1. Introduction
Tomatoes (Solanum lycopersicum L.), a vital horticultural crop ranking second only to potatoes in economic value, belong to the Solanaceae family and are extensively utilized fresh or in various processed forms (Mohan et al., 2016). With a rich nutritional profile, including vitamins, minerals, and bioactive compounds, tomatoes are associated with preventing chronic degenerative diseases (Perveen et al., 2015), making tomatoes widely acknowledged as healthful food, resulting in a substantial global increase in the cultivation acreage, production, and consumption of these nutritious fruits (Kim et al., 2016a). Globally, tomato production reached 182 million metric tons in 2018 (Ritchie et al., 2023), increasing to 186 million metric tons in 2020 as cited by Collins et al. (2022). Despite their economic importance, tomato cultivation faces challenges such as cultivar selection, management practices, pests, diseases, and abiotic stresses (Bita & Gerats, 2013; Roy et al., 2011; Shaheen et al., 2016; Zhou et al., 2018). Bacterial wilt (BW) caused by the soilborne bacterium Ralstonia solanacearum pathogen, stands out as a significant threat to tomato crops, causing significant loss of tomato fruit worldwide (Hong et al., 2018). The selection and cultivation of tomatoes with inherent genetic resistance to BW are advocated to enhance the quality and longevity of tomato crop yields (Kim et al., 2016a; Lee et al., 2015).
Ralstonia solanacearum ranks among the top 10 bacterial species in terms of scientific and economic importance in plant diseases (Yuliar et al., 2015). It is a soil-borne pathogen (Singh et al., 2015) causing vascular wilt disease by entering plants through wounds or root tips (Bindal & Srivastava, 2019). Classified into four phylogenic groups, this heterogeneous species complex includes strains with diverse geographical origins, host ranges, and pathogenic behaviors (Álvarez et al., 2007; Prior, 2005). The primary source of infection is soil, where the pathogen can survive for several years in the absence of a host plant and establishes latent infections in native weeds, adding to the challenge of eliminating the pathogen (Lebeau et al., 2011; Wang et al., 2019). Additionally, the pathogen can survive within a large temperature range of 10°C to 41°C and in varied environments (Muthoni et al., 2012). Spread occurs through contaminated water, infested soil, and contaminated farming equipment (Singh et al., 2015). R. solanacearum affects over 200 plant species, causing diseases like potato brown rot and bacterial wilt in tomatoes, tobacco, eggplants, and ornamentals, as well as Moko disease in bananas (Bindal & Srivastava, 2019). Symptoms include wilting during the day and recovery at night, progressing to plant death under severe conditions (Bindal & Srivastava, 2019). Bacterial wilt disease results in significant yield losses of approximately 90.62% (Aslam et al., 2017), impacting global crop productivity and causing annual losses exceeding US $1 billion (Champoiseau et al., 2009; Hong et al., 2012; Yuliar et al., 2015). Tomato bacterial wilt, induced by the soil-dwelling bacterium R. solanacearum, is prevalent in the southeastern United States. Under optimal conditions for disease development, it results in significant economic losses (Ji et al., 2007).
Several management strategies for BW have been suggested, including chemical fumigants, soil drainage, tillage practices, external nutrient supplements, and the use of resistant cultivars, transgenic plants, and microbes. However, each strategy has its limitations, and reliance on a single method often proves ineffective (Du et al., 2019). Chemical use, while temporarily effective, poses threats to beneficial microorganisms and human health (Aslam et al., 2017; Singh et al., 2015). Strategies like soil fumigation may not be economically feasible on a large scale as quoted by Mcavoy et al. (2012). As a result, it is essential to formulate strategies that are both effective and environmentally friendly to decrease the incidence of this disease (Lu et al., 2016). The development of resistant varieties emerges as the most effective, economical, and environmentally friendly approach to BW control (Du et al., 2019; Huet, 2014; Singh et al., 2015).
Breeding for resistance involves identifying resistant sources and ongoing screening against the pathogen, providing a sustainable and long-term solution to mitigate the impact of BW (Aslam et al., 2017; Yadav et al., 2023). The initial crucial step in any breeding program is germplasm collection, vital for crop improvement and diversity (Barik et al., 2020; Fock et al., 2000). Genetic resources, mainly found in the wild due to natural selection, provide valuable materials for enhancing crops. In the case of BW resistance, wild species like S. torvum, S. sisymbrifolium, and S. khasianum contribute resistance resources for Solanum species like eggplants (Barik et al., 2020). Additionally, some wild potato species exhibit BW tolerance (Patil et al., 2012). Regarding tomatoes, potential sources of resistance to BW have been identified but are not extensively utilized, mainly due to their small fruit sizes and poor horticultural characteristics, which may not align with commercial tomato industry standards. These sources include L285 and selections from wild tomato accessions PI 263722, PI 126408, PI 196298 (Lycopersicon esculentum), and PI 251323 (Lycopersicon pimpinellifolium) (Hanson & Licardo, 1998). While specific BW-resistant genotypes have been identified and released, they often exhibit region-specific resistance, and their effectiveness may decline outside the region of development due to environmental factors and variability among R solanacearum strains (Chellemi et al., 1994).
The identification of resistance varieties against BW is imperative (Zohoungbogbo et al., 2021), and it involves evaluating diverse genotypes from various regions for their response to the pathogen. Different studies have utilized various methods and tomato genotypes to discover strains resistant to BW. Screening methods include the sick plot method and artificial inoculation of tomato plants with R solanacearum strains (Barik et al., 2020). In the sick plot method, genotypes are planted in soil containing R. solanacearum, but uneven pathogen distribution across the soil necessitates the use of artificial inoculation methods. These methods encompass soil drenching, where bacterial suspension is injected into root incisions, leaf clipping, involving clipping leaves with bacterial suspension, and axil puncturing, where sterile needles are used to prick plant axils with the inoculum. Soil drenching has been found to significantly record the greatest BW incidences in screening tomatoes, brinjal, and chilies. Wilting symptoms are assessed using a BW score to measure resistance. In a study, thirteen genotypes from the USA, Taiwan, and Ghana were screened using the root dip technique, showing stable resistance in genotypes H7996, LA0442, and LA0443 (Newton B-Mensah et al., 2021). Another study evaluated fifty-five tomato genotypes and found seven greatly resistant genotypes, including RIL-118, Indam-1004, Arka Samrat, PKM-1, PED, EC-802390, and EC-816105, using the root-sectioned seedling dipping method (Kumar et al., 2018). This study aimed to identify tomato genotypes resistant to Bacterial wilt (BW) pathogen, providing valuable resources for future breeding initiatives. The goal is to incorporate these resistant genotypes into breeding programs, including genome-wide association studies and genomic selection, to facilitate the development of BW-resistant hybrids or cultivars. The specific objective was to evaluate the BW resistance of tomato germplasm sourced from the United States Department of Agriculture (USDA).
