Submitted:
27 December 2023
Posted:
28 December 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
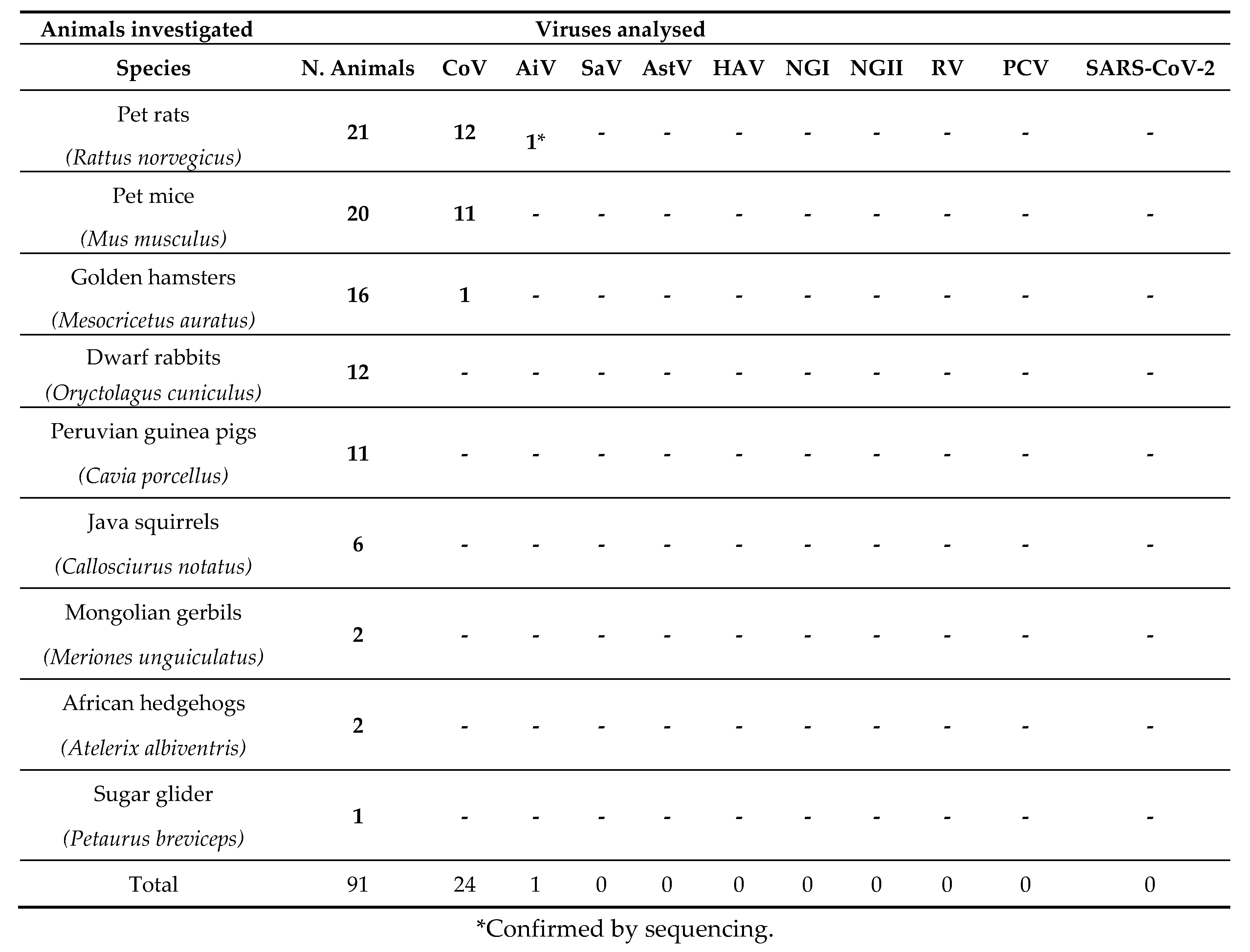
Animals investigated
Necropsy analysis
Nucleic acids extraction
Biomolecular analysis
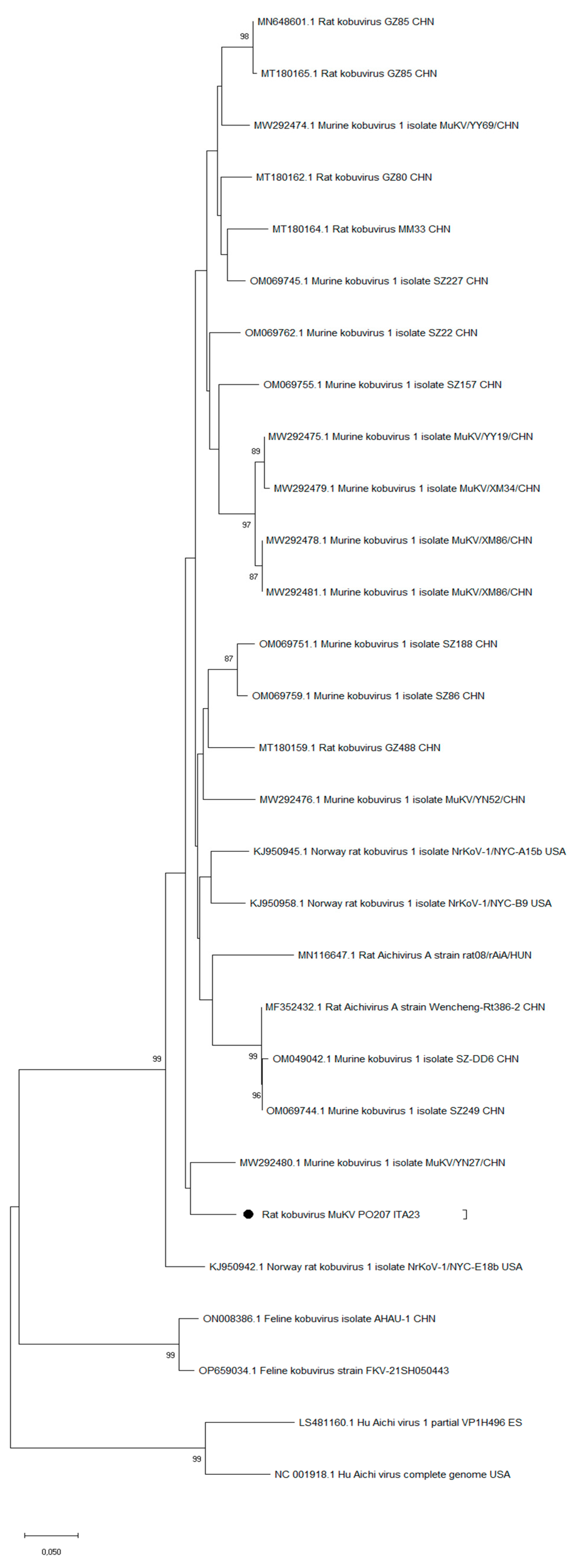
Aichivirus characterization
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abi, K.M.; Yang, C.; Tang, C.; Jing, Z.Z. Aichivirus C isolate is a diarrhoea-causing pathogen in goats. Transbound Emerg Dis 2022, 69, e2268-e2275. Epub 2022 May 11. PMID: 35502695. [CrossRef]
- Albano, F.; Bruzzese, E.; Bella, A.; Cascio, A.; Titone, L.; Arista, S.; Izzi, G.; Virdis, R.; Pecco, P.; Principi, N.; Fontana, M.; Guarino, A. Rotavirus and not age determines gastroenteritis severity in children: a hospital-based study. Eur J Pediatr 2007, 166, 241-7. Epub 2006 Aug 29. PMID: 16941130. [CrossRef]
- Alfano, F.; Dowgier, G.; Valentino, M.P.; Galiero, G.; Tinelli, A.; Decaro, N.; Fusco, G. Identification of Pantropic Canine Coronavirus in a Wolf (Canis lupus italicus) in Italy. J Wildl Dis 2019, 55, 504-508. [CrossRef]
- Alfano, F.; Fusco, G.; Mari, V.; Occhiogrosso, L.; Miletti, G.; Brunetti, R.; Galiero, G.; Desario, C.; Cirilli, M.; Decaro, N. Circulation of pantropic canine coronavirus in autochthonous and imported dogs, Italy. Transbound Emerg Dis 2020, 67, 1991–9. [CrossRef]
- Amoroso, MG.; Lucifora, G.; Degli Uberti, B.; Serra, F.; De Luca, G.; Borriello, G.; De Domenico, A.; Brandi, S.; Cuomo, MC.; Bove, F.; Riccardi, MG.; Galiero, G.; Fusco, G. Fatal Interstitial Pneumonia Associated with Bovine Coronavirus in Cows from Southern Italy. Viruses 2020, 12, 1331. PMID: 33228210; PMCID: PMC7699522. [CrossRef]
