientific Reports 2020, 10, 18572. [Google Scholar] [CrossRef]
- Cheng-Guang, H.; Gualerzi, C.O. The ribosome as a switchboard for bacterial Stress response. Frontiers in Microbiology 2021, 11, 619038. [Google Scholar] [CrossRef]
- Cheslock, M.A.; Embers, M.E. Human bartonellosis: an underappreciated public health problem. Tropical Medicine and Infectious Disease 2019, 4, 69. [Google Scholar] [CrossRef] [PubMed]
- Choo, Y.M.; Xu, P.; Hwang, J.K.; Leal, W.S. Reverse chemical ecology approach for the identification of an oviposition attractant for Culex qinquefasciatus. Proceedings of the National Academy of Sciences of USA 2018, 115, 714–719. [Google Scholar] [CrossRef]
- Cramer, P. Organization and regulation of gene transcription. Nature 2019, 573, 45–54. [Google Scholar] [CrossRef]
- Cronmiller, E.; Toor, D.; Shao, N.C.; Kariyawasam, T.; Wang, M.H.; Lee, J.H. Cell wall integrity signaling regulates cell wall-related gene expression in Chlamydomonas reinhardtii. Scientific Reports 2019, 9, 12204. [Google Scholar] [CrossRef] [PubMed]
- Cui, N.; Lu, H.; Wang, T.; Zhang, W.; Kang, L.; Cui, F. Armet, an aphid effector protein, induces pathogen resistance in plants by promoting the accumulation of salicylic acid. Philosophical Transactions B of the Royal Society B: Biological Sciences 2019, 374, 20180314. [Google Scholar] [CrossRef] [PubMed]
- Damude, H.G.; Zhang, H.; Farrall, L.; Ripp, K.G.; Tomb, J.F.; Hollerbach, D.; Yadav, N.S. Identification of bifunctional delta12/omega3 fatty acid desaturases for improving the ratio of omega3 to omega6 fatty acids in microbes and plants. Proceedings of the National Academy of Sciences USA 2006, 103, 9446–9451. [Google Scholar] [CrossRef] [PubMed]
- de Carvalho, C.C.C.R.; Caramujo, M.J. The various roles of fatty acids. Molecules 2018, 23, 2583. [Google Scholar] [CrossRef] [PubMed]
- Delrue, C.; Speeckaert, M.M. Vitamin D and vitamin D-binding protein in health and disease. International Journal of Molecular Sciences 2023, 24, 4642. [Google Scholar] [CrossRef] [PubMed]
- den Hartig, L.J. Conjugated linoleic acid effects on cancer, obesity, and atherosclerosis: a review of pre-clinical and human trials with current perspectives. Nutrients 2019, 11, 370. [Google Scholar] [CrossRef]
- Dias, R.O.; Cardoso, C.; Pimentel, A.C.; Damasceno, T.F.; Ferreira, C.; Terra, W.R. The roles of mucus-forming mucins, peritrophins and peritrophins with mucin domains in the insect midgut. Insect Molecular Biology 2018, 27, 46–60. [Google Scholar] [CrossRef] [PubMed]
- Dibrova, D.V.; Galperin, M.Y.; Koonin, E.V.; Mulkidjanian, A.Y. Ancient systems of sodium/potassium homeostasis as predecessors of membrane bioenergetics. Biochemistry (Mosc) 2015, 80, 495–516. [Google Scholar] [CrossRef] [PubMed]
- Diez-Hermano, S.; Ganfornina, M.D.; Skerra, A.; Guttiérez, G.; Sanchez, D. An evolutionary perspective of the lipocalin protein family. Frontiers in Physiology 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Dillon, R.J.; Vennard, C.T.; Charnley, A.K. Pheromones: exploitation of gut bacteria in the locust. Nature 2000, 403, 851. [Google Scholar] [CrossRef] [PubMed]
- Diomandé, S.E.; Nguyen-The, C.; Guinebretière, M.H.; Broussole, V.; Brillard, J. Role of fatty acids in Bacillus environmental adaptation. Frontiers in Microbiology 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- DiRusso, C.C.; Black, P.N. Bacterial long chain fatty acid transport: gateway to a fatty acid-responsive signaling system. Journal of Biological Chemistry 2004, 279, 49563–49566. [Google Scholar] [CrossRef]
- Doudna, J.A.; Batey, R.T. Structural insights into the signal recognition particle. Annual Review of Biochemistry 2004, 73, 539–557. [Google Scholar] [CrossRef] [PubMed]
- Doré, H.; Guyet, U.; Leconte, J.; Farrant, G.K.; Alric, B.; Ratin, M.; Ostrowski, M.; Ferrieux, M.; Brillet-Guéguen, L.; Hoebeke, M.; Siltanen, J.; Le Corguillé, G.; Corre, E.; Wincker, P.; Scanlan, D.J.; Eveillard, D.; Partensky, F.; Garczarek, L. Differential global distribution of marine picocyanobacteria gene clusters reveals distinct niche-related adaptive strategies. The ISME journal 2023, 17, 720–732. [Google Scholar] [CrossRef]
- Dyanov, H.M., Lyozin, G.I., Dzitoeva, S.G. and Korochkin, L.I. (1994) A cDNA of Drosophila melanogaster ejaculatory bulb specific protein III (PEBme III). #AAA87058, DMU08281 (RNA source: imago).
- Einhorn, E. and Imler, J.L. (2019) Insect immunity; from systemic to chemosensory organs protection. Olfactory Concepts of Insect Control-Alternative to Insecticides (ed. J.F. Picimbon), pp. 205–229. Vol. 2 Springer Nature, Switzerland. [CrossRef]
- Elzinga, D.A.; De Vos, M.; Jander, G. Suppression of plant defenses by a Myzus persicae (green peach aphid) salivary effector protein. Molecular Plant Microbe Interactions 2014, 27, 747–756. [Google Scholar] [CrossRef] [PubMed]
- Farvis, M.S.; Ding, M.; Pan, A.; Sun, Q.; Chiuve, S.E.; Steffen, L.M.; Willett, W.C.; Hu, F.B. Dietary linoleic acid and risk of coronary heart disease: a systematic review and meta-analysis of prospective cohort studies. Circulation 2014, 130, 1568–1578. [Google Scholar] [CrossRef] [PubMed]
- Forêt, S.; Wanner, K.W.; Maleszka, R. Chemosensory proteins in the honeybee: Insights from the annotated genome, comparative analysis and expression profiling. Insect Biochemistry and Molecular Biology 2007, 37, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, Y.; Wada, K.; Kabuta, T. Lysosomal degradation of intracellular nucleic acids—multiple autophagic pathways. Journal of Biochemistry 2017, 161, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Georgescauld, F.; Song, Y.; Dautant, A. Structure, folding and stability of nucleoside diphosphate kinases. International Journal of Molecular Sciences 2020, 21, 6779. [Google Scholar] [CrossRef] [PubMed]
- Gestwicki, J.E.; Lamanna, A.C.; Harshey, R.M.; McCarter, L.L.; Kiessling, L.L.; Adler, J. Evolutionary conservation of Methyl-Accepting Chemotaxis Protein location in bacteria and Archaea. Journal of Bacteriology 2017, 182, 6499–6502. [Google Scholar] [CrossRef] [PubMed]
- Ghai, R.; Mizuno, C.M.; Picazo, A.; Camacho, A.; Rodriguez-Valera, F. Metagenomics uncovers a new group of low GC and ultra-small marine Actinobacteria. Scientific Reports 2013, 3, 2471. [Google Scholar] [CrossRef] [PubMed]
- González-Caballero, N.; Valenzuela, J.G.; Ribeiro, J.M.C.; Cuervo, P.; Brazil, R.P. Transcriptome exploration of the sex pheromone gland of Lutzomyia longipalpis (Diptera: Psychodidae: Phlebotominae). Parasites and Vectors 2013, 6, 56. [Google Scholar] [CrossRef] [PubMed]
- Gopikrishnan, V., Radhakrishnan, M., Krupakar, P., Manigundan, K., Abirami, B., and Reshma, S. (2023) Chapter 36 – Endosymbiotic interactions of actinobacteria with the insects. Microbial Symbionts: Function and Molecular Interactions on Host. (ed. D. Dharumadurai), pp. 645–658, Academic Press. [CrossRef]
- Guo, X.; Xuan, N.; Liu, G.X.; Xie, H.Y.; Lou, Q.N.; Arnaud, P.; Offmann, B.; Picimbon, J.F. An expanded survey of the moth PBP/GOBP clade in Bombyx mori: new insight into expression and functional roles. Frontiers in Physiology 2021. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Wang, X.; Ma, Z.; Xue, L.; Han, J.; Yu, D.; Kang, L. CSP and Takeout genes modulate the switch between attraction and repulsion during behavioral phase change in the migratory locust. PLoS Genetics 2011, 7, e1001291. [Google Scholar] [CrossRef]
- Hammoud, A.; Louni, M.; Baldé, M.C.; Beavogui, A.H.; Gautret, P.; Raoult, D.; Fenollar, F.; Misse, D.; Mediannikov, O. Molecular characterization and genetic diversity of haplogroup E human lice in Guinea. Microorganisms 2021, 9, 257. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Luan, Y.S. Horizontal transfer of small RNAs to and from plants. Frontiers in Plant Science 2015, 6, 1113. [Google Scholar] [CrossRef] [PubMed]
- Han, R.; Feng, X.Q.; Vollmer, W.; Stoodley, P.; Chen, J. Deciphering the adaption of bacterial cell wall mechanical integrity and turgor to different chemical or mechanical environments. Journal of Colloid and Interface Science 2023, 640, 510–520. [Google Scholar] [CrossRef] [PubMed]
- Harrison, E.; Brockhurst, M.A. Plasmid-mediated horizontal gene transfer is a coevolutionary process. Trends in Microbiology 2012, 20, 262–267. [Google Scholar] [CrossRef] [PubMed]
- Hastings, P.J.; Lupski, J.R.; Rosenberg, S.M.; Ira, G. Mechanisms of change in gene copy number. Nature Review Genetics 2009, 10, 551–564. [Google Scholar] [CrossRef] [PubMed]
- Heaton, N.S.; Randall, G. Multifaceted roles for lipids in viral infection. Trends in Microbiology 2011, 19, 368–375. [Google Scholar] [CrossRef] [PubMed]
- Heinisch, J.J.; Rodicio, R. Protein kinase C in fungi—more than just cell wall integrity. FEMS Microbiology Reviews 2018, 42, fux051. [Google Scholar] [CrossRef] [PubMed]
- Hilgemann, D.W. From a pump to a pore: How palytoxin opens the gates. Proceedings of the National Academy of Sciences 2003, 100, 386–388. [Google Scholar] [CrossRef] [PubMed]
- Ho, S. The molecular clock and estimating species divergence. Nature Education 2008, 1, 168. [Google Scholar]
- Hou, S.; Guo, L.; Xu, B.; Dong, H.; Zhang, S.; Fu, Y.; Shi, J.; Li, L.; Fu, J.; Shi, F.; Meng, Y.; Jin, Y. Trans-splicing facilitated by RNA pairing greatly expands sDscam isoform diversity but not hemophilic binding specificity. Science Advances 2022, 8. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Huang, H.; Hui, X.; Cheng, X.; White, A.P.; Zhao, Z.; Wang, Y. Distribution and evolution of Yersinia Leucine-Rich Repeat proteins. Infection and Immunity 2016, 84, 2243–2254. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Li, L.; Rong, Y.S. JiangShi (僵尸): a widely distributed Mucin-like protein essential for Drosophila development. G3 (Bethesda) 2022, 12, jkac126. [Google Scholar] [CrossRef]
- Hunter, D. (2019) Locusts in the world. Olfactory Concepts of Insect Control-Alternative to Insecticides. (ed. J.F. Picimbon), pp. 29–49. Vol. 1, Springer Nature, Switzerland. [CrossRef]
- Hunter, W., Martinez-Torres, D., Rahbe, Y., Sabater-Munoz, B., Stern, D., Tagu, D. and Wincker, P. (2004) An expressed sequence sequence tags database for the pea aphid Acyrthosiphum pisum #CN760202.
