Introduction
SARS-CoV-2, known as the RNA virus responsible for the COVID-19 pandemic, has been identified to affect a number of cellular processes (Gioia et al., 2023), not just at the metabolic level but also extending to the genomic level. However, the virus’s impact on DNA integrity and related processes remains unclear and is a topic of debate in the scientific community.
A study elucidates that exposure to SARS-CoV-2 has long-term detrimental effects, marked by the progression towards organ damage and immunity impairment (Meagher, 2021). The most commonly reported impacted organs are the heart and blood vessels (Dennis et al., 2021), as well as the kidneys (Wang et al., 2022). This phenomenon is of serious concern, considering that organ damage invariably begins with cellular and tissue-level impairment. Furthermore, understanding the virus’s characteristic of "hijacking" host cells in its life cycle, it is highly plausible that damage also occurs at the genomic level. Reports have also shown an increase in cell membrane damage, indicated by elevated levels of malondialdehyde (MDA), a marker of cellular damage, in COVID-19 patients (Gonçalves et al., 2022).
Analyses of the impact of COVID-19 on DNA damage, genomic instability, and cell deregulation were reviewed by Publo and team in 2021, including supporting evidence related to these destructive hypotheses (Pánico et al., 2022). Another study clarified the role of DNA damage response (DDR)-dependent telomere dysfunction through the expression of angiotensin-converting enzyme 2 (ACE2) (Sepe et al., 2022). Unfortunately, a comprehensive mapping in this domain of study was not conducted, potentially overlooking significant topics. Science mapping could allow us to analyze and evaluate the development and evidence of research quickly. Therefore, this study aims to provide evidence-based proof through bibliometric mapping to analyze the relationship between the variables of DNA damage, genomic instability, and cell deregulation due to SARS-CoV-2 exposure in COVID-19.
Research Question
Is there a relationship between COVID-19 and induced DNA damage?
Is there a relationship between COVID-19 and Genomic Instability?
Is there a relationship between COVID-19 and Cell Cycle Deregulation during their replication in mammalian cells?
Method
This bibliometric study is designed to explore the impact of the virus causing COVID-19 at the genomic level. The initial phase involves the development of research questions aimed at understanding the correlation between the virus and DNA alterations. Based on the framework developed by Donthu (Donthu et al., 2021), we established a set of control vocabularies and relevant keywords, which were then utilized to filter and gather data from the available literature. This process was followed by stringent data selection and analysis, including the removal of outliers and distortions, as well as mathematical computations to gain deeper insights related to the research questions.
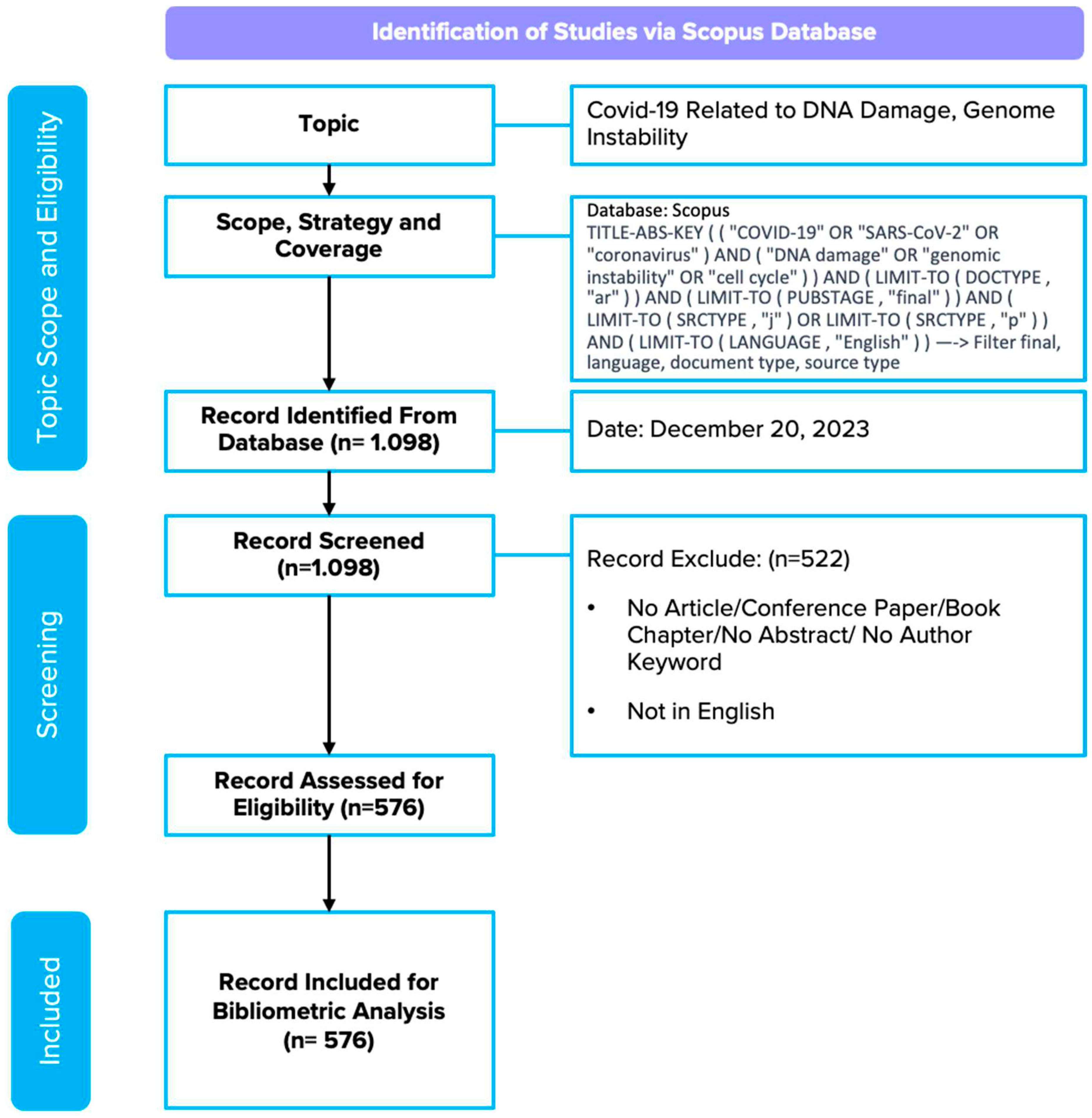
The literature search strategy involved two rounds using a combination of specific keywords in the Scopus database. The first round utilized the query "TITLE-ABS-KEY (('COVID-19' OR 'SARS-CoV-2' OR 'coronavirus') AND ('DNA damage' OR 'genomic instability' OR 'cell cycle'))", which yielded 1098 documents. The second round involved filtering to limit the results to research articles ('ar'), final publication stages ('final'), journal ('j') or proceedings ('p') source types, and documents in English, resulting in 576 relevant documents. This literature search was conducted on December 20, 2023, applying these filters to ensure the accuracy and relevance of the retrieved documents. This phase is summarized in a PRISMA flow diagram adapted from (Page et al., 2021).
Ronde 1: TITLE-ABS-KEY ( ( "COVID-19" OR "SARS-CoV-2" OR "coronavirus" ) AND ( "DNA damage" OR "genomic instability" OR "cell cycle" ) )
Ronde 2: TITLE-ABS-KEY ( ( "COVID-19" OR "SARS-CoV-2" OR "coronavirus" ) AND ( "DNA damage" OR "genomic instability" OR "cell cycle" ) ) AND ( LIMIT-TO ( DOCTYPE , "ar" ) ) AND ( LIMIT-TO ( PUBSTAGE , "final" ) ) AND ( LIMIT-TO ( SRCTYPE , "j" ) OR LIMIT-TO ( SRCTYPE , "p" ) ) AND ( LIMIT-TO ( LANGUAGE , "English" ) )
Following data collection, the next phase is data visualization using various bibliometric methods to map the relationship between COVID-19 and genomic changes. This visualization includes the creation of citation maps, co-citation analysis, and network analysis, all aiming to identify patterns, trends, and key relationships in the existing literature. The interpretation of these analytical results will provide critical insights into the impact of COVID-19 at the genomic level, as well as future directions for related research.
Figure 1.
Document Selection Stages Adapted from the PRISMA Flow Diagram (Page et al., 2021).
Figure 1.
Document Selection Stages Adapted from the PRISMA Flow Diagram (Page et al., 2021).
Result and Discussion
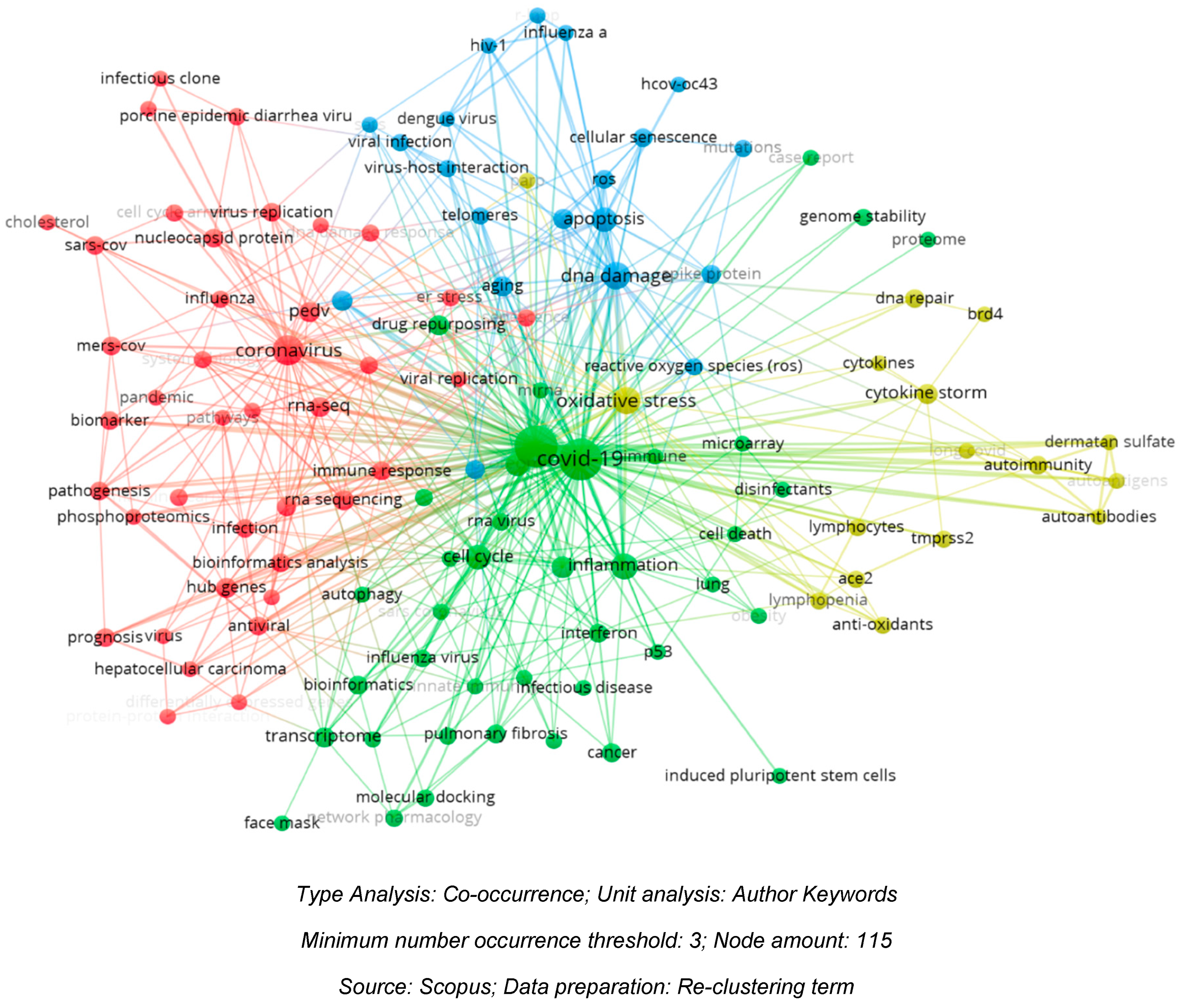
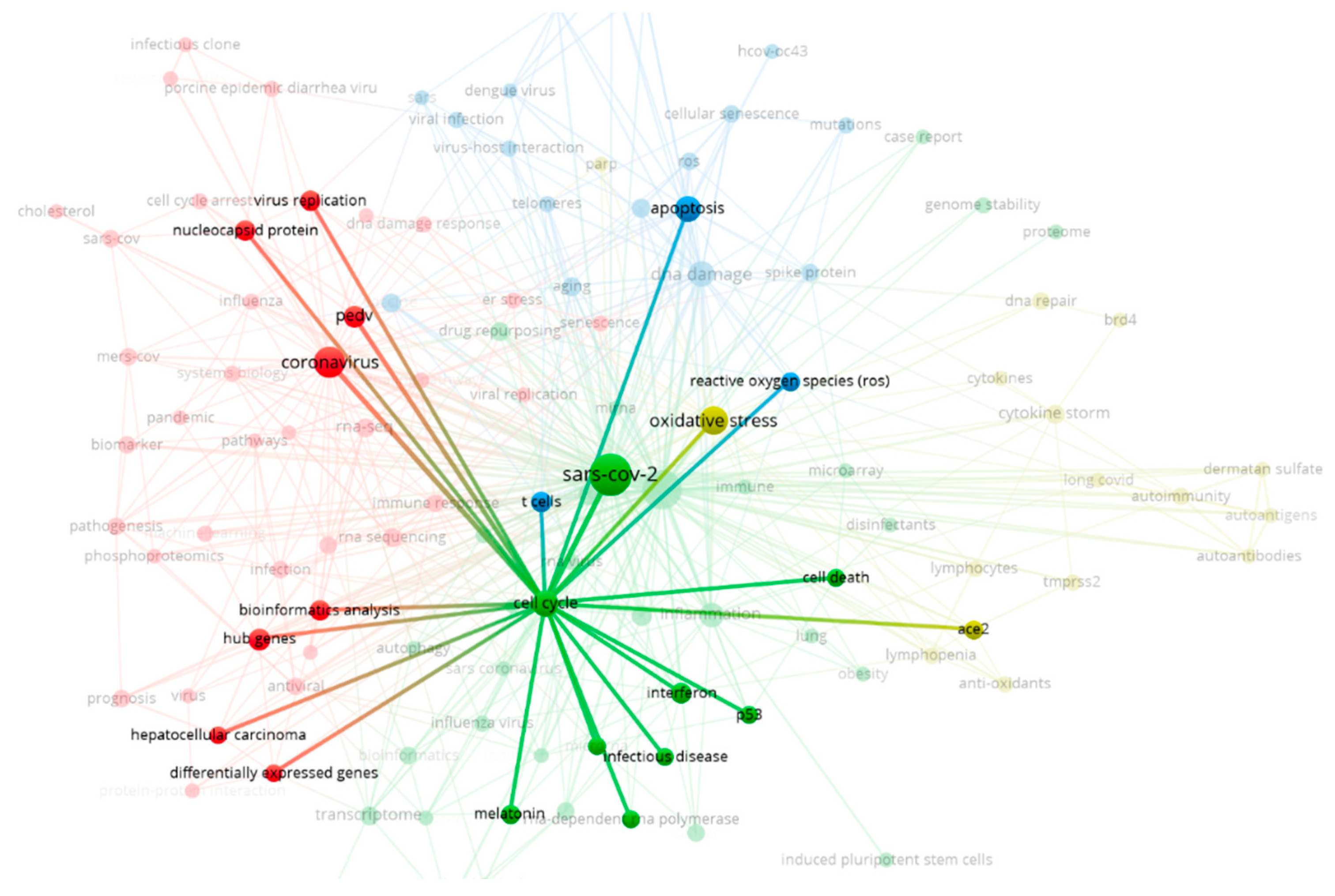
The mapping of the 576 articles included in the bibliometric analysis reveals four main clusters: coronavirus (red), COVID-19 (green), DNA damage (blue), and oxidative stress (yellow), as shown in
Figure 2 below. Each node represents a term, and nodes vary in size. The largest nodes, as seen in the figure below, indicate centrality, signifying a multitude of related topics. The distance between nodes also reflects the closeness of the studies. The closer the distance between nodes, the more they share in urgency. This can be observed in the proximity between apoptosis and DNA damage.
Figure 2.
Factors Involved in DNA Damage and Genomic Instability in COVID-19.
Figure 2.
Factors Involved in DNA Damage and Genomic Instability in COVID-19.
The illustration above displays the factors involved during the occurrence of an infection. The four clusters that have formed show interconnected factors. We can observe the proximity between the COVID-19 node and the nodes for DNA damage and oxidative stress, which are subsequently linked to apoptosis. This relationship provides an insight into the worst impact on the infected host cells, namely apoptosis.
3.1. Induce DNA damage
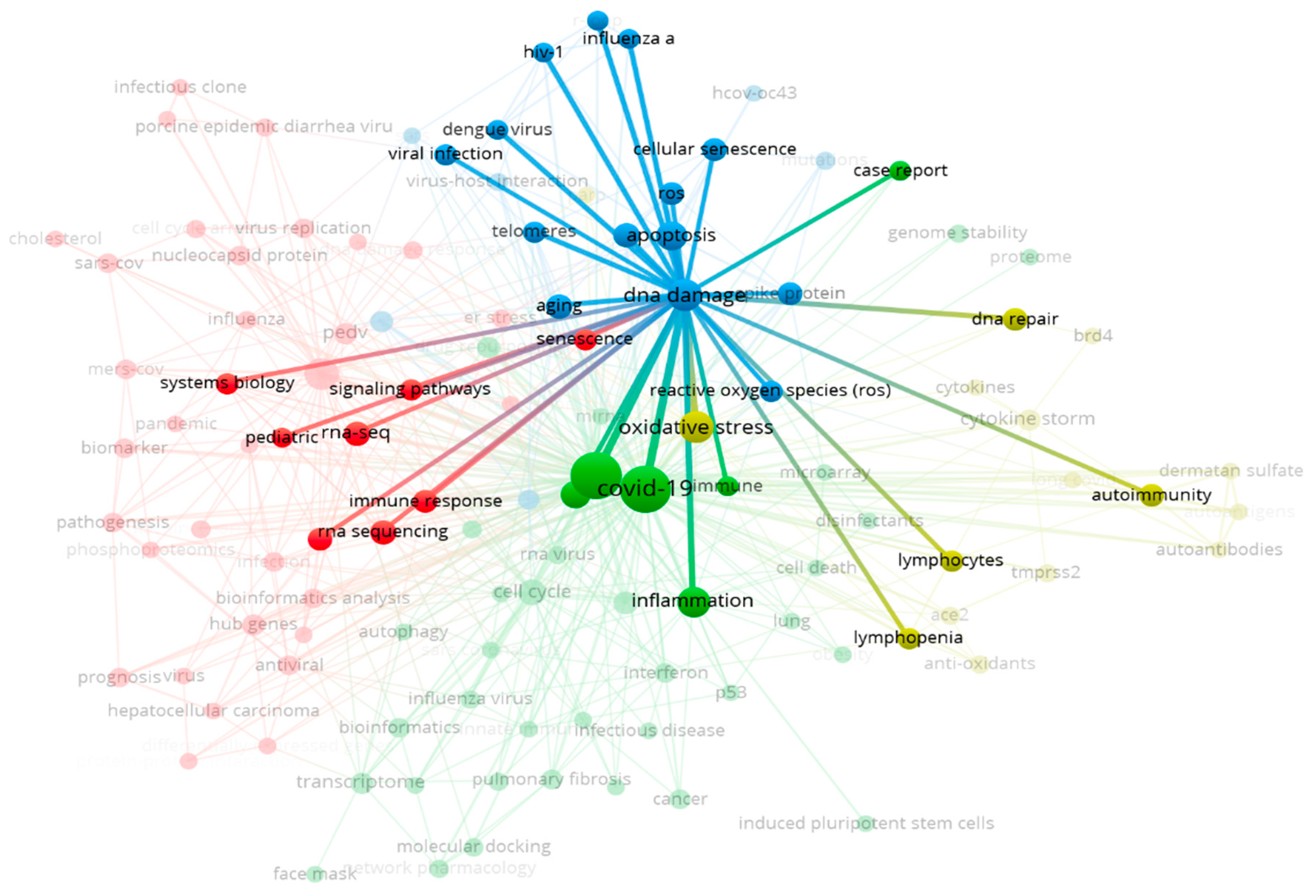
Errors in DNA synthesis, sequence reading, and DNA damage are common occurrences in biological systems. During the cell cycle, there are checkpoint control mechanisms in place to recognize and repair these errors (Rogan et al., 2021), akin to the balance between oxidants and antioxidants. This indicates that damage is tolerable within reasonable limits.
Unfortunately, the induction (intervention) of DNA damage beyond these thresholds leads to a state of imbalance. For instance, viral infection attacks by COVID-19 trigger the production of Reactive Oxygen Species (ROS) (Ming et al., 2022). This event can result in chromosomal damage (Rogan et al., 2021), cell apoptosis, and tissue damage. We analyzed this urgency through the study of scientific document evidence with term analysis. In the frequency analysis for the term “DNA damage,” 33 out of 575 documents (6%) were found. To reinforce this analysis, node distribution mapping in the form of science mapping was conducted. The proximity of the “DNA damage” and “apoptosis” nodes indicates a high level of urgency and association (
Figure 2). Furthermore, COVID-19 can lead to the depletion of host RNA binding proteins (RBPs) (Rogan et al., 2021).
Figure 3.
The Relationship with Factors Triggering DNA Damage by COVID-19.
Figure 3.
The Relationship with Factors Triggering DNA Damage by COVID-19.
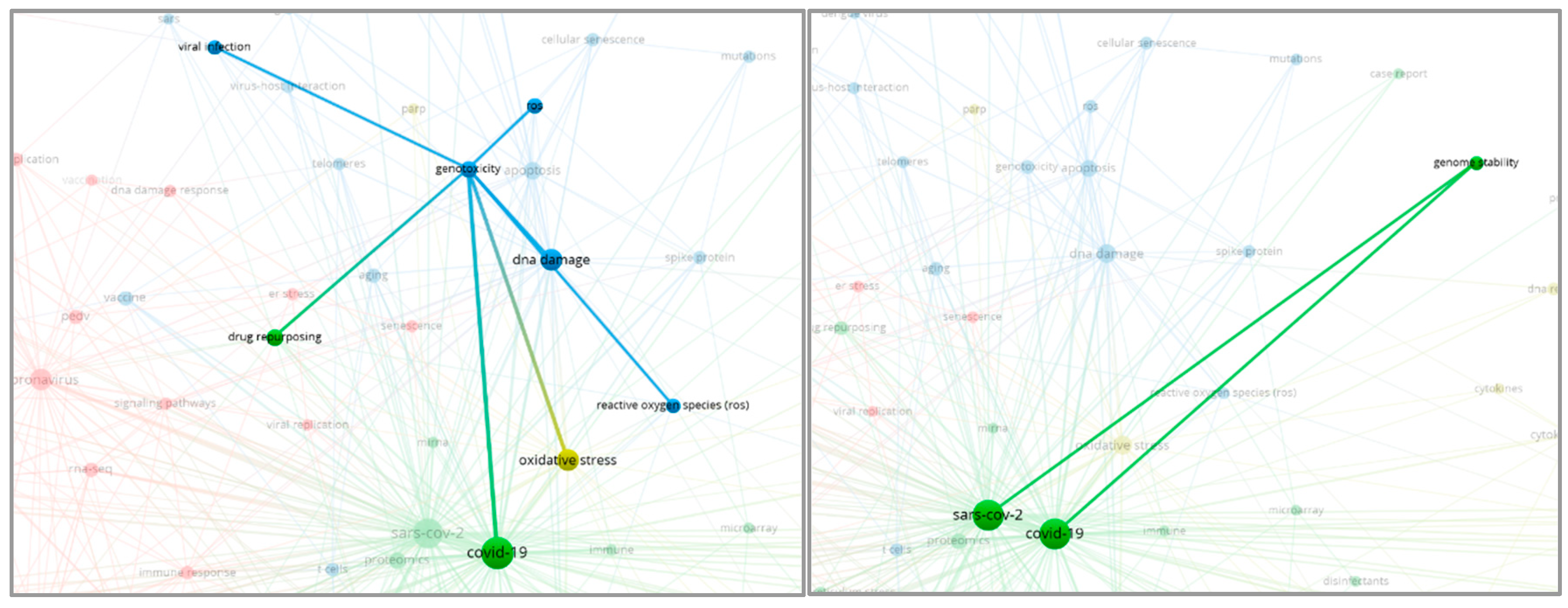
3.2. Genomic instability
It is widely recognized that COVID-19 infection poses numerous problems for the human genome, particularly in terms of genomic stability (Iourov & Vorsanova, 2022). One aspect of genomic instability is the issue of telomere regulation (telomere dysregulation), which can lead to cell death (Sepe et al., 2022).
Figure 4.
The Relationship with Factors Triggering Genomic Instability by COVID-19.
Figure 4.
The Relationship with Factors Triggering Genomic Instability by COVID-19.
3.3. Cell cycle deregulation during their replication in mammalian cells
Figure 5.
The Relationship with Factors Triggering Genomic Instability by COVID-19.
Figure 5.
The Relationship with Factors Triggering Genomic Instability by COVID-19.
The integrity of the cell membrane plays a crucial role. If the cell membrane is damaged, cells can undergo lysis and apoptosis. Research by Goncalves has demonstrated the dangers posed by COVID-19 protein fragments to the integrity of the cell membrane (Gonçalves et al., 2022). If this intervention is extensive, it can lead to DNA damage and cell death.
3.4. Technology Support for Identification
Identification at the genomic level is a complex task. There are a multitude of target genes that need to be identified, including the identification of genomic processes occurring with or without intervention. Marcus and his team (2007) discuss the DNA checkpoint. They emphasize the significance of understanding quantitative phospho-proteomics for identifying in vivo kinase substrates in DNA damage checkpoint kinases Mec1, Tel1, and Rad53 (orthologs of human ATR, ATM, and CHK2, respectively) (Smolka et al., 2007).
Similarly, with the impact of COVID-19 on DNA damage, there is a need for the design of biomarkers and the identification of appropriate target genes.
Conclusion
In the mapping of 576 scientific documents, four main clusters were identified: coronavirus, COVID-19, DNA damage, and oxidative stress. The analysis of the relationships and occurrences between these nodes reveals a close association between DNA damage and ROS, leading to oxidative stress, and potentially culminating in apoptosis. The bibliometric approach clearly demonstrates that COVID-19 is strongly linked to occurrences of DNA damage, genomic instability, and cell cycle deregulation.
References
- Dennis, A., Wamil, M., Alberts, J., Oben, J., Cuthbertson, D.J., Wootton, D., Crooks, M., Gabbay, M., Brady, M., Hishmeh, L., Attree, E., Heightman, M., Banerjee, R., & Banerjee, A. Multiorgan impairment in low-risk individuals with post-COVID-19 syndrome: A prospective, community-based study. BMJ Open 2021, 11, e048391. [CrossRef]
- Donthu, N., Kumar, S., Mukherjee, D., Pandey, N., & Lim, W.M. How to conduct a bibliometric analysis: An overview and guidelines. Journal of Business Research 2021, 133, 285–296. [CrossRef]
- Gioia, U., Tavella, S., Martínez-Orellana, P., Cicio, G., Colliva, A., Ceccon, M., Cabrini, M., Henriques, A.C., Fumagalli, V., Paldino, A., Presot, E., Rajasekharan, S., Iacomino, N., Pisati, F., Matti, V., Sepe, S., Conte, M.I., Barozzi, S., Lavagnino, Z., … d’Adda di Fagagna, F. SARS-CoV-2 infection induces DNA damage, through CHK1 degradation and impaired 53BP1 recruitment, and cellular senescence. Nature Cell Biology 2023, 25, Article 4. [CrossRef]
- Gonçalves, S. de O., Luz, T.M. da, Silva, A.M., de Souza, S.S., Montalvão, M.F., Guimarães, A.T.B., Ahmed, M.A.I., Araújo, A.P. da C., Karthi, S., & Malafaia, G. Can spike fragments of SARS-CoV-2 induce genomic instability and DNA damage in the guppy, Poecilia reticulate? An unexpected effect of the COVID-19 pandemic. Science of The Total Environment 2022, 825, 153988. [CrossRef]
- Iourov, I.Y., & Vorsanova, S.G. COVID-19 and Aging-Related Genome (Chromosome) Instability in the Brain: Another Possible Time-Bomb of SARS-CoV-2 Infection. Frontiers in Aging Neuroscience 2022, 14. Available online: https://www.frontiersin.org/articles/10.3389/fnagi.2022.786264.
- Meagher, T. Long COVID - An Early Perspective. Journal of Insurance Medicine 2021, 49, 19–23. [CrossRef]
- Ming, X., Chen, H., Yang, Y., Zhao, P., Sun, L., Zhang, C., Shin, H.-J., Lee, J.-S., Jung, Y.-S., & Qian, Y. Porcine Enteric Coronavirus PEDV Induces the ROS-ATM and Caspase7-CAD-γH2AX Signaling Pathways to Foster Its Replication. Viruses 2022, 14, 1782. [CrossRef]
- Page, M.J., McKenzie, J.E., Bossuyt, P.M., Boutron, I., Hoffmann, T.C., Mulrow, C.D., Shamseer, L., Tetzlaff, J.M., Akl, E.A., Brennan, S.E., Chou, R., Glanville, J., Grimshaw, J.M., Hróbjartsson, A., Lalu, M.M., Li, T., Loder, E.W., Mayo-Wilson, E., McDonald, S., … Moher, D. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [CrossRef]
- Pánico, P., Ostrosky-Wegman, P., & Salazar, A.M. The potential role of COVID-19 in the induction of DNA damage. Mutation Research/Reviews in Mutation Research 2022, 789, 108411. [CrossRef]
- Rogan, P.K., Mucaki, E.J., & Shirley, B.C. A proposed molecular mechanism for pathogenesis of severe RNA-viral pulmonary infections. F1000Research 2021, 9, 943. [CrossRef]
- Sepe, S., Rossiello, F., Cancila, V., Iannelli, F., Matti, V., Cicio, G., Cabrini, M., Marinelli, E., Alabi, B.R., di Lillo, A., Di Napoli, A., Shay, J.W., Tripodo, C., & d’Adda di Fagagna, F. DNA damage response at telomeres boosts the transcription of SARS-CoV-2 receptor ACE2 during aging. EMBO Reports 2022, 23, e53658. [CrossRef]
- