1. Introduction
Cell-free DNA (cfDNA) is known to be immunologically active, like lipopolysaccharides, and binds to Toll-like receptor (TLR) to release inflammatory cytokines such as tumor necrosis factor-alpha (TNF-α) and interleukin-6 (IL-6) into the peripheral blood and trigger inflammatory responses and tissue damage [
1]. Sepsis is a serious infection with a 30-day mortality rate of 24.4% according to a recent study [
2]. Accuracy and rapidity are essential for the diagnosis and treatment of sepsis, and predicting the prognosis of sepsis in advance is of utmost importance, so various biomarkers have been developed to determine the diagnosis, treatment response, and prognosis of sepsis, among which cfDNA is known to be a useful biomarker [
3].
It has also been found that in sepsis, the administration of nucleases lowers the blood levels of cfDNAs and, as a result, an improved prognosis for sepsis patients [
4]. The origin of these cfDNA is known to be from infected bacteria or cell necrosis due to various diseases or apoptosis [
5]. However, its exact origin has not been proven to date. In addition, previous studies have shown that synthesized cytosine-phosphate-guanosine oligodeoxynucleotides (CpG-ODNs) trigger inflammatory responses and sepsis, confirming that cfDNA triggers the same response and that the mechanism occurs by binding to TLRs [
6]. No animal studies have been conducted to determine whether sepsis-derived cfDNA can have biological effects when administered to normal hosts, and if so, the mechanisms are shrouded in mystery. Increased oxidative stress, which results in inflammatory responses and tissue damage, leads to the production of myriad oxidative species in the mitochondria of infected tissues, resulting in immunoredox imbalance [
7]. In this regard, this could be an important mechanism to explain prognosis. However, there is little documentation on whether sepsis-derived cfDNA administered to macrophages (RAW 264.7 cell) and healthy mice elicits an inflammatory response, and whether the mechanism relies solely on immune responses or immune redox imbalances. To address this, we hypothesized that cfDNA from septic mice and healthy humans would exhibit differential immune redox responses via immunoredox-related gene pathways.
Although cell line experiments have shown that cfDNA can induce immune responses by acting on TLRs [
8], there are few reports of biological effects in animal experiments. Furthermore, nothing is known about the biological effect of cfDNA in vitro. It is known that there are clear differences in inflammatory responses and redox balance between healthy individuals and septic groups [
9]. Given these, we speculated that cfDNA from septic mice and healthy humans would exhibit different immune redox responses via immuno-redox-related gene pathways.
The objectives of the study are the following:
(1) To demonstrate the proinflammatory action and oxidant effects of cfDNA from septic mice in macrophage cell lines and animal models.
(2) To evaluate the increased production of reactive oxygen species (ROS) and nitric oxide (NO) and the decrease of glutathione peroxidase to demonstrate the effect of sepsis-derived cfDNA as an oxidant in the macrophage cell line RAW 264.7 and healthy ICR mice.
(3) To evaluate the increase in TNF-α in the macrophage cell line RAW 264.7 and healthy ICR mice to assess the inflammatory effect of cfDNA derived from septic mice.
3. Discussion
This investigation substantiates the discernible variance in immunoredox response within cell-free DNA (cfDNA) extracted from septic murine subjects and non-septic human individuals, elucidating distinct patterns along immunoredox-associated genetic pathways. The affirmative outcomes are derived from a comprehensive analysis encompassing both in vitro experimentation and in vivo observations, collectively providing four lines of compelling evidence.
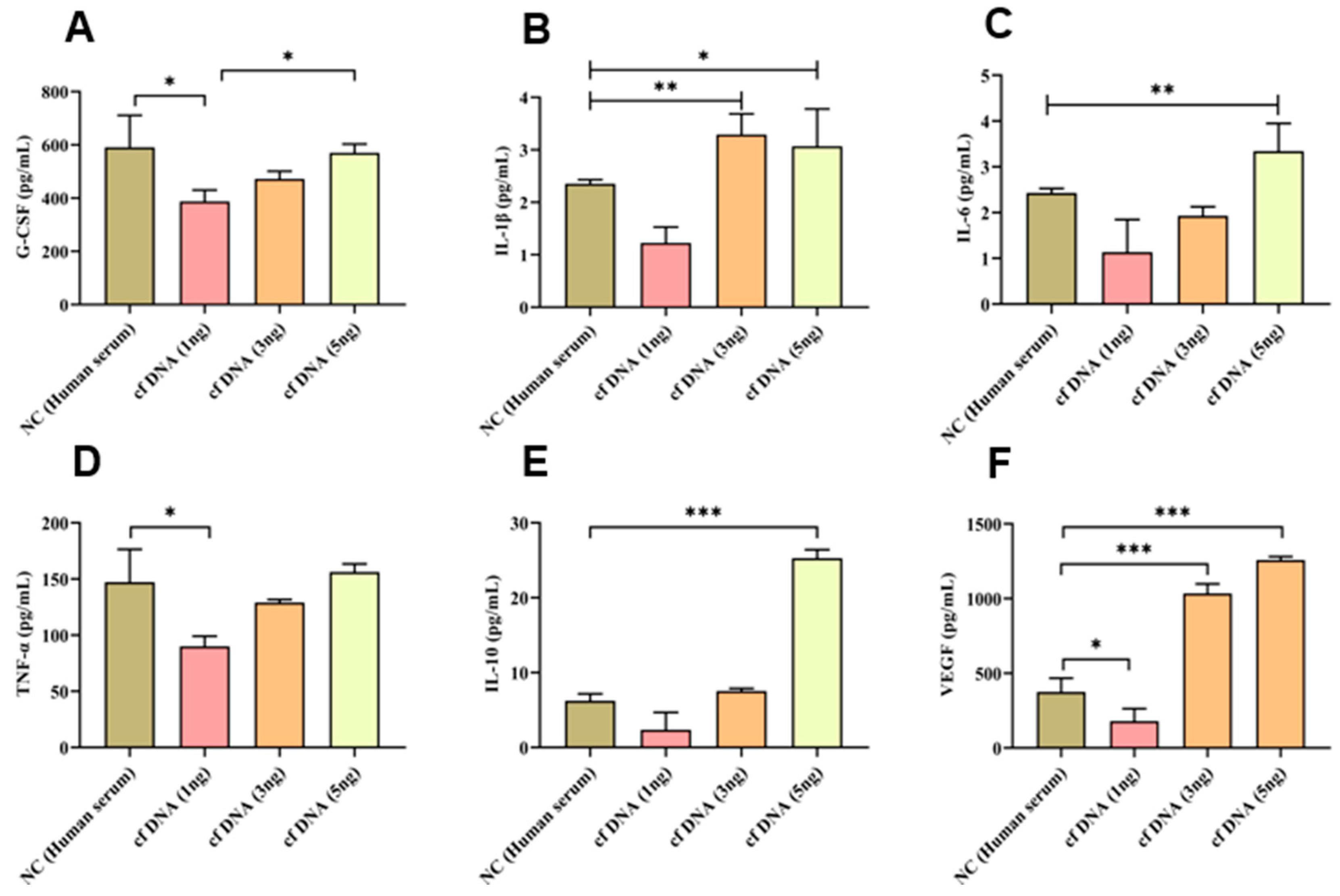
First, of the cytokines measured after administration of sepsis-derived cfDNA to macrophages 264.7, G-CSF, TNF-α, IL-10, VEGF, IL-1β, and IL-6 all increased exponentially in proportion to the concentration of cfDNA administered, with G-CSF, TNF-α, IL-10, and VEGF showing significant differences. cfDNA refers to DNA fragments that are present in body fluids such as blood, urine, and saliva [
11]. These fragments are released into the bloodstream by cells undergoing apoptosis (programmed cell death) or necrosis (cell death due to injury or disease). cfDNA can be used for a variety of diagnostic and research purposes, including detecting gene mutations, monitoring disease progression, and identifying fetal DNA from maternal blood during pregnancy [
12]. cfDNA can activate the innate immune system, the body's first line of defense against pathogens and foreign substances. Once released into the bloodstream, cfDNA can be recognized by immune cells, such as macrophages and dendritic cells, and trigger an inflammatory response [
13]. This response can help fight infections or cancer, but if it is improperly regulated, it can lead to the development of autoimmune diseases or other inflammatory conditions. "DAMP" stands for "damage-associated molecular pattern," and refers to molecules that are released from damaged or dying cells and can trigger an immune response or inflammation. cfDNA can act as a DAMP, activating the innate immune system and causing inflammation [
14]. However, the immune response can also be regulated by other factors in the body, such as anti-inflammatory cytokines, to prevent excessive inflammation. The balance between pro- and anti-inflammatory signals is essential for maintaining immune homeostasis and preventing the development of chronic inflammatory diseases. Our research will continue to better understand the mechanisms by which cfDNA interacts with the innate immune system and to develop new theragnostic based on this knowledge.