- Ibarra, A.; Hetzer, M.W. Nuclear pore proteins and the control of genome functions. Genes Development 2015, 29, 337–349. [Google Scholar] [CrossRef] [PubMed]
- Ikegami, A.; Honma, K.; Sharma, A.; Kuramitsu, H.K. Multiple functions of the leucine-rich repeat protein LrrA of Treponema denticola. Infection and Immunity 2004, 72, 4619–4627. [Google Scholar] [CrossRef] [PubMed]
- Ingham, V.A.; Anthousi, A.; Douris, V.; Harding, N.J.; Lycett, G.; Morris, M.; Vontas, J.; Ranson, H. A sensory appendage protein protects malaria vectors from pyrethroids. Nature 2020, 577, 376–380. [Google Scholar] [CrossRef] [PubMed]
- Ishii, E.; Eguchi, Y. Diversity in sensing and signaling of bacterial sensor histidine kinases. Biomolecules 2021, 11, 1524. [Google Scholar] [CrossRef] [PubMed]
- Jansen, S.; Zídek, L.; Löfstedt, C.; Picimbon, J.F.; Sklenar, V. 1H, 13C, and 15N resonance assignment of Bom- byxmori chemosensory protein 1 (BmorCSP1). Journal of Biomolecular NMR 2006, 36, 47. [Google Scholar] [CrossRef] [PubMed]
- Jansen, S.; Chmelik, J.; Zídek, L.; Padrta, P.; Novak, P.; Zdrahal, Z.; Picimbon, J.F.; Löfstedt, C.; Sklenar, V. Structure of Bombyx mori Chemosensory Protein 1 in solution. Archives of Insect Biochemistry and Physiology 2007, 66, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Jia, Q.; Zeng, H.; Zhang, J.; Gao, S.; Xiao, N.; Tang, J.; Dong, X.; Xie, W. The crystal structure of the Spodoptera litura Chemosensory Protein CSP8. Insects 2021, 12, 602. [Google Scholar] [CrossRef] [PubMed]
- Jian, Z.; Zeng, L.; Xu, T.; Sun, S.; Yan, S.; Zhao, S.; Su, Z.; Ge, C.; Zhang, Y.; Jia, J.; Dou, T. The intestinal microbiome associated with lipid metabolism and obesity in humans and animals. Journal of Applied Microbiology 2022, 133, 2915–2930. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Brandazza, A.; Navarrini, A.; Ban, L.; Zhang, S.; Steinbrecht, R.A.; Zhang, L.; Pelosi, P. Expression and immunolocalization of odorant-binding and chemosensory proteins in locusts. Cellular and Molecular Life Sciences 2005, 62, 1156–1166. [Google Scholar] [CrossRef] [PubMed]
- Jindra, M.; Palli, S.R.; Riddiford, L.M. The juvenile hormone signaling pathway in insect development. Annual Review of Entomology 2013, 58, 181–204. [Google Scholar] [CrossRef]
- Kazaz, S.; Miray, R.; Baud, S. Acyl-acyl carrier protein desaturases and plant biotic interactions. Cells 2021, 10, 674. [Google Scholar] [CrossRef] [PubMed]
- Kenis, S.; Istiban, M.N.; Van Damme, S.; Vandewyer, E.; Watteyne, J.; Schoofs, L.; Beets, I. Ancestral glycoprotein hormone-receptor pathway controls growth in C. elegans. Frontiers in Endocrinology (Lausanne) 2023, 14, 1200407. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.H.; Pham, V.; Jablonka, W.; Goodman, W.G.; Ribeiro, J.M.C.; Andersen, J.F. A mosquito hemolymph odorant-binding protein family member specifically binds juvenile hormone. Journal of Biological Chemistry 2017, 292, 15329–15339. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.D.; Pahuja, K.B.; Ravazzola, M.; Yoon, J.; Boyadjiev, S.A.; Hammamoto, S.; Schekman, R.; Orci, L.; Kim, J. The SEC23-SEC31 interface plays critical role for export of procollagen from the endoplasmic reticulum. Journal of Biological Chemistry 2012, 287, 10134–10144. [Google Scholar] [CrossRef] [PubMed]
- Kirkness, E.F.; et al. Genome sequences of the human body louse and its primary endosymbiont provide insights into the permanent parasitic life. Proceedings of the National Academy of Sciences USA 2010, 107, 12168–12173. [Google Scholar] [CrossRef] [PubMed]
- Kornblihtt, A.R.; de la Mata, M.; Fededa, J.P.; Muñoz, M.J.; Nogués, G. Multiple links between transcription and splicing. RNA 2004, 10, 1489–1498. [Google Scholar] [CrossRef] [PubMed]
- Ko, J.Y.; Oh, S.; Yoo, K.H. Functional enhancers as master regulators of tissue-specific gene regulation and cancer development. Molecules and Cells 2017, 40, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Kohlmeier, M.; Andreas, S.; Jorg, S.; Shearer, M.J. Transport of vitamin K to bone in humans. The Journal of Nutrition 1996, 126, 1192S–1196S. [Google Scholar] [CrossRef] [PubMed]
- Kramer, J.; Kang, R.; Grimm, L.M.; De Cola, L.; Picchetti, P.; Biedermann, F. Molecular probes, chemosensors, and nanosensors for optical detection of biorelevant molecules and ions in aqueous media and biofluids. Chemical Reviews 2022, 122, 3459–3636. [Google Scholar] [CrossRef]
- Kulmuni, J.; Wurm, Y.; Pamilo, P. Comparative genomics and chemosensory protein genes reveals rapid evolution and positive selection in ant-specific duplicates. Heredity 2013, 110, 538–547. [Google Scholar] [CrossRef] [PubMed]
- Lacroix, B.; Citovsky, V. Pathways of DNA transfer to plants from Agrobacterium tumefaciens and related bacterial species. Annual Review of Phytopathology 2019, 57, 231–251. [Google Scholar] [CrossRef] [PubMed]
- Lamichhane, S.; Sen, P.; Alves, M.A.; Ribeiro, H.C.; Raunioniemi, P.; Hyötyläine, T.; Orešič, M. Linking gut microbiome and lipid metabolism: moving beyond associations. Metabolites 2021, 11, 55. [Google Scholar] [CrossRef] [PubMed]
- Larkin, K.; Toloza, A.C.; Gabrie, J.A.; Rodriguez, C.A.; Rueda, M.M.; Matamoros, G.; Palacio, O.; Jamani, S.; Fontecha, G.; Sanchez, A.L. First detection of Acinetobacter baumannii in Pediculus humanus capitis from Latin America. Tropical Medicine and Infectious Disease 2023, 8, 345. [Google Scholar] [CrossRef] [PubMed]
- Lartigue, A.; Campanacci, V.; Roussel, A.; Larsson, A.M.; Jones, T.A.; Tegoni, M.; Cambillau, C. X-ray structure and ligand binding study of a moth chemosensory protein. Journal of Biological Chemistry 2002, 277, 32094–32098. [Google Scholar] [CrossRef] [PubMed]
- Laufer, H.; Borst, D.; Baker, F.C.; Reuter, C.C.; Tsai, L.W.; Schooley, D.A.; Carrasco, C.; Sinkus, M. Identification of a juvenile-hormone-like compound in a Crustacean. Science 1987, 235, 202–205. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.H.; Choi, K.C. Adversive effects of pesticides on the functions of immune system. Comparative Biochemistry and Physiology Part C: Toxicology & Pharmacology 2020, 235, 108789. [Google Scholar] [CrossRef]
- Le Goff, G. and Giraudo, M. (2019) Effects of pesticide on the environment and insecticide resistance. Olfactory Concepts of Insect Control-Alternative to Insecticides. (ed. J.F. Picimbon), pp. 51–78. Vol. 1, Springer Nature, Switzerland. [CrossRef]
- Lin, D.H.; Hoelz, A. The structure of the Nuclear Pore Complex (an update). Annual Review of Biochemistry 2019, 88, 725–783. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.X.; et al. Biotype expression and insecticide response of Bemisia tabaci chemosensory protein-1. Archives of Insect Biochemistry and Physiology 2014, 85, 137–151. [Google Scholar] [CrossRef]
- Liu, G.X.; Ma, H.M.; Xie, H.Y.; Xuan, N.; Picimbon, J.F. Sequence variation of Bemisia tabaci Chemosensory protein 2 in cryptic species B and Q: new DNA markers for whitefly recognition. Gene 2016a, 576, 284–291. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.X.; Ma, H.M.; Xie, Y.N.; Xuan, N.; Guo, X.; Fan, Z.X.; Rajashekar, B.; Arnaud, P.; Offmann, B.; Picimbon, J.F. Biotype characterization, developmental profiling, insecticide response and binding property of Bemisia tabaci chemosensory proteins: role of CSP in insect defense. PLoS ONE 2016b, 11, e0154706. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.X.; Picimbon, J.F. Bacterial origin of insect chemosensory odor-binding proteins. Gene & Translational Bioinformatics 2017, 3, e1548. [Google Scholar]
- Liu, G.X.; Arnaud, P.; Offmann, B.; Picimbon, J.F. Genotyping and bio-sensing chemosensory proteins in insects. Sensors 2017, 17, 1801. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.X.; Yue, S.; Rajashekar, B.; Picimbon, J.F. Expression of chemosensory protein (CSP) structures in Pediculus humanis corporis and Acinetobacter baumannii. SOJ Microbiology and Infectious Diseases 2019, 7, 1–17. [Google Scholar] [CrossRef]
- Liu, G.X.; Xuan, N.; Rajashekar, B.; Arnaud, P.; Offmann, B.; Picimbon, J.F. Comprehensive history of CSP genes: evolution, phylogenetic distribution, and functions. Genes 2020, 11, 413. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Liu, H.; Wang, L.; Song, L.; Jiang, G.; Lu, Q.; Yang, T.; Peng, H.; Cai, R.; Zhao, X.; Zhao, T.; Wu, H. Cochlear transcript diversity and its role in auditory functions implied by an otoferlin short isoform. Nature Communications 2023, 14, 3085. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.L.; Guo, H.; Huang, L.Q.; Pelosi, P.; Wang, C.Z. Unique function of a chemosensory protein in the proboscis of two Helicoverpa species. Journal of Experimental Biology 2014, 217, 1821–1826. [Google Scholar] [CrossRef] [PubMed]
- Loftus, B.J., Utterback, T., Pertea, G., Koo, H., Mori, A., Schneider, J., Lovin, D., de Bruyn, B., Song, Z., Raikhel, A., de Fatima, B.M., Casavant, T., Soares, B. and Severson, D. Aedes aegypti cDNA sequencing. #DV263125, DV263127, DV289920, DV289921, DV297938, DV314711, DV314712, DV316589, DV316619, DV334734, DV334735, DV335057, DV335058, DV344048, DV347318, DV347319, DV365747, DV365766, DV357763, DV368339, DV368340, DV393559, DV400785, DV400787, “…”.
- Los, D.A. and Murata, N. (2002) Chapter 10 – Sensing and responses to low temperature in cyanobacteria. Cell and Molecular Responses to Stress (K.B. Storey and J.M. Storey), Vol. 3, pp. 139–153. Elsevier, ScienceDirect, Amsterdam, Netherlands.
