Submitted:
27 December 2023
Posted:
28 December 2023
You are already at the latest version
Abstract
Keywords:
1. Anthracnose: A Foremost Devastating Threat in Lupins
2. Colletotrichum lupini: The Causative Agent for Anthracnose in Lupins
2.1. C. lupini Belongs to the Colletotrichum Acutatum Species Complex (CAsc)
2.2. Colletotrichum lupini Reproduces Clonally, Exhibiting Distinct Morphologies and Virulence Patterns Both between and within Clonal Lineages

2.3. Recently Unveiled Complete Genome Sequences May Provide Substantial Potential for Uncovering the Pathogenicity Factors of This Devastating Plant Pathogen
3. Pathogenesis and Epidemiology of Anthracnose in Lupins: Insights into Primary and Secondary Infection Mechanisms
3.1. Dissemination of Anthracnose through Primary Infection: Asymptomatic Infected Seeds as the Principal Mode of Spread
3.2. Colletotrichum lupini Adopts a Hemibiotrophic Lifestyle with an Initial Brief Biotrophic Phase Succeeded by a Highly Destructive Necrotrophic Development
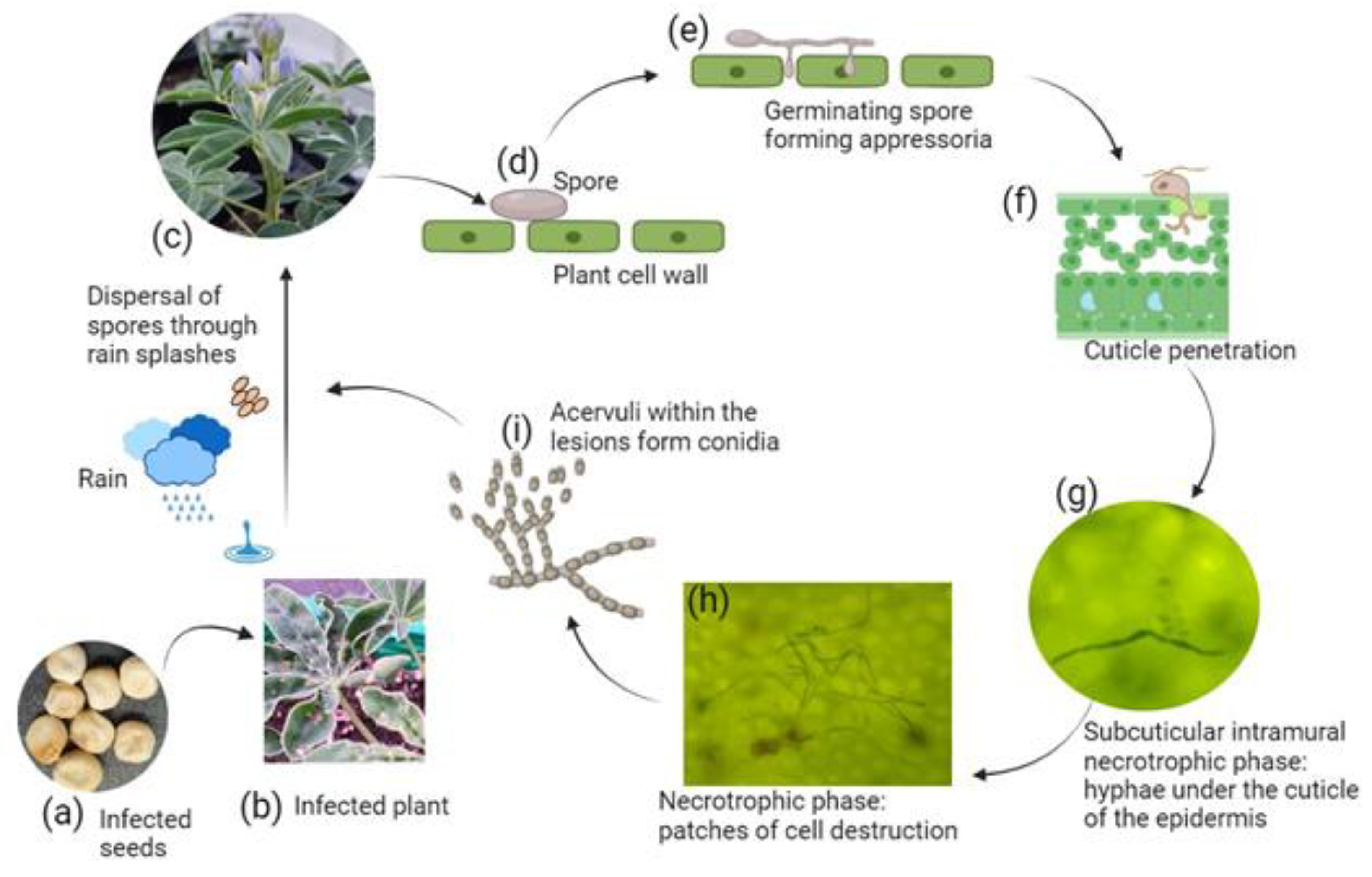
3.3. Colletotrichum lupini Exhibits a Wide Temperature Tolerance Range
4. Molecular Mechanisms Governing the Interaction between C. lupini and Lupins
4.1. Unveiling the Main Molecular Regulatory Pathways in the Interaction between C. lupini and lupins: Contributions from -Omics Analysis
4.1.1. Omics Analysis of the Hemibiotrophic Lifestyle of C. lupini to Uncover Putative Key Virulence Factors
4.1.2. –Omics Analysis of the Host Plant Response to Highlight Major Defense Pathways Involved
4.2. Plant and Fungal Secondary Metabolites as Key Regulators of the Pathogenic Interaction
5. Disease Management through Prophylaxis
5.1. The Utilization of Disease-Free Seeds as a Primary Prophylactic Measure for Mitigating Anthracnose Spread
5.2. Enhancing Precision and Efficiency in C. lupini Detection: Advances in Molecular Methodologies and Tools
6. Insights into the Genetics of Anthracnose Resistance in Domesticated Lupin Species and Implications for Crop Improvement
7. Addressing Technical Challenges to Improve Lupin Biotechnologies for Facilitating Anthracnose-Resistant Variety Breeding
8. Conclusions & Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- FAOSTAT Food and Agricultural Organization of the United Nations; 2021;
