Submitted:
27 December 2023
Posted:
28 December 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Clinicopathologic characteristics ofAdult Diffuse Glioma Patients
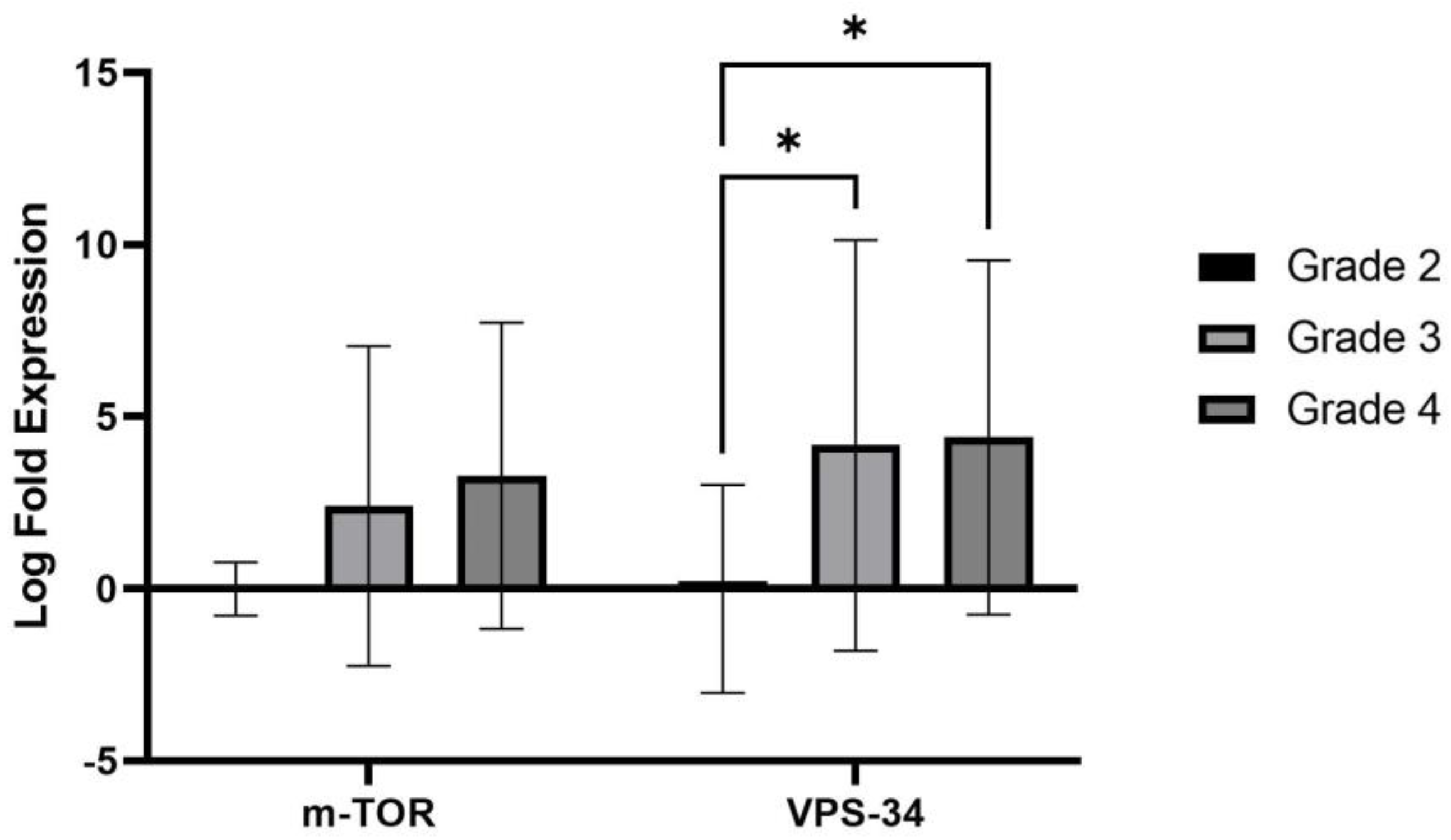
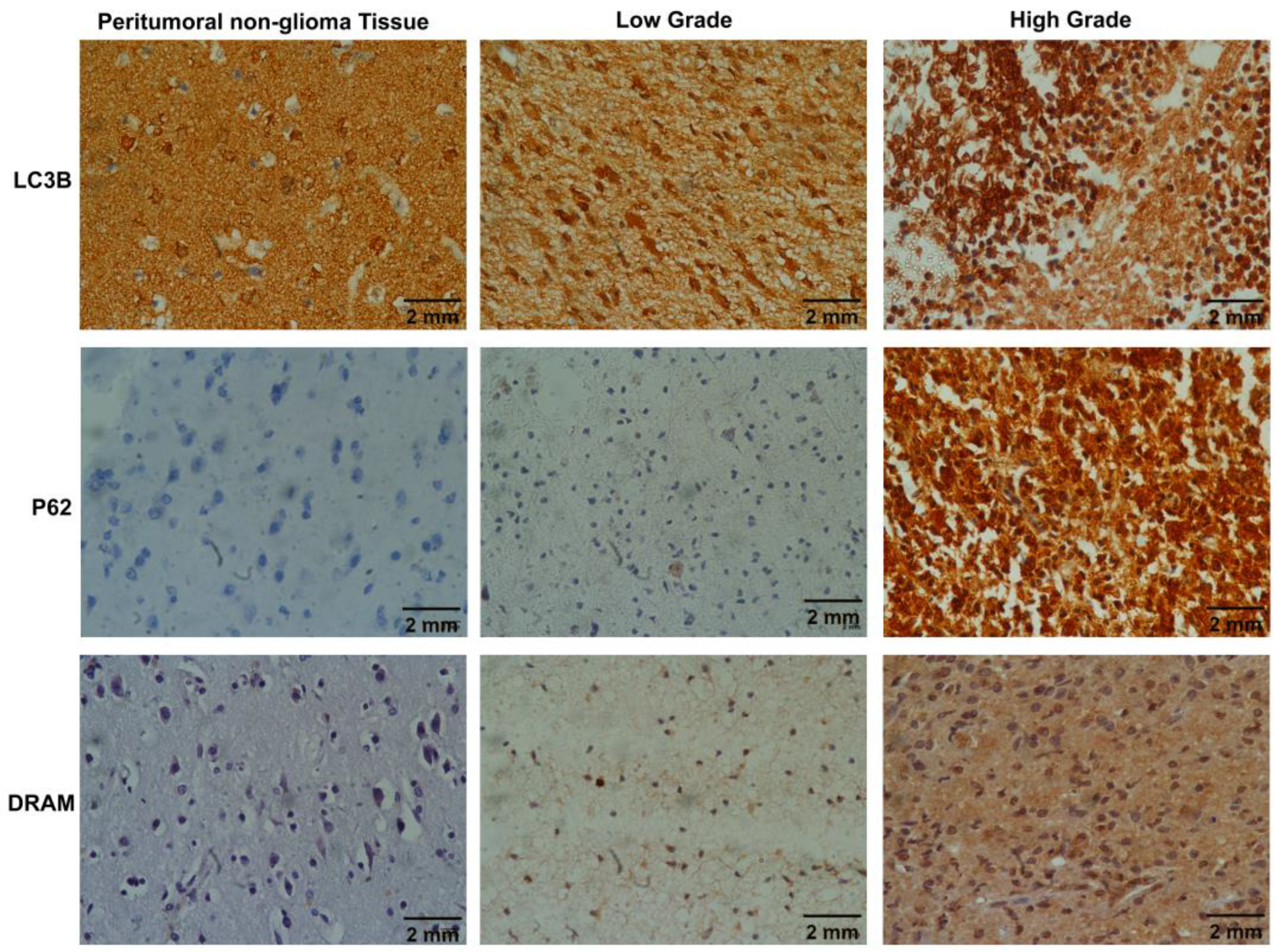
2.2. Evaluation of autophagy status using immunohistochemical analysis of Molecular markers (LC3B, SQSTM1/p62 and DRAM)
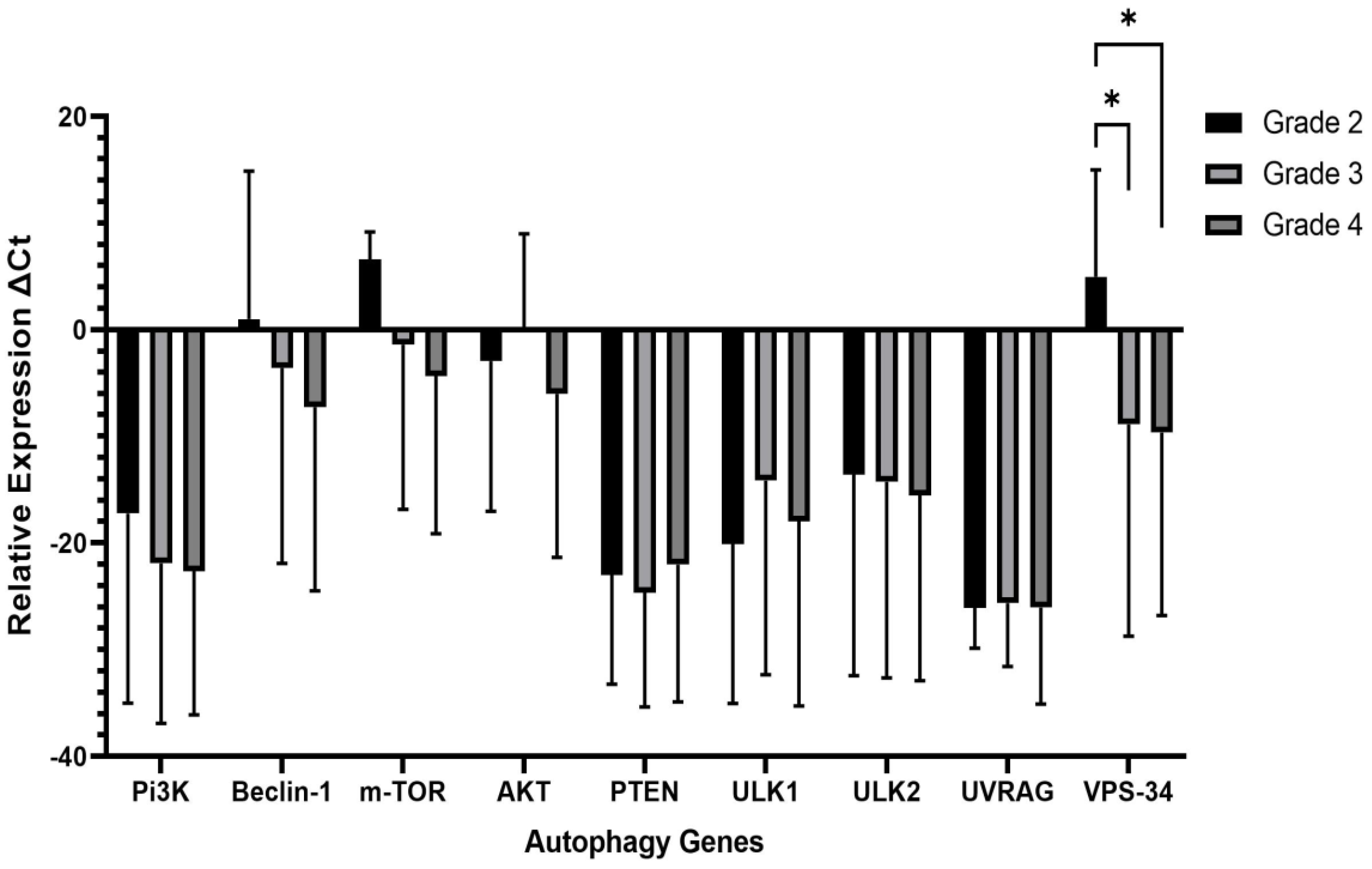
2.3. Correlation of clinicopathological features and autophagy markers
3. Discussion
4. Materials and Methods
4.1. Patients Selection
4.2. Tissue processing for histopathological analysis
4.3. Tissue processing for immunohistochemical examination
4.4. Immunohistochemical evaluation of autophagy markers
4.5. RNA extraction and cDNA synthesis
4.6. Gene expression analysis via quantitative real time (qPCR)
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ferlay, J.; Soerjomataram, I.; Ervik, M.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. GLOBOCAN 2012 v1. 0, cancer incidence and mortality worldwide. Iarc Cancerbase 2013, 11. [Google Scholar]
- Ostrom, Q.T.; Gittleman, H.; Xu, J.; Kromer, C.; Wolinsky, Y.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2009–2013. Neuro-oncology 2016, 18 (Suppl. 5), v1–v75. [Google Scholar] [CrossRef] [PubMed]
- Ostrom, Q.T.; Price, M.; Neff, C.; Cioffi, G.; Waite, K.A.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS statistical report: Primary brain and other central nervous system tumors diagnosed in the United States in 2015–2019. Neuro-oncology 2022, 24 (Suppl. 5), v1–v95. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.; Pfister, S.M.; Reifenberger, G. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro-oncology 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Fisher, J.; Schwartzbaum, J.; Wrensch, M.; Wiemels, J.L. Epidemiology of brain tumors. Neurol. Clin. 2007, 25, 867–890. [Google Scholar] [CrossRef]
- Krakstad, C.; Chekenya, M. Survival signalling and apoptosis resistance in glioblastomas: opportunities for targeted therapeutics. Molecular cancer 2010, 9, 1–14. [Google Scholar] [CrossRef]
- Miyashita, T.; Krajewski, S.; Krajewska, M.; Wang, H.G.; Lin, H.; Liebermann, D.A.; Hoffman, B.; Reed, J.C. Tumor suppressor p53 is a regulator of bcl-2 and bax gene expression in vitro and in vivo. Oncogene 1994, 9, 1799–1805. [Google Scholar] [PubMed]