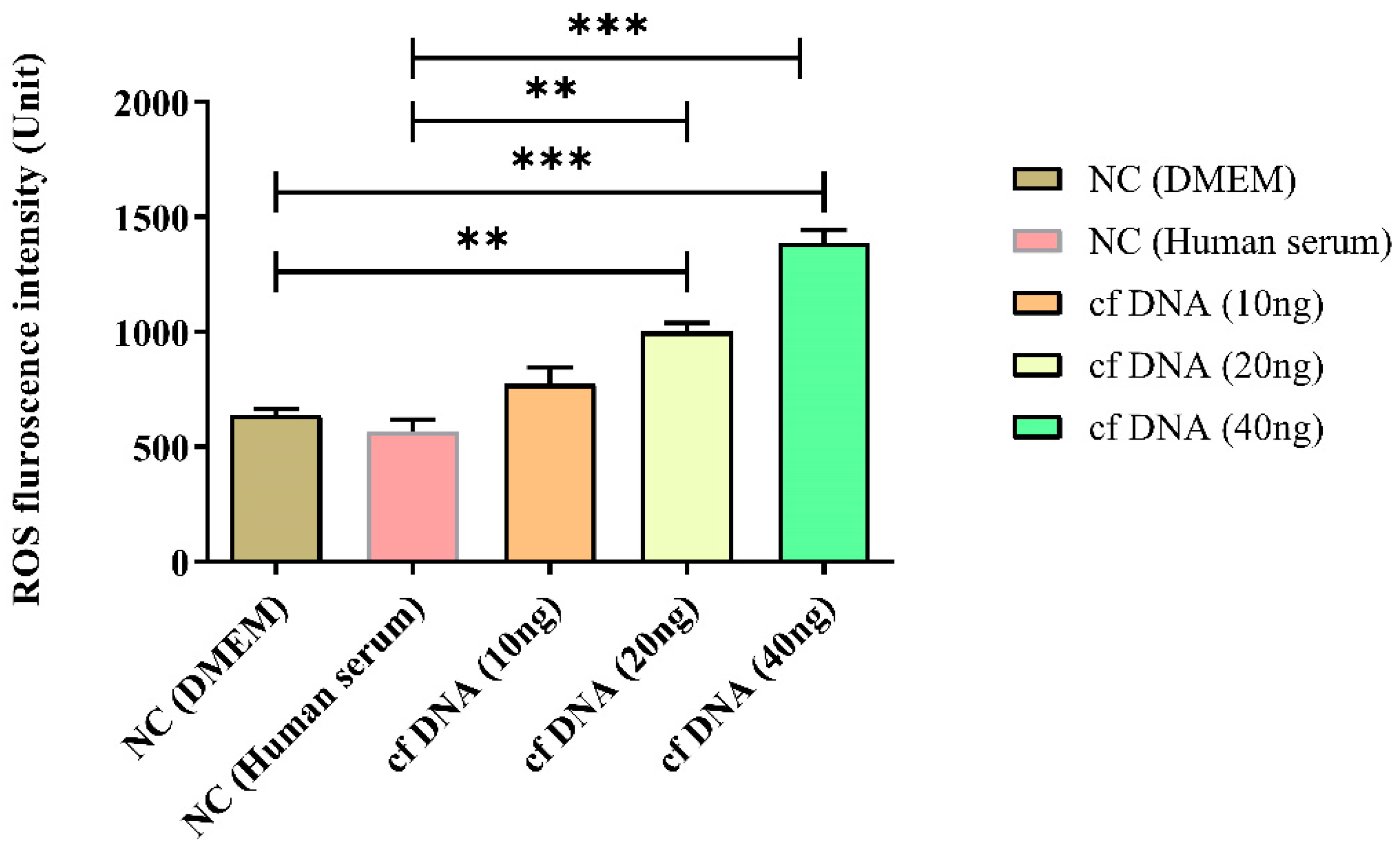
Second, we examined the impact of the immune redox system after administration of sepsis-derived cfDNA to a macrophage line and found a significant increase in total ROS, NO, and catalase concentrations. In addition, cell viability assays showed a significant decrease in viability proportional to the concentration of cfDNA administered. The redox response of cfDNA refers to the ability of cfDNA to induce oxidative stress in cells and tissues. Oxidative stress occurs when there is an imbalance between the production of ROS and the ability of cells to detoxify them. ROS can damage cellular components such as DNA, proteins, and lipids, leading to inflammation and cell death. cfDNA can induce oxidative stress by activating signaling transduction pathways that lead to ROS production, such as the NADPH oxidase pathway [
15]. The redox response of cfDNA is thought to play an important role in the pathogenesis of a variety of diseases, including cancer, autoimmune diseases, and cardiovascular disease [
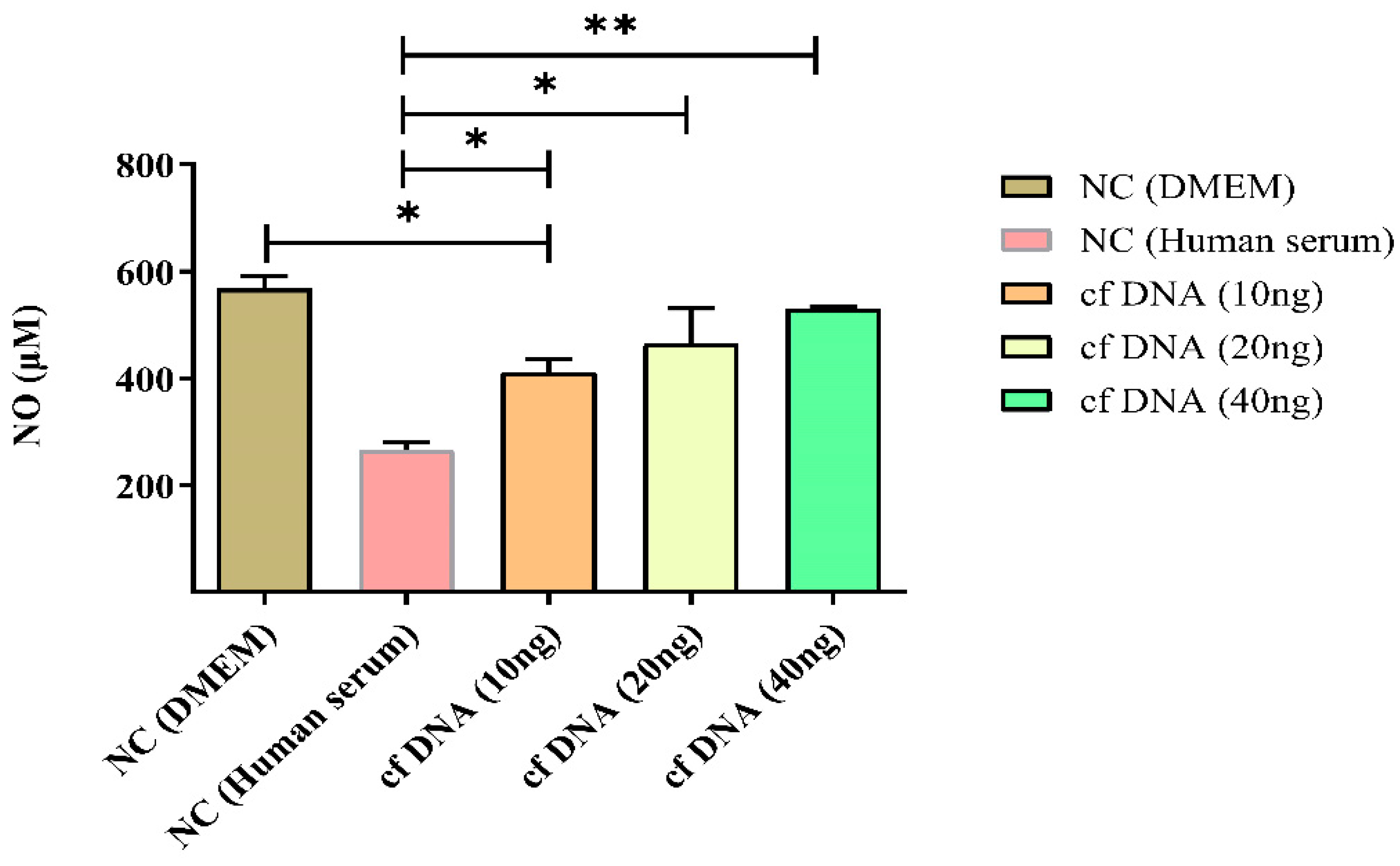
16]. Nitrosative response refers to the production of NO and other reactive nitrogen species (RNS) in response to various stimuli, including cfDNA. NO is a signaling molecule that plays a key role in regulating blood flow, immune function, and neurotransmission. However, excessive production of NO and other RNS can lead to oxidative and nitrosative stress, which can damage cellular components and lead to various diseases. cfDNA can activate signaling pathways that lead to the expression of inducible NO synthase (iNOS), the enzyme that catalyzes NO production, leading to the production of NO and other RNS.