- Guilengue, N.; Silva, M. do C.; Talhinhas, P.; Neves-Martins, J.; Loureiro, A. Subcuticular–Intracellular Hemibiotrophy of Colletotrichum Lupini in Lupinus Mutabilis. Plants 2022, 11. [CrossRef]
- Peres, N.A.; Timmer, L.W.; Adaskaveg, J.E.; Correll, J.C. Lifestyles of Colletotrichum Acutatum. Plant Disease 2005, 89, 784–796. [CrossRef]
- Sweetingham, M.W.; Cowling, W.A.; Buirchell, B.J.; Brown, A.G.P.; Shivas, R.G. Anthracnose of Lupins in Western Australia. Australasian Plant Pathology 1995, 24, 271–271. [CrossRef]
- Shea, G.; Thomas, G.; Buirchell, B.; Salam, M.; McKirdy, S.; Sweetingham, M. Case Study: Industry Response to the Lupin Anthracnose Incursion in Western Australia.; Palta, J.A., Berger, J.B., Ed.; International Lupin Association: Fremantle, Western Australia, September 2008; pp. 425–431.
- Talhinhas, P.; Baroncelli, R.; Le Floch, G. Anthracnose of Lupins Caused by Colletotrichum Lupini : A Recent Disease and a Successful Worldwide Pathogen. Journal of Plant Pathology 2016, 98, 5–14. [CrossRef]
- Książkiewicz, M.; Rychel-Bielska, S.; Plewiński, P.; Bielski, W.; Nuc, M.; Kozak, B.; Krajewski, P.; Jędryczka, M. A Successful Defense of the Narrow-Leafed Lupin against Anthracnose Involves Quick and Orchestrated Reprogramming of Oxidation–Reduction, Photosynthesis and Pathogenesis-Related Genes. Scientific Reports 2022, 12, 8164–8164. [CrossRef]
- Pandey, A.K.; Kumar, A.; Mbeyagala, E.K.; Barbetti, M.J.; Basandrai, A.; Basandrai, D.; Nair, R.M.; Lamichhane, J.R. Anthracnose Resistance in Legumes for Cropping System Diversification. Critical Reviews in Plant Sciences 2023, 42, 177–216. [CrossRef]
- Nirenberg, H.I.; Feiler, U.; Hagedorn, G. Description of Colletotrichum Lupini Comb. Nov. in Modern Terms. Mycologia 2002, 94, 307–320. [CrossRef]
- Sreenivasaprasad, S.; Talhinhas, P. Genotypic and Phenotypic Diversity in Colletotrichum Acutatum, a Cosmopolitan Pathogen Causing Anthracnose on a Wide Range of Hosts. Molecular Plant Pathology 2005, 6, 361–378. [CrossRef]
- Dean, R.; Van Kan, J.A.L.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The Top 10 Fungal Pathogens in Molecular Plant Pathology. Molecular Plant Pathology 2012, 13, 414–430. [CrossRef]
- Gresta, F.; Wink, M.; Prins, U.; Abberton, M.; Caprato, J.; Scarafoni, A.; Hill, G. Lupins in European Cropping Systems. In Legumes in cropping systems; Murphy-Bokern, D., Stoddard, F.L., Watson, C.A., Eds.; CABI International 2017, 2017; Vol. Chapter 6, pp. 88–108.
- Abreha, K.B.; Ortiz, R.; Carlsson, A.S.; Geleta, M. Understanding the Sorghum–Colletotrichum Sublineola Interactions for Enhanced Host Resistance. Frontiers in Plant Science 2021, 12. [CrossRef]
- Baroncelli, R.; Pensec, F.; Lio, D.D.; Boufleur, T.; Vicente, I.; Sarrocco, S.; Picot, A.; Baraldi, E.; Sukno, S.; Thon, M.; et al. Complete Genome Sequence of the Plant-Pathogenic Fungus Colletotrichum Lupini. Molecular Plant-Microbe Interactions 2021, 34, 1461–1464. [CrossRef]
- Talhinhas, P.; Baroncelli, R. Colletotrichum Species and Complexes: Geographic Distribution, Host Range and Conservation Status; Fungal Diversity; Springer Netherlands, 2021; Vol. 110, p. 198; ISBN 1322502100491.
- Msairi, S.; Chliyeh, M.; Touhami, A.O.; Abdelaziz, M.; Alaoui, E. FIRST REPORT OF Colletotrichum Lupini CAUSING ANTHRACNOSE DISEASE ON THE OLIVE FRUITS IN MOROCCO; Article in PLANT CELL BIOTECHNOLOGY AND MOLECULAR BIOLOGY; 2020;
- Dubrulle, G.; Pensec, F.; Picot, A.; Rigalma, K.; Pawtowski, A.; Nicolleau, S.; Harzic, N.; Nodet, P.; Baroncelli, R.; Le Floch, G. Phylogenetic Diversity and Effect of Temperature on Pathogenicity of Colletotrichum Lupini. Plant Disease 2020, 104, 938–950. [CrossRef]
- Talhinhas, P.; Sreenivasaprasad, S.; Neves-Martins, J.; Oliveira, H. Genetic and Morphological Characterization of Colletotrichum Acutatum Causing Anthracnose of Lupins. Phytopathology® 2002, 92, 986–996. [CrossRef]
- Lotter, H.C.; Berger, D.K. Anthracnose of Lupins in South Africa Is Caused by Colletotrichum Lupini Var. Setosum. Australasian Plant Pathology 2005, 34, 385–392. [CrossRef]
- Jayawardena, R.S.; Hyde, K.D.; Damm, U.; Cai, L.; Liu, M.; Li, X.H.; Zhang, W.; Zhao, W.S.; Yan, J.Y. Notes on Currently Accepted Species of Colletotrichum. Mycosphere 2016, 7, 1192–1260. [CrossRef]
- Masi, M.; Nocera, P.; Boari, A.; Zonno, M.C.; Pescitelli, G.; Sarrocco, S.; Baroncelli, R.; Vannacci, G.; Vurro, M.; Evidente, A. Secondary Metabolites Produced by Colletotrichum Lupini, the Causal Agent of Anthachnose of Lupin (Lupinus Spp.). Mycologia 2020, 112, 533–542. [CrossRef]
- Baroncelli, R.; Talhinhas, P.; Pensec, F.; Sukno, S.A.; Floch, G.L.; Thon, M.R. The Colletotrichum Acutatum Species Complex as a Model System to Study Evolution and Host Specialization in Plant Pathogens. Frontiers in Microbiology 2017, 8. [CrossRef]
- Damm, U.; Cannon, P.F.; Woudenberg, J.H.C.; Crous, P.W. The Colletotrichum Acutatum Species Complex. Studies in Mycology 2012, 73, 37–113. [CrossRef]
- Bhunjun, C.S.; Phukhamsakda, C.; Jayawardena, R.S.; Jeewon, R.; Promputtha, I.; Hyde, K.D. Investigating Species Boundaries in Colletotrichum. Fungal Diversity 2021, 107, 107–127. [CrossRef]
- Chen, Y. peng; Wu, T.; Tian, W.H.; Ilyukhin, E.; Hyde, K.D.; Maharachchikumbura S. S. N. Comparative Genomics Provides New Insights into the Evolution of Colletotrichum. Mycosphere 2022, 13, 134–187. [CrossRef]
- Lardner, R.; Johnston, P.R.; Plummer, K.M.; Pearson, M.N. Morphological and Molecular Analysis of Colletotrichum Acutatum Sensu Lato. Mycological Research 1999, 103, 275–285. [CrossRef]
- Alkemade, J.A.; Baroncelli, R.; Messmer, M.M.; Hohmann, P. Attack of the Clones: Population Genetics Reveals Clonality of Colletotrichum Lupini, the Causal Agent of Lupin Anthracnose. Molecular Plant Pathology 2023. [CrossRef]
- Yang, H.A.; Sweetingham, M.W. The Taxonomy of Colletotrichum Isolates Associated with Lupin Anthracnose. Australian Journal of Agricultural Research 1998, 49, 1213–1213. [CrossRef]
- Riegel, R.; Véliz, D.; Von Baer, I.; Quitral, Y.; Muñoz, M. Genetic Diversity and Virulence of Colletotrichum Lupini Isolates Collected in Chile. Tropical Plant Pathology 2010, 35, 144–152. [CrossRef]