2. Materials and Methods
2.2. Plant materials and phenotyping
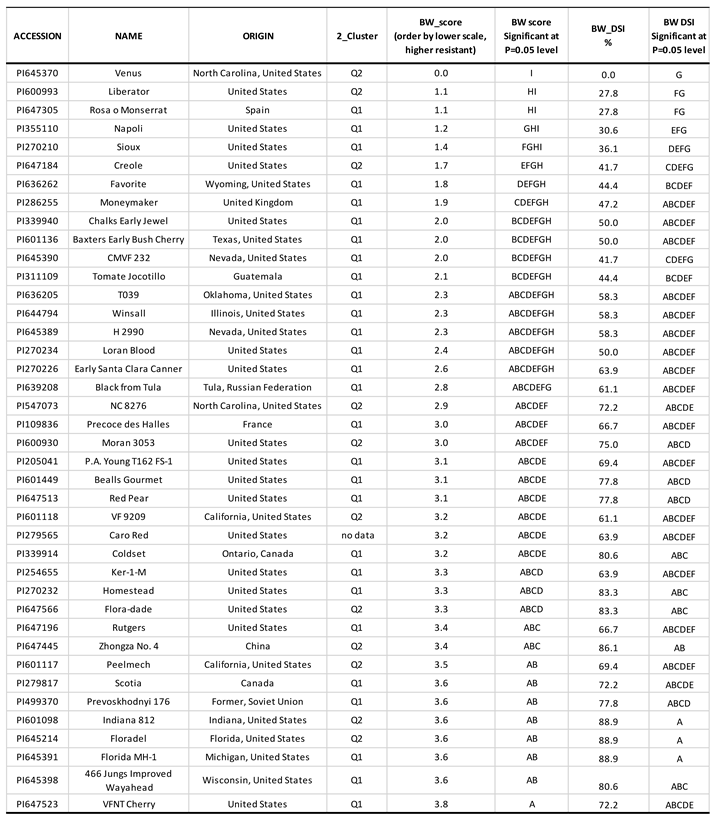
40 tomato accessions were obtained from the United States Department of Agriculture (USDA) Agricultural Research Service (ARS) Germplasm Resources Information Network (GRIN) (
Table 1). The majority (77.5%), 31 of the 40 accessions were originally from the United States; 2 were from Canada, and 1 from China, France, Guatemala, the Russian Federation, Former Soviet Union, Spain, and the United Kingdom, respectively.
Ralstonia solanacearum Strains P822 phylotype II sequevar 7 was used to test tomato accessions for BW resistance. The R. solanacearum Strains P822 was originally isolated from blueberry in Florida (Bocsanczy et al., 2019) and it was provided by Ana Maria Bocsanczy and David J. Norman at the University of Florida and stored at Alejandro Rojas’s laboratory of University of Arkansas, Fayetteville, AR for the bacterial growth, propagation, and preservation.
2.3. Greenhouse experiment for pathogen tests
The tomato accessions were planted in a commercial potting mix with 3 plants per pot arranged in a completely randomized design (CRD) with three replicates. Seedlings were watered daily and fertilized once after 7 days following seedling emergence. Five weeks after planting, seedlings were inoculated with R. solanacearum strain P822. The pathogen was tested for its virulence before being applied in this experiment. The pathogen was grown on Casamino acid-Peptone-Glucose (CPG) media maintained at 30oC (Allen et al., 2007), where the CPG medium constituents were 1 g casamino acid (casein hydrolysate), 10 g peptone, 5 g glucose, and 17 agar for solid media (plates).
A soil-drenching inoculation method was followed, where a knife was used to injure plant roots by cutting through the soil 1-2 cm away from the stem base before inoculation (
Figure 1). Before inoculation, the seedlings were kept without irrigation for a day, and 150 mL of the bacterial suspension was poured into the soil where the cut was made (Kalpage & De Costa, 2014). Following inoculation, the plants were watered with 150 mL of tap water daily.
The degree of wilt in seedlings was evaluated on a scale of 0 to 4, with each seedling being assessed individually (
Figure 2). A score of 0 indicated the absence of symptoms (no wilting), while a score of 1 indicated that 25% of the leaves were wilted. A score of 2 indicated that 50% of the leaves were wilted, and a score of 3 indicated that 75% of the leaves were wilted. A score of 4 indicated that all leaves were wilted, or the plant was dead (Bi-Hao et al., 2009; Kwon et al., 2021).
Disease severity index (DSI) was used for BW reaction in each of the 40 tomato accessions and was calculated as DSI = 100 * ∑ (frequency × score of rating) / [(Total number of observations) × (maximal disease index)] (Chiang et al., 2017).
3. Data analysis
3.1. Statistical model
The statistical model for ANOVA analysis was the following: Yij =µ + Gj +ℇij, where i= 1, 2, 3 and j=1.….40, with µ representing the overall mean, and Yij representing the response from the jth accession (Gj) (fixed effect) and eij representing the random error associated with the ijth observation.
3.2. ANOVA, distribution, descriptive statistics, Pearson’s correlation, and broad-sense heritability
The data were analyzed using JMP PRO 17. Variance variance (ANOVA) was analyzed using the general linear model (GLM) procedure. The distribution of the data was visualized using the ‘Distribution’; Descriptive statistics were estimated using the ‘Tabulate’; and the Person’s correlation coefficients (r-value) and their P-values were calculated by the ‘Multivariate Methods’ options of JMP PRO 17, respectively.
The broad-sense heritability (H2) was estimated as H2 = 100 * σ2g / [σ2g + (σ2e/r)] (Holland, 2003) with σ2g being the total genetic variance, σ2e being the residual variance, and r was the number of replicates. The estimates for σ2g and σ2e were [EMS(G)-Var(Residual)]/ r and Var(Residual), respectively. EMS(G) and Var(Residual) were obtained from the ANOVA table.
3.3. DNA extraction: genotyping by sequencing (GBS) and SNP discovery.
DNA (genome) was extracted from fresh leaves of tomato plants using the CTAB/SDS method. DNA sequencing was conducted using genotyping by sequencing (GBS) approach (Elshire et al., 2011) in Pair-end sequencing and libraries were sequenced by Illumina NovaSeq in the Biotechnology Center at the University of Wisconsin-Madison (
https://biotech.wisc.edu/). The short-read sequences data are aligned to tomato genome reference, Solanum lycopersicum, ITAG_4.0 (
https://phytozome-next.jgi.doe.gov/info/Slycopersicum_ITAG4_0) and SNPs were postulated in a pipeline using TASSE_GBS (Glaubitz et al., 2014) and Stacks 2 (Rochette et al., 2019;
https://catchenlab.life.illinois.edu/stacks/). A total of 392,496 single nucleotide polymorphism (SNP) markers were discovered across 287 tomato genotypes distributed on 12 chromosomes of tomato and provided by UWBC.
3.4. Principal Component Analysis (PCA) and Genetic Diversity
The principal component analysis (PCA) and genetic diversity analysis were performed in 39 of the 40 tomato accessions (except PI 279565 listed in
Table 1) in GAPIT 3 (Genomic Association and Prediction Integration Tool version 3) by setting PCA = 2 to 10 and NJ tree = 2 to 10 (Huang et al., 2019). The phylogenetic trees were drawn using the neighbor-joining (NJ) method in GAPIT 3 and also was drawn by the maximum likelihood (ML) method in MEGA 7 (Kumar et al., 2016) based on 4,847 single nucleotide polymorphism (SNP) markers distributed on 12 chromosomes. The SNP set consisted of 4,847 SNPs across the 39 accessions, after filtering and keeping the SNPs with minor allele frequency (MAF) >3.5%, missing allele <15%, and heterogeneous rate <=30% in this study.