The involvement of cfDNA's nitrosative response is believed to be significant in the development of different illnesses, such as cancer, autoimmune diseases, and cardiovascular disease [
17]. Various oxidant and antioxidant enzymes may be involved in the redox imbalances induced by cfDNA. For example, cfDNA can activate the NADPH oxidase pathway, leading to the production of ROS such as superoxide anion (O
2-) and hydrogen peroxide (H
2O
2) [
18]. However, ROS has the ability to react with biological components such as DNA, proteins, and lipids, causing oxidative stress and inflammation. Cells can create several antioxidant enzymes, such as superoxide dismutase (SOD), catalase, and GPx, to neutralize ROS and avoid oxidative damage to counteract the effects of ROS [
19]. Our research is underway to better understand the mechanisms by which cfDNA induces oxidative stress of ROS and NO and to develop new diagnostic and therapeutic approaches based on this knowledge. The exact mechanisms by which cfDNA induces redox imbalances and the involvement of specific oxidant and antioxidant enzymes are still under investigation.
Third, we evaluated the impact of sepsis-derived cfDNA on the immune system after injection in healthy mice. The results showed that in healthy ICR mice, blood TNF-α levels increased in proportion to cfDNA concentration, with substantial differences, but in C3H/HeJ animals, whose function is hindered by mutations in the receptor-4 gene, TNF-α levels did not differ despite variances in cfDNA concentration. The inflammasome is a multiprotein complex that plays a key role in the regulation of inflammation and immune responses. The inflammasome is activated in response to a variety of stimuli, including pathogens, cellular stress, and DAMPs such as cfDNA. The inflammasome is made up of numerous proteins, including NLRP3, ASC, and caspase-1. When activated, the inflammasome can convert pro-inflammatory cytokines like IL-1β and IL-18 into active versions, resulting in an inflammatory response. Inflammasome activation can also result in pyroptosis, a type of programmed cell death associated with inflammation [
20]. The exact mechanisms by which cfDNA activates the inflammasome and the downstream effects on immune function are still under investigation.
The cfDNA can activate the inflammasome, a multi-protein complex that plays an important role in regulating inflammation and immune responses [
21]. The inflammasome can be activated, resulting in pyroptosis, a type of programmed cell death associated with inflammation and the release of pro-inflammatory cytokines. These cytokines can enhance the inflammatory response and contribute to the development of a variety of inflammatory diseases. The exact mechanisms by which cfDNA activates the inflammasome and triggers pyroptosis are still under investigation.
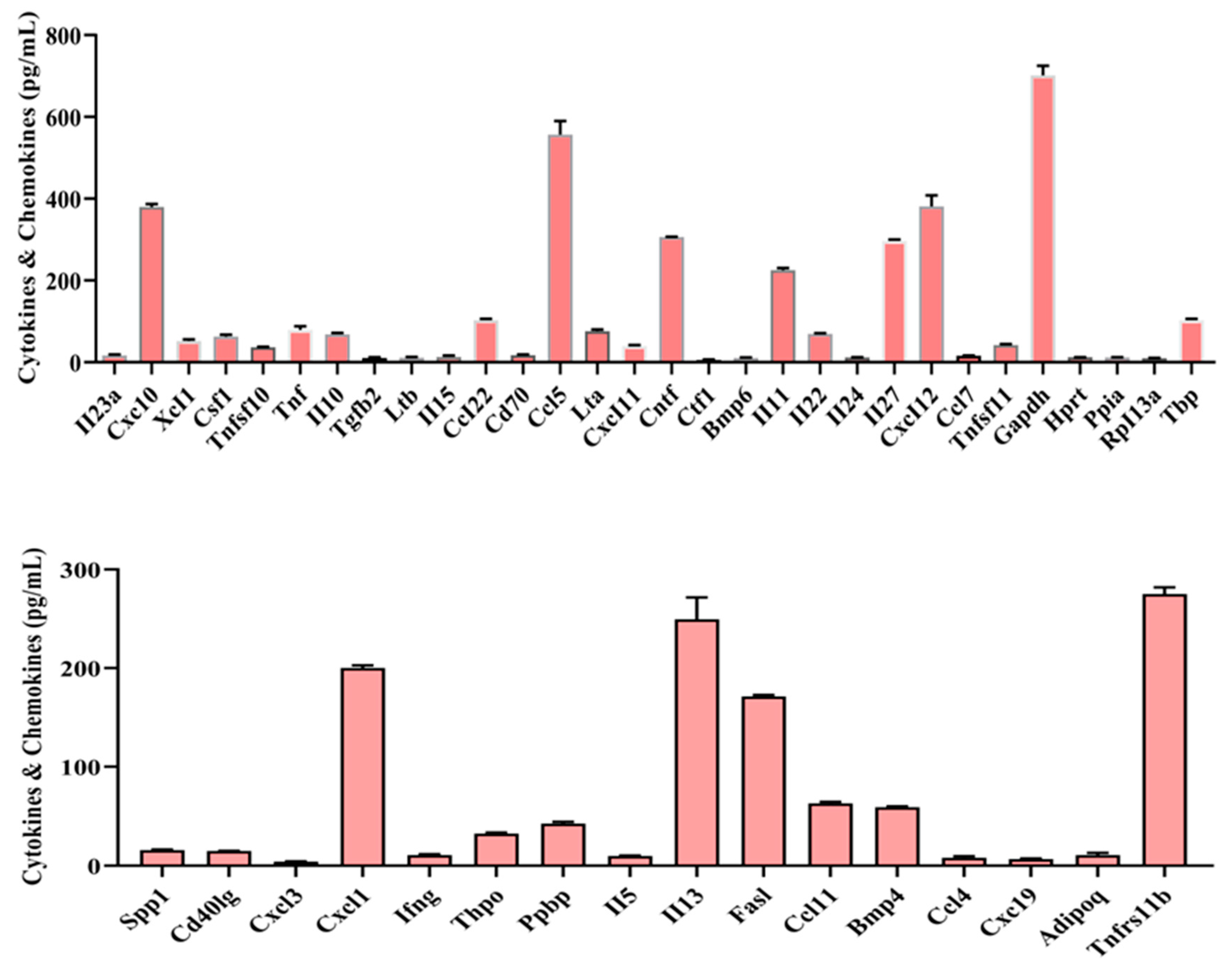
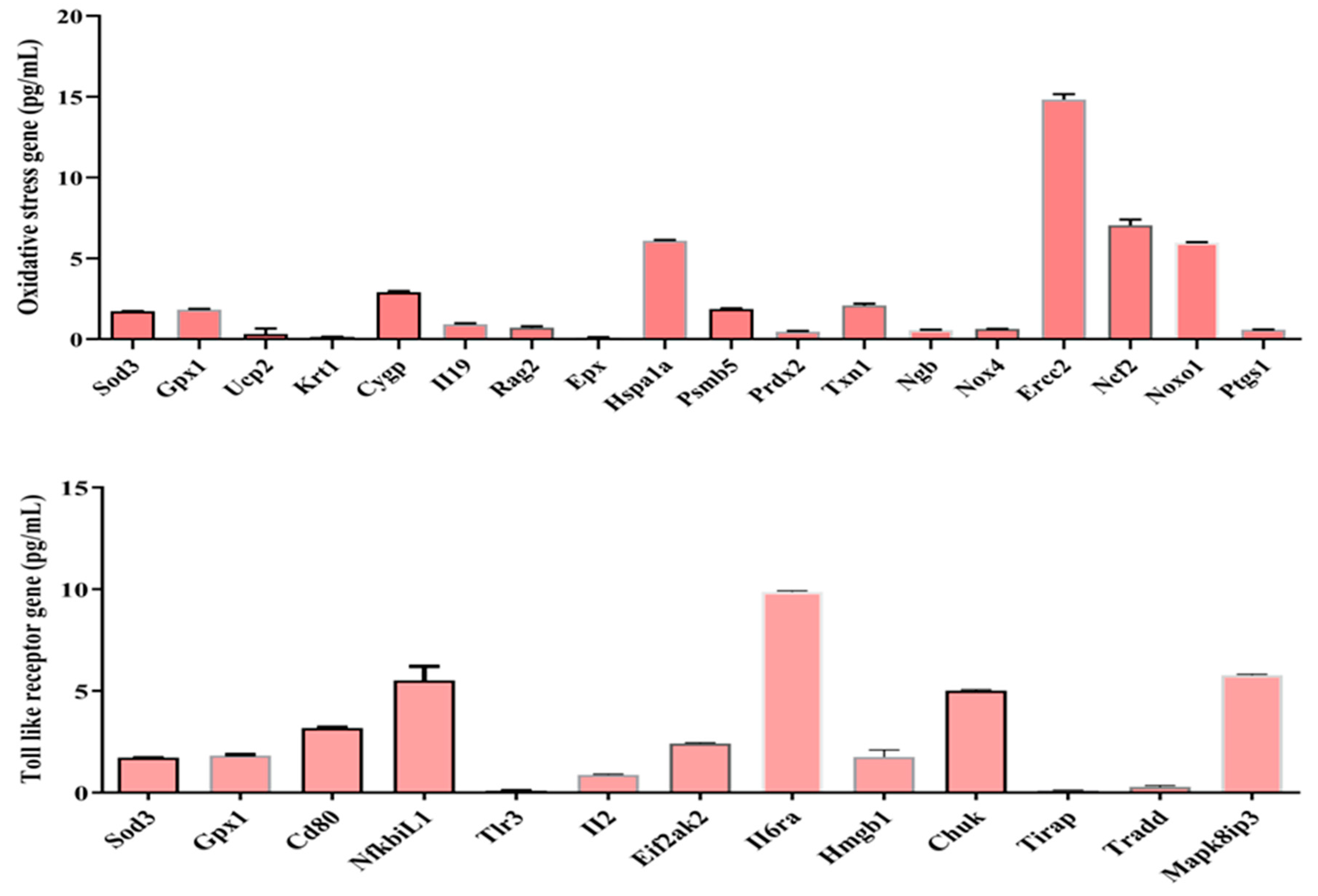
