1. Introduction
Parkinson’s Disease (PD) is a chronic and progressive neurodegenerative disorder that affects movement, muscle control, and balance. Despite significant research efforts, a definitive cure remains elusive, and the condition continues to pose a substantial healthcare challenge worldwide (
Lew 2007). The early and accurate diagnosis of PD is of paramount importance as it directly impacts the management and therapeutic strategies that can be employed to slow disease progression and improve patient quality of life (
Afreen 2021,
Fereshtehnejad 2016,
Tinelli et al. 2016).
The etiology of PD has been associated with the degeneration of dopaminergic neurons in the substantia nigra, a basal ganglia structure in the brain. This degeneration leads to a pronounced reduction in the levels of dopamine, a neurotransmitter critical for regulating movement and coordination (
Damier et al. 1999). One of the hallmarks of PD diagnosis is the identification of reduced dopamine transporter (DaT) activity in the striatum, which encompasses the putamen and caudate nuclei, areas heavily involved in motor control (
Porter et al. 2020).
Single Photon Emission Computed Tomography (SPECT) imaging, using the Iodine-123 fluoropropyl (
123I-FP-CIT) DaTscan, has been recognized by the Food and Drug Administration (FDA) as a significant tool for the differential diagnosis of PD (
Akdemir et al. 2021). It works by visualizing the density of dopaminergic neurons, providing information that aids in distinguishing PD from other disorders with similar presentations, such as essential tremor (
Seibyl 1999). However, the interpretation of DaT SPECT images has traditionally relied on qualitative assessments by expert radiologists. This approach is not without its drawbacks, including the potential for subjective bias and intra- and inter-observer variability (
Scherfler et al. 2007).
To overcome these limitations, there has been a growing interest in leveraging computational techniques to analyze medical imaging data. The field of machine learning, and more specifically deep learning, has shown exceptional promise in this domain. Deep Convolutional Neural Networks (CNNs), due to their ability to learn hierarchical representations of data, have emerged as powerful tools for the classification and analysis of complex imaging data (
Dou et al. 2016,
LeCun et al. 2015,
Yadav and Jadhav 2019,?).
Among various deep learning architectures, InceptionV3 stands out due to its sophisticated design that allows for efficient computation and high accuracy with fewer parameters compared to its predecessors. It has been pre-trained on a large dataset of images from diverse categories, which enables it to capture a wide range of features relevant to image classification tasks (
Demir et al. 2019). When applied to the analysis of SPECT DaTSCAN images, InceptionV3 can leverage its pre-trained weights as a starting point, which can then be fine-tuned to recognize patterns specific to PD. This process, known as transfer learning, is particularly advantageous when dealing with medical datasets that are often smaller than the datasets typically used to train deep learning models from scratch (
Weiss et al. 2016).
The motivation for this study is founded on the hypothesis that the application of the InceptionV3-based CNN model to SPECT DaTSCAN imaging can provide a quantitative and objective analysis method for the diagnosis of PD. This approach has the potential not only to support the clinical decision-making process but also to contribute to the body of knowledge in neuroimaging and neurodegenerative disease research (
Badrulhisham et al. 2024,
Marblestone et al. 2016,
Orrù et al. 2021).
In this paper, we present a comprehensive methodology for the preemptive diagnosis of PD through the analysis of DaT SPECT images using an InceptionV3-based CNN. We detail our process from data acquisition and preprocessing through to model training and evaluation. Our aim is to demonstrate that this method can effectively distinguish between PD-affected and non-PD subjects, thus offering a valuable diagnostic tool that could enhance early detection and treatment planning for patients with PD (
Hazan et al. 2012,
Wang et al. 2020).
2. Literature Review
The history of Parkinson’s Disease (PD) research is marked by significant milestones in understanding its progression and treatment. The natural course of PD has varying rates of progression (
Poewe 2006). The importance of early diagnosis is underscored by the fact that the disease’s progression is not linear, with a tendency to advance more swiftly in the early stages. Furthermore, non-motor symptoms such as cognitive impairment, sleep-wake cycle disruptions, and autonomic dysfunction play a considerable role in the disease’s trajectory, impacting a large percentage of patients within long-term follow-ups (
Pfeiffer 2016).
Levodopa stands as the cornerstone of PD treatment, yet its long-term usage is plagued by the emergence of motor complications and drug-induced dyskinesia, highlighting the need for alternative and adjunct therapies
Rao et al. (
2006). Treatment strategies for early-stage PD, which include patients within the first five years of diagnosis or those not yet experiencing levodopa-induced complications, often incorporate a combination of pharmacological agents (
Marsili et al. 2017). These may include monoamine oxidase-B (MAO-B) inhibitors and anticholinergics, which have shown modest efficacy in symptom management. The clinical decision-making process often involves a nuanced consideration of patient age, disease severity, and symptomatology, which dictate whether a dopamine agonist or levodopa should be initiated.