- Alkemade, J.A.; Messmer, M.M.; Voegele, R.T.; Finckh, M.R.; Hohmann, P. Genetic Diversity of Colletotrichum Lupini and Its Virulence on White and Andean Lupin. Scientific Reports 2021, 11. [CrossRef]
- Pszczółkowska, A.; Okorski, A.; Jastrzębski, J.P.; Paukszto; Fordoński, G. The Complete Mitogenome of Colletotrichum Lupini Var. Setosum. Mitochondrial DNA Part B: Resources 2016, 1, 37–38. [CrossRef]
- Thomas, G.J.; Sweetingham, M.W.; Yang, H.A.; Speijers, J. Effect of Temperature on Growth of Colletotrichum Lupini and on Anthracnose Infection and Resistance in Lupins. Australasian Plant Pathology 2008, 37, 35–35. [CrossRef]
- Falconí, C.E.; Yánez–Mendizábal, V. Dry Heat Treatment of Andean Lupin Seed to Reduce Anthracnose Infection. Crop Protection 2016, 89, 178–183. [CrossRef]
- Falconí, C.E.; Yánez-Mendizábal, V. Available Strategies for the Management of Andean Lupin Anthracnose. Plants 2022, 11. [CrossRef]
- Falconí, C.E.; Visser, R.G.F.; Van Heusden, S. Influence of Plant Growth Stage on Resistance to Anthracnose in Andean Lupin (Lupinus Mutabilis). Crop and Pasture Science 2015, 66, 729–734. [CrossRef]
- Diggle, A.J.; Salam, M.U.; Thomas, G.J.; Yang, H.A.; O’connell, M.; Sweetingham, M.W. Analytical and Theoretical Plant Pathology AnthracnoseTracer: A Spatiotemporal Model for Simulating the Spread of Anthracnose in a Lupin Field; 2002;
- Semaškienė, R.; Brazauskienė, I.; Lisova, R.; Liepienė, N.; Maknickienė, Z. The Incidence of Anthracnose (Colletotrichum Spp.) on Lupine Seed. Zemdirbyste-Agriculture 2008, 144–150.
- Thomas, G.J.; Sweetingham, M.W. Cultivar and Environment Influence the Development of Lupin Anthracnose Caused by Colletotrichum Lupini. Australasian Plant Pathology 2004, 33, 571–577. [CrossRef]
- Perfect, S.E.; Bleddyn Hughes, H.; O’connell, R.J.; Green, J.R. Colletotrichum: A Model Genus for Studies on Pathology and Fungal-Plant Interactions; 1999. [CrossRef]
- Cannon, P.F.; Damm, U.; Johnston, P.R.; Weir, B.S. Colletotrichum - Current Status and Future Directions. Studies in Mycology 2012, 73, 181–213. [CrossRef]
- O’Connell, R.J.; Thon, M.R.; Hacquard, S.; Amyotte, S.G.; Kleemann, J.; Torres, M.F.; Damm, U.; Buiate, E.A.; Epstein, L.; Alkan, N.; et al. Lifestyle Transitions in Plant Pathogenic Colletotrichum Fungi Deciphered by Genome and Transcriptome Analyses. Nature Genetics 2012, 44, 1060–1065. [CrossRef]
- De Silva, D.D.; Crous, P.W.; Ades, P.K.; Hyde, K.D.; Taylor, P.W.J. Life Styles of Colletotrichum Species and Implications for Plant Biosecurity. Fungal Biology Reviews 2017, 31, 155–168. [CrossRef]
- Dubrulle, G.; Picot, A.; Madec, S.; Corre, E.; Pawtowski, A.; Baroncelli, R.; Zivy, M.; Balliau, T.; Le Floch, G.; Pensec, F. Deciphering the Infectious Process of Colletotrichum Lupini in Lupin through Transcriptomic and Proteomic Analysis. Microorganisms 2020, 8, 1621–1621. [CrossRef]
- Diéguez-Uribeondo, J.; Förster, H.; Adaskaveg, J.E. Digital Image Analysis of Internal Light Spots of Appressoria of Colletotrichum Acutatum; 2003; Vol. 93, pp. 923–923;.
- Diéguez-Uribeondo, J.; Förster, H.; Soto-Estrada, A.; Adaskaveg, J.E. Subcuticular-Intracellular Hemibiotrophic and Intercellular Necrotrophic Development of Colletotrichum Acutatum on Almond. Phytopathology® 2005, 95, 751–758. [CrossRef]
- You, M.; Buirchell, B.; M Siddique, K.H.; Sweetingham, M. A PCR-Based Molecular Marker Applicable for Marker-Assisted Selection for Anthracnose Disease Resistance in Lupin Breeding. Cellular & Molecular Biology Letters 2005, 10, 123–134.