Fourth, in macrophage line experiments, we examined oxidative stress gene and TLR gene expression after administration of sepsis-derived cfDNA to macrophage RAW 264.7. 46 cytokines & and chemokines genes, 18 oxidative stress genes, and 13 TLR genes were expressed at varying concentrations. These results may support the immunoredox imbalance at the gene level mentioned earlier. To my knowledge, this is the first identification of these genes, so further studies are needed to clarify their roles and functions. The signaling pathway by which cfDNA activates the innate immune system involves the recognition of cfDNA by TLRs [
22], which are expressed on the surface of immune cells such as macrophages and dendritic cells. TLRs can recognize specific patterns in cfDNA, such as unmethylated CpG motifs [
23], which are commonly found in bacterial DNA but are rarely in mammalian DNA. This recognition sets off a chain of events that results in the activation of transcription factors like NF-κB and the generation of pro-inflammatory cytokines such as IL-6 and TNF-α [
24]. Activation of these signaling pathways can also recruit other immune cells such as neutrophils and natural killer cells, to the site of inflammation. The exact mechanisms by which cfDNA activates these signaling pathways and their downstream effects on immune function are still under investigation. Our findings on oxidative stress and receptor gene expression suggest which genes are primarily involved in oxidative stress and which genes are involved in the activation of signaling pathways by which receptors recognize cfDNA and cfDNA produces inflammatory cytokines and directs other immune cells to the site of inflammation. Taken together, cfDNA can induce redox imbalance by activating various signaling pathways that lead to the production of ROS and other reactive species. For example, cfDNA can activate the NADPH oxidase pathway, which is a major source of ROS in cells. This pathway involves the activation of enzymes such as NOX2 and NOX4, which produce superoxide anion (O2-) and hydrogen peroxide (H
2O
2). These ROS can react with cellular components such as DNA, proteins, and lipids, leading to oxidative stress and inflammation.
The exact mechanisms by which cfDNA induces redox imbalances and the involvement of specific oxidant and antioxidant enzymes are still under investigation. However, my findings suggest that targeting these pathways may be a promising approach to developing new theragnostic for intractable human diseases.
4. Materials and Methods
4.1. Reagents
Hyclone Laboratories, Inc. (South Logan, USA) provided Dulbecco's Modified Eagle Medium (DMEM) and 10% fetal bovine serum (FBS). GibcoTM, Invitrogen Corporation (Auckland, N.Z., USA) provided a 1% penicillin/streptomycin solution. Dojindo Molecular Technologies, Inc. (Rockville, MD, USA) provided the Cell Counting Kit-8 (CCK-8), and Sigma-Aldrich (St. Louis, MO, USA) provided the 2'7'-dichlorodihydrofluorescin diacetate (DCFH-DA). iNtRON Biotechnology (Sungnam, Gyeonggi-do, South Korea) provided the NO reagent (Griess reagent kit). The Catalase test kit and the GPx assay kit were provided by Biovision Inc. (Milpitas, CA, USA). Bio-Rad Laboratories (San Diego, CA, USA) provided the Bio-Plex® Multiple Bead Suspension Array kit for TNF- α and IL-6. DAEJUNG (Siheung-si, Gyeonggi-do, South Korea) provided 30% H2O2 solution, and cfDNA was measured using the MagListoTM cfDNA extraction kit from BIONEER (Daejeon, South Korea).