4. Results
4.1. Parameters and distributions of bacterial wilt resistance
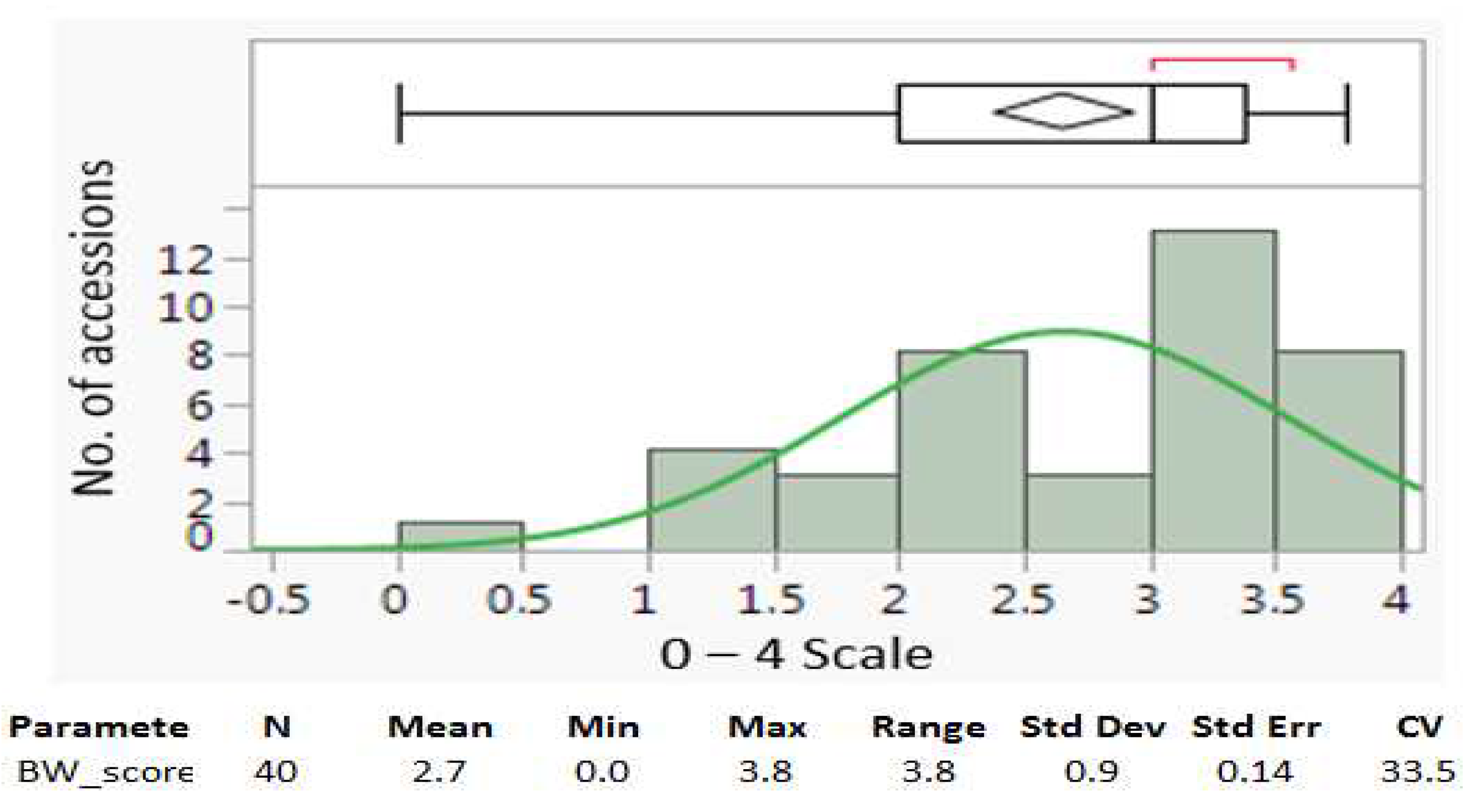
The investigation focused on evaluating BW symptoms in tomato plants, employing a 0-4 severity scale. Analysis of data from 40 tomato accessions on day 11 post-symptom onset revealed a rightward skew in the distribution, indicating increased susceptibility to BW (
Figure 3a). Among the 40 accessions assessed, the accession PI 645370 demonstrated the greatest resistance with a score of 0, while four other accessions PI 647305, PI 600993, PI 355110, and PI 270210 exhibited scores below 1.5 (
Table 1), suggesting resistance to BW pathogen
R. solanacearum Strains P822. These five accessions emerge as promising candidates for BW resistance, providing valuable insights for the prospective development of more robust tomato crops.
On the other hand, the seven accessions, PI 279817, PI 499370, PI 645391, PI 645398, PI 645214, PI 601098, and PI 647523 showed a score of 3.6 or higher on a scale of 4 (
Table 1), indicating their more susceptibility to BW and may be used as susceptible controls to screen BW resistant in tomato germplasm and as susceptible parents in genetic studies.
The recorded scores for BW resistance in the 40 tomato accessions had an average value of 2.7, a standard deviation (Std Dev) of 0.9, a standard error (Std Err) of 0.14, and a coefficient variation (CV) of 33.5% (
Figure 3a). These statistics suggest that the population had a large range (3.8) and variation ( 0.9 Std Dev & 33.5% CV) in BW resistance. Among the 40 genotypes, the top five genotypes with scores of 0 or <1.5 averaged mean demonstrated a great level of BW resistance, showing their potential to be used in the breeding programs targeting the development of new cultivars or lines with BW resistance.