- Wojakowska, A.; Muth, D.; Narozna, D.; Mądrzak, C.; Stobiecki, M.; Kachlicki, P. Changes of Phenolic Secondary Metabolite Profiles in the Reaction of Narrow Leaf Lupin (Lupinus Angustifolius) Plants to Infections with Colletotrichum Lupini Fungus or Treatment with Its Toxin. Metabolomics 2013, 9, 575–589. [CrossRef]
- Yang, H.; Tao, Y.; Zheng, Z.; Shao, D.; Li, Z.; Sweetingham, M.W.; Buirchell, B.J.; Li, C. Rapid Development of Molecular Markers by Next-Generation Sequencing Linked to a Gene Conferring Phomopsis Stem Blight Disease Resistance for Marker-Assisted Selection in Lupin (Lupinus Angustifolius L.) Breeding. Theoretical and Applied Genetics 2013, 126, 511–522. [CrossRef]
- Fischer, K.; Dieterich, R.; Nelson, M.N.; Kamphuis, L.G.; Singh, K.B.; Rotter, B.; Krezdorn, N.; Winter, P.; Wehling, P.; Ruge-Wehling, B. Characterization and Mapping of LanrBo: A Locus Conferring Anthracnose Resistance in Narrow-Leafed Lupin (Lupinus Angustifolius L.). Theoretical and Applied Genetics 2015, 128, 2121–2130. [CrossRef]
- Bitarishvili, S.; Samad, S.; Boldyrev, S.; Ben, C.; Volkova, P.; Shavarda, A.; Lukashevich, M.; Bondarenko, E. Metabolic Profiling Reveals Fumaric Acid and GABA as Possible Markers of Colletotrichum Lupini Infection of White Lupin. Physiological and Molecular Plant Pathology 2023, 128. [CrossRef]
- Moraga, J.; Gomes, W.; Pinedo, C.; Cantoral, J.M.; Hanson, J.R.; Carbú, M.; Garrido, C.; Durán-Patrón, R.; Collado, I.G. The Current Status on Secondary Metabolites Produced by Plant Pathogenic Colletotrichum Species. Phytochemistry Reviews 2019, 18, 215–239. [CrossRef]
- Anjali; Kumar, S.; Korra, T.; Thakur, R.; Arutselvan, R.; Kashyap, A.S.; Nehela, Y.; Chaplygin, V.; Minkina, T.; Keswani, C. Role of Plant Secondary Metabolites in Defence and Transcriptional Regulation in Response to Biotic Stress. Plant Stress 2023, 8, 100154. [CrossRef]
- Wojakowska, A.; Kułak, K.; Jasiński, M.; Kachlicki, P.; Stawiński, S.; Stobiecki, M. Metabolic Response of Narrow Leaf Lupine (Lupinus Angustifolius) Plants to Elicitation and Infection with Colletotrichum Lupini under Field Conditions. Acta Physiologiae Plantarum 2015, 37. [CrossRef]
- Pecchia, S.; Caggiano, B.; Da Lio, D.; Cafà, G.; Le Floch, G.; Baroncelli, R. Molecular Detection of the Seed-Borne Pathogen Colletotrichum Lupini Targeting the Hyper-Variable IGS Region of the Ribosomal Cluster. Plants 2019, 8, 4–6. [CrossRef]
- Kamber, T.; Malpica-López, N.; Messmer, M.M.; Oberhänsli, T.; Arncken, C.; Alkemade, J.A.; Hohmann, P. A Qpcr Assay for the Fast Detection and Quantification of Colletotrichum Lupini. Plants 2021, 10, 2–9. [CrossRef]
- Falconí, C.E.; Yánez-Mendizábal, V. Efficacy of UV-C Radiation to Reduce Seedborne Anthracnose (Colletotrichum Acutatum) from Andean Lupin (Lupinus Mutabilis). Plant Pathology 2018, 67, 831–838. [CrossRef]
- Alkemade, J.A.; Arncken, C.; Hirschvogel, C.; Messmer, M.M.; Leska, A.; Voegele, R.T.; Finckh, M.R.; Kölliker, R.; Groot, S.P.C.; Hohmann, P. The Potential of Alternative Seed Treatments to Control Anthracnose Disease in White Lupin. Crop Protection 2022, 158. [CrossRef]
- Phan, H.T.T.; Ellwood, S.R.; Adhikari, K.; Nelson, M.N.; Oliver, R.P. The First Genetic and Comparative Map of White Lupin (Lupinus Albus L.): Identification of QTLs for Anthracnose Resistance and Flowering Time, and a Locus for Alkaloid Content. DNA Research 2007, 14, 59–70. [CrossRef]
- Książkiewicz, M.; Nazzicari, N.; Yang, H.; Nelson, M.N.; Renshaw, D.; Rychel, S.; Ferrari, B.; Carelli, M.; Tomaszewska, M.; Stawiński, S.; et al. A High-Density Consensus Linkage Map of White Lupin Highlights Synteny with Narrow-Leafed Lupin and Provides Markers Tagging Key Agronomic Traits. Scientific Reports 2017, 7, 15335–15335. [CrossRef]