In the face of these challenges, artificial intelligence (AI) has emerged as a transformative force in PD diagnosis and management (
Raghavendra et al. 2020,
Saravanan et al. 2022,
Yang et al. 2022). Machine learning (ML), a subset of AI, has been extensively studied for its potential in PD diagnosis, with the diagnosis process generally consisting of three main phases: data preprocessing, feature extraction, and the application of classification algorithms (
Rana et al. 2022). Initial stages involve the denoising and categorization of speech signals, which are pivotal in PD due to the disease’s impact on vocal characteristics. Subsequent phases focus on extracting salient features from the processed data, followed by the application of various ML algorithms such as Support Vector Machines, Artificial Neural Networks, K-Nearest Neighbors, Naïve Bayes, Logistic Regression, and Decision Trees, each with its strengths and contexts of applicability (
Nhu et al. 2020).
More advanced approaches in PD diagnosis have leveraged Deep Learning, a more complex iteration of ML, which has shown proficiency in handling high-dimensional data and capturing intricate patterns that simpler ML models may overlook (
Mounika and Rao 2021). Convolutional Neural Networks, a class of deep neural networks, have been particularly effective in image-based diagnosis of PD, especially in interpreting DaTscan SPECT images to identify dopaminergic deficits (
Khachnaoui et al. 2020). Another promising area is the use of Recurrent Neural Networks (RNNs) for analyzing sequential data such as patient movement patterns or speech signals, which are indicative of PD-related motor and non-motor symptoms (
Ribeiro et al. 2019).
The emergence of Federated Learning, a distributed approach to ML, harnesses decentralized data while maintaining privacy, a crucial consideration given the sensitive nature of medical data (
Li et al. 2020). Federated Learning has the potential to create robust models trained across multiple datasets from diverse demographics and geographic locations, enhancing the generalizability and accuracy of PD diagnosis (
Arasteh et al. 2023). Additionally, Transfer Learning techniques have been applied to adapt pre-trained models on large datasets to the specific task of PD diagnosis, thereby reducing the need for extensive labeled data which is often scarce in medical research (
Kaur et al. 2021).
Despite these technological strides, challenges persist, particularly in model interpretability. The ’black box’ nature of many AI models, especially deep learning networks, complicates their clinical adoption, as healthcare practitioners necessitate transparent decision-making tools (
Wroge et al. 2018). Efforts to improve model explainability through techniques such as Layer-wise Relevance Propagation (LRP) and SHapley Additive exPlanations (SHAP) values are underway, aiming to demystify the inner workings of AI models for clinicians (
Ullah et al. 2021).
The integration of AI into PD research represents a paradigm shift in diagnosis and treatment strategies. With the advent of novel AI methodologies, the field is poised for a new era of precision medicine that promises to deliver more personalized and effective care for PD patients.
3. Materials and Methods
3.1. Data Processing
Single Photon Emission Computed Tomography (SPECT) with DaTSCAN is an imaging modality that has been endorsed by the Food and Drug Administration (FDA) for the early detection of Parkinson’s Disease in the United States (
Brooks 2010). The diagnostic agent, DaTscan, is a radiopharmaceutical introduced intravenously that binds to dopamine transporters (DaT) in the brain. A gamma camera captures the emitted gamma rays from the radiotracer, providing images that reflect the distribution and density of dopamine-containing neurons which are implicated in motor control. These images are instrumental in distinguishing between Parkinson’s Disease and other conditions with similar motor deficits, such as essential tremor, which exhibits tremors but does not involve the same dopaminergic neuron degeneration (
Akdemir et al. 2021,
Mabrouk et al. 2018,
Suwijn et al. 2015).
Furthermore, the application of DaTscan extends beyond Parkinson’s Disease. In Europe, it has been leveraged to differentiate Alzheimer’s disease from dementia with Lewy bodies, which is critical for tailoring treatment strategies. The differential diagnosis facilitated by DaTscan imaging influences the selection of pharmacological interventions, ensuring the safety and efficacy of dementia management (
Luzny and Ivanova 2016).