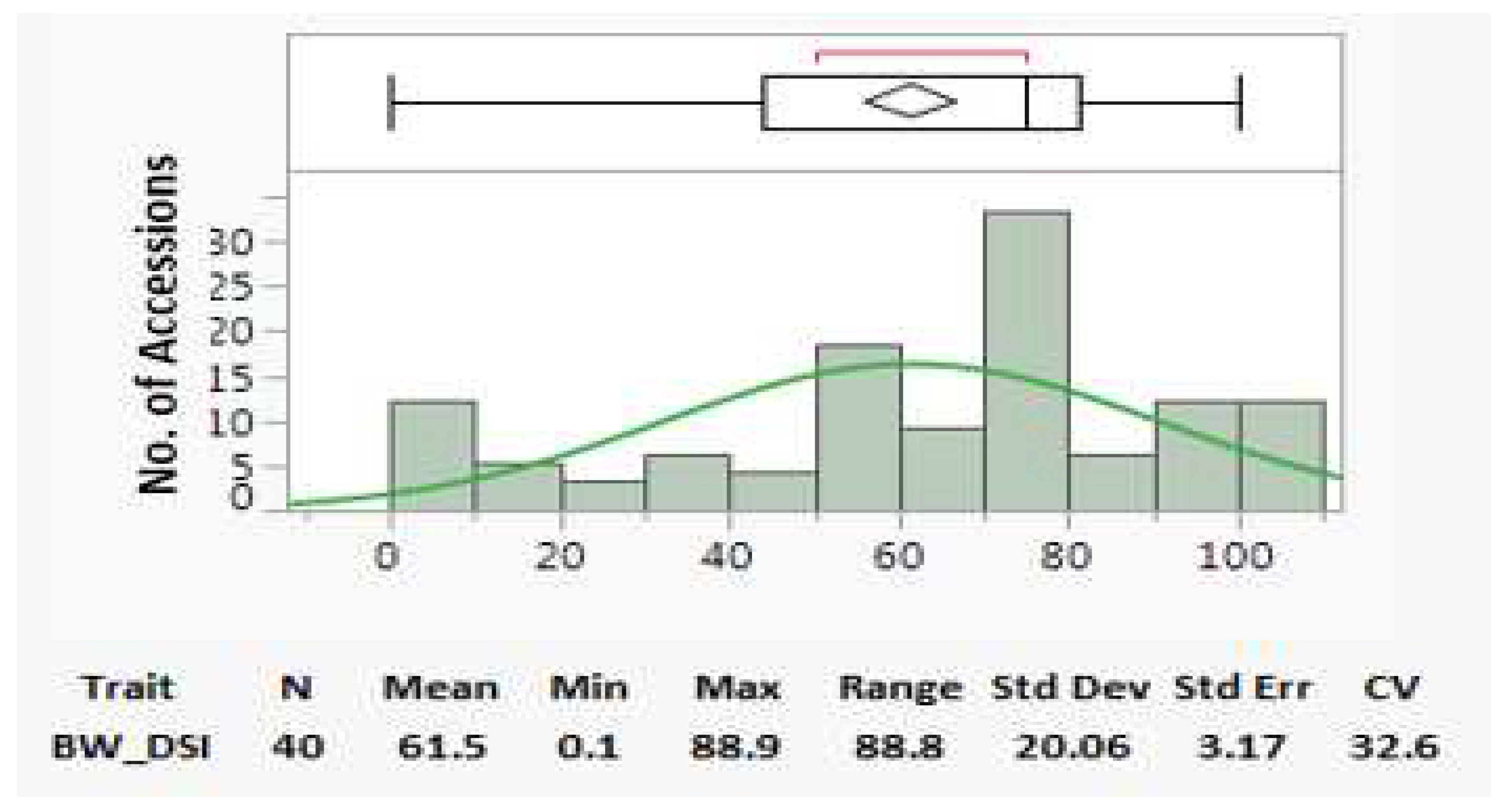
For the DSI, an average value of 61.5, a standard deviation (Std Dev) of 20.06, a standard error (Std Err) of 3.17, and a coefficient variation (CV) of 32.6% (
Figure 3b) were detected. These statistics suggest that the population had a large range (88.8) and variation (20.06 Std Dev & 32.6% CV) in BW DSI. Among the 40 genotypes, PI 645370 had a 0 DSI and demonstrated the greatest level of BW resistance and both PI 600993 and PI 647305 had 27.8% DSI, showing mediate resistance (
Table 1). The three accessions were also the top greatest resistant with 0 or 1.1 score based on the 0-4 score (
Table 1), thus indicating the potential to be used in breeding programs to develop new cultivars or lines with BW resistance.
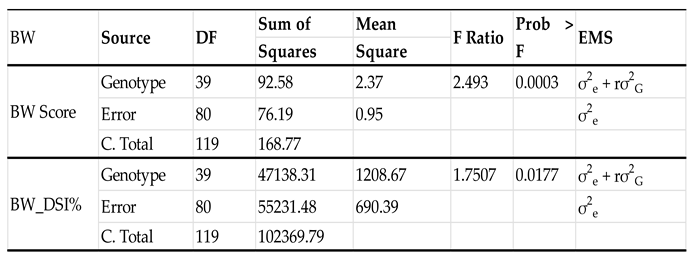
The ANOVA showed a signicant genotype (accession) effect (
P = 0.0003 < 0.001) for the disease scale and (
P = 0.0177 <0.05) (
Table 2), indicating there were significant differences for the BW resistance level (0-4 scale). Among the 40 accessions, accessions PI 645370, PI 647305, PI 600993, PI 355110, and PI 270210 had the lowest scores ranging from 0 to 1.5, showing the five accessions were BW-resistant.
The estimated broad-sense heritability (H
2) was 59.9% and 42.8% for the BW resistance scale and DSI, respectively, indicating that genetic factors accounted for a significant portion of the variation in BW resistance among the tomato genotypes (accessions) tested, demonstrating that the BW resistance is inheritable (
Table 2.).
4.2. Principal Component Analysis (PCA) and Genetic Diversity Analysis
PCA was analyzed in 39 of the 40 tomato accessions (except PI 279565 listed in
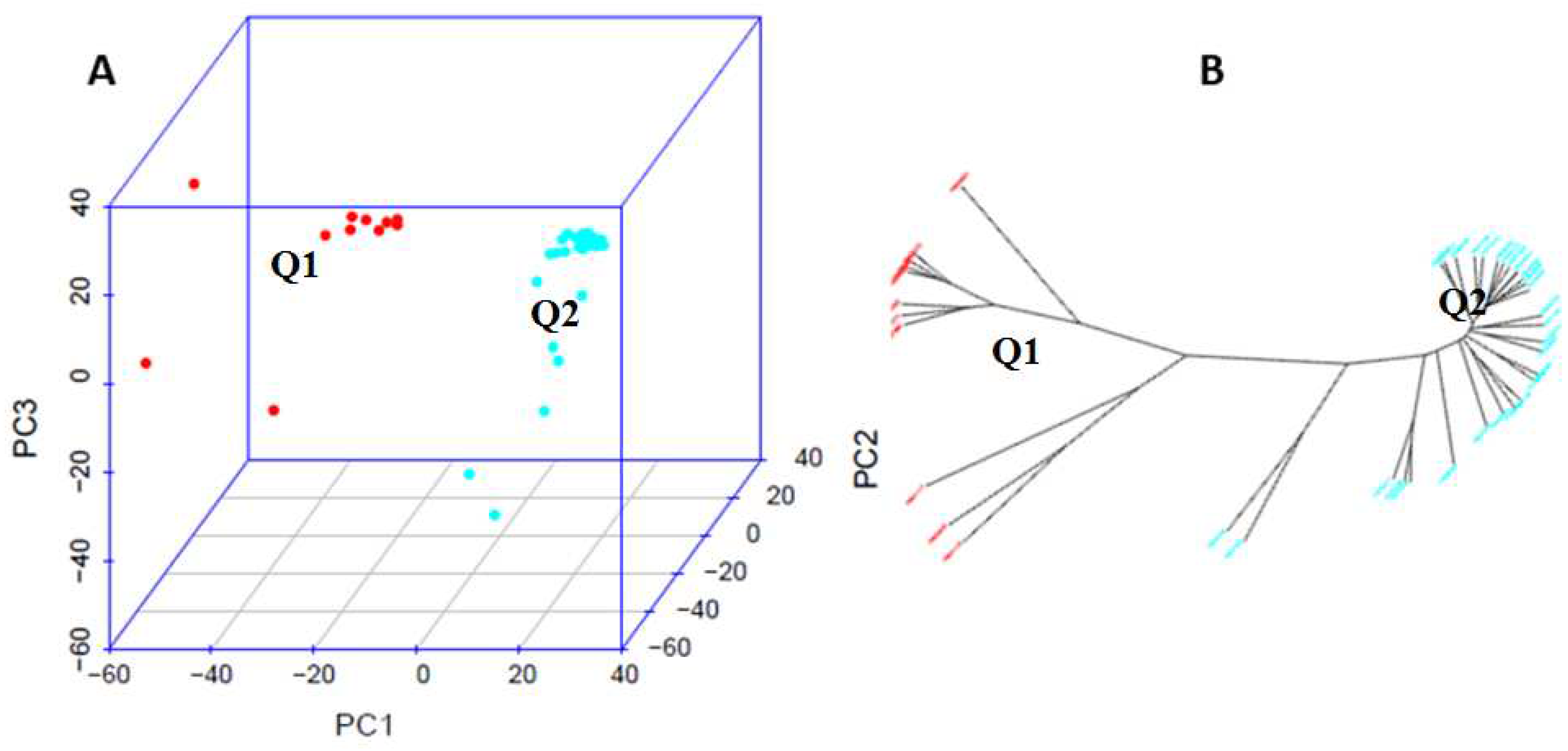
Table 1). The 39 accessions were divided into two distinct clusters or sub-populations, represented by red (Q1) and blue (Q2) colors in both the PCA plot (
Figure 4A) and phylogenetic tree (
Figure 4B) based on the neighbor-joining (NJ) algorithm in GAPIT 3. The Q2 sub-population was the majority with 28 accessions, accounting for 71.8% of the total population and the Q1 had 11 accessions (28.2%) (
Table 1,
Figure 4).