- Alkemade, J.A.; Nazzicari, N.; Messmer, M.M.; Annicchiarico, P.; Ferrari, B.; Voegele, R.T.; Finckh, M.R.; Arncken, C.; Hohmann, P. Genome-Wide Association Study Reveals White Lupin Candidate Gene Involved in Anthracnose Resistance. Theoretical and Applied Genetics 2022, 135, 1011–1024. [CrossRef]
- Yang, H.; Boersma, J.G.; You, M.; Buirchell, B.J.; Sweetingham, M.W. Development and Implementation of a Sequence-Specific PCR Marker Linked to a Gene Conferring Resistance to Anthracnose Disease in Narrow-Leafed Lupin (Lupinus Angustifolius L.). Molecular Breeding 2004, 14, 145–151. [CrossRef]
- Yang, H.; Renshaw, D.; Thomas, G.; Buirchell, B.; Sweetingham, M. A Strategy to Develop Molecular Markers Applicable to a Wide Range of Crosses for Marker Assisted Selection in Plant Breeding: A Case Study on Anthracnose Disease Resistance in Lupin (Lupinus Angustifolius L.). Molecular Breeding 2008, 21, 473–483. [CrossRef]
- Ruge-Wehling, B.; Dieterich, R.; Thiele, C.; Eickmeyer, F.; Wehling, P. Resistance to Anthracnose in Narrow-Leafed Lupin (Lupinus Angustifolius L.): Sources of Resistance and Development of Molecular Markers. Journal Für Kulturpflanzen 2009, 61, 62–65.
- Lichtin, N.; Salvo-Garrido, H.; Till, B.; Caligari, P.D.S.; Rupayan, A.; Westermeyer, F.; Olivos, M. Genetic and Comparative Mapping of Lupinus Luteus L. Highlight Syntenic Regions with Major Orthologous Genes Controlling Anthracnose Resistance and Flowering Time. Scientific Reports 2020, 10. [CrossRef]
- Guilengue, N.; Neves-Martins, J.; Talhinhas, P. Response to Anthracnose in a Tarwi (Lupinus Mutabilis) Collection Is Influenced by Anthocyanin Pigmentation. Plants 2020, 9. [CrossRef]
- Boersma, J.G.; Pallotta, M.; Li, C.D.; Buirchell, B.J.; Sivasithamparam, K.; Yang Huaan Construction of a Genetic Linkage Map Using MFLP and Identification of Molecular Markers Linked to Domestication Genes in Narrow-Leafed Lupin (Lupinus Angustifolius L.). Cell. Mol. Biol. Lett. 2005, 10, 331–344.
- Nelson, M.N.; Phan, H.T.T.; Ellwood, S.R.; Moolhuijzen, P.M.; Hane, J.; Williams, A.; O’Lone, C.E.; Fosu-Nyarko, J.; Scobie, M.; Cakir, M.; et al. The First Gene-Based Map of Lupinus Angustifolius L. -Location of Domestication Genes and Conserved Synteny with Medicago Truncatula. Theoretical and Applied Genetics 2006, 113, 225–238. [CrossRef]
- Yang, H.; Tao, Y.; Zheng, Z.; Li, C.; Sweetingham, M.W.; Howieson, J.G. Application of Next-Generation Sequencing for Rapid Marker Development in Molecular Plant Breeding: A Case Study on Anthracnose Disease Resistance in Lupinus Angustifolius L. BMC Genomics 2012, 13, 318–318. [CrossRef]
- Kamphuis, L.G.; Hane, J.K.; Nelson, M.N.; Gao, L.; Atkins, C.A.; Singh, K.B. Transcriptome Sequencing of Different Narrow-leafed Lupin Tissue Types Provides a Comprehensive Uni-gene Assembly and Extensive Gene-based Molecular Markers. Plant Biotechnology Journal 2015, 13, 14–25. [CrossRef]
- Hane, J.K.; Ming, Y.; Kamphuis, L.G.; Nelson, M.N.; Garg, G.; Atkins, C.A.; Bayer, P.E.; Bravo, A.; Bringans, S.; Cannon, S.; et al. A Comprehensive Draft Genome Sequence for Lupin (Lupinus Angustifolius), an Emerging Health Food: Insights into Plant–Microbe Interactions and Legume Evolution. Plant Biotechnology Journal 2017, 15, 318–330. [CrossRef]
- Plewiński, P.; Książkiewicz, M.; Rychel-Bielska, S.; Rudy, E.; Wolko, B. Candidate Domestication-Related Genes Revealed by Expression Quantitative Trait Loci Mapping of Narrow-Leafed Lupin (Lupinus Angustifolius L.). International Journal of Molecular Sciences 2019, 20. [CrossRef]