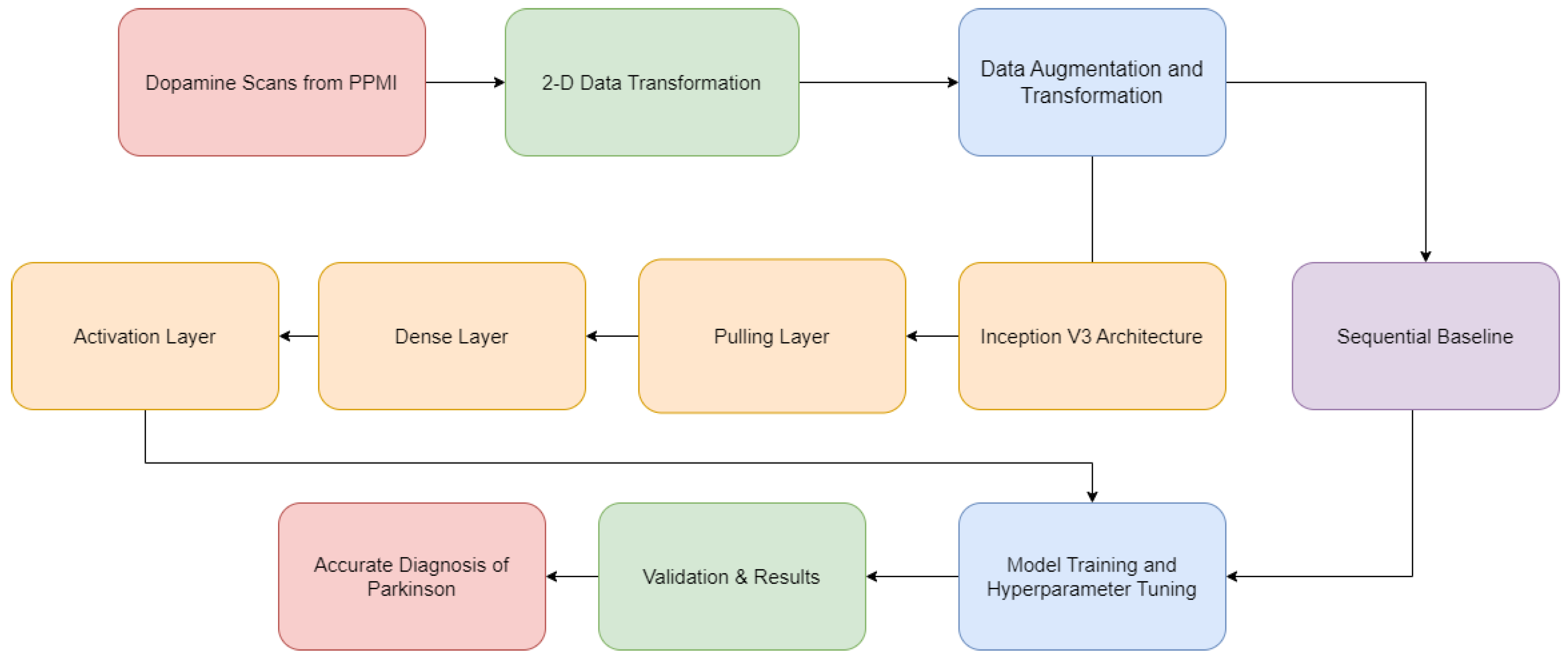
The dataset for this study was sourced from the Parkinson’s Progression Markers Initiative (PPMI), which is an extensive collection aimed at identifying biomarkers for Parkinson’s Disease. The PPMI’s database includes clinical, imaging, and biosample data from individuals with and without Parkinson’s disease. For the current investigation, a subset of the database consisting of 708 DaT SPECT images from unique patients was used. This subset was categorized into two groups based on clinical diagnosis: 384 patients diagnosed with PD and 324 controls.
Table 1 shows the data distribution of the dataset. To maintain the integrity of the training process, the dataset was meticulously curated to exclude any follow-up images, ensuring that each patient contributed only a single image to the dataset. This precaution was taken to prevent data leakage and to validate the model’s performance on independent patient data.
In preparing the imaging data for analysis, raw
123I-FP-CIT SPECT images were processed in accordance with the standardized protocols of the PPMI (
Marek et al. 2011). The images underwent reconstruction and were transformed into a standardized format within the Montreal Neurological Institute (MNI) coordinate system, which is widely accepted for brain imaging analyses. This standardization involved the conversion of images into three-dimensional volumes with matrix sizes of 91 x 109 x 91, which is compatible with the spatial resolution commonly used in neuroimaging studies.
Due to the constraints imposed by system memory, the ImageDataGenerator class from the Keras library was employed to manage the dynamic loading of image batches during model training. The original images, which were in the DICOM format typical for medical imaging, were converted into PNG files to facilitate the use of the ImageDataGenerator. This tool cannot process volumetric data directly; hence, the conversion to a compatible format was necessary. The representative slices of the striatum, specifically slices numbered 40-42, were selected based on their anatomical relevance to PD. These slices were then combined to create composite RGB images, conforming to the three-channel input requirement of the InceptionV3 model (
Liu et al. 2020). One such 2D-Processed DaT Scan is shown in
Figure 1.