- Lu, Y.; Li, H.; Zhuang, S.; Zhang, D.; Zhang, Q.; Zhou, J.; Dong, S.; Liu, Q.; Wang, P. Olfactory biosensor using odorant-binding proteins from honeybee: ligands of floral odors and pheromones detection by electrochemical impedance. Sensors and Actuators B Chemical 2014, 193, 420–427. [Google Scholar] [CrossRef]
- Ma, C.; Cui, S.; Tian, Z.; Zhang, Y.; Chen, G.; Gao, X.; Tian, Z.; Chen, H.; Guo, J.; Zhou, Z. OcomCSP12, a chemosensory protein expressed specifically by ovary, mediates reproduction in Ophraella communa (Coleoptera: Chrysomelidae). Frontiers Physiology 2019, 10, 1290. [Google Scholar] [CrossRef] [PubMed]
- Ma, J. Transcriptional activators and activation mechanisms. Protein Cells 2011, 2, 879–888. [Google Scholar] [CrossRef] [PubMed]
- Macabuhay, A.; Arsova, B.; Walker, R.; Johnson, A.; Watt, M.; Roessner, U. Modulators or facilitators? Roles of lipids in plant root-microbe interactions. Trends in Plant Science 2021, 27, P180–P190. [Google Scholar] [CrossRef] [PubMed]
- Malcicka, M.; Visser, B.; Ellers, J. An evolutionary perspective on linoleic acid synthesis in animals. Evolutionary Biology 2018, 45, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Maleszka, J.; Forêt, S.; Saint, R.; Maleszka, R. RNAi-induced phenotypes suggest a novel role for a chemosensory protein CSP5 in the development of embryonic integument in the honeybee (Apis mellifera). Development Genes and Evolution 2007, 217, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Mandrioli, M.; Melchiori, G.; Panini, M.; Chiesa, O.; Giordano, R.; Mazzoni, E.; Manicardi, G.C. Analysis of the extent of synteny and conservation in the gene order in aphids: a first glimpse from the Aphis glycines genome. Insect Biochemistry and Molecular Biology 2019, 113, 103228. [Google Scholar] [CrossRef] [PubMed]
- Martín-Blázquez, R.; Chen, B.; Kang, L.; Bakkali, M. Evolution, expression and association of the chemosensory protein genes with the outbreak phase of the two main pest locusts. Scientific Reports 2018, 7, 6653. [Google Scholar] [CrossRef] [PubMed]
- Matlin, K.S. The strange case of the signal recognition particle. Nature Review Molecular Cell Biology 2002, 3, 538–542. [Google Scholar] [CrossRef] [PubMed]
- McKenna, M.P.; Hekmat-Scafe, D.S.; Gaines, P.; Carlson, J.R. Putative Drosophila pheromone-binding-proteins expressed in a subregion of the olfactory system. Journal of Biological Chemistry 1994, 269, 16340–16347. [Google Scholar] [CrossRef] [PubMed]
- Mei, T.; Fu, W.B.; Li, B.; He, Z.B.; Chen, B. Comparative genomics of chemosensory protein genes (CSPs) in twenty-two species (Diptera: Culicidae): identification, characterization, and evolution. PLoS ONE 2018, 13, e0190412. [Google Scholar] [CrossRef] [PubMed]
- Michaelson, D.; Silletti, J.; Murphy, G.; D’Eustachio, P.; Rush, M.; Philips, M.R. Differential localization of Rho Gtpases in live cells. Journal of Cell Biology 2001, 152, 111–126. [Google Scholar] [CrossRef] [PubMed]
- Moelling, K.; Broecker, F. Viruses and Evolution – Viruses First? A Personal Perspective. Frontiers in Microbiology 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- https://doi.org/10.3389/fmicb.2019.00523. [CrossRef]
- Moon, S.Y.; Zheng, Y. Rho GTPase-activating proteins in cell regulation. Trends in Cell Biology 2003, 13, P13–P22. [Google Scholar] [CrossRef] [PubMed]
- Moloney, F.; Yeow, T.P.; Mullen, A.; Nolan, J.J.; Roche, H.M. Conjugated linoleic acid supplementation, insulin sensitivity, and lipoprotein metabolism in patients with diabetes mellitus. American Journal of Clinical Nutrition 2004, 80, 887–895. [Google Scholar] [CrossRef] [PubMed]
- Monturiol-Gross, L.; Villalta-Romero, F.; Flores-Díaz, M.; Alape-Girón, A. Bacterial phospholipases C with dual activity: phosphatidylcholinesterase and sphingomyelinase. FEBS OpenBio 2021, 11, 3262–3275. [Google Scholar] [CrossRef] [PubMed]
- Morris, F.C.; Dexter, C.; Kostoulias, X.; Uddin, M.I.; Peleg, A.Y. The mechanisms of disease caused by Acinetobacter baumannii. Frontiers in Microbiology 2019, 10, 1601. [Google Scholar] [CrossRef] [PubMed]
- Mosbah, A.; Campanacci, V.; Lartigue, A.; Tegoni, M.; Cambillau, C.; Darbon, H. Solution structure of a chemosensory protein from the moth Mamestra brassicae. Biochemical Journal 2003, 369, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Moto, K.; Suzuki, M.G.; Hull, J.J.; Kurata, R.; Takahashi, S.; Yamamoto, M.; Okano, K.; Imai, K.; Ando, T.; Matsumoto, S. Involvement of a bifunctional fatty-acyl desaturase in the biosynthesis of the silkmoth, Bombyx mori, sex pheromone. Proceedings of the National Academy of Sciences USA 2004, 101, 8631–8636. [Google Scholar] [CrossRef] [PubMed]
- Nachtschatt, M.; Okada, S.; Speight, R. Integral membrane fatty acid desaturases: a review of biochemical, structural, and biotechnological advances. European Journal of Lipid Science and Technology 2020, 122, 20000181. [Google Scholar] [CrossRef]
- Nene, V.; Worthman, J.R.; Lawson, D.; et al. Genome sequence of Aedes aegypti, a major arbovirus vector. Science 2007, 22, 1718–1723. [Google Scholar] [CrossRef] [PubMed]
- Neves, G.; Zucker, J.; Daly, M.; Chess, A. Stochastic yet biased expression of multiple Dscam splice variants by individual cells. Nature Genetics 2004, 36, 240–246. [Google Scholar] [CrossRef] [PubMed]
- Njenga, R.; Boele, J.; Öztürk, Y.; Koch, H.G. Coping with stress: How bacteria fine-tune protein synthesis and protein transport. Journal of Biological Chemistry 2023, 299, 105163. [Google Scholar] [CrossRef] [PubMed]
- Nomura, A.; Kawasaki, K.; Kubo, T.; Natori, S. Purification and localization of p10, a novel protein that increases in nymphal regenerating legs of Periplaneta americana (American cockroach). International Journal of Developmental Biology 1992, 36, 391–398. [Google Scholar] [PubMed]
- Noriega, F.G.; Ribeiro, J.M.C.; Koener, J.F.; Valenzuela, J.G.; Hernandez-Martinez, S.; Pham, V.M.; Feyereisen, R. Comparative genomics of insect juvenile hormone biosynthesis. Insect Biochemistry and Molecular Biology 2006, 36, 366–374. [Google Scholar] [CrossRef] [PubMed]
- Ohnishi, A.; Hashimoto, K.; Imai, K.; Matsumoto, S. Functional characterization of the Bombyx mori fatty acid transport protein (BmFATP) within the silkmoth pheromone gland. Journal of Biological Chemistry 2009, 284, 5128–5136. [Google Scholar] [CrossRef] [PubMed]
- Ollivier, M.; Legeai, F.; Rispe, C. Comparative analysis of the Acyrthosiphon pisum genome and expressed sequence tag-based gene sets from other aphid species. Insect Molecular Biology 2019, 19, 33–45. [Google Scholar] [CrossRef] [PubMed]
- O’Neal, A.J.; Butler, L.R.; Rolandelli, A.; Gilk, S.D.; Pedra, J.H.F. Lipid hijacking: A unifying theme in vector-borne diseases. eLife 2020, 9, e61675. [Google Scholar] [CrossRef] [PubMed]
- Ozaki, K.; Utoguchi, A.; Yamada, A.; Yoshikawa, H. Identification and genomic structure of chemosensory proteins (CSP) and odorant binding proteins (OBP) genes expressed in foreleg tarsi of the swallowtail butterfly Papilio xuthus. Insect Biochemistry and Molecular Biology 2008, 38, 969–976. [Google Scholar] [CrossRef] [PubMed]
- Ozaki, M.; Wada-Katsumata, A.; Fujikawa, K.; Iwasaki, M.; Yokohari, F.; Satoji, Y.; Nisimura, T.; Yamaoka, R. Ant nestmate and non-nestmate discrimination by a chemosensory sensillum. Science 2005, 309, 311–314. [Google Scholar] [CrossRef] [PubMed]
- Pedra, J.H.; Brandt, A.; Li, H.M.; Westerman, R.; Romero-Serverson, J.; Pollack, R.J.; Murdock, L.L.; Pittendrigh, B.R. Transcriptome identification of putative genes involved in protein catabolism and innate immune response in human body louse (Pediculicidae: Pediculus humanus). Insect Biochemistry and Molecular Biology 2003, 33, 1135–1143. [Google Scholar] [CrossRef]
- Pereira, S.L.; Leonard, A.E.; Mukerji, P. Recent advances in the study of fatty acid desaturases from animals and lower eukaryotes. Prostaglandins, Leukotrienes & Essential Fatty Acids 2003, 68, 97–106. [Google Scholar] [CrossRef]
- Perkin, L.C.; Friesen, K.S.; Flinn, P.W.; Oppert, B. Venom gland components of the ectoparasitoid wasp, Anisopteromalus calandrae. Journal of Venom Research 2015, 6, 19–37. [Google Scholar] [PubMed]
- Persson, T.; Battenberg, K.; Demina, I.V.; Vigil-Stenman, T.; Vanden Heuvel, B.; Pujic, P.; Faccioti, M.T.; Wilbanks, E.G.; O’Brien, A.; Fournier, P.; Cruz Hernandez, M.A.; Mendoza Herrera, A.; Médigue, C.; Normand, P.; Pawlowski, K.; Berry, A.M. Candidatus Frankia Datiscae Dg1, the actinobacterial microsymbiont of Datisca glomerata, expresses the canonical nod genes nodABC in symbiosis with its host plant. PLoS One 2015, 10, e0127630. [Google Scholar] [CrossRef] [PubMed]
- Picimbon, J.F. Les périrécepteurs chimiosensoriels des insectes. Médecine et Science M/S (INSERM Journal) 2002, 18, 1089–1094. [Google Scholar] [CrossRef]
- Picimbon, J.F. (2003) Biochemistry and evolution of CSP and OBP proteins. Insect Pheromone Biochemistry and Molecular Biology, The Biosynthesis and Detection of Pheromones and Plant Volatiles (eds. G.J. Blomquist and R.G. Vogt), pp. 539–566, Elsevier Academic Press, London, San Diego, UK, USA. [CrossRef]
- Picimbon, J.F. (2005) Synthesis of odorant reception–suppressing agents: odorant binding proteins (OBPs) and Chemosensory Proteins (CSPs) as molecular targets for pest management. Biopesticides of plant origin (eds. C. Regnault-Roger, B. Philogène and C. Vincent), pp. 245–266, Intercept Ltd., Hampshire, UK.