- Adhikari, K.N.; Thomas, G.; Buirchell, B.J.; Sweetingham, M.W. Identification of Anthracnose Resistance in Yellow Lupin (Lupinus Luteus L.) and Its Incorporation into Breeding Lines. Plant Breeding 2011, 130, 660–664. [CrossRef]
- French, R.J.; Sweetingham, M.W.; Shea, G.G. A Comparison of the Adaptation of Yellow Lupin (Lupinus Luteus L.) and Narrow-Leafed Lupin (L. Angustifolius L.) to Acid Sandplain Soils in Low Rainfall Agricultural Areas of Western Australia. Australian Journal of Agricultural Research 2001, 52, 945–954. [CrossRef]
- Adhikari, K.N.; Buirchell, B.J.; Thomas, G.J.; Sweetingham, M.W.; Yang, H. Identification of Anthracnose Resistance in Lupinus Albus L. and Its Transfer from Landraces to Modern Cultivars. Crop and Pasture Science 2009, 60, 472–479. [CrossRef]
- Raman, R.; Cowley, R.B.; Raman, H.; Luckett, D.J. Analyses Using SSR and DArT Molecular Markers Reveal That Ethiopian Accessions of White Lupin (Lupinus Albus L.) Represent a Unique Genepool. Open Journal of Genetics 2014, 04, 87–98. [CrossRef]
- Alkemade, J.A.; Messmer, M.M.; Arncken, C.; Leska, A.; Annicchiarico, P.; Nazzicari, N.; Książkiewicz, M.; Voegele, R.T.; Finckh, M.R.; Hohmann, P. A High-Throughput Phenotyping Tool to Identify Field-Relevant Anthracnose Resistance in White Lupin. Plant Disease 2021, 105, 1719–1727. [CrossRef]
- Mulin, M.; Bellio-Spataru, A. Organogenesis from Hypocotyl Thin Cell Layers of Lupinus Mutabilis and Lupinus Albus; Plant Growth Regulation; 2000; Vol. 30, pp. 177–183;.
- Nguyen, A.H.; Hodgson, L.M.; Erskine, W.; Barker, S.J. An Approach to Overcoming Regeneration Recalcitrance in Genetic Transformation of Lupins and Other Legumes. Plant Cell, Tissue and Organ Culture 2016, 127, 623–635. [CrossRef]
- Aslam, M.M.; Karanja, J.K.; Zhang, Q.; Lin, H.; Xia, T.; Akhtar, K.; Liu, J.; Miao, R.; Xu, F.; Xu, W. In Vitro Regeneration Potential of White Lupin (Lupinus Albus) from Cotyledonary Nodes. Plants 2020, 9, 1–14. [CrossRef]
- Molvig, L.; Tabe, L.M.; Eggum, B.O.; Moore, A.E.; Craig, S.; Spencer, D.; Higgins, T.J.V. Enhanced Methionine Levels and Increased Nutritive Value of Seeds of Transgenic Lupins ( Lupinus Angustifolius L.) Expressing a Sunflower Seed Albumin Gene. Proceedings of the National Academy of Sciences 1997, 94, 8393–8398. [CrossRef]
- Pigeaire, A.; Abernethy, D.; Smith, P.M.; Simpson, K.; Fletcher, N.; Lu, C.-Y.; Atkins, C.A.; Cornish, E. Transformation of a Grain Legume (Lupinus Angustifolius L.) via Agrobacterium Tumefaciens-Mediated Gene Transfer to Shoot Apices; Molecular Breeding; Kluwer Academic Publishers, 1997; Vol. 3, pp. 341–349;.
- Barker, S.J.; Si, P.; Hodgson, L.; Ferguson-Hunt, M.; Khentry, Y.; Krishnamurthy, P.; Averis, S.; Mebus, K.; O’Lone, C.; Dalugoda, D.; et al. Regeneration Selection Improves Transformation Efficiency in Narrow-Leaf Lupin. Plant Cell, Tissue and Organ Culture 2016, 126, 219–228. [CrossRef]
- Li, H.; Wylie, S.J.; Jones, M.G.K. Transgenic Yellow Lupin (Lupinus Luteus); Plant Cell Reports; Springer-Verlag, 2000; Vol. 19, pp. 634–637;.
- Pniewski, T.; Kapusta, J.; Płucienniczak, A. Agrobacterium-Mediated Transformation of Yellow Lupin to Generate Callus Tissue Producing HBV Surface Antigen in a Long-Term Culture. Journal of Applied Genetics 2006, 47, 309–318. [CrossRef]
- Babaoglu, M.; Mccabe, M.S.; Power, J.B.; Dave, M.R. Agrobacterium-Mediated Transformation of Lupinus Mutabilis L. Using Shoot Apical Explants; ACTA PHYSIOLOGIAE PLANTARUM; 2000; Vol. 22, pp. 111–119;.