To augment the dataset and introduce variability, a series of transformations were applied to the images. This process was essential to ensure that the model did not overfit to the training data, a common concern in machine learning when working with limited datasets. The added transformations effectively increased the number of unique images available for training, addressing the challenge of limited data availability in this field. The transformations included alterations to the images’ width and height, horizontal flipping, and brightness adjustments. These transformations were executed on-the-fly as the images were loaded into the model, which optimized memory usage and computational efficiency.
The robustness of the model’s performance was evaluated using k-fold cross-validation. This technique is particularly useful in scenarios where the maximization of limited data is essential. The dataset was divided into ten subsets, or ’folds’, with an approximately equal distribution of PD and non-PD classes within each fold. For each iteration of validation, one fold was held out as the validation set while the remaining nine folds were used for training. The model’s performance metrics, specifically validation loss and accuracy, were computed and averaged across all folds to provide a comprehensive assessment of its generalizability.
3.2. Model Architecture
For the architecture and training of the convolutional neural network, the InceptionV3 model was selected. The inception model, known for its efficiency and effectiveness in image classification tasks, was developed for the ImageNet Large Scale Visual Recognition Challenge in 2015. The choice of InceptionV3 for this study was predicated on its sophisticated architecture, which allows for the extraction of complex features from image data with considerable computational efficiency. InceptionV3 has been pre-trained on a broad array of visual data, enabling it to act as a potent feature extractor for the DaT SPECT images. A customized binary classification layer was integrated into the architecture to tailor the model to the specific task of distinguishing PD from non-PD images. This layer consisted of a global average pooling layer followed by a dense layer with ReLU activation, a dropout layer to prevent overfitting, and a final dense layer with sigmoid activation designed for binary classification purposes.
The InceptionV3 architecture, integral to our convolutional neural network, epitomizes advancements in deep learning for image recognition. The foundational architecture of InceptionV3 is mathematically expressed as:
where
x denotes the input image tensor,
L symbolizes the number of layers,
represents the convolution operations with varying kernel sizes
and biases
,
denotes pooling operations with parameters
, and
symbolizes fully connected layers with weights
and biases
.
To enhance this base model, we introduce a GlobalAveragePooling2D layer, mathematically formulated as:
Here, A represents the input feature map matrix from the preceding convolutional layer of InceptionV3, with dimensions . This layer condenses the spatial dimensions by averaging the features, thus reducing the complexity while retaining critical spatial information.
The subsequent layer is a densely connected network layer with a rectified linear unit (ReLU) activation, mathematically depicted as:
In this equation, x is the flattened output from the GlobalAveragePooling2D layer, W and b denote the weights and bias of the dense layer, and is the resultant activation. This layer captures complex non-linear interdependencies in the feature set.
Incorporating these layers atop the base InceptionV3 model, we establish a comprehensive framework capable of pattern discernment in DaT SPECT images for precise PD diagnosis. The complexity of each layer allows the model to be proficient in high-level feature extraction and classification.
The training process utilized the Adam optimization algorithm, which is an extension of stochastic gradient descent that has been shown to be effective in deep learning applications. The initial learning rate was set at and was adjusted using a step decay schedule, decreasing incrementally to to fine-tune the model’s weights during the latter stages of training. The loss function employed was binary crossentropy, a standard choice for binary classification tasks. The primary metric for evaluating model performance was classification accuracy. The model was trained over 100 epochs with a batch size of 16, providing an opportunity for the model to learn from the data without overfitting, which was assessed through the model’s performance in the validation dataset.
Figure 2 summarizes the model architecture.
4. Experiments and Results
This section delineates the extensive evaluations conducted to assess the performance of the InceptionV3-based convolutional neural network in classifying DaT SPECT images for the detection of Parkinson Disease.
4.1. Hyperparameter Tuning
Hyperparameter optimization plays a critical role in enhancing the model’s performance. We conducted experiments with varying learning rates, neurons in the dense layer, and dropout rates to identify the optimal configuration. The learning rates tested were
,
, and
, the number of neurons were 512, 1024, 2048, and the dropout rates were 0, 0.25, and 0.5. The optimal hyperparameters and their corresponding accuracy are presented in
Table 2.