- Amoroso, MG.; Serra, F.; Esposito, C.; D'Alessio, N.; Ferrara, G.; Cioffi, B.; Anzalone, A.; Pagnini, U.; De Carlo, E.; Fusco, G.; Montagnaro, S. Prevalence of Infection with Porcine Circovirus Types 2 and 3 in the Wild Boar Population in the Campania Region (Southern Italy). Animals (Basel) 2021, 11,3215. 10: ,. [CrossRef]
- Amoroso, M.G.; Corrado, F.; De Carlo, E.; Lucibelli, M.G.; Martucciello, A.; Guarino, A.; Galiero, G. Bubaline Herpesvirus 1 Associated with Abortion in a Mediterranean Water Buffalo. Res Vet Sci 2013, 94, 813–816. [CrossRef]
- Amoroso, MG.; Russo, D.; Lanave, G.; et al. Detection and phylogenetic characterization of astroviruses in insectivorous bats from Central-Southern Italy. Zoonoses Public Health 2018, 65, 702–710. [CrossRef]
- Baaert, L.; Wobus, C.E.; Van Coillie, E.; Thackray, L.B.; Debevere, J.; Uyttendaele, M. Detection of murine norovirus 1 by using plaque assay, transfection assay, and real-time reverse transcription-PCR before and after heat exposure. Appl Environ Microbiol 2008, 74, 543–546. [CrossRef]
- Bányai, K.; Papp, H.; Dandár, E.; Molnár, P.; Mihály, I.; Van Ranst, M.; Martella, V.; Matthijnssens, J. Whole genome sequencing and phylogenetic analysis of a zoonotic human G8P[14] rotavirus strain. Infect Genet Evol 2010, 10, 1140-4. 1016. [CrossRef]
- Bányai, K.; Kemenesi, G.; Budinski, I.; Földes, F.; Zana, B.; Marton, S.; Varga-Kugler, R.; Oldal, M.; Kurucz, K.; Jakab, F. Candidate new rotavirus species in Schreiber's bats, Serbia. Infect Genet Evol 2017, 48, 19-26. Epub 2016 Dec 6. PMID: 27932285; PMCID: PMC7106153. 1016. [CrossRef]