- Picimbon, J.F. RNA mutations in the moth pheromone gland. RNA Diseases 2014a, 1, e240. [Google Scholar] [CrossRef]
- Picimbon, J.F. RNA mutations: source of life. Gene Technology 2014b, 3, 2. [Google Scholar] [CrossRef]
- Picimbon, J.F. Mutations in the insect transcriptome. International Journal of Clinical and Experimental Pathology 2016, 6, e122. [Google Scholar] [CrossRef]
- Picimbon, J.F. A new view of genetic mutations. Australasian Medical Journal 2017, 10, 701–715. [Google Scholar] [CrossRef]
- Picimbon, J.F. (2019) Evolution of protein physical structures in insect chemosensory systems. Olfactory Concepts of Insect Control-Alternative to Insecticides. (ed. J.F. Picimbon), pp. 231–263. Vol. 2, Springer Nature, Switzerland.
- Picimbon, J.F. Chapter three—bioinformatic, genomic and evolutionary analysis of genes: a case study in Dipteran CSPs. Methods in Enzymology 2020, 642, 35–79. [Google Scholar] [CrossRef] [PubMed]
- Picimbon, J.F. (2023a) RNA + peptide editing in chemosensory proteins (CSPs), a new theory for the origin of life on Earth’s crust. Gene, submitted.
- Picimbon, J.F. Insect OBPs, CSPs and NPC2s for control of SARS-CoV-2 and COVID-19: transport/degradation of viral capsid lipids. Revue Médicale de Bruxelles 2023b, 44, 111–122. [Google Scholar] [CrossRef]
- Picimbon, J.F.; Leal, W.S. Olfactory soluble proteins of cockroaches. Insect Biochemistry and Molecular Biology 1999, 30, 973–978. [Google Scholar] [CrossRef]
- Picimbon, J.F. and Regnault-Roger, C. (2008) Composés sémiochimiques volatils, phytoprotection et olfaction: cibles moléculaires de la lutte intégrée. Biopesticides d’Origine Végétale (eds. C. Regnault-Roger, B. Philogène and C. Vincent), pp. 383–415. Lavoisier Tech & Doc, Paris.
- Picimbon, J.F.; Dietrich, K.; Breer, H.; Krieger, J. Chemosensory proteins of Locusta migratoria (Orthoptera: Acrididae). Insect Biochemistry and Molecular Biology 2000a, 30, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Picimbon, J.F.; Dietrich, K.; Angeli, S.; Scaloni, A.; Krieger, J.; Breer, H.; Pelosi, P. Purification and molecular cloning of chemosensory proteins from Bombyx mori. Archives of Insect Biochemistry and Physiology 2000b, 44, 120–129. [Google Scholar] [CrossRef] [PubMed]
- Picimbon, J.F.; Dietrich, K.; Krieger, J.; Breer, H. Identity and expression pattern of chemosensory proteins in Heliothis virescens (Lepidoptera, Noctuidae). Insect Biochemistry and Molecular Biology 2001, 31, 1173–1181. [Google Scholar] [CrossRef] [PubMed]
- Pikielny, C.W.; Hasan, G.; Rouyer, F.; Rosbach, M. Members of a family of Drosophila putative odorant-binding proteins are expressed in different subsets of olfactory hairs. Neuron 1994, 12, 35–49. [Google Scholar] [CrossRef] [PubMed]
- Pirtilla, A.M.; Brusila, V.; Koskimaki, J.J.; Wali, P.R.; Ruotsalainen, A.L.; Mutanen, M.; Markkola, A.M. Exchange of microbiomes in plant-insect herbivore interactions. mBio 2023, 14. [Google Scholar] [CrossRef] [PubMed]
- Pittendrigh, B.R., Clark, J.M., Lee, S.H., Yoon, K.S., Sun, W., Steele, L.D. and Seong, K.M. (2015) Body lice: from the genome project to functional genomics and reverse genetics. Short views on insect genomics and proteomics, Entomology in focus (eds: C. Raman, M. Goldsmith & T. Agunbiade), pp. 1–18. Springer Cham, Switzerland.
- Poirel, L.; Madec, J.Y.; Lupo, A.; Schink, A.K.; Kieffer, N.; Nordmann, P.; Schwarts, S. Antimicrobial resistance in Escherichia coli. Microbiology Spectrum-American Society for Microbiology Press 2018, 6, ARBA-0026-2017. [Google Scholar] [CrossRef]
- Rai, K.S.; Black, W.C. Mosquito genomes: structure, organization and evolution. Advances in Genetics 1999, 41, 1–32. [Google Scholar] [CrossRef]
- Ray, S.; Casteel, C.L. Effector-mediated plant-virus-vector interactions. Plant Cell 2022, 34, 1514–1531. [Google Scholar] [CrossRef] [PubMed]
- Redondo-Salvo, S.; Fernández-López, R.; Ruiz, R.; Vielva, L.; de Toro, M.; Rocha, E.P.C.; Garcillán-Barcia, M.P.; de la Cruz, F. Pathways for horizontal gene transfer in bacteria revealed by a global map of their plasmids. Nature Communications 2020, 11, 3602. [Google Scholar] [CrossRef]
- Richards, S., Moran, N., Morioka, M., Fukatsu, T. and Nakabachi, A. (2007) Acyrthosiphon pisum ESTs. #EX608974.
- Robertson, H.M.; Martos, R.; Sears, C.R.; Todres, E.Z.; Walden, K.K.; Nardi, J.B. Diversity of odourant binding proteins revealed by an expressed sequence tag project on male Manduca sexta moth antennae. Insect Molecular Biology 1999, 8, 501–518. [Google Scholar] [CrossRef] [PubMed]
- Rock, C.O. (2008) Chapter 3 – Fatty acid and phospholipid metabolism in prokaryotes. Biochemistry of Lipids, Lipoproteins and Membranes Fifth Edition (eds. D.E. Vance and J.E. Vance), pp. 59–96, Elsevier Science, Amsterdam, Netherlands.
- Rodriguez, P.A.; Stam, R.; Warbroek, T.; Bos, J.I. Mp10 and Mp42 from the aphid species Myzus persicae trigger plant defenses in Nicotiana benthamiana through different activities. Molecular Plant Microbe Interactions 2014, 27, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, M.F.; Sánchez-García, A.; Salas, J.J.; Garcés, R.; Martínez-Force, E. Characterization of soluble acyl-ACP desaturases from Camelina sativa, Macadamia tetraphylla and Dolichandra unguis-cati. Journal of Plant Physiology 2015, 178, 35–42. [Google Scholar] [CrossRef]
- Rooney, A.P., Jackson, M.A., Dunlap, C.A., Behle, R.W. and Muturi, E.J. (2019) Discovery and development of microbial biological control agents. Olfactory Concepts of Insect Control-Alternative to Insecticides. (ed. J.F. Picimbon), pp. 79–91. Vol. 1, Springer Nature, Switzerland.
- Rosonina, E.; Blencowe, B.J. Gene expression: the close coupling of transcription and splicing. Current Biology 2002, 12, R319–R321. [Google Scholar] [CrossRef]
- Rupawate, P.S.; Roylawar, P.; Khandagale, K.; Gawande, S.; Ade, A.B.; Jaiswal, D.K.; Borgave, S. Role of gut symbionts of insect pests: A novel target for insect-pest control. Frontiers in Microbiology 2023, 14, 1146390. [Google Scholar] [CrossRef] [PubMed]
- Sabatier, L.; Jouanguy, E.; Dostert, C.; Zachary, D.; Dimarcq, J.L.; Bulet, P.; Imler, J.C. Pherokine-2 and -3: Two Drosophila molecules related to pheromone/odor-binding proteins induced by viral and bacterial infections. European Journal of Biology 2003, 270, 3398–3407. [Google Scholar] [CrossRef]
- Salah Ud-Din, A.I.M.; Roujeinikova, A. Methyl-accepting chemotaxis proteins: a core sensing element in prokaryotes and archaea. Cellular and Molecular Life Sciences 2017, 74, 3293–3303. [Google Scholar] [CrossRef]
- Sánchez-Gracia, A.; Vieira, F.G.; Rozas, J. Molecular evolution of the major chemosensory gene families in insects. Heredity 2009, 103, 208–216. [Google Scholar] [CrossRef]
- Sasaki, K.; Yoshida, H. Organelle autoregulation—stress responses in the ER, Golgi, mitochondria and lysosome. Journal of Biochemistry 2015, 157, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Savarit, F.; Sureau, G.; Cobb, M.; Ferveur, J.F. Genetic elimination of known pheromones reveals the fundamental chemical bases of mating and isolation in Drosophila. Proceedings of the National Academy of Sciences USA 1999, 96, 9015–9020. [Google Scholar] [CrossRef] [PubMed]
- Schachat, S.R.; Goldstein, P.Z.; Desalle, R.; Bobo, D.M.; Boyce, K.; Payne, J.L.; Labandeira, C.C. Illusion of flight? Absence, evidence and the age of winged insects. Biological Journal of the Linnean Society 2023, 138, 143–168. [Google Scholar] [CrossRef]
- Scheffers, D.J.; Pinho, M.G. Bacterial cell wall synthesis: new insights from localization studies. Microbiology and Molecular Biology Reviews 2005, 69, 585–607. [Google Scholar] [CrossRef]
- Schneewind, O.; Missiakas, D. Sec-secretion and sortase-mediated anchoring of proteins in Gram-positive bacteria. Biochimica et Biophysica Acta – Molecular Cell Research 2014, 1843, 1687–1697. [Google Scholar] [CrossRef] [PubMed]
- Schneider, E.S.; Kleineidam, C.J.; Leitinger, G.; Römer, H. Ultrastructure and electrophysiology of thermosensitive sensilla coeloconica in a tropical katydid of the genus Mecopoda (Orthoptera, Tettigoniidae). Arthropod Structure & Development 2018, 47, 482–497. [Google Scholar] [CrossRef]
- Schultz, D.J.; Suh, M.C.; Ohlrogge, J.B. Stearoyl-acyl carrier protein and unusual acyl-acyl carrier protein desaturase activities are differentially influenced by ferredoxin. Plant Physiology 2000, 124, 681–692. [Google Scholar] [CrossRef] [PubMed]
- Seshasayee, A.S.; Sivaraman, K.; Luscombe, N.M. An overview of prokaryotic transcription factors: a summary of function and occurrence in bacterial genomes. Subcell Biochemistry 2011, 52, 7–23. [Google Scholar] [CrossRef]
- Shalaeva, D.N.; Galperin, M.Y.; Mulkidjanian, A.Y. Eukaryotic G protein-coupled receptors as descendants of prokaryotic sodium-translocating rhodopsins. Biology Direct 2015, 10, 63. [Google Scholar] [CrossRef]
- Shangguan, X.; Zhang, J.; Liu, B.; Zhao, Y.; Wang, H.; Wang, Z.; Guo, J.; Rao, W.; Jing, S.; Guan, W.; Ma, Y.; Wu, Y.; Hu, L.; Chen, R.; Du, B.; Zhu, L.; Yu, D.; He, G. A mucin-like protein of planthopper is required for feeding and induces immunity responses in plants. Plant Physiology 2018, 176, 552–565. [Google Scholar] [CrossRef] [PubMed]
- Shigenobu, S.; Richards, S.; Cree, A.G.; Morioka, M.; Fukatsu, T.; Kudo, T.; Miyagishima, S.; Gibbs, R.A.; Stern, D.L.; Nakabashi, A. A full-length cDNA resource for the pea aphid, Acyrtosiphon pisum. Insect Molecular biology 2010, 19, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, J.A.; Khan, M.M.; Bamisile, B.S.; Hafeez, M.; Qasim, M.; Rasheed, M.T.; Rasheed, M.A.; Ahmad, S.; Shahid, M.I.; Xu, Y. Role of insect gut microbiota in pesticide degradation: a review. Frontiers in Microbiology 2022, 13. [Google Scholar] [CrossRef] [PubMed]
- Siebers, M.; Brands, M.; Wever, V.; Duan, Y.; Hölzl, G.; Dörmann, P. Lipids in plant-microbe interactions. Biochimica et Biophysica Acta 2016, 1861, 1379–1395. [Google Scholar] [CrossRef] [PubMed]
- Silva-Junior, E.A.; Ruzzini, A.C.; Paludo, C.R.; Nascimento, F.S.; Currie, C.R.; Clardy, J.; Pupo, M.T. Pyrazines from bacteria and ants: convergent chemistry within an ecological niche. Scientific Reports 2018, 8, 2595. [Google Scholar] [CrossRef] [PubMed]
- Spealman, P.; Burrelli, J.; Gresham, D. Inverted duplicate DNA sequences increase translocation rates through sequencing nanopores resulting in reduced base calling accuracy. Nucleic Acids Research 2020, 48, 4940–4945. [Google Scholar] [CrossRef] [PubMed]
- Sperling, P.; Ternes, P.; Zank, T.K.; Heinz, E. The evolution of desaturases. Prostaglandins, Leukotrienes & Essential Fatty Acids 2003, 68, 73–95. [Google Scholar] [CrossRef]
- Steinbrecht, R.A. (2003) What can we learn from localizing lipoclistins? European Symposium For Insect Taste and Olfaction (ESITO VIII), July 2-7th, Harstad, Norway.