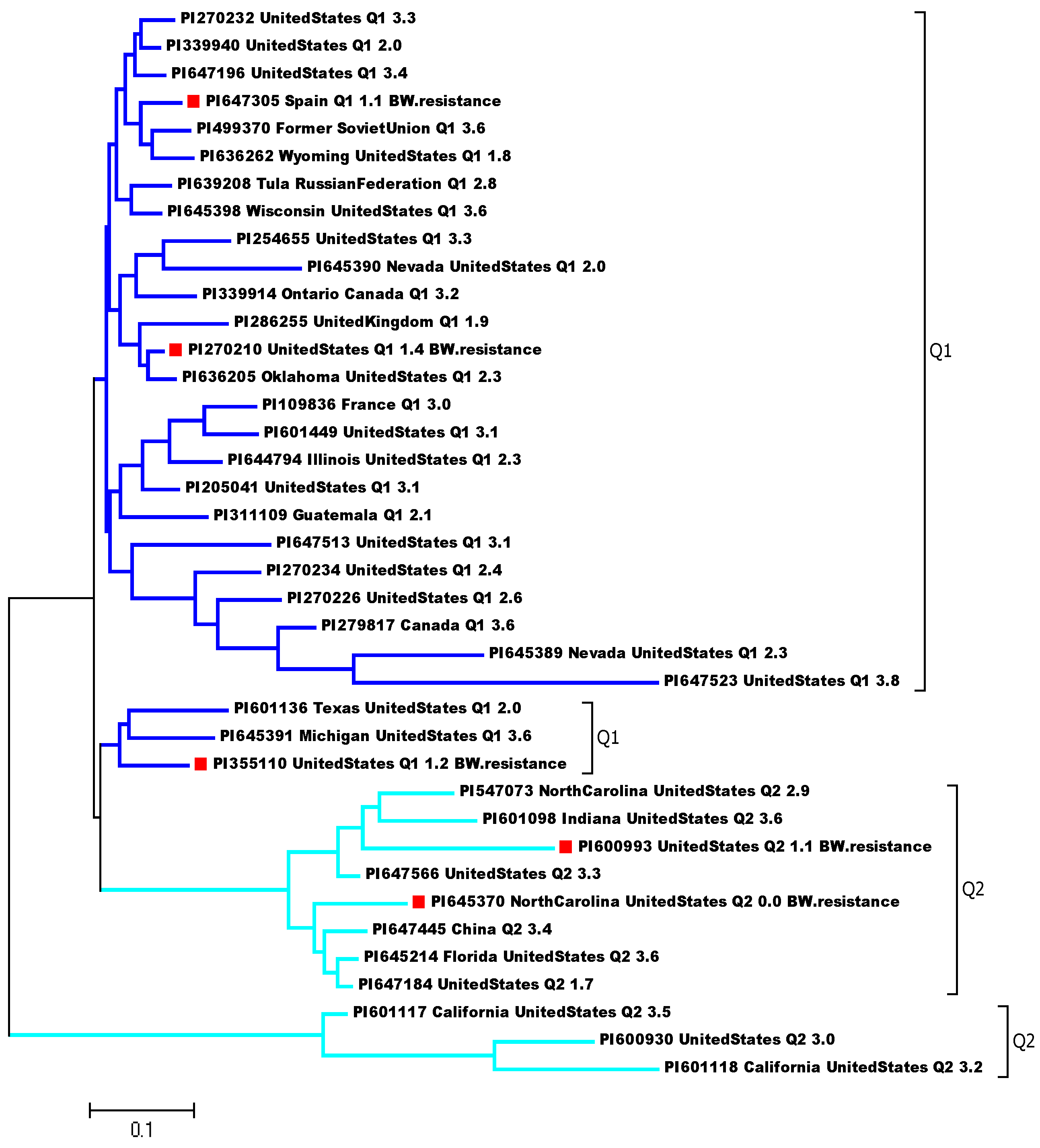
Two distinct clusters were also observed among the 39 tomato accessions in the phylogenetic tree postulated by the Maximum Likelihood (ML) method in MEGA 7 (
Figure 5). The top five greatly BW-resistant accessions, PI 645370, PI 647305, PI 600993, PI 355110, and PI 270210 were arranged into the two clusters (Q1 and Q2) marked red and squared shape in
Figure 5 and
Table 1 with the accession information, indicating different genetic base (background). Interestingly, despite most of the accessions in the study originating from the USA, the genotype diversity analysis revealed diverse genetic differences among the U.S. accessions, different countries, and States of U.S. (
Figure 5). Four of the five resistant accessions were originally collected from the United States of America, while one was collected from Spain (
Table 1 and
Figure 5). This highlights the availability of resistant genotypes to
R. solanacearum isolate P822, which is common in the USA. The sup-population Q1 contains three BW-resistant accessions, two from the USA and one from Spain; and the Q2 had two BW-resistant accessions from the USA. This information can be used to identify and utilize BW-resistant genotypes in breeding programs to develop more resilient tomato crops.