- Bergallo, M.; Galliano, I.; Montanari, P.; Rassu, M.; Daprà, V. Aichivirus in Children with Diarrhea in Northern Italy. Intervirology 2017, 60, 196-200. Epub 2018 Mar 2. PMID: 29502122. [CrossRef]
- Bergallo, M.; Galliano, I.; Daprà, V.; Rassu, M.; Montanari, P.; Tovo, PA. Molecular Detection of Human Astrovirus in Children With Gastroenteritis, Northern Italy. Pediatr Infect Dis J 2018, 37, 738-742. PMID: 30004391. [CrossRef]
- Biscaro, V.; Piccinelli, G.; Gargiulo, F.; Ianiro, G.; Caruso, A.; Caccuri, F.; De Francesco, M.A. Detection and molecular characterization of enteric viruses in children with acute gastroenteritis in Northern Italy. Infect Genet Evol 2018, 60, 35-41. doi: 10.1016/j.meegid.2018.02.011. Epub 2018 Feb 10. PMID: 29438743. [CrossRef]
- Bonadonna, L.; Briancesco, R.; Suffredini, E.; Coccia, A.; Della Libera, S.; Carducci, A.; Verani, M.; Federigi, I.; Iaconelli, M.; Bonanno Ferraro, G.; Mancini, P.; Veneri, C.; Ferretti, E.; Lucentini, L.; Gramaccioni, L.; La Rosa, G. Enteric viruses, somatic coliphages and Vibrio species in marine bathing and non-bathing waters in Italy. Mar Pollut Bull 2019, 149, 110570. Epub 2019 Sep 8. PMID: 31542593. [CrossRef]
- Boros, Á.; Orlovácz, K.; Pankovics, P.; Szekeres, S.; Földvári, G.; Fahsbender, E.; Delwart, E.; Reuter, G. Diverse picornaviruses are prevalent among free-living and laboratory rats (Rattus norvegicus) in Hungary and can cause disseminated infections. Infect Genet Evol 2019, 75, 103988. Epub 2019 Aug 1. PMID: 31377399. [CrossRef]
- Centers for Disease Control and Prevention. Diseases from rodents. www.cdc.gov/rodents/diseases/index.html (2010).