- Steinbrecht, R.A. Fine structure immunocytochemistry—An important tool for research on odorant-binding proteins. Methods in Enzymology 2020, 642, 259–278. [Google Scholar] [CrossRef] [PubMed]
- Stern, D.L., Wincker, P. and Tagu, D. (2005) Large-scale gene discovery in the pea aphid Acyrthosiphon pisum (Hemiptera). #DY230371.
- Suenami, S.; Koto, A.; Miyazaki, R. Basic structures of gut bacterial communities in eusocial insects. Insects 2023, 14, 444. [Google Scholar] [CrossRef] [PubMed]
- Taga, M.E.; Bassler, B.L. Chemical communication among bacteria. Proceedings of the National Academy of Sciences USA 2003, 100, 14549–14554. [Google Scholar] [CrossRef] [PubMed]
- Teng, O.; Ang, C.K.E.; Guan, X.L. Macrophage-bacteria interactions—A lipid-centric relationship. Frontiers in Immunology 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Thakur, S.; Chauhan, V. State of the Globe: Re-emergence of the louse-borne infections. Journal of Global Infectious Diseases 2022, 14, 45–46. [Google Scholar] [CrossRef]
- The International Aphid Genomics Consortium. Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biology 2010, 8, e1000313. [Google Scholar] [CrossRef]
- Titball, R.W. Bacterial phospholipases C. Microbiology Reviews 1993, 57, 347–366. [Google Scholar] [CrossRef] [PubMed]
- Toledo, A.; Benach, J.L. Hijacking and use of host lipids by intracellular pathogens. Microbiology Spectrum 2015, 3. [Google Scholar] [CrossRef] [PubMed]
- doi:10.1128/microbiolspec.VMBF-0001-2014. [CrossRef]
- Tomaselli, S.; Crescenzi, O.; Sanfelice, D.; Ab, E.; Wechsel-berger, R.; Angeli, S.; Scaloni, A.; Boelens, R.; Tancredi, T.; Pelosi, P.; Picone, D. Solution structure of a chemosensory protein from the desert locust Schistocerca gregaria. Biochemistry 2006, 45, 1606–1613. [Google Scholar] [CrossRef] [PubMed]
- Turcotte, S.; Letellier, J.; Lippé, R. Herpes simplex virus type 1 capsids transit by the trans-Golgi network, where viral glycoproteins accumulate independently of capsid egress. Journal of Virology 2005, 79, 8847–8860. [Google Scholar] [CrossRef] [PubMed]
- van der Meer-Janssen, Y.P.M.; van Galen, J.; Batenburg, J.J.; Helms, J.B. Lipids in host-pathogen interactions: Pathogens exploit the complexity of the host cell lipidome. Progress in Lipid Research 2010, 49, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Vega Rúa, A. and Okech, B.A. (2019) The spread of mosquito-borne diseases: a major and global public health problem. Olfactory Concepts of Insect Control-Alternative to Insecticides. (ed. J.F. Picimbon), pp.1-27. Vol. 1, Springer Nature, Switzerland.
- Vela, J.; Montiel, E.E.; Mora, P.; Lorite, P.; Palomeque, T. Aphids and ants, mutualistic species, share a mariner element with an unusual location on aphid chromosomes. Genes 2020, 12, 1966. [Google Scholar] [CrossRef] [PubMed]
- Verjovski-Almeida, S., Eiglmeier, K., El-Dorry, H., Gomes, S.L., Menck, S.F.M., Nascimento, A.L. and Roth, C.W. (2005) FAPESP and Institut Pasteur/AMSUD Network Aedes aegypti cDNA sequencing project. #EG001037.
- Vogt, R.G.; Riddiford, L.M. Pheromone binding and inactivation by moth antennae. Nature 1981, 293, 161–163. [Google Scholar] [CrossRef] [PubMed]
- Vogt, R.G.; Rogers, M.E.; Franco, M.D.; Sun, M. A comparative study of odorant binding protein genes: differential expression of the PBP1-GOBP2 gene cluster in Manduca sexta (Lepidoptera) and the organization of OBP genes in Drosophila melanogaster (Diptera). Journal of Experimental Biology 2002, 205, 719–744. [Google Scholar] [CrossRef] [PubMed]
- Wada-Katsumata, A.; Zurek, L.; Nalyanya, G.; Roelofs, W.L.; Zhang, A.; Schal, C. Gut bacteria mediate aggregation in the German cockroach. Proceedings of the National Academy of Sciences USA 2015, 112, 15678–15683. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.E.; Delannay, C.; Goindin, D.; Deng, L.; Guan, S.; Lu, X.; Fouque, F.; Vega-Rúa, A.; Picimbon, J.F. Cartography of odor chemicals in the dengue vector mosquito (Aedes aegypti L., Diptera/Culicidae). Scientific Reports 2019, 9, 8510. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Granados, R.R. An intestinal mucin is the target substrate for a baculovirus enhancin. Proceedings of the National Academy of Sciences USA 1997, 94, 6977–6982. [Google Scholar] [CrossRef] [PubMed]
- Wanner, K.W.; Willis, L.G.; Theilmann, D.A.; Isman, M.B.; Feng, Q.; Plettner, E. Analysis of the insect os-d-like gene family. Journal of Chemical Ecology 2004, 30, 889–911. [Google Scholar] [CrossRef] [PubMed]
- Wanner, K.W.; Isman, M.B.; Feng, Q.; Plettner, E.; Theilmann, D.A. Developmental expression patterns of four chemosensory protein genes from the Eastern spruce budworm, Choristoneura fumiferana. Insect Molecular Biology 2005, 14, 289–300. [Google Scholar] [CrossRef] [PubMed]
- Wari, D.; Kabir, M.A.; Mujiono, K.; Hojo, Y.; Shinya, T.; Tani, A.; Nakatani, H.; Galis, I. Honeydew-associated microbes elicit defense responses against brown planthopper in rice. Journal of Experimental Botany 2019, 70, 1683–1696. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; Lepore, R.; Schwede, T. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Research 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.; Feng, K.; Qin, J.; Wei, P.; Cao, P.; Zhang, Y.; Yuchi, Z.; He, L. A detoxification pathway initiated by a nuclear receptor TcHR96h in Tetranychus cinnabarinus (Boisduval). PLoS Genetics 2023, 19, e1010911. [Google Scholar] [CrossRef] [PubMed]
- Wicker-Thomas, C.; Garrido, D.; Bontonou, G.; Napal, L.; Mazuras, N.; Denis, B.; Rubin, T.; Parvy, J.P.; Montagne, J. Flexible origin of hydrocarbon/pheromone precursors in Drosophila melanogaster (S). Journal of Lipid Research 2015, 56, 2094–2102. [Google Scholar] [CrossRef] [PubMed]
- Wirth, R.; Muscholl, A.; Wanner, G. The role of pheromones in bacterial interactions. Trends in Microbiology 1996, 4, 96–103. [Google Scholar] [CrossRef] [PubMed]
- Xuan, N.; Bu, X.; Liu, Y.Y.; Yang, X.; Liu, G.X.; Fan, Z.X.; Bi, Y.P.; Yang, L.Q.; Lu, Q.N.; Rajashekar, B.; Leppik, G.; Kasvandik, S.; Picimbon, J.F. Molecular evidence of RNA editing in the Bombyx chemosensory protein family. PLoS ONE 2014, 9, e86932. [Google Scholar] [CrossRef] [PubMed]
- Xuan, N.; Guo, X.; Xie, H.Y.; Lou, Q.N.; Bo, L.X.; Liu, G.X.; Picimbon, J.F. Increased expression of CSP and CYP genes in adult silkworm females exposed to avermectins. Insect Science 2015, 22, 203–219. [Google Scholar] [CrossRef]
- Xuan, N.; Rajashekar, B.; Kasvandik, S.; Picimbon, J.F. Structural components of chemosensory protein mutations in the silkworm moth, Bombyx mori. Agri Gene 2016, 2, 53–58. [Google Scholar] [CrossRef]
- Xuan, N.; Rajashekar, B.; Picimbon, J.F. DNA and RNA-dependent polymerization in editing of Bombyx chemosensory protein (CSP) gene family. Agri Gene 2019, 12, 100087. [Google Scholar] [CrossRef]
- Ye, Z.; Bishop, T.; Wang, Y.; Shahriari, R.; Lynch, M. Evolution of sex determination in crustaceans. Marine Life Science & Technology 2023, 5, 1–11. [Google Scholar] [CrossRef]
- Yew, J.Y.; Chung, H. Insect pheromones: An overview of function, form, and discovery. Progress in Lipid Research 2015, 59, 88–105. [Google Scholar] [CrossRef] [PubMed]
- Yue, S.; He, Q.X.; Picimbon, J.F. Lactobacillus for ribosome peptide editing cancer. Clinical Translational Oncology 2023, 25, 1522–1544. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Shi, Q.; Li, T.; Cheng, P.; Guo, X.; Song, X.; Gong, M. Comparative proteomics reveals mechanisms that underlie insecticide resistance in Culex pipiens pallens Coquillett. PLoS Neglected Tropical Diseases 2021, 15, e0009237. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Lin, X.; Guo, X. The role of insect symbiotic bacteria in metabolizing phytochemicals and agrochemicals. Insects 2022, 13, 583. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Gonsior, M.; Schmitt-Kopplin, P.; Zhan, Y.; Zhang, R.; Jiao, N.; Chen, F. Microbial transformation of virus-induced dissolved organic matter from picocyanobacteria: coupling of bacterial diversity and DOM chemodiversity. The ISME Journal 2019, 13, 2551–2565. [Google Scholar] [CrossRef]
- Zaremska, V.; Maria Fisher, I.; Renzone, G.; Arena, S.; Scaloni, A.; Knoll, W.; Pelosi, P. Reverse chemical ecology suggests putative primate pheromones. Molecular Biology and Evolution 2022, 39, msab338. [Google Scholar] [CrossRef]
- Zhu, J.; Wang, G.; Pelosi, P. Plant transcriptomes reveal hidden guests. Biochemical Biophysical Research Communications 2016a, 474, 497–502. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Iovinella, I.; Dani, F.R.; Liu, Y.L.; Huang, L.Q.; Liu, Y.; Wang, C.Z.; Pelosi, P.; Wang, G. Conserved chemosensory proteins in the proboscis and eyes of Lepidoptera. International Journal of Biological Sciences 2016b, 12, 1394–1404. [Google Scholar] [CrossRef]
- Zhu, J.; Arena, S.; Spinelli, S.; Liu, D.; Zhang, Q.; Wei, R.; Cambillau, C.; Scaloni, A.; Wang, G.; Pelosi, P. Reverse chemical ecology: Olfactory proteins from the giant panda and their interactions with putative pheromones and bamboo volatiles. Proceedings of the National Academy of Sciences USA 2017, 114, E9802–E9810. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J., Iovinella, I., Dani, F.R., Pelosi, P. and Wang, G. (2019) Chemosensory proteins: A versatile binding family. Olfactory Concepts of Insect Control-Alternative to Insecticides (ed. J.F. Picimbon), pp. 147–169. Vol. 2, Springer Nature, Switzerland.