- Polowick, P.L.; Loukanina, N.N.; Doshi, K.M. Agrobacterium-Mediated Transformation of Tarwi (Lupinus Mutabilis Sweet), a Potential Platform for the Production of Plant-Made Proteins. In Vitro Cellular & Developmental Biology - Plant 2014, 50, 401–411. [CrossRef]
- Xu, W.; Zhang, Q.; Yuan, W.; Xu, F.; Muhammad Aslam, M.; Miao, R.; Li, Y.; Wang, Q.; Li, X.; Zhang, X.; et al. The Genome Evolution and Low-Phosphorus Adaptation in White Lupin. Nature Communications 2020, 11. [CrossRef]
- Aslam, M.M.; Waseem, M.; Zhang, Q.; Ke, W.; Zhang, J.; Xu, W. Identification of ABC Transporter G Subfamily in White Lupin and Functional Characterization of L.albABGC29 in Phosphorus Use. BMC Genomics 2021, 22. [CrossRef]
- Aslam, M.M.; Fritschi, F.B.; Zhang, D.; Wang, G.; Li, H.; Lam, H.; Waseem, M.; Xu, W.; Zhang, J. Overexpression of LaGRAS Enhances Phosphorus Acquisition via Increased Root Growth of Phosphorus-deficient White Lupin. Physiologia Plantarum 2023. [CrossRef]
- Zhou, Y.; Olt, P.; Neuhäuser, B.; Moradtalab, N.; Bautista, W.; Uhde-Stone, C.; Neumann, G.; Ludewig, U. Loss of LaMATE Impairs Isoflavonoid Release from Cluster Roots of Phosphorus-Deficient White Lupin. Physiologia Plantarum 2021, 173, 1207–1220. [CrossRef]
- Zhou, Y.; Neuhäuser, B.; Neumann, G.; Ludewig, U. LaALMT1 Mediates Malate Release from Phosphorus-Deficient White Lupin Root Tips and Metal Root to Shoot Translocation. Plant Cell and Environment 2020, 43, 1691–1706. [CrossRef]
- Zhu, X.; Xu, W.; Liu, B.; Zhan, Y.; Xia, T. Adaptation of High-Efficiency CRISPR/Cas9-Based Multiplex Genome Editing System in White Lupin by Using Endogenous Promoters. Physiologia Plantarum 2023, 175. [CrossRef]
- Monteiro, A.; Miranda, C.; Trindade, H. Mediterranean Lupines as an Alternative Protein Source to Soybean.; MDPI: Basel Switzerland, May 2021; pp. 38–38.
- Szczepański, A.; Adamek-Urbańska, D.; Kasprzak, R.; Szudrowicz, H.; Śliwiński, J.; Kamaszewski, M. Lupin: A Promising Alternative Protein Source for Aquaculture Feeds? Aquaculture Reports 2022, 26. [CrossRef]
- Gresta, F.; Oteri, M.; Scordia, D.; Costale, A.; Armone, R.; Meineri, G.; Chiofalo, B. White Lupin (Lupinus Albus L.), an Alternative Legume for Animal Feeding in the Mediterranean Area. Agriculture (Switzerland) 2023, 13. [CrossRef]
- Foyer, C.H.; Lam, H.-M.; Nguyen, H.T.; Siddique, K.H.M.; Varshney, R.K.; Colmer, T.D.; Cowling, W.; Bramley, H.; Mori, T.A.; Hodgson, J.M.; et al. Neglecting Legumes Has Compromised Human Health and Sustainable Food Production. Nature Plants 2016, 2, 16112. [CrossRef]
- Adhikari, K.N.; Thomas, G.; Diepeveen, D.; Trethowan, R. Overcoming the Barriers of Combining Early Flowering and Anthracnose Resistance in White Lupin (Lupinus Albus L.) for the Northern Agricultural Region of Western Australia. Crop and Pasture Science 2013, 64, 914–921. [CrossRef]

| Lupin species | Source for resistance (Cultivar / Line) | Origin | Locus/ candidate gene | Reference |
|
Lupinus albus (White lupin) |
P27174 P27175 P27178 Blu-25 |
Ethiopia Ethiopia Ethiopia Chile |
2 QTLs on ALB02 and ALB04 linkage groups NA NA Lalb_Chr05_g0216161 |
[58,59,60] |
| Rumbo Baer(a) | Chile | “ | ||
| Rumbo Baer(b) | Chile | “ | ||
|
Lupinus angustifolius (Narrow-leaved lupine) |
Tanjil | Australia | Lanr1 | [43,61,62,63] |
| Wonga Mandelup Bo7212 |
Australia Australia Germany |
Lanr1 AnMan LanrBo |
||
|
Lupinus luteus (Yellow lupin) |
Core 98 | Chile | Lanr1 homolog | [64,72] |
| Gyulatanya P20856 | Hungary | NA | ||
| P28716 | Portugese | NA | ||
|
Lupinus mutabilis (Andean lupin) |
LM34 | Chile | NA | [65] |
| I82 | Chile | NA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