- Chhabra, P.; de Graaf, M.; Parra, G.I.; Chan, M.C.; Green, K.; Martella, V.; Wang, Q.; White, P.A.; Katayama, K.; Vennema, H.; Koopmans, M.P.G.; Vinjé, J. Updated classification of norovirus genogroups and genotypes. J Gen Virol 2019, 100, 1393-1406. Erratum in: J Gen Virol 2020, 101, 893. PMID: 31483239; PMCID: PMC7011714. [CrossRef]
- Chafekar, A.; Fielding, B.C. MERS-CoV: Understanding the Latest Human Coronavirus Threat. Viruses 2018, 10, 93. PMID: 29495250; PMCID: PMC5850400. Viruses 2018, 10. [CrossRef]
- Chan, J.F.; Yuan, S.; Kok, K.H.; To, K.K.; Chu, H.; Yang, J.; Xing, F.; Liu, J.; Yip, C.C.; Poon, R.W.; Tsoi, H.W.; Lo, S.K.; Chan, K.H.; Poon, V.K.; Chan, W.M.; Ip, J.D.; Cai, J.P.; Cheng, V.C.; Chen, H.; Hui, C.K.; Yuen, K.Y. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet 2020, 395, 514-523. Epub 2020 Jan 24. PMID: 31986261; PMCID: PMC7159286. 1016. [CrossRef]
- Chomel, B.B.; Belotto, A.; Meslin, F.X. Wildlife, exotic pets, and emerging zoonoses. Emerg Infect Dis 2007, 13, 6-11.
- Cioffi, B.; Ianiro, G.; Iaccarino, D.; D'Apice, F.; Ferraro, A.; Race, M.; Spasiano, D.; Esposito, E.; Monini, M.; Serra, F.; Cozza, D.; Di Nocera, F.; De Maio, L.; Amoroso, M.G.; De Carlo E.; Fusco, G. A potential risk assessment tool to monitor pathogens circulation in coastal waters. Environ Res 2021, 200, 111748. Epub 2021 Jul 22. PMID: 34303676. [CrossRef]
- Cooney, M.A.; Gorrell, R.J.; Palombo, E.A. Characterisation and phylogenetic analysis of the VP7 proteins of serotype G6 and G8 human rotaviruses. J Med Microbiol 2001, 50, 462-467. PMID: 11339255. [CrossRef]
- Costafreda, M.I.; Bosch, A.; Pintó, R.M. Development, evaluation, and standardization of a real-time TaqMan reverse transcription-PCR assay for quantification of hepatitis A virus in clinical and shellfish samples. Appl Environ Microbiol 2006, 72, 3846-55. PMID: 16751488; PMCID: PMC1489592. [CrossRef]
- Dakroub, H.; Russo, D.; Cistrone, L.; Serra, F.; Fusco, G.; De Carlo E.; Amoroso, M.G. A First Assessment of SARS-CoV-2 Circulation in Bats of Central-Southern Italy. Pathogens. 2022, 11, 742. PMID: 35889988 Free PMC article. [CrossRef]
- Dance, A. Beyond coronavirus: The virus discoveries transforming biology. Nature 2021, 595, 22–25.