4.2. Macrophage RAW 264.7 cells culture and cfDNA hit into cells
Sigma-Aldrich (catalog number 91062702) and the American Type Culture Collection (ATCC® TIB-71TM) supplied the RAW 264.7 cell lines. They were cultivated in DMEM high glucose media supplemented with 10% FBS (consistently used batch) and 1% penicillin/streptomycin, under a 37°C, 5% CO2, and 95% humidity culture environment. Cells were detached using a cell scraper after reaching 90% confluence, passaged at a 1:6 ratio onto T-75 flasks, and all cell culture-related products came from the same batch.
Cell culture continued up to passage 10, with cells progressing from the third passage. Before phenotypic and functional studies, cells were frozen and underwent the next two passages (e.g., cells from passage 3 were thawed, cultured to passage 5, and then subjected to further investigations). Regular testing for Mycoplasma contamination was conducted on the cells. Cells were cultivated by only one person to prevent variations in culture methods. Subsequently, 96-well plate were seeded with 3 x 106 cells, and septic mice-derived cfDNA was added at various concentrations, including 1ng/mL, 5ng/mL, and 10ng/mL, and incubated for 24 hours at 37°C. As a control experiment, the same cells were seeded in a 96-well plate and normal control (NC)-derived cfDNA extracted using Pioneer cfDNA synthesis kit was added and incubated at the same temperature and time course. After the incubation period, we measured cell viability at 450nm using a microplate reader. We also used a microplate reader to measure assays such as ROS, NO, GPx, and catalase (CAT) at various wavelengths according to the manufacturer’s protocol instructions.
4.3. Cell viability assay
In this study, we used the cell counting Kit-8 (CCK-8) reagent from Quanti-MAXTM (Seoul, Korea) to evaluate cell viability according to the manufacturer's instructions. To begin the method, RAW 264.7 cells were seeded at a density of 3 x 106 in DMEM media in a 96-well plate. After that, the plate was placed in an incubator at 37°C with 5% CO2 for 24 hours. Following a wash with 1X-PBS, the cells were exposed to varying concentrations of cfDNA and treated with various experimental waters in DMEM media for 24 hours. Following treatment, the cells were washed three times with 1X-PBS, and each well was given 10 µL of CCK-8 reagent coupled with DMEM medium. After 3 hours of incubation at 37°C, cell viability was measured at 450nm using a SpectraMax® ABS Plus absorbance microplate reader (Molecular Devices, San Jose, CA, USA).
4.4. Measurement of total ROS
The manufacturer's instructions were followed while measuring ROS levels in RAW 264.7 macrophage cells with the 2′-7′-dichloro-dihydro-fluorescein diacetate (DCFHDA) reagent for ROS detection (Abcam, Cambridge, MA, USA) as previously described [
25].
4.5. Measurement of NO levels
NO levels in RAW 264.7 macrophage cells were also analyzed using the Griess reagent (Promega Corp., Madison, WI, USA) per the manufacturer's instructions as previously described [
25,
26].
4.6. Measurement of antioxidant activities (Catalase)
Catalase levels, an inherent antioxidant enzyme, were assessed 24 hours post-treatment with various concentrations of cfDNA using a BioVision kit (BioVision, Inc., Milpitas, CA, USA) following the provided guidelines. In summary, 78µL of samples (10µL from the stock and 68µL of assay buffer) for the catalase assay were introduced to a 96-well microplate and left to incubate for 30 minutes. The optical density of catalase at 570nm was gauged using SpectraMax® ABS Plus (Molecular Devices, San Jose, CA, USA). Catalase activity outcomes were quantified in nmol/min/mL.
4.7. Extraction and purification of cfDNA, along with the quantification of genes related to cytokines & and chemokines, TLR, and oxidative stress using qPCR DNA.
The entirety of cfDNA from the samples was extracted employing the bead-beating technique. Assessment of cfDNA quality and quantity ensured using a NanoDrop 1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). Subsequently, the total DNA was diluted to attain final concentrations of 10 ng/μL, which were then employed to generate amplicons for pyrosequencing. Various qPCR screening kits targeting genes related to oxidative stress, TLR, and cytokines & and chemokines were employed for analysis. The cfDNA fragment of the vector was amplified, utilizing oligonucleotide primers acquired from Bioneer Co. Ltd. qPCR was executed in 96-well PCR plates with a final volume of 20μL, employing GreenstarTM master mix (Bioneer Co. Ltd.), cfDNA templates, and qPCR primers. For each primer/cfDNA pair, duplicate reactions were established, with Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) serving as the internal control. The qPCR reactions were started in a QuantGene 9600 (Bioneer Co. Ltd.) with a 15-minute heating stage, followed by 40-45 cycles of 95°C (15s) and 58°C (30s), with an extension at 72°C (30s). The threshold cycle (Ct) approach was used to do relative measurement of gene expression.
4.8. Measurement of cytokines
The Milliplex® MAP Mouse Cytokine/Chemokine Magnetic Bead Panel 96-well plate test by Millipore Corporation in Billerica, MA, USA, is a Luminex-based multiplex technology used for cytokine and chemokine profiling. G-CSF, IL-1, IL-6, IL-10, TNF-α, and VEGF were assessed using a multiplex immunoassay according to the manufacturer's instructions as previously described [
26] [
27].
4.9. Experimental design
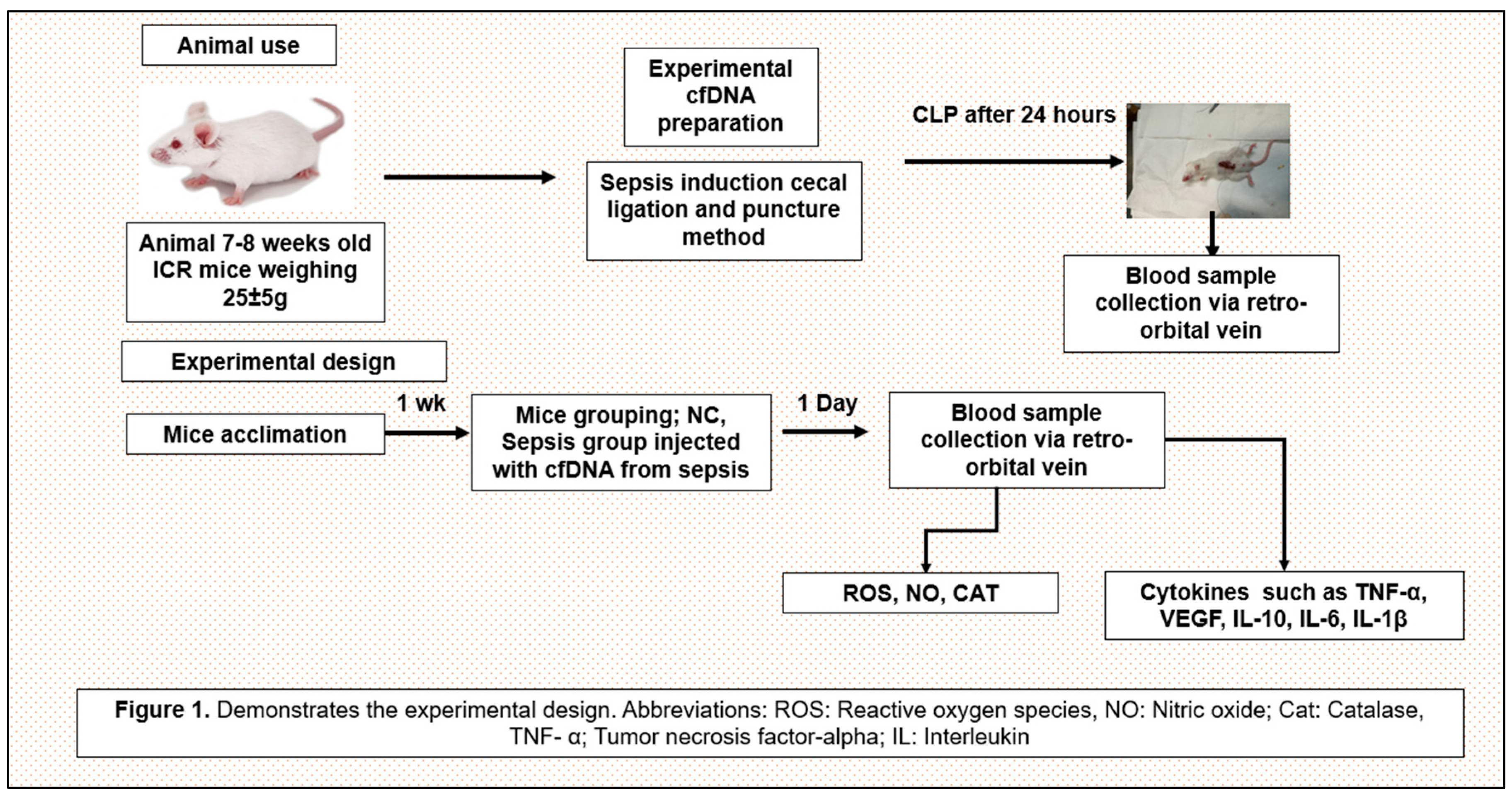
A total of 295 healthy ICR mice used for animal experiments were purchased from KOATECH (Pyeongtaek-si, Gyeonggi-do, South Korea) and DBL. Co., Ltd (Eumseong-gun, Chungcheongbuk-do, South Korea). Mice were 7-9 weeks old, male, and weighed 25-30g. Animal experiments were conducted by national ethical guidelines and were submitted to and approved by the Institutional Animal Care and Use Committee (R1(2019-6).