- Liu, F.; Liu, D.; Yang, Y.; Zhao, S. Effect of autophagy inhibition on chemotherapy-induced apoptosis in A549 lung cancer cells. Oncology letters 2013, 5, 1261–1265. [Google Scholar] [CrossRef]
- Condello, M.; Mancini, G.; Meschini, S. The exploitation of liposomes in the inhibition of autophagy to defeat drug resistance. Frontiers in Pharmacology 2020, 11, 787. [Google Scholar] [CrossRef]
- Levine, B.; Klionsky, D.J. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Developmental cell 2004, 6, 463–477. [Google Scholar] [CrossRef]
- Murrow, L.; Debnath, J. Autophagy as a stress-response and quality-control mechanism: implications for cell injury and human disease. Annual Review of Pathology: Mechanisms of Disease 2013, 8, 105–137. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Bao, S.; Wu, Q.; Wang, H.; Eyler, C.; Sathornsumetee, S.; Shi, Q.; Cao, Y.; Lathia, J.; McLendon, R.E. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer cell 2009, 15, 501–513. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Xing, X.; Liu, Q.; Wang, Z.; Xin, Y.; Zhang, P.; Hu, C.; Liu, Y. Hypoxia-induced autophagy reduces radiosensitivity by the HIF-1α/miR-210/Bcl-2 pathway in colon cancer cells. International journal of oncology 2015, 46, 750–756. [Google Scholar] [CrossRef]
- Wu, H.-M.; Jiang, Z.-F.; Ding, P.-S.; Shao, L.-J.; Liu, R.-Y. Hypoxia-induced autophagy mediates cisplatin resistance in lung cancer cells. Scientific reports 2015, 5, 12291. [Google Scholar] [CrossRef] [PubMed]
- Denton, D.; Nicolson, S.; Kumar, S. Cell death by autophagy: facts and apparent artefacts. Cell Death & Differentiation 2012, 19, 87–95. [Google Scholar] [CrossRef]
- Lu, N.; Li, X.; Tan, R.; An, J.; Cai, Z.; Hu, X.; Wang, F.; Wang, H.; Lu, C.; Lu, H. HIF-1α/Beclin1-mediated autophagy is involved in neuroprotection induced by hypoxic preconditioning. Journal of Molecular Neuroscience 2018, 66, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Menon, M.B.; Dhamija, S. Beclin 1 phosphorylation–at the center of autophagy regulation. Frontiers in cell and developmental biology 2018, 6, 137. [Google Scholar] [CrossRef] [PubMed]
- Pirtoli, L.; Cevenini, G.; Tini, P.; Vannini, M.; Oliveri, G.; Marsili, S.; Mourmouras, V.; Rubino, G.; Miracco, C. The prognostic role of Beclin 1 protein expression in high-grade gliomas. Autophagy 2009, 5, 930–936. [Google Scholar] [CrossRef] [PubMed]