- Di Martino, B.; Di Felice, E.; Ceci, C.; Di Profio, F.; Marsilio, F. Canine kobuviruses in diarrhoeic dogs in Italy. Vet Microbiol 2013, 166, 246-249. Epub 2013 Jun 6. PMID: 23806200; PMCID: PMC7117211. [CrossRef]
- Di Martino, B.; Di Profio, F.; Melegari, I.; Marsilio, F.; Martella, V. Detection of feline kobuviruses in diarrhoeic cats, Italy. Vet Microbiol 2015, 176, 186-189. Epub 2015 Jan 17. PMID: 25631253; PMCID: PMC7117564. [CrossRef]
- Di Martino, B.; Di Profio, F.; Robetto, S.; Fruci, P.; Sarchese, V.; Palombieri, A.; Melegari, I.; Orusa, R.; Martella, V.; Marsilio, F. Molecular Survey on Kobuviruses in Domestic and Wild Ungulates From Northwestern Italian Alps. Front Vet Sci 2021, 8, 679337. PMID: 34195249; PMCID: PMC8237713. [CrossRef]
- Diakoudi, G.; Jamnikar-Ciglenečki, U.; Lanave, G.; Lelli, D.; Martella, V.; Kuhar, U. Genome sequence of an aichivirus detected in a common pipistrelle bat (Pipistrellus pipistrellus). Arch Virol 2020, 165, 1019-1022. Epub 2020 Feb 13. PMID: 32056001. 32056001. [CrossRef]
- d'Ovidio, D.; Santoro, D. Zoonotic Dermatoses of Exotic Companion Mammals. Vet Clin North Am Exot Anim Pract 2023, 26, 511-523. [CrossRef]
- Drosten, C.; Gunther, S.; Preiser, W.; van der Werf, S.; Brodt, H.R.; Becker, S.; Rabenau, H.; Panning, M.; Kolesnikova, L.; Fouchier, R.A.; et al. Identification of a Novel Coronavirus in Patients with Severe Acute Respiratory Syndrome. N. Engl. J. Med 2003, 348, 1967–1976. [CrossRef]
- Firth, C.; Bhat, M.; Firth, M.A.; Williams, S.H.; Frye M.J.; Simmonds, P.; Conte, J.M.; Ng, J.; Garcia, J.; Bhuva, N.P.; Lee, B.; Che, X.; Quan, P.L.; Lipkin, W.I. Detection of zoonotic pathogens and characterization of novel viruses carried by commensal Rattus norvegicus in New York City. mBio 2014, 5, e01933-14. PMID: 25316698; PMCID: PMC4205793. [CrossRef]
- Fusco, G.; Di Bartolo, I.; Cioffi, B.; Ianiro, G.; Palermo, P.; Monini, M.; Amoroso, M.G. Prevalence of Foodborne Viruses in Mussels in Southern Italy. Food Environ Virol 2017, 9, 187-194. Epub 2017 Jan 4. PMID: 28054332. [CrossRef]
- Fusco, G.; Anastasio, A.; Kingsley, D.H.; Amoroso, M.G.; Pepe, T.; Fratamico, P.M.; Cioffi, B.; Rossi, R.; La Rosa, G.; Boccia, F. Detection of Hepatitis A Virus and Other Enteric Viruses in Shellfish Collected in the Gulf of Naples, Italy. Int J Environ Res Public Health 2019, 16, 2588. PMID: 31331104; PMCID: PMC6678136. [CrossRef]
- Gao, Y.; He, W.; Fu, J.; Li, Y.; He, H.; Chen, Q. Epidemiological Evidence for Fecal-Oral Transmission of Murine Kobuvirus. Front Public Health 2022, 10, 865605. PMID: 35517645; PMCID: PMC9062591. [CrossRef]
- Haake, C.; Cook, S.; Pusterla, N.; Murphy, B. Coronavirus Infections in Companion Animals: Virology, Epidemiology, Clinical and Pathologic Features. Viruses 2020, 12, 1023.
- Han, B.A.; Schmidt, J.P.; Bowden, S.E. & Drake, J.M. Rodentreser-voirsoffuture zoonotic diseases. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112, 7039–7044. [CrossRef]
- Isegawa, Y.; Nakagomi, O.; Nakagomi, T.; Ueda, S. A. VP4 sequence highly conserved in human rotavirus strain AU-1 and feline rotavirus strain FRV-1. J Gen Virol 1992, 73, 1939-46. PMID: 1322955. [CrossRef]
- Johnson, C.K.; Hitchens, P.L.; Pandit, P.S.; Rushmore, J.; Evans, T.S.; Young, C.C.W.; Doyle, M.M. Global shifts in mammalian population trends reveal key predictors of virus spillover risk. Proc Biol Sci 2020, 287, 20192736. Epub 2020 Apr 8. PMID: 32259475; PMCID: PMC7209068. [CrossRef]
- Kaikkonen, S.; Rasanen, S.; Ramet, M.; et al: Aichivirus infection in children with acute gastroenteritis in Finland. Epidemiol Infect 2010, 138, 1166–1171.