- Zhu, L.; Zhang, Y.; Zhang, W.; Yang, S.; Chen, J.Q.; Tian, D. Patterns of exon-intron architecture variation of genes in eukaryotic genomes. BMC Genomics 2009, 10, 47. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Dai, X. Bacterial stress defense: the crucial role of ribosome speed. Cellular and Molecular Life Sciences 2020, 77, 853–858. [Google Scholar] [CrossRef] [PubMed]
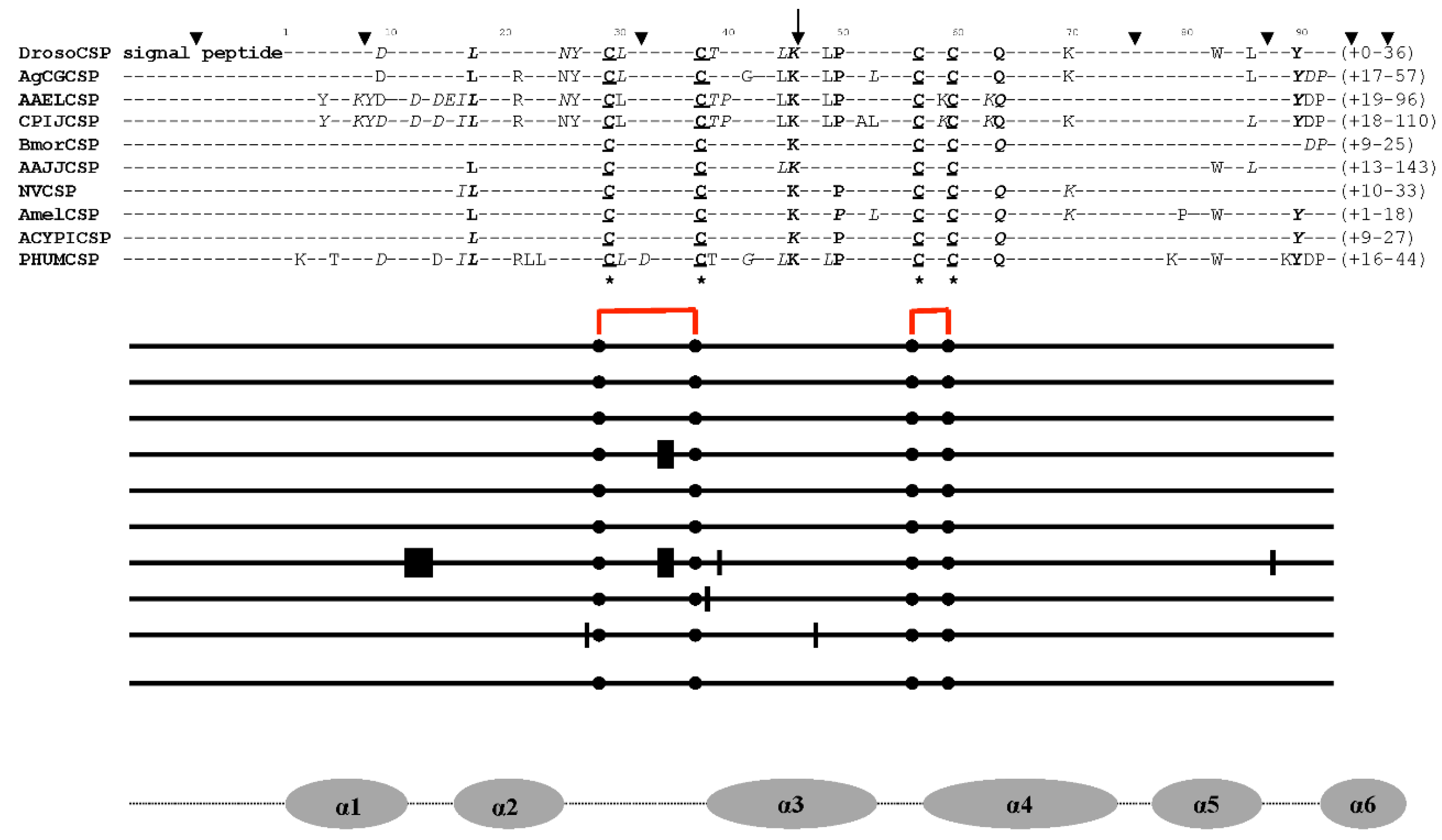
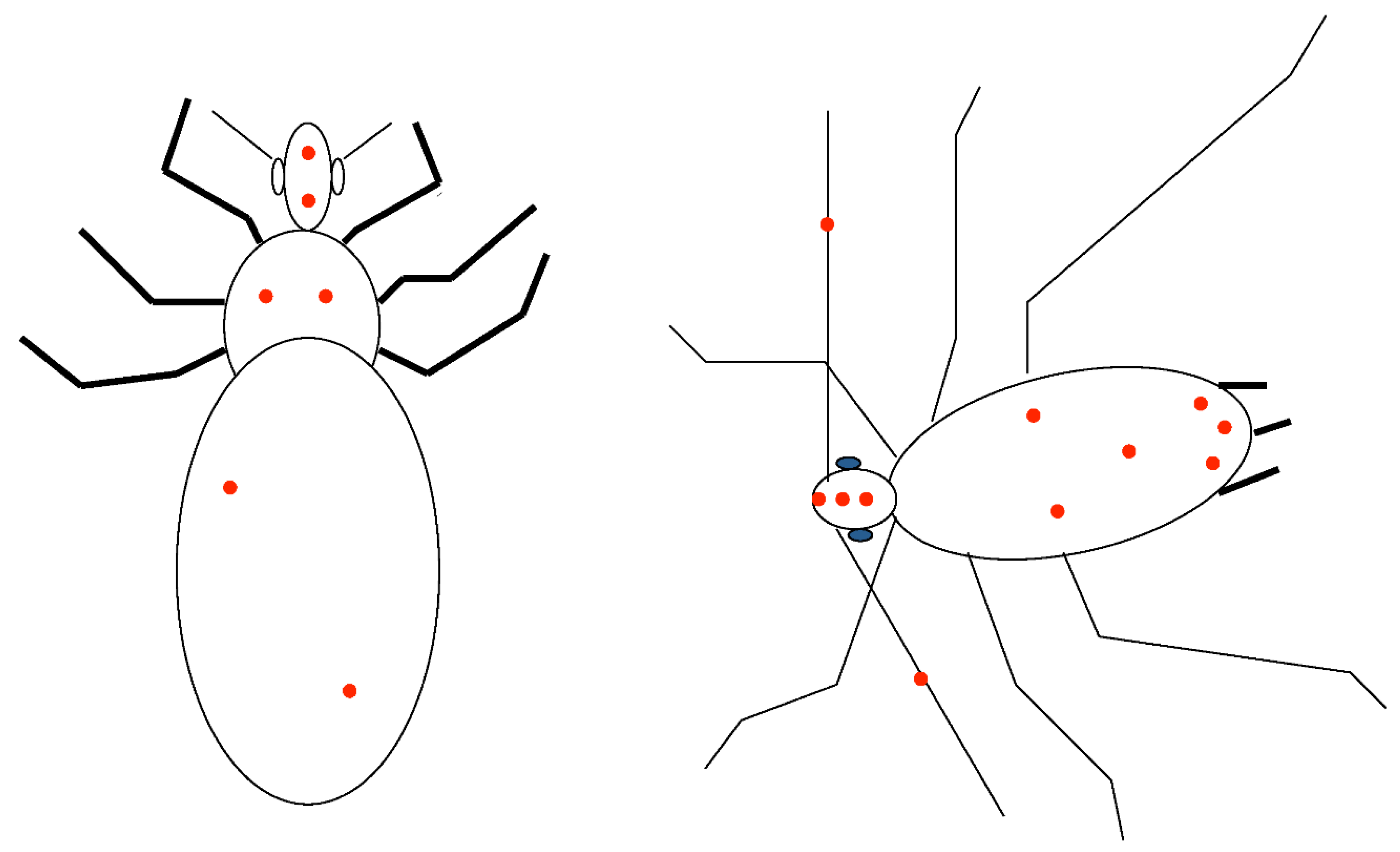
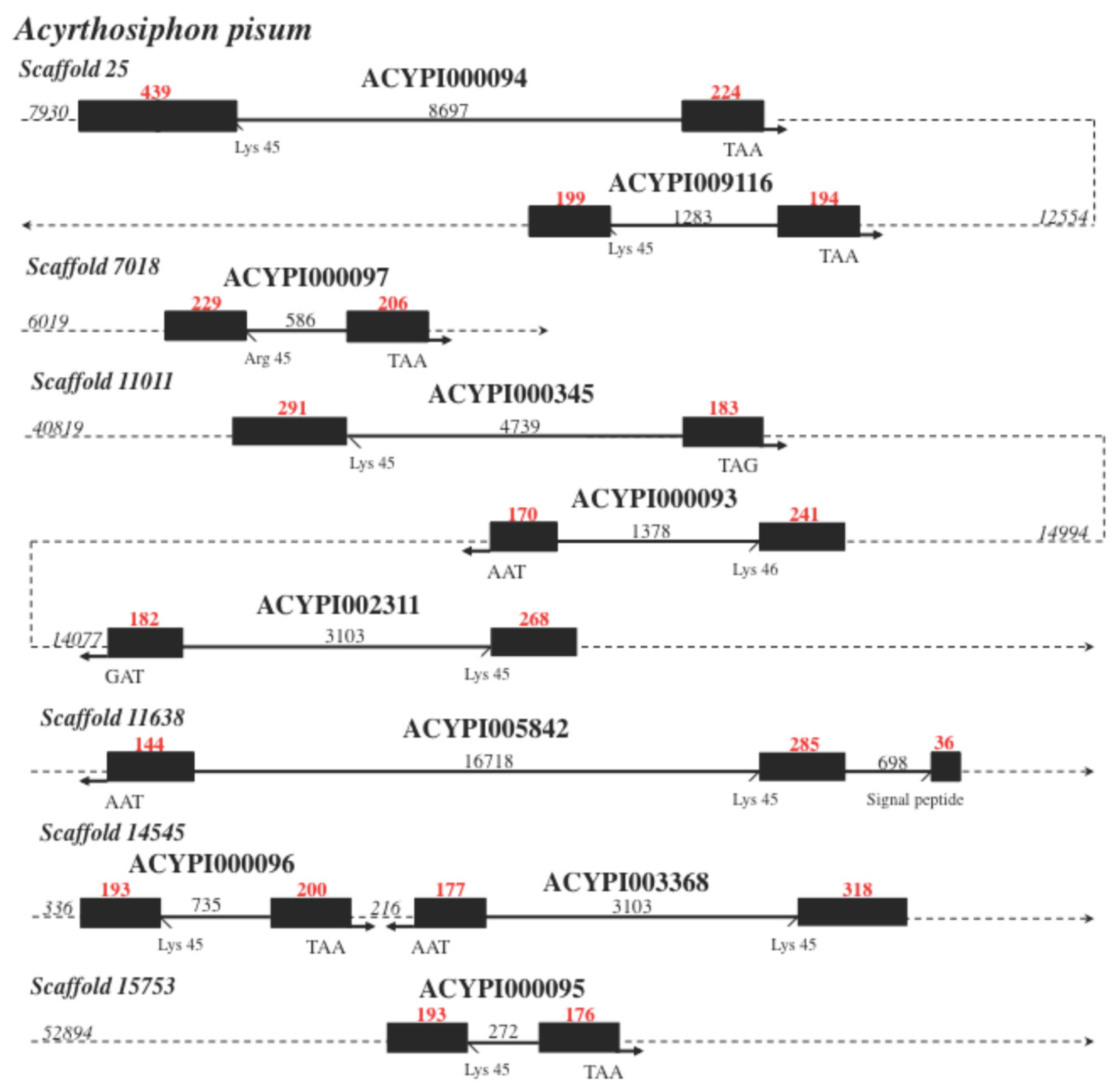
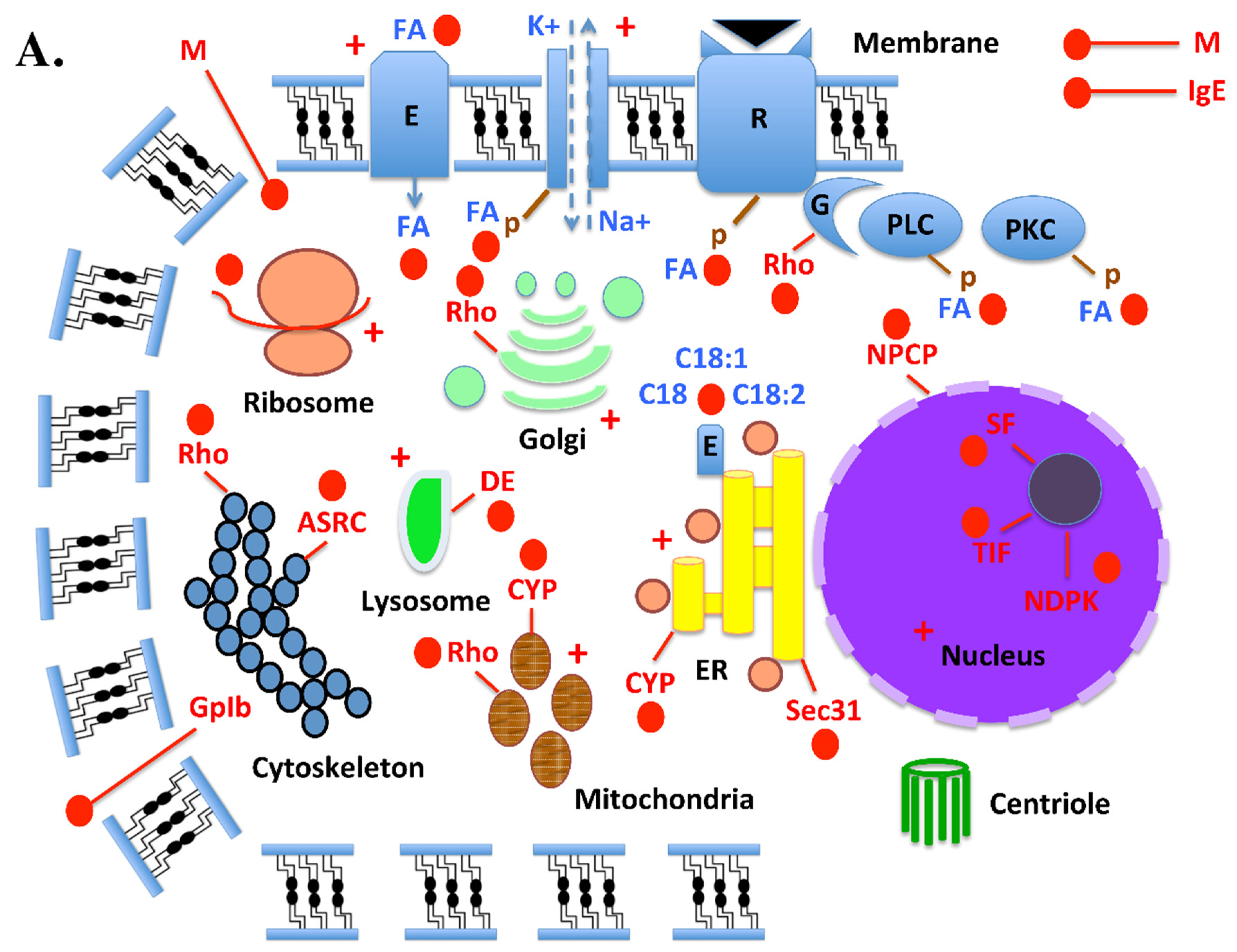
| Locus | mRNA bps | Access Number (R) |
Protein aa (kDa) |
Access Number (P) |
Species | Identity | Tissue | Function |
|---|---|---|---|---|---|---|---|---|
| LOC111036095 | 459 | XM_022317999 |
153 (17.2) | XP_022173691 |
Myzus persicae |
Mp10 “CSP4” Ebsp-3 |
Whole body Asexual adult |
Innate Immunity induction |
| BABH01021709 | 633 | NM_001043604 | 211 (24.6) | AK368835 |
Bombyx mori |
“BmorCSP10” |
Antennae Legs Fat body Gut Epidermisƒ |
Immune responseƒ |
| LOC126581753 | 2492 | XM_050245629 | 351 (37.7) | XP_050101586 |
Anopheles aquasalis |
Mucin-2 Variant X6 |
Translation initiation factor | |
| LOC109621340 | 2802 | XM_029870982 | 408 (43.4) | XP_029726842 | Aedes albopictus | SamkC Variant X9 |
All body Pupae |
Serine-Threonine protein kinase |
| LOC129745337 | 1986 | XM_055738352 | 364 (38.8) | XP_055594327 | Uranotaenia lowii | Mucin C.1 Variant X6 |
Integument Whole body |
Translation initiation factor |
| LOC115260417 | 1257 | XM_029861367 | 418 (45.8) | XP_029717227 | Aedes albopictus | Mucin-2 | All body Pupae |
Translation initiation factor |
| LOC126581753 | 2693 | XM_050245628 | 418 (45.6) | XP_050101585 |
Anopheles aquasalis |
Mucin-2 Variant X5 |
WAS-WASL-interacting protein | |
| LOC118459415 | 2720 | XM_035922756 | 418 (46.2) | XP_035778649 | Anopheles albimanus | Extensin-like Variant X5 |
Whole body Pupae |
Cell wall component Defense |
| LOC118459415 | 2909 | XM_035922754 | 418 (52.7) | XP_035778647 | Anopheles albimanus | Extensin-like Variant X7 |
Whole body Pupae |
Leucine-rich repeat Extensin-like |
| LOC129952650 | 2022 | XM_056065396 | 361 (39.4) | XP_055921371 | Eupeodes corollae | Rho GTPase-activating protein X3 | Rho GTPase-activator | |
| LOC109621340 | 3093 | XM_029870976 | 505 (54.4) | XP_029726836 | Aedes albopictus | Mucin-2 Variant X3 | All body Pupae |
Nuclear pore protein complex |
| LOC126564444 | 1245 | XM_050221501 | 401 (44.3) | XP_050077458 | Aedes maculipalpis | Mucin-2 Variant X4 |
Translation initiation factor | |
| LOC118513797 | 2611 | XM_036059988 | 400 (43.9) | XP_035915881 | Aedes stephensi | Mucin-2 Variant X3 |
Whole body | Translation initiation factor |
| LOC118459415 | 2461 | XM_035922752 | 547 (56.2) | XP_035778645 | Aedes albimanus | Mucin-5AC Variant X2 |
Whole body Pupae |
Extensin-like |
| LOC109621340 | 3084 | XM_029870977 | 502 (54.8) | XP_029726837 | Aedes albopictus | Mucin-2 Variant X4 |
All body Pupae |
Nuclear pore protein complex |
| LOC129744111 | 2632 | XM_055736495 | 496 (50.1) | XP_055592470 | Uranotaenia lowii | Mucin-2 Variant X3 |
Whole body |
Nuclear pore protein complex |
| LOC109621340 | 3318 | XM_029870961 | 580 (62.3) | XP_029726821 | Aedes albopictus | Mucin-2 Variant X1 |
All body Pupae |
Nuclear pore protein complex |
| LOC120904750 | 2951 | XM_040315040 | 493 (54.4) | XP_040170974 | Aedes arabiensis | Extensin-like Variant X5 | Extensin-like | |
| LOC118459415 | 2921 | XM_035922753 | 493 (51.4) | XP_035778646 | Aedes albimanus | Mucin-2 Variant X3 |
Whole body Pupae |
Extensin-like |
| LOC121596701 | 3034 | XM_041921881 | 452 (48.7) | XP_041777815 | Anopheles merus | Mucin-5AC Variant X7 |
Whole body |
Protein transport protein Sec31-like |
| LOC121596701 | 3371 | XM_041921880 | 564 (59.6) | XP_041777814 | Anopheles merus | Mucin-2 Variant X2 |
Whole body |
Mucin5AC-like |
| LOC121596701 | 3377 | XM_041921874 | 566 (59.8) | XP_041777808 | Anopheles merus | Mucin-2 Variant X1 |
Whole body |
Mucin5AC-like |
| LOC121596701 | 3022 | XM_041921883 | 448 (47.8) | XP_041777817 | Anopheles merus | Mucin-2 Variant X5 |
Whole body |
Mucin5AC-like |
| LOC121596701 | 3028 | XM_041921882 | 450 (47.9) | XP_041777816 | Anopheles merus | Mucin-5AC Variant X8 |
Whole body |
Mucin5AC-like |
| LOC120904750 | 3137 | XM_040315039 | 555 (60.8) | XP_040170973 | Anopheles arabiensis | Extensin-like Variant X4 |
Male |
Extensin-like |
| LOC129745337 | 2346 | XM_055738346 | 483 (51.0) | XP_055594321 | Uranotaenia lowii | Mucin-C1 Variant X1 |
Integument Whole body |
Mucin-C1-like |