Twenty-four hours after inducing sepsis in specific pathogen-free ICR mice via cecal ligation and puncture, blood was collected from the orbital venous plexus of the mice, centrifuged, and the resulting plasma cfDNA was extracted and purified. Sepsis-derived cfDNA at concentrations of 100ng/mL, 200ng/mL, and 400ng/mL was injected into a vein of a healthy ICR mouse and the cfDNA remaining in the blood was measured 24 hours later. TNF-α was also measured 24 hours after the same concentrations of sepsis-derived cfDNA was injected into healthy ICR mice. TNF-α and cfDNA were measured using the ELISA kit described above.
Cecal ligation and puncture (CLP) procedure
(a) After the mouse is anesthetized, the hairs on the mouse's skin are shaved with a razor and sterilized.
(b) The mouse's abdomen is exposed and a midline laparotomy is performed.
(c) The cecum in the abdomen is identified and exposed, and then the cecum is tied distally with a silk thread.
(d) Puncture the distal portion of the tied cecum twice with a 23-gauge needle. Then gently press on the cecum to squeeze out the contents of the cecum.
(e) Then, after placing the cecum into the abdominal cavity, close the incised abdominal muscles with running sutures.
(f) The abdominal skin incision is closed by applying a metal clip.
(g) 1 ml of normal saline is injected intraperitoneally and the mouse is placed on an electric pad to recover from anaesthesia [
28].
Figure 10 shows the experimental design.
4.10. Data management and statistical analysis
Graph Pad Prism version 8.0 software (Graph Pad Software, La Jolla, CA, USA) was used for statistical analysis. The comparisons were performed using one-way ANOVA and multiple comparison tests (Tukey). A P value of less than 0.05 was considered statistically significant.
Figure 1.
(A) G-CSF, (B) IL-1β, (C), IL-6 (D) TNF-α, VEGF, (E) IL-10 (F) VEGF release at 24 hours after inducing different concentrations of cfDNA, Data are shown as mean ± standard deviation (S.D.). n=5* p < 0.05, ** p < 0.01, *** p < 0.001, ****p < 0.0001 (one-way ANOVA, Tukey’s post-hoc test).
Figure 1.
(A) G-CSF, (B) IL-1β, (C), IL-6 (D) TNF-α, VEGF, (E) IL-10 (F) VEGF release at 24 hours after inducing different concentrations of cfDNA, Data are shown as mean ± standard deviation (S.D.). n=5* p < 0.05, ** p < 0.01, *** p < 0.001, ****p < 0.0001 (one-way ANOVA, Tukey’s post-hoc test).
Figure 2.
ROS intensity at 24 hours after inducing different concentrations of cfDNA. Data were shown as mean ± standard deviation (S.D.). n=5* p < 0.05, ** p < 0.01, *** p < 0.001 (one-way ANOVA, Tukey’s post-hoc test).
Figure 2.
ROS intensity at 24 hours after inducing different concentrations of cfDNA. Data were shown as mean ± standard deviation (S.D.). n=5* p < 0.05, ** p < 0.01, *** p < 0.001 (one-way ANOVA, Tukey’s post-hoc test).
Figure 3.
NO intensity at 24 hours after inducing different concentrations of cfDNA. NC: normal control, cf: cell-free. Data were shown as mean ± standard deviation (S.D.). n=5* p < 0.05, ** p < 0.01 (one-way ANOVA, Tukey’s post-hoc test).
Figure 3.
NO intensity at 24 hours after inducing different concentrations of cfDNA. NC: normal control, cf: cell-free. Data were shown as mean ± standard deviation (S.D.). n=5* p < 0.05, ** p < 0.01 (one-way ANOVA, Tukey’s post-hoc test).
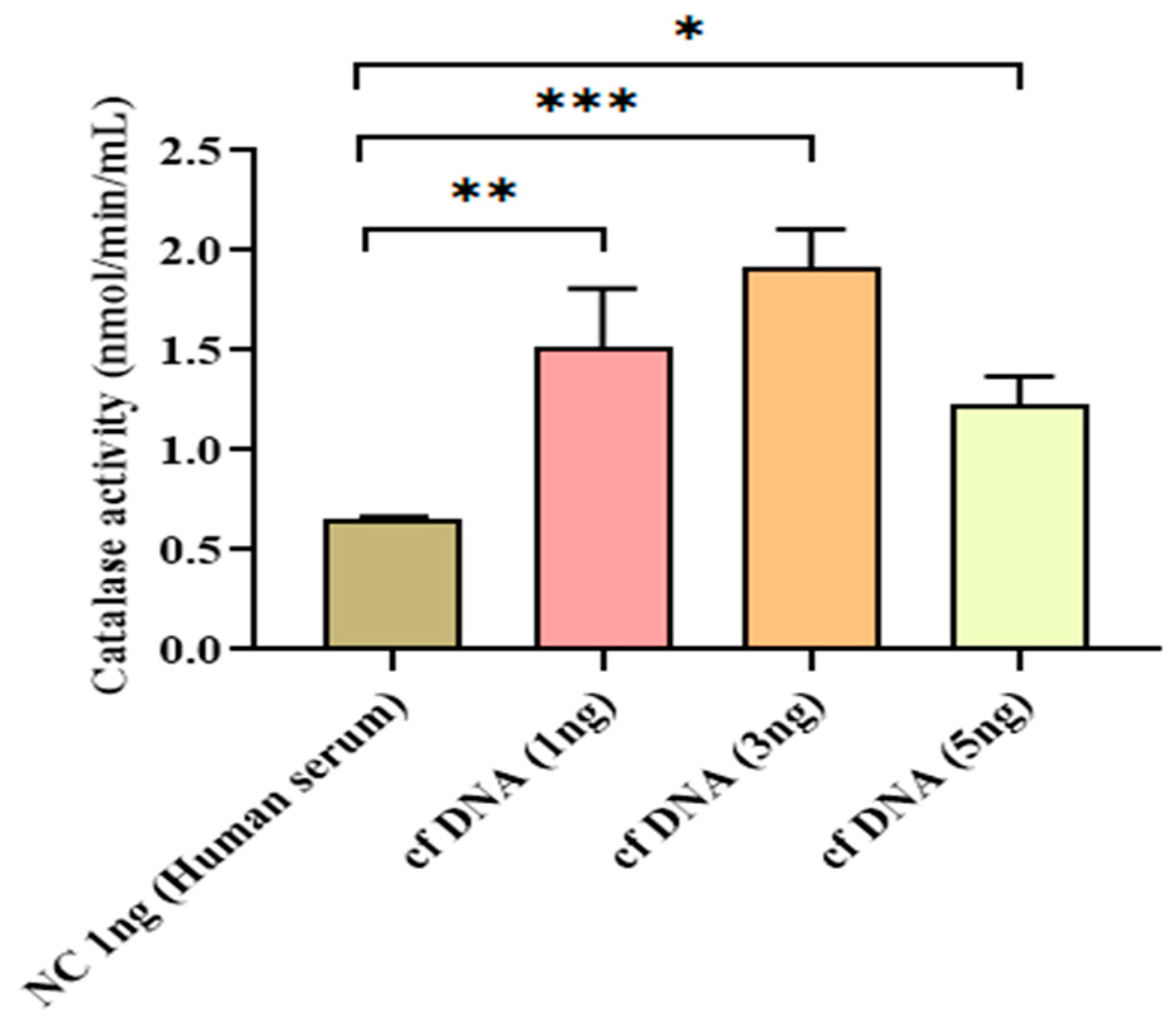
Figure 4.