- Kapoor, A.; Simmonds, P.; Dubovi, E.J.; Qaisar, N.; Henriquez, JA.; Medina, J.; Shields, S.; Lipkin, W.I. Characterization of a canine homolog of human Aichivirus. J Virol 2011, 85, 11520-5. Epub 2011 Aug 31. PMID: 21880761; PMCID: PMC3194941. [CrossRef]
- Khamrin, P.; Maneekarn, N.; Peerakome, S.; Yagyu, F.; Okitsu, S.; Ushijima, H. Molecular characterization of a rare G3P[3] human rotavirus reassortant strain reveals evidence for multiple human-animal interspecies transmissions. J Med Virol 2006, 78, 986-94. PMID: 16721863. [CrossRef]
- Kim, H.R.; Park, Y.R.; Lim, D.R.; Park, M.J.; Park, J.Y.; Kim, S.H.; Lee, K.K.; Lyoo, Y.S.; Park, C.K. Multiplex real-time poly-merase chain reaction for the differential detection of porcine circovirus 2 and 3. J. Virol. Methods 2017, 250, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Kitajima, M.; Hata, A.; Yamashita, T.; Haramoto, E.; Minagawa, H.; Katayama, H. Development of a reverse transcription-quantitative PCR system for detection and genotyping of aichi viruses in clinical and environmental samples. Appl Environ Microbiol 2013, 79, 3952-8. Epub 2013 Apr 19. PMID: 23603673; PMCID: PMC3697579. [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Le Cann, P.; Ranarijaona, S.; Monpoeho, S.; Le Guyader, F.; Ferré, V. Quantification of human astroviruses in sewage using real-time RT-PCR. Res Microbiol 2004, 155, 11-5. PMID: 14759703. [CrossRef]
- Lodder, W.J., Rutjes, S.A.; Takumi, K.; de Roda Husman, A.M. Aichi virus in sewage and surface water, the Netherlands. Emerg Infect Dis 2013, 19, 1222-30. [CrossRef]
- Lu, L.; Van Dung, N.; Ivens, A.; Bogaardt, C.; O'Toole, A.; Bryant, J.E.; Carrique-Mas, J.; Van Cuong, N.; Anh, P.H.; Rabaa, M.A., et al. Genetic diversity and cross-species transmission of kobuviruses in Vietnam. Virus Evol 2018, 4, vey002. [CrossRef]
- Malasao, R.; Saito, M.; Suzuki, A.; Imagawa, T.; Nukiwa-Soma, N.; Tohma, K.; Liu, X.; Okamoto, M.; Chaimongkol, N.; Dapat, C.; Kawamura, K.; Kayama, Y.; Masago, Y.; Omura, T.; Oshitani, H. Human G3P[4] rotavirus obtained in Japan, 2013, possibly emerged through a human-equine rotavirus reassortment event. Virus Genes 2015, 50, 129-33. Epub 2014 Oct 29. PMID: 25352228; PMCID: PMC4349953. [CrossRef]
- Maneekarn, N.; Khamrin, P.; Chan-it, W.; Peerakome, S.; Sukchai, S.; Pringprao, K.; Ushijima, H. Detection of rare G3P[19] porcine rotavirus strains in Chiang Mai, Thailand, provides evidence for origin of the VP4 genes of Mc323 and Mc345 human rotaviruses. J Clin Microbiol 2006, 44, 4113-9. Epub 2006 Sep 20. PMID: 16988014; PMCID: PMC1698310. [CrossRef]
- Matthijnssens, J.; Rahman, M.; Martella, V.; Xuelei, Y.; De Vos, S.; De Leener, K.; Ciarlet, M.; Buonavoglia, C.; Van Ranst, M. Full genomic analysis of human rotavirus strain B4106 and lapine rotavirus strain 30/96 provides evidence for interspecies transmission. J Virol 2006, 80, 3801-10. PMID: 16571797; PMCID: PMC1440464. [CrossRef]