| LOC131210824 | 2638 | XM_058204117 | 386 (42.7) | XP_058060100 | Anopheles bellator | Mucin-2 Variant X1 |
Tegument Whole body |
Protein UL36 |
| LOC124638351 | 1860 | XM_047175293 | 292 (32.1) | XP_047031249 | Helicoverpa zea | PAN1-like | Whole body Pupae |
Actin cytoskeleton-regulatory complex |
| LOC128303246 | 2787 | XM_053040137 | 472 (51.2) | XP_052896097 | Anopheles moucheti | Mucin-2 Variant X1 |
Mucin-like | |
| LOC118513797 | 2824 | XM_036059983 | 470 (51.2) | XP_035915876 | Anopheles stephensi | Extensin-like Variant X1 |
Whole body | Leucine-rich repeat Extensin-like |
| Scaffold00198 | 878 | JH668476 | 288 (32.1) | KAG6454812 | Manduca sexta | Hypothetical | ||
| LOC118513797 | 2774 | XM_036059986 | 454 (49.5) | XP_035915879 | Anopheles stephensi | Mucin-like Variant X2 |
Whole body | Mucin-2-like |
| LOC126374415 | 3912 | XM_050021059 | 298 (32.7) | XP_049877016 | Pectinophora gossypiella | YLP motif- protein 1 | Whole body Larvae |
Nuclear nucleoside kinase |
| LOC126564444 | 2774 | XM_050221498 | 554 (59.7) | XP_050077455 | Anopheles macupalpis | Mucin-2 Variant X1 |
Mucin-2-like | |
| LOC118459415 | 3326 | XM_035922748 | 628 (66.0) | XP_035778641 | Anopheles albimanus | Mucin-2 Variant X1 |
Whole body Pupae |
Mucin-2-like |
| LOC126564444 | 1410 | XM_050221500 | 456 (50.0) | XP_050077457 | Anopheles maculipalpis | Mucin-like Variant X2 |
Mucin-2-like | |
| Taxon93504 | 651 | LC027715 | 217 (24.0) | BAV56819 | Ostrinia furnacalis | “CSP15” | Antennae* | |
| LOC128271871 | 1721 | XM_053009541 | 396 (43.6) | XP_052865501 | Anopheles cruzii | WAS/WASL |
Actin skeleton-regulatory complex | |
| LOC128677457 | 1206 | XM_053758324 | 301 (33.6) | XP_053614299 | Plodia interpunctella | Formin-1-like |
Whole body Larvae |
Actin-microtubule association |
| LOC120904750 | 3488 | XM_040315029 | 672 (72.6) | XP_040170963 | Anopheles arabiensis | Extensin-like Variant X1 |
Extensin-like | |
| LOC111349097 | 1268 | XM_022960083 | 287 (31.8) | XP_022815851 | Spodoptera litura | PAN-1 | Whole body | Actin skeleton-regulatory complex |
| LOC5573135 | 2771 | XM_021847563 | 482 (51.4) | XP_021703255 |
Aedes aegypti |
DAN4 Variant X4 |
Whole body Pupae |
Nuclear pore protein complex |
| LOC126581753 | 2885 | XM_050245624 | 482 (52.0) | XP_050101581 | Anopheles aquasalis | Mucin-2 Variant X1 |
Mucin-like | |
| LOC120904750 | 3470 | XM_040315038 | 666 (71.5) | XP_040170972 | Anopheles arabiensis | Extensin-like Variant X9 |
Leucine-rich protein Extensin-like |
|
| LOC120904750 | 3476 | XM_040315037 | 668 (71.7) | XP_040170971 | Anopheles arabiensis | Mucin-2 Variant X2 |
Male | Mucin-like |
| LOC5573135 | 3059 | XM_021847553 | 576 (71.5) | XP_021703245 |
Aedes aegypti |
NPCP Variant X1 |
Whole body Pupae |
Nuclear pore protein complex |
| LOC120955660 | 3533 | XM_040376732 | 667 (71.4) | XP_040232666 | Anopheles coluzzii | Mucin-2 Variant X1 |
Mucin-like | |
| LOC106137243 | 1976 | XM_021847553 | 305 (34.0) | XP_013193496 | Amyelois transitella | WASP-2 | Larvae | Actin skeleton-regulatory complex |
| LOC6034358 | 2296 | XM_038253956 | 402 (43.3) | XP_038109884 | Culex quinquefasciatus | DAN4 Variant X4 |
Whole body | Cell wall protein |
| LOC106102595 | 1190 | XM_013282121 | 294 (32.9) | XP_013137575 |
Papilio polites |
GP Ib-like α-chain |
Outer surface membrane protein | |
| LOC120427056 | 2533 | XM_052707633 | 461 (49.5) | XP_052563593 |
Culex pipiens pallens |
Mucin-2 Variant X5 |
Whole body |
Mucin-like |
| M_scaff_1411 | 820 | CAKXAJ010025123 | 269 (30.3) | CAH2235359 |
Pararge aegeria aegeria |
Jg5928 | Major outer envelope protein | |
| LOC108904830 | 1520 | XM_018707509 | 576 (32.8) | XP_018563025 | Anoplophora glabripennis | CWA-3 Variant X1 |
Larvae | Cell wall integrity Stress response |
| LOC125234523 | 2861 | XM_048140790 | 287 (32.0) | XP_047996747 | Leguminivora glycinivorella | RickA-like Variant X2 |
Whole body Larvae |
Arp2/3 complex-activating protein |
| LOC128271871 | 2177 | XM_053009537 | 548 (59.0) | XP_052865497 |
Anopheles cruzii |
Mucin-2 Variant X1 |
Tegument | Virus-tegument cross-linker |
| LOC6034358 | 2697 | XM_038253951 | 535 (58.9) | XP_038109879 | Culex quinquefasciatus | Mucin-2 Variant X1 |
Whole body |
Cell wall protein |
| LOC123011164 | 1400 | XM_044408485 | 239 (26.3) | XP_044264420 | Tribolium maedens | Allergen Tha p1 Variant X2 |
Whole body |
IgE binding protein |
| LOC116772069 | 1195 | XM_032664103 | 297 (32.9) | XP_032519994 |
Danaus plexipus plexipus |
WAS/WASL |
Actin skeleton-regulatory complex | |