5. Discussion
Bacterial diseases triggered by R. solanacearum, a devastating pathogen causing a threat to many plants worldwide (Hai et al., 2008a), are among the major phytopathological problems of solanaceous crops. Information on the sources of resistance and inheritance is necessary to devise efficient and successful breeding strategies for developing resistant cultivars. Resistant tomato accessions have been found in other studies that are specific to different isolates of the pathogen. In this study, the use of a 0-4 scale and DSI to assess disease severity is a widely accepted method for evaluating the resistance of tomato genotypes to BW (Morel et al., 2018). Out of the forty genotypes (accessions) tested in this study, five genotypes were found to be resistant to R. solanacearum isolate P822, with many genotypes being susceptible to the bacteria isolate. The discovery of many genotypes susceptible to BW is consistent with previous studies that have reported a high degree of variability in resistance to the disease among tomato genotypes. In a previous study conducted by Hai et al. (2008), 252 wild tomato accessions were evaluated with Taiwanese race 1 strains, and five accessions were identified to be resistant to strain Pss186. In another study, 285 tomato accessions were screened and four out of all accessions showed high resistance against the pathogen (Kim et al., 2016). Consistent with prior research, including the current study, several tomato genotypes (accessions) resistant to BW were identified. This aligns with Hai et al. (2008b) report, which highlighted the limited frequency of discovering resistance resources in tomato germplasm. The identification of PI 645370 as an extremely resistant genotype and promising candidate genotypes PI 647306, PI 600993, PI 355110, and PI 270210 for BW resistance is particularly important, as it suggests that these genotypes may possess genetic traits that make them more resilient to the disease. Previous studies have documented the existence of BW-resistant genes in tomatoes, including the RRS1-R gene (Deslandes et al., 2002). As such, similar genes could probably be present in the current genotypes under investigation. If the presence of the resistance genes is verified in these genotypes through further investigations, they would represent a significant addition to the collection of identified resistant tomato genotypes, such as Hawaii 7996 (Grimault et al., 1995). This would expand the selection of tomato cultivars with desirable traits, potentially enabling breeders to develop more effective and sustainable strategies for controlling disease outbreaks and enhancing crop productivity. The confirmation of resistance genes in this genotype would also contribute to a deeper understanding of the molecular mechanisms underlying tomato-pathogen interactions, which could inform the development of novel approaches for enhancing crop resilience and ensuring food security. Future studies should strive to confirm the presence of resistance genes in this genotype and to elucidate their functional significance. This is particularly important considering the increasing prevalence of BW in many tomato-growing regions worldwide.
Broad-sense heritability is an important concept in quantitative genetics that describes the proportion of phenotypic variance that is attributed to genetic variation (Kaushik, 2011). In this study, the broad-sense heritability for BW resistance was estimated to be 59.9% based on a 0-4 scale and 42.8% based on DSI. These results suggest that BW resistance is a transmissible trait in tomatoes, and that selection for resistance can be effective in the breeding programs. These results are consistent with a previous report indicating a high broad-sense heritability for BW incidence (Boakye-Mensah, 2020). However, it is important to note that heritability estimates can be influenced by several factors, including the genetic architecture of the trait, the environmental conditions where the plants are grown, and the methods used to estimate heritability (Schmidt et al., 2019). Overall, the estimated broad-sense heritability of BW resistance in tomatoes provides important information for plant breeding programs, indicating that the selection for resistance can be an effective strategy for developing tomato cultivars with improved resistance to this devastating disease.
The clustering of genotypes in principal component analysis (PCA) and in phylogenetic tree can have important implications for plant breeding and conservation (Maji, 2012a). For example, by identifying genetically diverse genotypes, breeders can develop tomato cultivars with improved resistance to diseases and environmental stressors. Similarly, conservation efforts can focus on preserving the genetic diversity represented by both sub-populations to ensure the long-term viability of tomato production systems (Maji, 2012). In this study, PCA and phylogenetic analysis identified two distinct sub-populations of tomato genotypes (accessions) with different levels of genetic diversity and the five BW-resistant accessions distributed in the two sub-populations provide different genetic bases for breeders to choose from. These findings can inform breeding and conservation efforts to develop more resilient and sustainable tomato production systems.
6. Conclusion
This study evaluated bacterial wilt (BW) resistance in 40 USDA tomato germplasm accessions. Five accessions PI 645370, PI 647306, PI 600993, PI 355110, and PI 270210 were observed as BW resistance with PI 645370 as the greatest resistance. The broad-sense heritability was estimated as 59.9% based on a 0-4 scale of disease incidence and 42.8% based on the DSI of disease severity for BW resistance. Two distinct clusters (sub-populations) were shown among 39 of the 40 accessions, consisting of 3 and 2 BW resistant accessions in each cluster, respectively, suggesting the presence of different genetic bases in the five resistance accessions. The identification of resistant tomato genotypes for BW resistance provides valuable information for plant breeding programs to develop BW-resistant tomato cultivars.
Author Contributions
TMP, KEC, GB, and HX conducted a phenotypic evaluation of Bacteria wilt resistance in tomato accessions. AS performed the DNA sequencing and provided the SNP data. TMP, GB, HX, and AS performed phenotypic and genotypic data analysis. TMP, GB, HX, and AS are the Project Investigators. KEC, YC, IA, RD, and NJ collaborated in the study. TMP, KEC, and GB wrote the draft of the manuscript. AS, GB, and HX revised the manuscript. All authors reviewed and approved the manuscript.
Funding
The research was partially supported by USDA Crop Germplasm Evaluation grant 58-8060-1-008; USDA ARS Agreement Number/FAIN 58-6080-3-012; University of Arkansas Provost’s Collaborative Research Grant; USDA NIFA Hatch project ARK0VG2018 and ARK02440; and a scholarship from the Agricultural Transformation Initiative Fellowship and Scholarship Fund (ATI FSF). The ATI FSF is funded by The Foundation for a Smoke-Free World.
Data Availability Statement
The original information presented in the study is available in the Tables and Figures of the article text body and in the Supplementary Material.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Álvarez, B., López, M. M., & Biosca, E. G. (2007). Influence of native microbiota on survival of Ralstonia solanacearum phylotype II in river water microcosms. Applied and Environmental Microbiology, 73(22), 7210–7217. [CrossRef]
- Aslam, M. N., Mukhtar, T., Hussain, M. A., & Raheel, M. (2017). Assessment of resistance to bacterial wilt incited by Ralstonia solanacearum in tomato germplasm. Journal of Plant Diseases and Protection, 124(6), 585–590. [CrossRef]
- Barik, S., Reddy, A. C., Ponnam, N., Kumari, M., C, A. G., Reddy D C, L., Petikam, S., & Gs, S. (2020). Breeding for bacterial wilt resistance in eggplant (Solanum melongena L.): Progress and prospects. Crop Protection, 137, 105270. [CrossRef]
- Bi-Hao, C., Jian-Jun, L., Yong, W., & Guo-Ju, C. (2009). Inheritance and identification of SCAR marker linked to bacterial wilt-resistance in eggplant. African Journal of Biotechnology, 8(20), 5201–5207. http://www.academicjournals.org/AJB.