- Mira, F.; Purpari, G.; Lorusso, E.; Di Bella, S.; Gucciardi, F.; Desario, C.; Macaluso, G.; Decaro, N.; Guercio, A. Introduction of Asian canine parvovirus in Europe through dog importation. Transbound Emerg Dis 2018, 65, 16-21. [CrossRef]
- Oh, D.Y.; Silva, P.A.; Hauroeder, B.; et al: Molecular characterization of the first Aichi viruses isolated in Europe and in South America. Arch Virol 2006, 151, 1199–1206. [CrossRef]
- Pham, N.T.; Khamrin, P.; Nguyen, T.A.; et al: Isolation and molecular characterization of Aichi viruses from fecal specimens collected in Japan.; Bangladesh, Thailand, and Vietnam. J Clin Microbiol 2007, 45, 2287–2288. [CrossRef]
- Plyusnin, I.; Kant, R.; Jääskeläinen, A.J.; Sironen, T.; Holm, L.; Vapalahti, O.; Smura, T. Novel NGS pipeline for virus discovery from a wide spectrum of hosts and sample types. Virus Evol 2020, 6, veaa091. [CrossRef]
- Reperant, L.A.; Brown, I.H.; Haenen, O.L.; de Jong, M.D.; Osterhaus, A.D.; Papa, A.; Rimstad, E.; Valarcher, J.F.; Kuiken, T. Companion Animals as a Source of Viruses for Human Beings and Food Production Animals. J Comp Pathol 2016, 155, S41-53. Epub 2016 Aug 10. PMID: 27522300. [CrossRef]
- Reuter, G.; Boldizsár, A.; Pankovics, P. Complete nucleotide and amino acid sequences and genetic organization of porcine kobuvirus.; a member of a new species in the genus Kobuvirus.; family Picornaviridae. Arch Virol. 2009, 154, 101-108. Epub 2008 Dec 19. PMID: 19096904. [CrossRef]
- Reuter, G.; Boros, A. and Pankovics, P. 'Kobuviruses - a comprehensive review'. Rev Med Virol 2011, 21, 32-41. [CrossRef]
- Riley, P.Y, Chomel, B.B. Hedgehog zoonoses. Emerg Infect Dis 2005, 11, 1-5.
- Rivadulla, E.; Romalde, J.L. A Comprehensive Review on Human Aichi Virus. Virol Sin 2020, 35, 501-516. Epub 2020 Apr 27. Erratum in: Virol Sin. 2020 Dec 2;: PMID: 32342286; PMCID: PMC7223127. [CrossRef]
- Sdiri-Loulizi, K.; Hassine, M.; Aouni, Z.; et al: First molecular detection of Aichi virus in sewage and shellfish samples in the Monastir region of Tunisia. Arch Virol 2010, 155, 1509–1513. [CrossRef]
- Tofani, S.; Ianiro, G.; De Sabato, L.; Monini, M.; Angeloni, G.; Ponterio, E.; D’Agostino, C.; Di Bari, M.A.; Valeri, M.; Di Bartolo, I. Detection and whole genome sequencing of murine norovirus in animal facility in Italy. Anim. Biotechnol 2021, 29, 1–8. 34. [CrossRef]
- UNI EN ISO 15216-2:2019.
- Varela, M.F.; Hooper, A.S.; Rivadulla, E.; Romalde, J.L. Human Sapovirus in Mussels from Ría do Burgo, A Coruña (Spain). Food Environ Virol 2016, 8, 187–193. Epub 2016 May 7. PMID: 27156175. [CrossRef]
- Vijgen, L.; Keyaerts, E.; Moës, E.; Thoelen, I.; Wollants, E.; Lemey, P.; Vandamme, A.M.; Van Ranst, M. Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J Virol 2005, 79, 1595-1604. PMID: 15650185; PMCID: PMC544107. [CrossRef]