Catalase activity at 24 hours after inducing different concentrations of cfDNA. Data were shown as mean ± standard deviation (S.D.). n=5* p < 0.05, ** p < 0.01, *** p < 0.001 (one-way ANOVA, Tukey’s post-hoc test).
Figure 4.
Catalase activity at 24 hours after inducing different concentrations of cfDNA. Data were shown as mean ± standard deviation (S.D.). n=5* p < 0.05, ** p < 0.01, *** p < 0.001 (one-way ANOVA, Tukey’s post-hoc test).
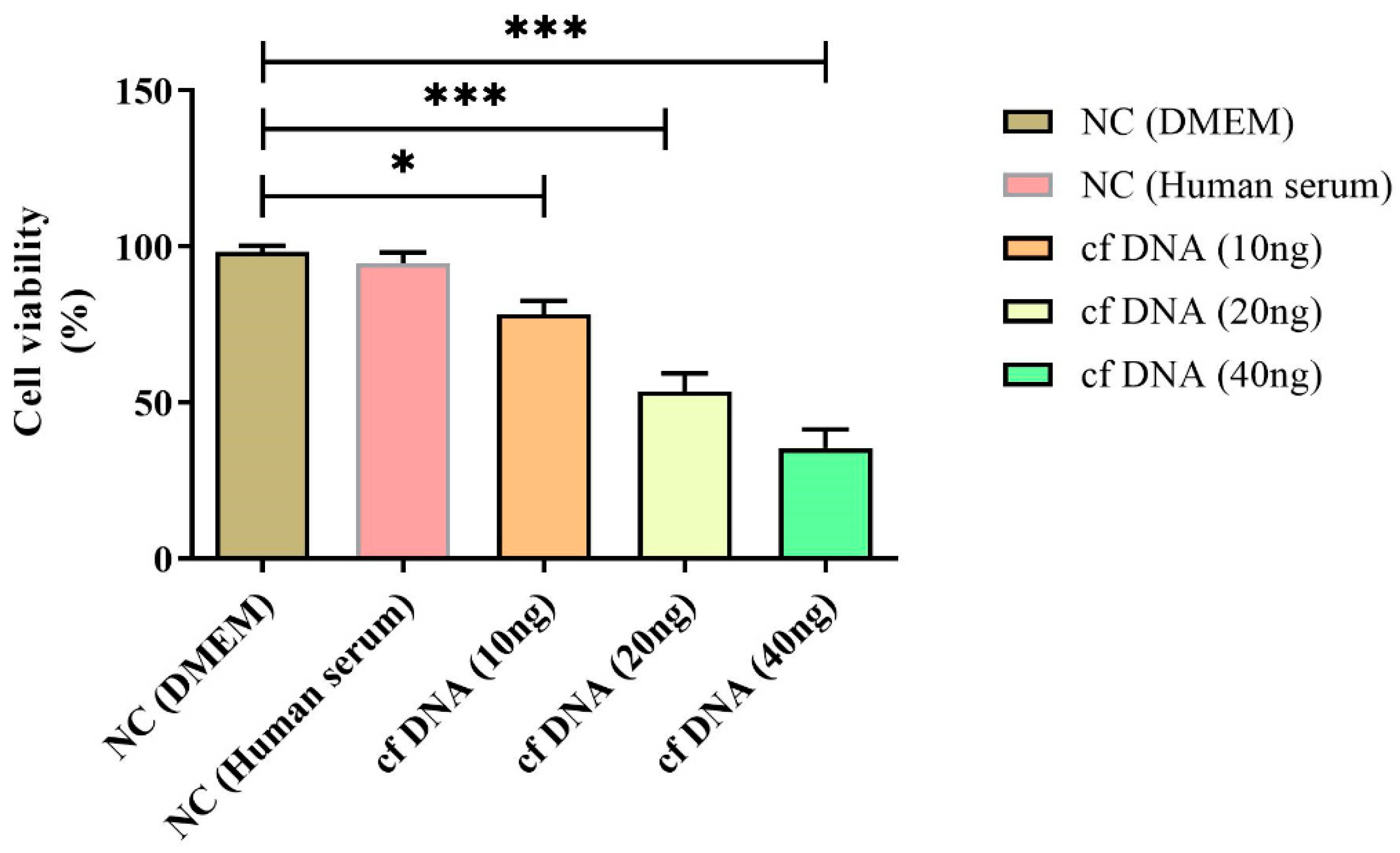
Figure 5.
Cell viability at 24 hours after inducing different concentrations of cfDNA. Data are shown as mean ± standard deviation (S.D.). n=5* p < 0.05, ** p < 0.01, *** p < 0.001, ****p < 0.0001 (one-way ANOVA, Tukey’s post-hoc test).
Figure 5.
Cell viability at 24 hours after inducing different concentrations of cfDNA. Data are shown as mean ± standard deviation (S.D.). n=5* p < 0.05, ** p < 0.01, *** p < 0.001, ****p < 0.0001 (one-way ANOVA, Tukey’s post-hoc test).