- Bindal, S., & Srivastava, S. (2019). Bacterial wilt of solanaceous crops: Sign, symptoms and management. Agrica, 8(2), 134. [CrossRef]
- Bita, C. E., & Gerats, T. (2013). Plant tolerance to high temperature in a changing environment: Scientific fundamentals and production of heat stress-tolerant crops. Frontiers in Plant Science (Vol. 4, Issue JUL). Frontiers Research Foundation. [CrossRef]
- Bocsanczy, A. M., Espindola, A. S., & Norman, D. J. (2019). Whole-genome sequences of Ralstonia solanacearum strains, emerging pathogens of blueberry in Florida. [CrossRef]
- Champoiseau, P. G., Jones, J. B., & Allen, C. (2009). Ralstonia solanacearum Race 3 Biovar 2 causes tropical losses and temperate anxieties. Plant Health Progress, 10(1). [CrossRef]
- Chellemi, D. O., Dankers, H. A., Olson, S. M., Hodge, N. C., & Scott, J. W. (1994). Evaluating bacterial wilt-resistant tomato genotypes using a regional approach. J. AMER. SOC. HORT. SCI (Vol. 119, Issue 2).
- Chiang, K. S., Liu, H. I., & Bock, C. H. (2017). A discussion on disease severity index values. Part I: warning on inherent errors and suggestions to maximize accuracy. Annals of Applied Biology, 171(2), 139–154. [CrossRef]
- Collins, E. J., Bowyer, C., Tsouza, A., & Chopra, M. (2022). Tomatoes: an extensive review of the associated health impacts of tomatoes and factors that can affect their cultivation. Biology (Vol. 11, Issue 2). MDPI. [CrossRef]
- Deslandes, L., Olivier, J., Theulières, F., Hirsch, J., Feng, D. X., Bittner-Eddy, P., Beynon, J., & Marco, Y. (2002). Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive rrs1-r gene, a member of a novel family of resistance genes. Proceedings of the National Academy of Sciences of the United States of America, 99(4), 2404. [CrossRef]
- Du, H., Wen, C., Zhang, X., Xu, X., Yang, J., Chen, B., & Geng, S. (2019). Identification of a major qtl (Qrrs-10.1) that confers resistance to Ralstonia solanacearum in pepper (capsicum annuum) using slaf-bsa and qtl mapping. International Journal of Molecular Sciences, 20(23). [CrossRef]
- Elshire, R. J., Glaubitz, J. C., Sun, Q., Poland, J. A., Kawamoto, K., Buckler, E. S., & Mitchell, S. E. (2011). A robust, simple genotyping-by-sequencing (GBS) approach for high-diversity species. PLOS ONE, 6(5). [CrossRef]
- Fock, I., Collonnier, C., Purwito, A., Luisetti, J., Souvannavong, V., Vedel, F., Servaes, A., Ambroise, A., Kodja, H., Ducreux, G., & Sihachakr, D. (2000). Resistance to bacterial wilt in somatic hybrids between Solanum tuberosum and Solanum phureja. Plant Science, 160(1), 165–176. [CrossRef]
- Glaubitz, J. C., Casstevens, T. M., Lu, F., Harriman, J., Elshire, R. J., Sun, Q., & Buckler, E. S. (2014). TASSEL-GBS: A high-capacity genotyping by sequencing analysis pipeline. PLOS ONE, 9(2). [CrossRef]
- Grimault, V., Prior, P., & Anaïs, G. (1995). A monogenic dominant resistance of tomato to bacterial wilt in Hawaii 7996 is associated with plant colonization by pseudomonas solanacearum. Journal of Phytopathology, 143(6), 349–352. [CrossRef]
- Hong Hai, T. T., Esch, E., & Wang, J. F. (2008a). Resistance to Taiwanese race 1 strains of Ralstonia solanacearum in wild tomato germplasm. European Journal of Plant Pathology, 122(4), 471–479. [CrossRef]
- Hong Hai, T. T., Esch, E., & Wang, J. F. (2008b). Resistance to Taiwanese race 1 strains of Ralstonia solanacearum in wild tomato germplasm. European Journal of Plant Pathology, 122(4), 471–479. [CrossRef]
- Hong, J. C., Norman, D. J., Reed, D. L., Timur Momol, M., & Jones, J. B. (2012). Diversity among Ralstonia solanacearum strains isolated from the southeastern United States. Phytopathology, 102(10), 924–936. [CrossRef]
- Hong, J. K., Jang, S. J., Lee, Y. H., Jo, Y. S., Yun, J. G., Jo, H., Park, C. J., & Kim, H. J. (2018). Reduced bacterial wilt in tomato plants by bactericidal peroxyacetic acid mixture treatment. Plant Pathology Journal, 34(1), 78–84. [CrossRef]
- Huang, M., Liu, X., Zhou, Y., Summers, R. M., & Zhang, Z. (2019). BLINK: A package for the next level of genome-wide association studies with both individuals and markers in the millions. GigaScience, 8(2). [CrossRef]
- Huet, G. (2014). Breeding for resistance to Ralstonia solanacearum. Frontiers in Plant Science (Vol. 5, Issue DEC). Frontiers Media S.A. [CrossRef]
- Ji, P., Allen, C., Sanchez-Perez, A., Yao, J., Elphinstone, J. G., Jones, J. B., & Momol, M. T. (2007). New diversity of Ralstonia solanacearum strains associated with vegetable and ornamental crops in Florida. Plant Disease, 91(2), 195–203. [CrossRef]
- Jung, E. J., Joo, H. J., Choi, S. Y., Lee, S. Y., Jung, Y. H., Lee, M. H., Kong, H. G., & Lee, S.-W. (2014). Resistance evaluation of tomato germplasm against bacterial wilt by Ralstonia solanacearum. Research in Plant Disease, 20(4), 253–258. [CrossRef]
- Kalpage, M. D., & De Costa, D. M. (2014). Isolation of bacteriophages and determination of their efficiency in controlling Ralstonia solanacearum causing bacterial wilt of tomato. Tropical Agricultural Research (Vol. 26, Issue 1).
- Kim, S. G., Hur, O. S., Ro, N. Y., Ko, H. C., Rhee, J. H., Sung, J. S., Ryu, K. Y., Lee, S. Y., & Baek, H. J. (2016a). Evaluation of resistance to Ralstonia solanacearum in tomato genetic resources at seedling stage. Plant Pathology Journal, 32(1), 58–64. [CrossRef]
- Kim, S. G., Hur, O. S., Ro, N. Y., Ko, H. C., Rhee, J. H., Sung, J. S., Ryu, K. Y., Lee, S. Y., & Baek, H. J. (2016b). Evaluation of resistance to Ralstonia solanacearum in tomato genetic resources at seedling stage. Plant Pathology Journal, 32(1), 58–64. [CrossRef]
- Kumar, S., Ramanjini Gowda, P. H., Saikia, B., Debbarma, J., Velmurugan, N., & Chikkaputtaiah, C. (2018). Screening of tomato genotypes against bacterial wilt (Ralstonia solanacearum) and validation of resistance-linked DNA markers. Australasian Plant Pathology, 47(4), 365–374. [CrossRef]
- Kumar, S., Stecher, G., & Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33(7), 1870–1874. [CrossRef]
- Kwon, J. S., Nam, J. Y., Yeom, S. I., & Kang, W. H. (2021). Leaf-to-whole plant spread bioassay for pepper and Ralstonia solanacearum interaction determines the inheritance of resistance to bacterial wilt for further breeding. International Journal of Molecular Sciences, 22(5), 1–14. [CrossRef]
- Lebeau, A., Daunay, M. C., Frary, A., Palloix, A., Wang, J. F., Dintinger, J., Chiroleu, F., Wicker, E., & Prior, P. (2011). Bacterial wilt resistance in tomato, pepper, and eggplant: Genetic resources respond to diverse strains in the Ralstonia solanacearum species complex. Phytopathology, 101(1), 154–165. [CrossRef]
- Lee, J. H., Jang, K. S., Choi, Y. H., Kim, J.-C., & Choi, G. J. (2015). Development of an efficient screening system for resistance of tomato cultivars to Ralstonia solanacearum. Research in Plant Disease, 21(4), 290–296. [CrossRef]
- Lu, Y., Rao, S., Huang, F., Cai, Y., Wang, G., & Cai, K. (2016). Effects of biochar amendment on tomato bacterial wilt resistance and soil microbial amount and activity. International Journal of Agronomy, 2016. [CrossRef]
- Maji A. T. (2012a). Application of principal component analysis for rice germplasm characterization and evaluation. Journal of Plant Breeding and Crop Science, 4(6). [CrossRef]
- Maji A. T. (2012b). Application of principal component analysis for rice germplasm characterization and evaluation. Journal of Plant Breeding and Crop Science, 4(6). [CrossRef]
- Mcavoy, T., Freeman, J. H., Rideout, S. L., Olson, S. M., & Paret, M. L. (2012). Evaluation of grafting using hybrid rootstocks for management of bacterial wilt in field tomato production. HORTSCIENCE (Vol. 47, Issue 5).