- Apel, A.; Herr, I.; Schwarz, H.; Rodemann, H.P.; Mayer, A. Blocked autophagy sensitizes resistant carcinoma cells to radiation therapy. Cancer research 2008, 68, 1485–1494. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Lei, Z.; Yu, J. Hypoxia induces autophagy in human vascular endothelial cells in a hypoxia-inducible factor 1-dependent manner. Molecular Medicine Reports 2015, 11, 2677–2682. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Zhu, K.; Yang, Z.; Zhou, Y.; Xia, Z.; Ren, J.; Zhao, Y.; Wu, G.; Liu, C. Hypoxia-induced autophagy is involved in radioresistance via HIF1A-associated beclin-1 in glioblastoma multiforme. Heliyon 2023, 9. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Sun, E.G.; Lee, Y.; Kim, M.S.; Kim, J.H.; Kim, W.J.; Jung, J.Y. Autophagy induction plays a protective role against hypoxic stress in human dental pulp cells. Journal of Cellular Biochemistry 2018, 119, 1992–2002. [Google Scholar] [CrossRef] [PubMed]
- White, E. Deconvoluting the context-dependent role for autophagy in cancer. Nature reviews cancer 2012, 12, 401–410. [Google Scholar] [CrossRef] [PubMed]
- Mathew, R.; Karp, C.M.; Beaudoin, B.; Vuong, N.; Chen, G.; Chen, H.-Y.; Bray, K.; Reddy, A.; Bhanot, G.; Gelinas, C. Autophagy suppresses tumorigenesis through elimination of p62. Cell 2009, 137, 1062–1075. [Google Scholar] [CrossRef] [PubMed]
- Karantza-Wadsworth, V.; Patel, S.; Kravchuk, O.; Chen, G.; Mathew, R.; Jin, S.; White, E. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes & development 2007, 21, 1621. [Google Scholar] [CrossRef]
- Mathew, R.; Kongara, S.; Beaudoin, B.; Karp, C.M.; Bray, K.; Degenhardt, K.; Chen, G.; Jin, S.; White, E. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes & development 2007, 21, 1367–1381. [Google Scholar] [CrossRef]
- Azad, M.B.; Chen, Y.; Henson, E.S.; Cizeau, J.; McMillan-Ward, E.; Israels, S.J.; Gibson, S.B. Hypoxia induces autophagic cell death in apoptosis-competent cells through a mechanism involving BNIP3. Autophagy 2008, 4, 195–204. [Google Scholar] [CrossRef]
- Bellot, G.; Garcia-Medina, R.; Gounon, P.; Chiche, J.; Roux, D.; Pouysségur, J.; Mazure, N.M. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Molecular and cellular biology 2009, 29, 2570–2581. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, S.M.; Elesawy, Y.F.; Abd El Aziz, A.M.; Khairy, R.A. The Pathological Evaluation of Autophagy-Related Protein (LC3B) and Its Association with the Infiltration of Immune Cells in Glioma. Asian Pacific Journal of Cancer Prevention: APJCP 2022, 23, 1777. [Google Scholar] [CrossRef] [PubMed]
- Tamrakar, S.; Yashiro, M.; Kawashima, T.; Uda, T.; Terakawa, Y.; Kuwae, Y.; Ohsawa, M.; Ohata, K. Clinicopathological significance of autophagy-related proteins and its association with genetic alterations in gliomas. Anticancer Research 2019, 39, 1233–1242. [Google Scholar] [CrossRef]
- Mathew, R.; Karantza-Wadsworth, V.; White, E. Role of autophagy in cancer. Nature Reviews Cancer 2007, 7, 961–967. [Google Scholar] [CrossRef]
- Das, C.K.; Mandal, M.; Kögel, D. Pro-survival autophagy and cancer cell resistance to therapy. Cancer and Metastasis Reviews 2018, 37, 749–766. [Google Scholar] [CrossRef]
- Ivankovic, D.; Chau, K.Y.; Schapira, A.H.; Gegg, M.E. Mitochondrial and lysosomal biogenesis are activated following PINK 1/parkin-mediated mitophagy. Journal of neurochemistry 2016, 136, 388–402. [Google Scholar] [CrossRef]
- Deng, D.; Luo, K.; Liu, H.; Nie, X.; Xue, L.; Wang, R.; Xu, Y.; Cui, J.; Shao, N.; Zhi, F. p62 acts as an oncogene and is targeted by miR-124-3p in glioma. Cancer Cell International 2019, 19, 1–13. [Google Scholar] [CrossRef]
- Rzymski, T.; Milani, M.; Pike, L.; Buffa, F.; Mellor, H.; Winchester, L.; Pires, I.; Hammond, E.; Ragoussis, I.; Harris, A. Regulation of autophagy by ATF4 in response to severe hypoxia. Oncogene 2010, 29, 4424–4435. [Google Scholar] [CrossRef]
- Crighton, D.; Wilkinson, S.; O'Prey, J.; Syed, N.; Smith, P.; Harrison, P.R.; Gasco, M.; Garrone, O.; Crook, T.; Ryan, K.M. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell 2006, 126, 121–134. [Google Scholar] [CrossRef]
- Geng, J.; Zhang, R.; Yuan, X.; Xu, H.; Zhu, Z.; Wang, X.; Wang, Y.; Xu, G.; Guo, W.; Wu, J. DRAM1 plays a tumor suppressor role in NSCLC cells by promoting lysosomal degradation of EGFR. Cell Death & Disease 2020, 11, 768. [Google Scholar] [CrossRef]
- Galavotti, S.; Bartesaghi, S.; Faccenda, D.; Shaked-Rabi, M.; Sanzone, S.; McEvoy, A.; Dinsdale, D.; Condorelli, F.; Brandner, S.; Campanella, M. The autophagy-associated factors DRAM1 and p62 regulate cell migration and invasion in glioblastoma stem cells. Oncogene 2013, 32, 699–712. [Google Scholar] [CrossRef]
- Ronan, B.; Flamand, O.; Vescovi, L.; Dureuil, C.; Durand, L.; Fassy, F.; Bachelot, M.-F.; Lamberton, A.; Mathieu, M.; Bertrand, T. A highly potent and selective Vps34 inhibitor alters vesicle trafficking and autophagy. Nature chemical biology 2014, 10, 1013–1019. [Google Scholar] [CrossRef]
- Dyczynski, M.; Yu, Y.; Otrocka, M.; Parpal, S.; Braga, T.; Henley, A.B.; Zazzi, H.; Lerner, M.; Wennerberg, K.; Viklund, J. Targeting autophagy by small molecule inhibitors of vacuolar protein sorting 34 (Vps34) improves the sensitivity of breast cancer cells to Sunitinib. Cancer letters 2018, 435, 32–43. [Google Scholar] [CrossRef]
- Marsh, T.; Debnath, J. Ironing out VPS34 inhibition. Nature cell biology 2015, 17, 1–3. [Google Scholar] [CrossRef]
- Hennessy, B.T.; Smith, D.L.; Ram, P.T.; Lu, Y.; Mills, G.B. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nature reviews Drug discovery 2005, 4, 988–1004. [Google Scholar] [CrossRef]
- Hu, M.; Zhu, S.; Xiong, S.; Xue, X.; Zhou, X. MicroRNAs and the PTEN/PI3K/Akt pathway in gastric cancer. Oncology reports 2019, 41, 1439–1454. [Google Scholar] [CrossRef]
- Jiang, N.; Dai, Q.; Su, X.; Fu, J.; Feng, X.; Peng, J. Role of PI3K/AKT pathway in cancer: the framework of malignant behavior. Molecular biology reports 2020, 47, 4587–4629. [Google Scholar] [CrossRef]
- Brennan, C.W.; Verhaak, R.G.; McKenna, A.; Campos, B.; Noushmehr, H.; Salama, S.R.; Zheng, S.; Chakravarty, D.; Sanborn, J.Z.; Berman, S.H. The somatic genomic landscape of glioblastoma. Cell 2013, 155, 462–477. [Google Scholar] [CrossRef]
- Errafiy, R.; Aguado, C.; Ghislat, G.; Esteve, J.M.; Gil, A.; Loutfi, M.; Knecht, E. PTEN increases autophagy and inhibits the ubiquitin-proteasome pathway in glioma cells independently of its lipid phosphatase activity. PloS one 2013, 8, e83318. [Google Scholar] [CrossRef]
- Giotta Lucifero, A.; Luzzi, S. Immune landscape in PTEN-related glioma microenvironment: A bioinformatic analysis. Brain Sciences 2022, 12, 501. [Google Scholar] [CrossRef]
- Simpson, L.; Parsons, R. PTEN: life as a tumor suppressor. Experimental cell research 2001, 264, 29–41. [Google Scholar] [CrossRef]
- Morris, D.H.; Yip, C.K.; Shi, Y.; Chait, B.T.; Wang, Q.J. Beclin 1-Vps34 complex architecture: Understanding the nuts and bolts of therapeutic targets. Frontiers in biology 2015, 10, 398–426. [Google Scholar] [CrossRef]
- Schläfli, A.; Berezowska, S.; Adams, O.; Langer, R.; Tschan, M. Reliable LC3 and p62 autophagy marker detection in formalin fixed paraffin embedded human tissue by immunohistochemistry. European journal of histochemistry: EJH 2015, 59. [Google Scholar] [CrossRef]
- Ladoire, S.; Chaba, K.; Martins, I.; Sukkurwala, A.Q.; Adjemian, S.; Michaud, M.; Poirier-Colame, V.; Andreiuolo, F.; Galluzzi, L.; White, E. Immunohistochemical detection of cytoplasmic LC3 puncta in human cancer specimens. Autophagy 2012, 8, 1175–1184. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Wu, Z. Immunohistochemical assessment of autophagic protein LC3B and p62 levels in glioma patients. International Journal of Clinical and Experimental Pathology 2018, 11, 862. [Google Scholar]
- Wudu, M.; Ren, H.; Hui, L.; Jiang, J.; Zhang, S.; Xu, Y.; Wang, Q.; Su, H.; Jiang, X.; Dao, R. DRAM2 acts as an oncogene in non-small cell lung cancer and suppresses the expression of p53. Journal of Experimental & Clinical Cancer Research 2019, 38, 1–13. [Google Scholar] [CrossRef]
- Masuda, G.; Yashiro, M.; Kitayama, K.; Miki, Y.; Kasashima, H.; Kinoshita, H.; Morisaki, T.; Fukuoka, T.; Hasegawa, T.; Sakurai, K. Clinicopathological correlations of autophagy-related proteins LC3, Beclin 1 and p62 in gastric cancer. Anticancer research 2016, 36, 129–136. [Google Scholar] [PubMed]
- Ahmed, K.; Sheikh, A.; Fatima, S.; Haider, G.; Ghias, K.; Abbas, F.; Mughal, N.; Abidi, S.H. Detection and characterization of latency stage of EBV and histopathological analysis of prostatic adenocarcinoma tissues. Scientific Reports 2022, 12, 10399. [Google Scholar] [CrossRef] [PubMed]
| Variables | Values |
|---|---|
|
Gender Male Female |
23 (59%) 16 (41%) |
| Age in years Median (Range) | 43 (47) |
|
Histopathological Type Astrocytoma (Grade 2-4) Oligodendroglioma (2 and 3) Glioblastoma (Grade 4) |
19 (48.7%) 12 (30.8%) 08 (20.5%) |
|
Histopathological grade 2 3 4 |
11 (28.2%) 13 (33.3%) 15 (38.5%) |
|
Glioma Grade Group Low Grade High Grade |
11 (28.20%) 28 (71.8%) |
|
IDH 1Mutant, N (%) |
25 (64.1%) |
| TP53, N (%) Mutant | 16 (41%) |
| ATRX, N (%) Mutant | 21 (53.8%) |
| LC3B | P62 | DRAM | |||||
| Cases | Total | Punctate | Diffuse | High | Low | Positive | Negative |
| 39 | 25 (64.1%) | 14 (35.9%) | 20 (51.3%) | 19 (48.7%) | 11 (28.2%) | 28 (71.8%) | |
| Grades | G-2 (11) | 02 (8%) | 09 (64.2%) | 00 (0%) | 11 (58%) | 04 (36.3%) | 07 (25%) |
| G-3 (13) | 12 (48%) | 01 (7.1%) | 10 (50%) | 03 (15.7%) | 03 (27.2%) | 10 (35.7%) | |
| G-4 (15) | 11 (44%) | 04 (28.5%) | 10 (50%) | 05 (26.3%) | 04 (36.3%) | 11 (39.2%) | |
| Grade | LC3B | p-value | P62 | p-value | DRAM | p-value | |||
| Punctate | Diffuse | High | Low | Positive | Negative | ||||
| 2 | 2(18%) | 09(82%) | 0.001* | 0(0%) | 11(100%) | 0.001* | 4(36%) | 07(64%) | 0.760 |
| 3 | 12(92%) | 1(7%) | 10(77%) | 3(23) | 3(23%) | 10(77%) | |||
| 4 | 11(73%) | 4(27%) | 10(67%) | 5(33%) | 4(27%) | 11(73%) | |||
| Clinicopathologic Parameters | LC3B | p-value | P62 | p-value | DRAM | p-value | |||
| Punctate | Diffuse |
0.022 |
High | Low |
0.017 |
Present | Absent |
0.380 |
|
|
Age <45 >45 |
10(40%) 15(60%) |
11(79%) 03(21%) |
07(35%) 13(65%) |
14(74%) 05(26%) |
05(45%) 06(55%) |
16(57%) 12(43%) |
|||
|
Gender Male Female |
17(68%) 08(32%) |
06(43%) 08(57%) |
0.117 |
14(70%) 06(30%) |
09(47%) 10(53%) |
0.133 |
09(82%) 02(18%) |
14(50%) 14(50%) |
0.70 |
|
Histological type Oligodendroglioma Astrocytoma Glioblastoma |
09(75%) 12(63%) 04(50%) |
3(25%) 07(37%) 04(50%) |
0.517 |
09(75%) 08(42%) 03(38%) |
03(25%) 11(58%) 05(62%) |
0.139 |
04(33%) 05(26%) 02(25%) |
08(67%) 14(74%) 06(75%) |
0.891 |
|
Histological grade 2 3 4 |
02(18%) 12(92%) 11(73%) |
09(82%) 01(8%) 04(27%) |
0.001 |
0(0%) 10(77%) 10(67%) |
11(100%) 03(23%) 05(33%) |
<0.001 |
04(36%) 03(23%) 04(27%) |
07()64%) 10(77%) 11(73%) |
0.760 |
|
IDH1-R132 mut Present Absent |
18(72%) 07(50%%) |
07(28%) 07(50%) |
0.153 |
14(56%) 06(43%) |
11(44%) 08(57%) |
0.325 |
08(32%) 03(21%) |
17(68%) 11(79%) |
0.376 |
|
ATRX mut/Loss Present Absent |
12(57%) 13(72%) |
09(43%) 05(28%) |
0.261 |
07(33%) 13(72%) |
14(67%) 05(28%) |
0.17 |
05(24%) 06(33%) |
16(76%) 12(67%) |
0.380 |
|
Tp53 mut Present Absent |
09(56%) 16(70%) |
07(44%) 07(30%) |
0.303 |
06(38%) 14(61%) |
10(62%0 09(39%) |
0.133 |
06(38%) 05(22%) |
10(62%) 18(78%) |
0.237 |
| PI3K | BECLIN1 | mTOR | AKT | PTEN | ULKI | ULK2 | UVRAG | VPS34 | |
| PI3K | - | 0.495 | 0.863 | 0.505 | 0.824 | 0.995 | 0.731 | 0.978 | 0.560 |
| BECLIN1 | 0.495 | - | 0.615 | 0.110 | 0.670 | 0.505 | 0.462 | 0.495 | 0.275 |
| m-TOR | 0.863 | 0.615 | - | 0.357 | 0.835 | 0.846 | 0.604 | 0.896 | 0.473 |
| AKT | 0.505 | 0.110 | 0.357 | - | 0.434 | 0.500 | 0.247 | 0.489 | 0.055 |
| PTEN | 0.824 | 0.670 | 0.835 | 0.434 | - | 0.802 | 0.714 | 0.846 | 0.577 |
| ULK1 | 0.995 | 0.505 | 0.846 | 0.500 | 0.802 | - | 0.753 | 0.956 | 0.516 |
| ULK2 | 0.731 | 0.462 | 0.604 | 0.247 | 0.714 | 0.753 | - | 0.670 | 0.626 |
| UVRAG | 0.978 | 0.495 | 0.896 | 0.489 | 0.846 | 0.956 | 0.670 | - | 0.599 |
| VPS34 | 0.560 | 0.275 | 0.473 | 0.055 | 0.577 | 0.516 | 0.626 | 0.599 | - |
| PI3K | BECLIN1 | mTOR | AKT | PTEN | ULKI | ULK2 | UVRAG | VPS34 | |
| PI3K | - | 0.866 | 0.748 | -.094 | 0.988 | 0.674 | 0.781 | 0.747 | 0.192 |
| BECLIN1 | 0.886 | -- | 0.659 | 0.143 | 0.852 | 0.566 | 0.549 | 0.567 | 0.875 |
| mTOR | 0.748 | 0.659 | - | -.088 | 0.714 | 0.544 | 0.527 | 0.396 | 0.627 |
| AKT | -.o94 | 0.143 | -.088 | - | -.104 | -.346 | -.538 | -.380 | -.204 |
| PTEN | 0.988 | 0.852 | 0.714 | -.104 | - | 0.621 | 0.753 | 0.735 | 0.889 |
| ULK1 | 0.674 | 0.566 | 0.544 | -.346 | 0.621 | - | 0.742 | 0.784 | 0.652 |
| ULK2 | 0.781 | 0.549 | 0.527 | -.538 | 0.753 | 0.742 | - | 0.814 | 0.715 |
| UVRAG | 0.747 | 0.567 | 0.396 | -.380 | 0.735 | 0.784 | 0.814 | - | 0.802 |
| VPS34 | 0.912 | 0.875 | 0.627 | -.204 | 0.899 | 0.652 | 0.715 | 0.802 | - |
| PI3K | BECLIN1 | mTOR | AKT | PTEN | ULKI | ULK2 | UVRAG | VPS34 | |
| PI3K | - | 0.721 | 0.461 | 0.479 | 0.646 | 0.943 | 0.782 | 0.893 | 0.600 |
| BECLIN1 | 0.721 | - | 0.575 | 0.514 | 0.382 | 0.707 | 0.714 | 0.579 | 0.768 |
| mTOR | 0.461 | 0.575 | - | 0.504 | 0.557 | 0.496 | 0.386 | 0.432 | 0.411 |
| AKT | 0.479 | 0.514 | 0.504 | - | 0.443 | 0.571 | 0.486 | 0.361 | 0.464 |
| PTEN | 0.646 | 0.382 | 0.577 | 0.443 | - | 0.568 | 0.375 | 0.504 | 0.257 |
| ULK1 | 0.943 | 0.707 | 0.496 | 0.571 | 0.568 | - | 0.811 | 0.886 | 0.643 |
| ULK2 | 0.782 | 0.714 | 0.386 | 0.486 | 0.375 | 0.811 | - | 0.729 | 0.786 |
| UVRAG | 0.893 | 0.579 | 0.432 | 0.386 | 0.574 | 0.886 | 0.729 | - | 0.514 |
| VPS34 | 0.600 | 0.768 | 0.411 | 0.464 | 0.257 | 0.643 | 0.786 | 0.514 | - |
| Genes | Forward primer | Reverse primer |
| Beclin-1 | 5’ -AATGACTTTTTTCCTTAGGGGG-3’ | 5’ -GTGGCTTTTGTGGATTTTTTCT-3’ |
| m-TOR | 5’ -TGGGACAGCATGGAAGAATA-3’ | 5’- TGTTGTGCCAAGGAGAAGAG-3’ |
| UVRAG | 5′- CTGTTGCCCTTGGTTATACTGC -3′ | 5′- GATGATTTCTTCTGCTTGCTCC -3′ |
| VPS34 | 5′-GCT GTC CTG GAA GAC CCA AT-3′ | 5′-TTC TCA CTG GCA AGG CCA AA-3′ |
| PTEN | 5’-CCAAGCTTATGACAGCCATCATC-3’ | 5’-CGCGGATCCTCAGACTTTTGTAA-3’ |
| ULK1 | 5’-GGACACCATCAGGCTCTTCC-3’ | 5’-GAAGCCGAAGTCAGCGATCT-3’ |
| ULK2 | 5’-TTCCTGCTCTAAGGGTTTGCTT-3’ | 5’-CCAGCGAGGGAGAACAACTG-3’ |
| PI3K | 5’ - ATGCAAATTCAGTGCAAAGG-3’ | 5’ - CGTGTAAACAGGTCAATGGC-3’ |
| AKT | 5’ -GCAGCACGTGTACGAGAAGA-3’ | 5’ -GGTGTCAGTCTCCGACGTG-3’ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).