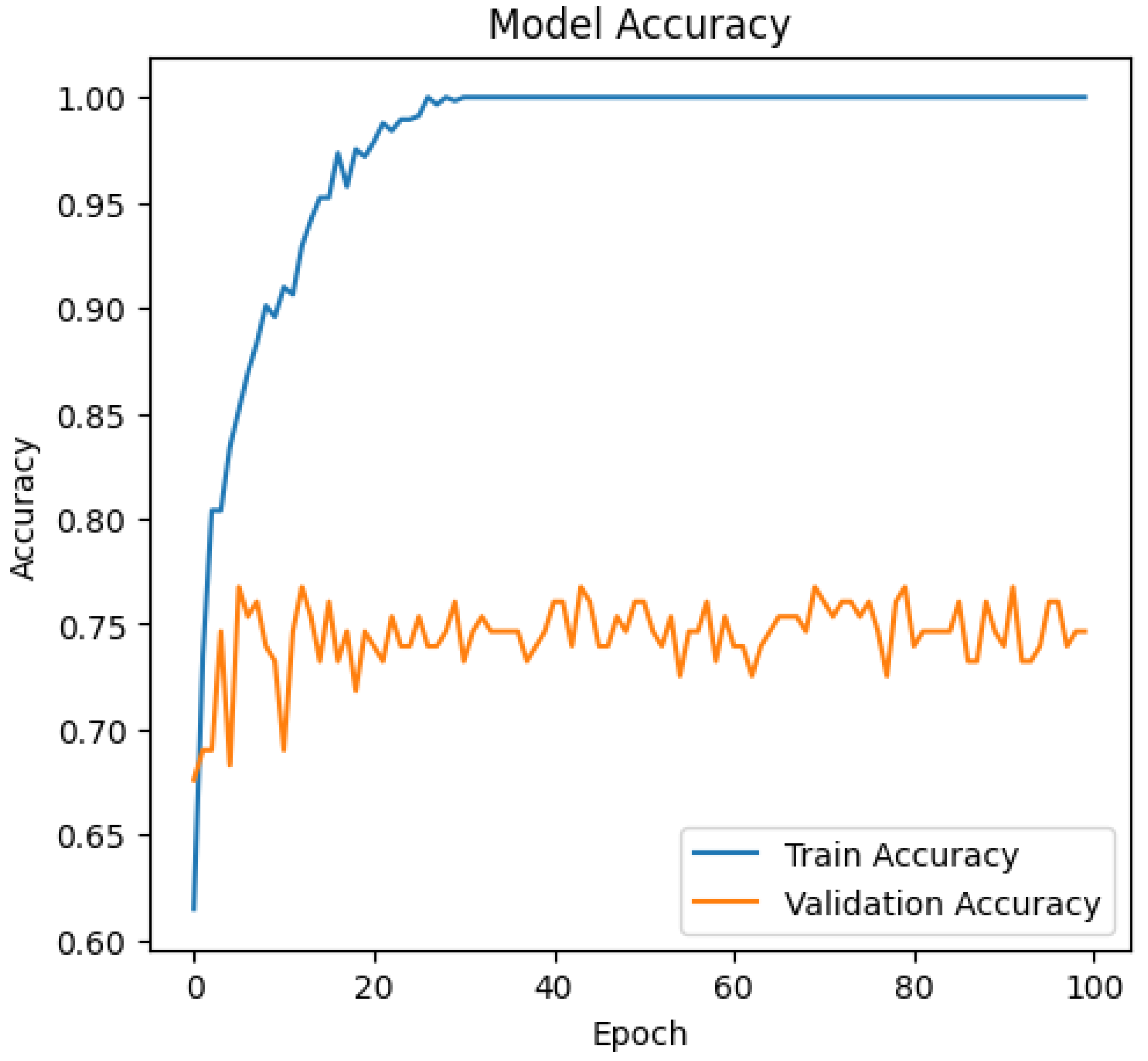
The InceptionV3 model, configured with these hyperparameters, was trained for 100 epochs, achieving an accuracy of 74.65%.
Figure 3 illustrates the model’s accuracy over the 100 training epochs.
The accuracy achieved by our InceptionV3 model indicates its effectiveness in distinguishing Parkinson’s Disease from controls, which has substantial implications in early diagnosis and personalized medicine.
To benchmark our model, we conducted a comparative analysis against a baseline Sequential Model, which is commonly used in similar applications. This comparison aims to highlight the relative improvement offered by the proposed model.
This comparison highlights the efficacy of our InceptionV3 model in classifying DaT SPECT images for Parkinson’s Disease detection, as it significantly outperforms the conventional Sequential Model approach.
In summary, our experiments and results demonstrate the proficiency of the InceptionV3-based model in classifying DaT SPECT images for Parkinson’s Disease detection. The model’s high accuracy during both training and validation testing highlights its potential as an effective tool in medical diagnostics. Future work will involve further expanding the dataset, exploring additional hyperparameters, and potentially integrating multimodal data to refine diagnostic capabilities.