- Vioque, F.; Dashti, A.; Santín, M.; Ruiz-Fons, F.; Köster, P.C.; Hernández-Castro, C.; García, J.T.; Bailo, B.; Ortega, S.; Olea, P.P.; Arce, F.; Chicharro, C.; Nieto, J.; González, F.; Viñuela, J.; Carmena, D.; González-Barrio, D. Wild micromammal host spectrum of zoonotic eukaryotic parasites in Spain. Occurrence and genetic characterisation. Transbound Emerg Dis 2022, 69, e2926-e2942. Epub 2022 Jul 8. PMID: 35752461. [CrossRef]
- Williams, S.H.; Che, X.; Garcia, J.A.; Klena, J.D.; Lee, B.; Muller, D.; Ulrich, W.; Corrigan, R.M.; Nichol, S.; Jain, K.; Lipkin, W.I. Viral Diversity of House Mice in New York City. mBio 2018, 9, e01354-17. PMID: 29666290; PMCID: PMC5904411. [CrossRef]
- You, F.F.; Zhang, M.Y.; He, H.; He, W.Q.; Li, Y.Z.; Chen, Q. Kobuviruses carried by Rattus norvegicus in Guangdong.; China. BMC Microbiol 2020, 20, 94. PMID: 32295529; PMCID: PMC7161169. [CrossRef]
- Zeng, S.Q.; Halkosalo, A.; Salminen, M.; Szakal, E.D.; Puustinen, L.; Vesikari, T. One-step quantitative RT-PCR for the detection of rotavirus in acute gastroenteritis. J Virol Methods 2008, 153, 238-40. Epub 2008 Sep 17. PMID: 18765254. [CrossRef]
- Zhang, M.; You, F.; Wu, F.; He, H.; Li, Q.; Chen Q. Epidemiology and genetic characteristics of murine kobuvirus from faecal samples of Rattus losea, Rattus tanezumi and Rattus norvegicus in southern China. J Gen Virol 2021, 102, 001646. PMID: 34486970; PMCID: PMC8567428. [CrossRef]
| Virus | Primers | Bibliography | |
|---|---|---|---|
| Astrovirus | Primer forward | 5’-CCGAGTAGGATCGAGGGT-3’ | Le Cann et al. 2004 |
| Primer reverse | 5’-GCTTCTGATTAAATCAATTTTAA-3’ | ||
| Probe | FAM: 5’-CTTTTCTGTCTCTGTTTAGATTATTTTAATCACC-3’ Tamra | ||
| Aichivirus | Primer forward | 5’-GTCTCCACHGACACYAAYTGGAC-3’ | Kitajima et al. 2013 |
| Primer reverse | 5’- GTTGTACATRGCAGCCCAGG-3’ | ||
| Probe | 5’-FAM-TTYTCCTTYGTGCGTGC- 3’NFQ (MGB) | ||
| Sapovirus | Primer forward | 5’-GAYCASGCTCTCGCYACCTAC-3’ | Varela et al. 2016 |
| Primer reverse | 5’-CCCTCCATYTCAAACACTA-3’ | ||
| Probe | 5’- FAM-CCCCTATRAACCA-3’NFQ (MGB) | ||
| Rotavirus | Primer forward | 5’-ACCATCTWCACRTRACCCTCTATGAG-3’ | Zeng et al. 2008 |
| Primer reverse | 5’-GGTCACATAACGCCCCTATAGC-3’ | ||
| Probe | 5’- FAM-AGTTAAAAGCTAACACTGTCAAA-3’(MGB) | ||
| Norovirus GI | Primer forward | 5’-CGCTGGATGCGNTTCCAT-3’ | ISO 15216 |
| Primer reverse | 5’-CCTTAGACGCCATCATCATTTAC-3’ | ||
| Probe | FAM-5’-TGGACAGGAGAYCGCRATCT-3’TAMRA | ||
| Norovirus GII | Primer forward | 5’-ATGTTCAGRTGGATGAGRTTCTCWGA-3’ | ISO 15216 |
| Primer reverse | 5’-TCGACGCCATCTTCATTCACA-3’ | ||
| Probe | FAM-5’-AGCACGTGGGAGGGCGATCG-3’-MGB/NFQ | ||
| Hepatitis A Virus | Primer forward | 5’-TCACCGCCGTTTGCCTAG-3’ | ISO 15216 |
| Primer reverse | 5’-GGAGAGCCCTGGAAGAAAG-3’ | ||
| Probe | FAM-CCTGAACCTGCAGGAATTAA-3’-MGB/NFQ | ||
| Porcine Circovirus | Primer forward | ACGTCCCTTTACTTTCAATTCACA | Kim et al. 2017 |
| Primer reverse | TATACTTGGTACACACATCCAGAGTCA | ||
| Probe | FAM-TGAGTTGATTACTGGCACGCCTAAACCAC-BHQ | ||
| Coronavirus | Primer forward | GGGTTGGGACTATCCTAAGTGTGA | Drosten et al. 2003 |
| Primer reverse | TAACACACAAACACCATCATCA | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