| LOC129727069 | 2801 | XM_055684473 | 625 (66.2) | XP_055540448 | Wyeomyia smithii | Mucin-2 Variant X1 |
Whole body | Mucin-like |
| LOC129727069 | 2580 | XM_055684478 | 553 (58.5) | XP_055540453 | Wyeomyia smithii | Mucin-2 Variant X3 |
Whole body | Mucin-like |
| LOC122855961 | 1243 | XM_044157672 | 128 (14.5) | XP_044013607 | Aphidius gifuensis | Allergen Tha p1 |
Adults |
IgE binding protein |
| LOC129727069 | 2476 | XM_055684480 | 518 (54.9) | XP_055540455 | Wyeomyia smithii | Mucin-2 Variant X5 |
Whole body |
Mucin-like |
| Scaffold_15245 | 372 | CH916367 | 122 (14.0) | GH22042 | Drosophila grimshawi | Pherokine-3-PC | Embryo Pupae |
Immunity |
| LOC6532152 | 607 | XM_002092892 | 239 (13.6) | XP_002092928 | Drosophila yakuba | Ebsp-3/PebIII | Ejaculatory bulb/duct | Sperm storage |
| LOC122859060 | 559 | XM_044162408 | 128 (14.2) | XP_044018343 | Aphidius gifuensus | Allergen Tha p1 |
Adults |
IgE binding protein |
| LOC125060609 | 487 | XM_047665563 | 121 (14.2) | XP_047521519 |
Pieris napi |
Allergen Tha p1 |
IgE binding protein | |
| LOC123307467 | 1891 | XM_044889794 | 457 (50.7) | XP_044745729 | Coccinella septempunctata | Eukaryotic TIF Variant 4 γ |
Translation initiation factor | |
| Taxon80765 | 387 | KJ451426 | 128 (14.4) | AHX71992 |
Aphis gossypii |
Acid trehalase “CSP3” |
Fat body |
Trehalose metabolism |
| LOC123712760 | 465 | XM_045666008 | 239 (26.3) | XP_045521964 |
Pieris brassicae |
Allergen Tha p1 |
IgE binding protein | |
| Scaffold00027 | 846 | JANFCV010000027 | 282 (31.4) | KAI5642933 | Phthorimaea operculella | A10/OS-D domain | Adults |
|
| Scaffold133 | 393 | JAANIC010002949 | 131 (14.7) | KAG5343447 | Acromyrmex charruanus | PEB3 | Whole body |
|
| LOC124166051 | 721 | XM_046543645 | 131 (14.5) | XP_046399601 |
Ischnura elegans |
Allergen Tha p1 Variant X2 |
IgE binding protein | |
| PHUM594540 | 423 | XM_002432550 | 140 (15.5) | XP_002432595 | Pediculus hum. corporis | Ebsp-3/PebIII | Larvae |
|
| LOC123010695 | 476 | XM_044407769 | 124 (14.4) | XP_044263704 | Tribolium maedens | Allergen Tha p1 |
Whole body Pupae |
IgE binding protein |
| Chromosome 2R | 339 | CP012524 | 113 (13.1) | ALC42649 | Drosophila busckii | PebIII | Whole body Larva |
|
| Dmel_CG9358 | 363 | NM_001299941 | 121 (13.4) | NP_001286870 | Drosophila melanogaster | Pherokine-3 Variant B |
Embryonic gonads |
Immunity |
| Scaffold_15245 | 366 | CH916367 | 122 (13.8) | EDW02527 | Drosophila grimshawi | GH22042/OS-D | ||
| LOC107225040 | 553 | XM_015665345 | 127 (14.1) | XP_015520831 | Neodiprion lecontei | Allergen Tha p1 Variant X1 |
Thorax Abdomen |
IgE binding protein |
| LOC124186858 | 574 | XM_046578909 | 127 (14.2) | XP_046434865 | Neodiprion fabricii | Allergen Tha p1 |
Thorax Abdomen |
IgE binding protein |
| LOC124166051 | 708 | XM_046543644 | 131 (14.2) | XP_046399600 |
Ischnura elegans |
Allergen Tha p1 Variant X1 |
IgE binding protein | |
| LOC124223445 | 658 | XM_046635425 | 127 (14.2) | XP_046491381 | Neodiprion pinetum | Allergen Tha p1 Variant X2 |
Thorax Abdomen |
IgE binding protein |
| LOC124308780 | 659 | XM_046771826 | 127 (14.2) | XP_046627782 | Neodiprion virginianus | Allergen Tha p1 Variant X2 |
Thorax Abdomen |
IgE binding protein |
| LOC110991536 | 755 | XM_022256909 | 121 (14.2) | XP_022112601 |
Pieris rapae |
Allergen Tha p1 |
IgE binding protein | |
| Contig_79 | 360 | LYFD01000069 | 120 (13.8) | OIB81003 |
Acinetobacter baumannii |
A10/OS-D |
Prokaryotic cell | |
| Contig_92 | 261 | LYGG01000091 | 87 (9.8) | OIC85870 |
Acinetobacter baumannii |
A10/OS-D |
Prokaryotic cell |
| Name | Organism | Authors | Year |
|---|---|---|---|
| p10 | Periplaneta americana | Nomura et al. | 1992 |
| Ebsp-3/PebIII | Drosophila melanogaster | Dyanov et al. | 1994 |
| A10 | Drosophila melanogaster | Pikielny et al. | 1994 |
| OS-D | Drosophila melanogaster | McKenna et al. | 1994 |
| Pam | Periplaneta americana | Picimbon and Leal | 1999 |
| CSP | Schistocerca gregaria | Angeli et al. | 1999 |
| SAP | Manduca sexta | Robertson et al. | 1999 |
| Pherokine | Drosophila melanogaster | Sabatier et al. | 2003 |
| Mp10 | Myzus persicae | Bos et al. | 2010 |
| LA-BP | Bemisia tabaci | Liu et al. | 2016 |
| Toxin-BP | Bemisia tabaci | Liu et al. | 2016 |
| B-CSP | Acinetobacter baumannii | Liu et al. | 2019 |
| Lipid-BP | Bemisia tabaci | Liu et al. | 2020 |
| JHRP | Aedes aegypti | Picimbon | 2020 |
| Mucin module | Aedes aegypti | Liu et al. | 2023 |
| TIF module | Aedes aegypti | Liu et al. | 2023 |
| DNA-BP | Aedes aegypti | Liu et al. | 2023 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