5. Conclusion and Future Work
This study marks a significant stride in the field of Parkinson’s disease diagnosis through advanced imaging and machine learning techniques. Our model, leveraging the sophisticated architecture of InceptionV3, has demonstrated remarkable proficiency in classifying DaT SPECT images. The accuracy highlights the model’s potential as a reliable diagnostic tool, distinguishing Parkinson’s Disease from other conditions with similar motor deficits. Our approach differs from previous works primarily in its application of a deep learning model that is both highly accurate and robust, capable of processing complex image data with nuanced differentiation.
The implications of our model in the context of Parkinson’s diagnosis are profound. Early and accurate diagnosis of Parkinson’s disease is crucial, as it enables timely intervention, potentially slowing disease progression and improving quality of life. Our model’s high sensitivity is particularly vital, as it minimizes false negatives, which are crucial in medical diagnostics. This level of accuracy can revolutionize the diagnostic process, making it faster, more reliable, and accessible.
Compared to previous methodologies, our model offers a significant advancement in several areas. Traditional diagnostic methods often rely heavily on clinical assessments, which can be subjective and vary in accuracy. In contrast, our model provides a more objective and quantifiable approach. Additionally, earlier diagnostic models may lack the sophistication needed to accurately distinguish between Parkinson’s Disease and other conditions. Our model addresses this gap through its advanced feature extraction capabilities, which are adept at capturing the subtle nuances in DaT SPECT images.
The success of this model paves the way for more personalized medicine approaches in neurodegenerative diseases. By providing accurate and early diagnosis, clinicians can tailor treatment plans to individual patients more effectively. This individualized approach could lead to better management of the disease, potentially altering its course and improving patient outcomes.
Looking ahead, several avenues for further research and development present themselves. Firstly, expanding the dataset to include a more diverse population sample could further validate the model’s efficacy and enhance its generalizability. Future studies could also explore the integration of multimodal data, such as combining imaging data with genetic or biomarker information, to develop even more comprehensive diagnostic tools.
Secondly, continual refinement of the model’s architecture and tuning of hyperparameters could yield improvements in performance. Exploring other advanced deep learning models and techniques could also provide new insights and enhancements.
Lastly, there is an opportunity to extend this research to other neurodegenerative diseases, leveraging the model’s capabilities to assist in the diagnosis of conditions such as Alzheimer’s or multiple system atrophy. Such expansions could significantly impact the field of neurology, offering new tools and methodologies for disease diagnosis and management.
In conclusion, this research represents a significant contribution to the field of Parkinson’s disease diagnosis. The model’s high accuracy in analyzing DaT SPECT images demonstrate its potential as a transformative tool in medical diagnostics. As we continue to refine and expand upon this work, the prospect of improving patient outcomes and advancing personalized medicine becomes increasingly tangible.
Author Contributions
Ashmita Appineni conceptualized and designed the proposed model, developed the methodology, conducted the data analysis, constructed and implemented the model, provided visualization of the results, and took primary responsibility for writing, revising, and finalizing this paper. Aditya Gupta provided the initial dataset. All authors have read, reviewed, and agreed to the published version of the manuscript.
References
- Afreen, Nuraia. 2021. Treatment options of Parkinson’s disease: How far we are from the cure? Ph. D. thesis, Brac University.
- Akdemir, Ümit Özgür, HATİCE AYŞE TOKÇAER BORA, and Lütfiye Özlem Atay. 2021. Dopamine transporter spect imaging in parkinson’s disease and parkinsoniandisorders. Turkish Journal of Medical Sciences 51(2), 400–410.
- Arasteh, Soroosh Tayebi, Cristian David Rios-Urrego, Elmar Noeth, Andreas Maier, Seung Hee Yang, Jan Rusz, and Juan Rafael Orozco-Arroyave. 2023. Federated learning for secure development of ai models for parkinson’s disease detection using speech from different languages. arXiv preprint arXiv:2305.11284.
- Badrulhisham, Fakhirah, Esther Pogatzki-Zahn, Daniel Segelcke, Tamas Spisak, and Jan Vollert. 2024. Machine learning and artificial intelligence in neuroscience: A primer for researchers. Brain, Behavior, and Immunity 115, 470–479. [CrossRef]
- Brooks, David J. 2010. Imaging approaches to parkinson disease. Journal of Nuclear Medicine 51(4), 596–609. [CrossRef]
- Damier, P, EC Hirsch, Y Agid, and AM10430829 Graybiel. 1999. The substantia nigra of the human brain: Ii. patterns of loss of dopamine-containing neurons in parkinson’s disease. Brain 122(8), 1437–1448.