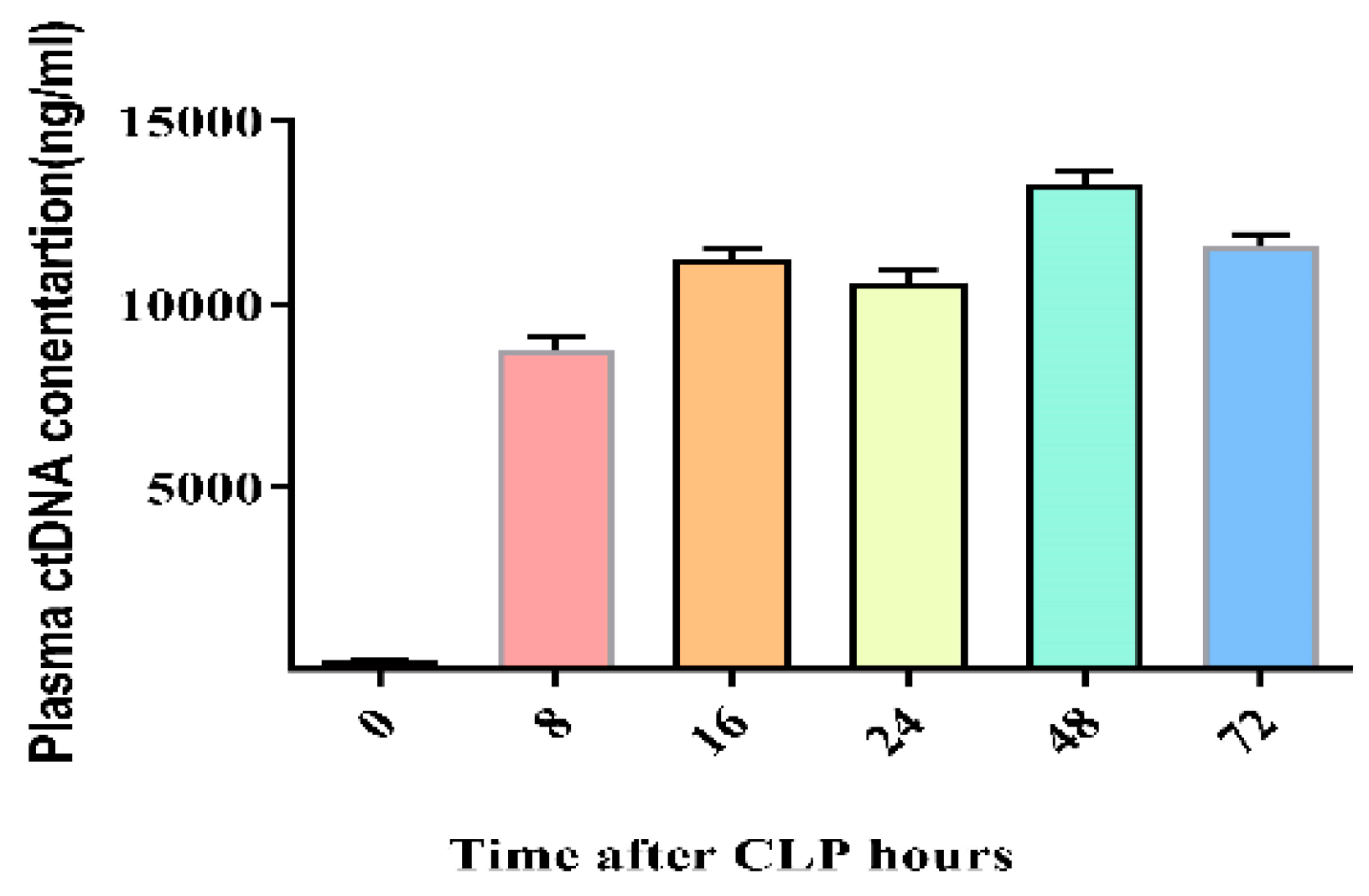
Figure 6.
Optimal Cecal ligation and puncture (CLP) time kinetics for cfDNA in CLP-induced mouse sepsis. Data were shown as mean ± standard deviation (S.D.). n=5* p < 0.05 (one-way ANOVA, Tukey’s post-hoc test).
Figure 6.
Optimal Cecal ligation and puncture (CLP) time kinetics for cfDNA in CLP-induced mouse sepsis. Data were shown as mean ± standard deviation (S.D.). n=5* p < 0.05 (one-way ANOVA, Tukey’s post-hoc test).
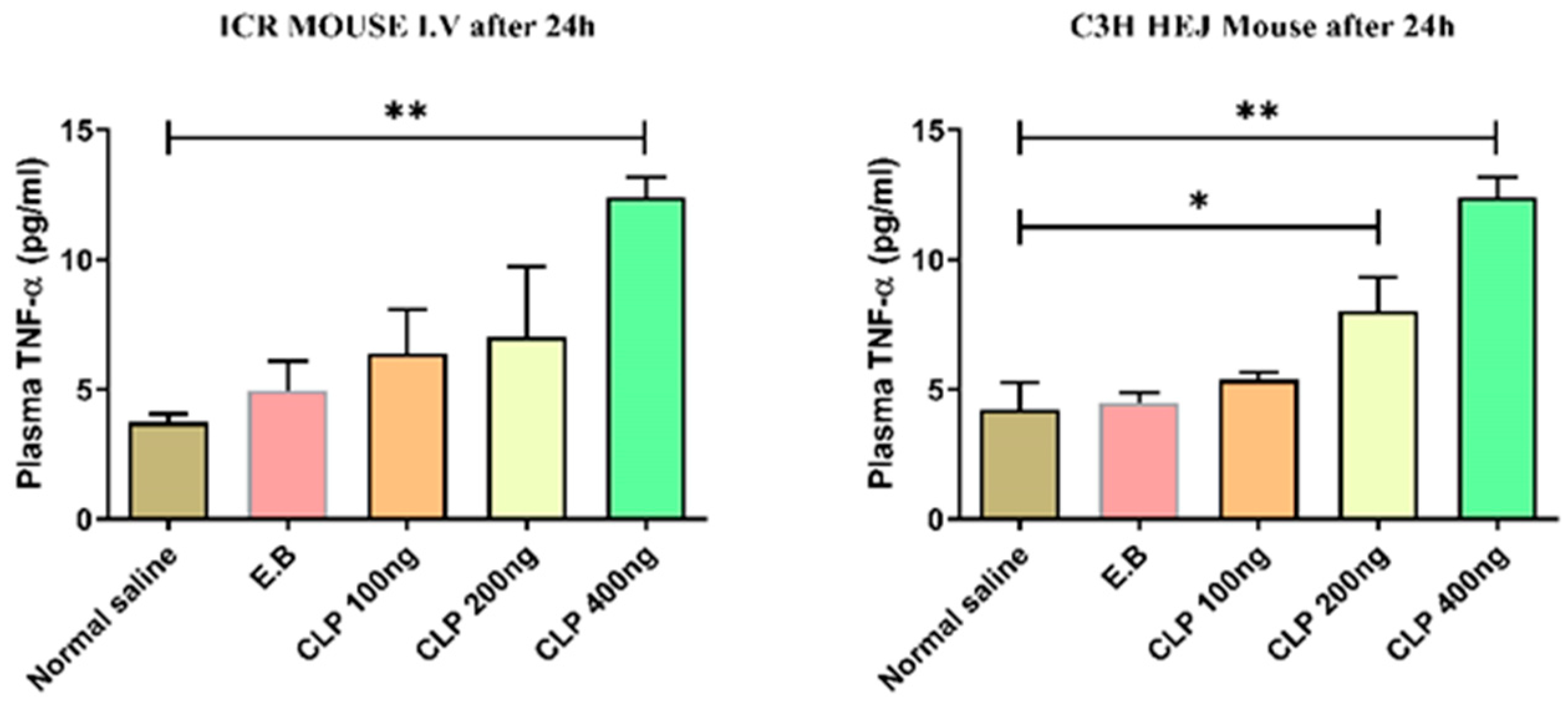
Figure 8.
Tumor necrosis factor-α (TNF-α) levels after intravenous injection of sepsis-derived cfDNA and control cfDNA into healthy ICR and C3H/HeJ mice. E.B: elution buffer. Data were shown as mean ± standard deviation (S.D.). n=5, * p < 0.05, ** p < 0.01 (one-way ANOVA, Tukey’s post-hoc test).
Figure 8.
Tumor necrosis factor-α (TNF-α) levels after intravenous injection of sepsis-derived cfDNA and control cfDNA into healthy ICR and C3H/HeJ mice. E.B: elution buffer. Data were shown as mean ± standard deviation (S.D.). n=5, * p < 0.05, ** p < 0.01 (one-way ANOVA, Tukey’s post-hoc test).
Figure 9.
Sepsis-derived cfDNA-induced cytokines & and chemokines, TLR, and oxidative stress gene expression in vitro.
Figure 9.
Sepsis-derived cfDNA-induced cytokines & and chemokines, TLR, and oxidative stress gene expression in vitro.
Figure 10.
Schematic diagram of the experimental design.
Figure 10.
Schematic diagram of the experimental design.