- Mohan, V., Gupta, S., Thomas, S., Mickey, H., Charakana, C., Chauhan, V. S., Sharma, K., Kumar, R., Tyagi, K., Sarma, S., Gupta, S. K., Kilambi, H. V., Nongmaithem, S., Kumari, A., Gupta, P., Sreelakshmi, Y., & Sharma, R. (2016). Tomato fruits show wide phenomic diversity but fruit developmental genes show low genomic diversity. PLOS ONE, 11(4). [CrossRef]
- Morel, A., Peeters, N., Vailleau, F., Barberis, P., Jiang, G., Berthomé, R., & Guidot, A. (2018). Plant pathogenicity phenotyping of Ralstonia solanacearum strains. Methods in Molecular Biology, 1734, 223–239. [CrossRef]
- Muthoni, J., Shimelis, H., & Melis, R. (2012). Management of bacterial wilt [Rhalstonia solanacearum Yabuuchi et al., 1995] of potatoes: An opportunity for host resistance in Kenya. Journal of Agricultural Science, 4(9). [CrossRef]
- Newton B-Mensah, I., Osei, K., & Prempeh, R. N. A. (2021). Screening tomato genotypes for bacterial wilt disease (Ralstonia solanacearum) resistance in Ghana. European Journal of Agriculture and Food Sciences, 3(5), 1–8. [CrossRef]
- Newton Boakye-Mensah, I. (2020). Evaluation of tomato (solanum lycopersicum l) genotypes for bacterial wilt (Ralstonia solanacearum) resistance in Ghana. https://ir.ucc.edu.gh/xmlui.
- Patil, V. U., Gopal, J., & Singh, B. P. (2012). Improvement for bacterial wilt resistance in potatoes by conventional and biotechnological approaches. Agricultural Research (Vol. 1, Issue 4, pp. 299–316). Springer. [CrossRef]
- Perveen, R., Suleria, H. A. R., Anjum, F. M., Butt, M. S., Pasha, I., & Ahmad, S. (2015). Tomato (solanum lycopersicum) carotenoids and lycopene chemistry; metabolism, absorption, nutrition, and allied health claims—a comprehensive review. Critical Reviews in Food Science and Nutrition, 55(7), 919–929. [CrossRef]
- Prior, P. (2005). How complex is the “Ralstonia solanacearum species complex? https://www.researchgate.net/publication/37628297.
- Ritchie, H., Rosado, P., & Roser, M. (2023). Agricultural production. Our World in Data. https://ourworldindata.org/agricultural-production.
- Rochette, N. C., Rivera-Colón, A. G., & Catchen, J. M. (2019). Stacks 2: Analytical methods for paired-end sequencing improve RADseq-based population genomics. Molecular Ecology, 28(21), 4737–4754. [CrossRef]
- Roy, S. J., Tucker, E. J., & Tester, M. (2011). Genetic analysis of abiotic stress tolerance in crops. Current Opinion in Plant Biology, 14(3), 232–239. [CrossRef]
- S. K. Kaushik. (2011). Genetics of fruit yield and its contributing characteristics in tomato (Solanum lycopersicom). Journal of Agricultural Biotechnology and Sustainable Development, 3(10). [CrossRef]
- Schmidt, P., Hartung, J., Bennewitz, J., & Hans-Peter, P. (2019). Heritability in plant breeding on a genotype-difference basis. Genetics, 212(4), 991–1008. [CrossRef]
- Shaheen, M. R., Ayyub, C. M., Amjad, M., & Waraich, E. A. (2016). Morpho-physiological evaluation of tomato genotypes under high-temperature stress conditions. Journal of the Science of Food and Agriculture, 96(8), 2698–2704. [CrossRef]
- Singh, S., Gautam, R. K., Singh, D. R., Sharma, T. V. R. S., Sakthivel, K., & Roy, S. D. (2015). Genetic approaches for mitigating losses caused by bacterial wilt of tomato in tropical islands. European Journal of Plant Pathology (Vol. 143, Issue 2, pp. 205–221). Kluwer Academic Publishers. [CrossRef]
- Wang, G., Kong, J., Cui, D., Zhao, H., Niu, Y., Xu, M., Jiang, G., Zhao, Y., & Wang, W. (2019). Resistance against Ralstonia solanacearum in tomatoes depends on the methionine cycle and the γ-aminobutyric acid metabolic pathway. Plant Journal, 97(6), 1032–1047. [CrossRef]
- Yadav, R. K., Tripathi, M. K., Tiwari, S., Tripathi, N., Asati, R., Patel, V., Sikarwar, R. S., & Payasi, D. K. (2023). Breeding and genomic approaches towards the development of fusarium wilt resistance in chickpea. Life (Vol. 13, Issue 4). MDPI. [CrossRef]
- Yuliar, Asi Nion, Y., & Toyota, K. (2015). Recent trends in control methods for bacterial wilt diseases caused by Ralstonia solanacearum. Microbes and Environments (Vol. 30, Issue 1, pp. 1–11). Japanese Society of Microbial Ecology. [CrossRef]
- Zhou, R., Wu, Z., Wang, X., Rosenqvist, E., Wang, Y., Zhao, T., & Ottosen, C. O. (2018). Evaluation of temperature stress tolerance in cultivated and wild tomatoes using photosynthesis and chlorophyll fluorescence. Horticulture Environment and Biotechnology, 59(4), 499–509. [CrossRef]
- Zohoungbogbo, H., Quenum, A., Honfoga, J., Chen, J. R., Achigan-Dako, E., Kenyon, L., & Hanson, P. (2021). Evaluation of resistance sources of tomato (Solanum lycopersicum L.) to phylotype I strain of Ralstonia solanacearum species complex in Benin. Agronomy, 11(8). [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).