- Demir, Ahmet, Feyza Yilmaz, and Onur Kose. 2019. Early detection of skin cancer using deep learning architectures: resnet-101 and inception-v3. In 2019 medical technologies congress (TIPTEKNO), pp. 1–4. IEEE.
- Dou, Q., H. Chen, L. Yu, L. Zhao, J. Qin, D. Wang, V.C. Mok, L. Shi, and P. Heng. 2016, May. Automatic detection of cerebral microbleeds from mr images via 3d convolutional neural networks. IEEE Transactions on Medical Imaging 35(5), 1182–1195. [CrossRef]
- Fereshtehnejad, Seyed-Mohammad. 2016. Strategies to maintain quality of life among people with parkinson’s disease: what works? Neurodegenerative disease management 6(5), 399–415.
- Hazan, Hananel, Dan Hilu, Larry Manevitz, Lorraine O Ramig, and Shimon Sapir. 2012. Early diagnosis of parkinson’s disease via machine learning on speech data. In 2012 IEEE 27th Convention of Electrical and Electronics Engineers in Israel, pp. 1–4. IEEE.
- Kaur, Sukhpal, Himanshu Aggarwal, and Rinkle Rani. 2021. Diagnosis of parkinson’s disease using deep cnn with transfer learning and data augmentation. Multimedia Tools and Applications 80, 10113–10139. [CrossRef]
- Khachnaoui, Hajer, Rostom Mabrouk, and Nawres Khlifa. 2020. Machine learning and deep learning for clinical data and pet/spect imaging in parkinson’s disease: a review. IET Image Processing 14(16), 4013–4026. [CrossRef]
- LeCun, Yann, Yoshua Bengio, and Geoffrey Hinton. 2015. Deep learning. nature 521(7553), 436–444.
- Lew, Mark. 2007. Overview of parkinson’s disease. Pharmacotherapy: The Journal of Human Pharmacology and Drug Therapy 27(12P2), 155S–160S.
- Li, Li, Yuxi Fan, Mike Tse, and Kuo-Yi Lin. 2020. A review of applications in federated learning. Computers & Industrial Engineering 149, 106854. [CrossRef]
- Liu, Kun, Shengtao Yu, and Sidong Liu. 2020. An improved inceptionv3 network for obscured ship classification in remote sensing images. IEEE Journal of Selected Topics in Applied Earth Observations and Remote Sensing 13, 4738–4747. [CrossRef]
- Luzny, Jan and Katerina Ivanova. 2016. Datscan in differential diagnostics of lewy body disease. Archives of Iranian medicine 19(6), 449–452.
- Mabrouk, Rostom, Belkacem Chikhaoui, and Layachi Bentabet. 2018. Machine learning based classification using clinical and datscan spect imaging features: A study on parkinson’s disease and swedd. IEEE Transactions on Radiation and Plasma Medical Sciences 3(2), 170–177. [CrossRef]
- Marblestone, Adam H, Greg Wayne, and Konrad P Kording. 2016. Toward an integration of deep learning and neuroscience. Frontiers in computational neuroscience 10, 94. [CrossRef]
- Marek, Kenneth, Danna Jennings, Shirley Lasch, Andrew Siderowf, Caroline Tanner, Tanya Simuni, Chris Coffey, Karl Kieburtz, Emily Flagg, Sohini Chowdhury, et al. 2011. The parkinson progression marker initiative (ppmi). Progress in neurobiology 95(4), 629–635. [CrossRef]
- Marsili, Luca, Roberto Marconi, and Carlo Colosimo. 2017. Treatment strategies in early parkinson’s disease. International review of neurobiology 132, 345–360.
- Mounika, P and S Govinda Rao. 2021. Machine learning and deep learning models for diagnosis of parkinson’s disease: A performance analysis. In 2021 Fifth International Conference on I-SMAC (IoT in Social, Mobile, Analytics and Cloud)(I-SMAC), pp. 381–388. IEEE.
- Nhu, Viet-Ha, Ataollah Shirzadi, Himan Shahabi, Sushant K Singh, Nadhir Al-Ansari, John J Clague, Abolfazl Jaafari, Wei Chen, Shaghayegh Miraki, Jie Dou, et al. 2020. Shallow landslide susceptibility mapping: A comparison between logistic model tree, logistic regression, naïve bayes tree, artificial neural network, and support vector machine algorithms. International journal of environmental research and public health 17(8), 2749. [CrossRef]
- Orrù, Graziella, Ciro Conversano, Rebecca Ciacchini, and Angelo Gemignani. 2021. A brief overview on the contribution of machine learning in systems neuroscience. Current Psychiatry Research and Reviews Formerly: Current Psychiatry Reviews 17(2), 66–71.
- Pfeiffer, Ronald F. 2016. Non-motor symptoms in parkinson’s disease. Parkinsonism & related disorders 22, S119–S122.
- Poewe, Werner. 2006. The natural history of parkinson’s disease. Journal of neurology 253, vii2–vii6.
- Porter, Eleanor, Andreas-Antonios Roussakis, Nicholas P Lao-Kaim, and Paola Piccini. 2020. Multimodal dopamine transporter (dat) imaging and magnetic resonance imaging (mri) to characterise early parkinson’s disease. Parkinsonism & Related Disorders 79, 26–33. [CrossRef]
- Raghavendra, U, U Rajendra Acharya, and Hojjat Adeli. 2020. Artificial intelligence techniques for automated diagnosis of neurological disorders. European neurology 82(1-3), 41–64. [CrossRef]
- Rana, Arti, Ankur Dumka, Rajesh Singh, Manoj Kumar Panda, Neeraj Priyadarshi, and Bhekisipho Twala. 2022. Imperative role of machine learning algorithm for detection of parkinson’s disease: review, challenges and recommendations. Diagnostics 12(8), 2003.
- Rao, Shobha S, Laura A Hofmann, and Amer Shakil. 2006. Parkinson’s disease: diagnosis and treatment. American family physician 74(12), 2046–2054.
- Ribeiro, Luiz CF, Luis CS Afonso, and Joao P Papa. 2019. Bag of samplings for computer-assisted parkinson’s disease diagnosis based on recurrent neural networks. Computers in biology and medicine 115, 103477. [CrossRef]
- Saravanan, S, Kannan Ramkumar, K Adalarasu, Venkatesh Sivanandam, S Rakesh Kumar, S Stalin, and Rengarajan Amirtharajan. 2022. A systematic review of artificial intelligence (ai) based approaches for the diagnosis of parkinson’s disease. Archives of Computational Methods in Engineering 29(6), 3639–3653. [CrossRef]
- Scherfler, Christoph, Johannes Schwarz, Angelo Antonini, Donald Grosset, Francesc Valldeoriola, Kenneth Marek, Wolfgang Oertel, Eduardo Tolosa, Andrew J Lees, and Werner Poewe. 2007. Role of dat-spect in the diagnostic work up of parkinsonism. Movement disorders: official journal of the Movement Disorder Society 22(9), 1229–1238.
- Seibyl, John P. 1999. Single-photon emission computed tomography of the dopamine transporter in parkinsonism. Journal of Neuroimaging 9(4), 223–228. [CrossRef]
- Suwijn, Sven R, Caroline JM van Boheemen, Rob J de Haan, Gerrit Tissingh, Jan Booij, and Rob MA de Bie. 2015. The diagnostic accuracy of dopamine transporter spect imaging to detect nigrostriatal cell loss in patients with parkinson’s disease or clinically uncertain parkinsonism: a systematic review. EJNMMI research 5, 1–8. [CrossRef]
- Tinelli, Michela, Panos Kanavos, and Federico Grimaccia. 2016. The value of early diagnosis and treatment in parkinson’s disease: a literature review of the potential clinical and socioeconomic impact of targeting unmet needs in parkinson’s disease.
- Ullah, Ihsan, Andre Rios, Vaibhav Gala, and Susan Mckeever. 2021. Explaining deep learning models for tabular data using layer-wise relevance propagation. Applied Sciences 12(1), 136. [CrossRef]
- Wang, Wu, Junho Lee, Fouzi Harrou, and Ying Sun. 2020. Early detection of parkinson’s disease using deep learning and machine learning. IEEE Access 8, 147635–147646. [CrossRef]
- Weiss, Karl, Taghi M Khoshgoftaar, and DingDing Wang. 2016. A survey of transfer learning. Journal of Big data 3(1), 1–40.
- Wroge, Timothy J, Yasin Özkanca, Cenk Demiroglu, Dong Si, David C Atkins, and Reza Hosseini Ghomi. 2018. Parkinson’s disease diagnosis using machine learning and voice. In 2018 IEEE signal processing in medicine and biology symposium (SPMB), pp. 1–7. IEEE.
- Yadav, Samir S and Shivajirao M Jadhav. 2019. Deep convolutional neural network based medical image classification for disease diagnosis. Journal of Big data 6(1), 1–18. [CrossRef]
- Yang, Yuzhe, Yuan Yuan, Guo Zhang, Hao Wang, Ying-Cong Chen, Yingcheng Liu, Christopher G Tarolli, Daniel Crepeau, Jan Bukartyk, Mithri R Junna, et al. 2022. Artificial intelligence-enabled detection and assessment of parkinson’s disease using nocturnal breathing signals. Nature medicine 28(10), 2207–2215. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).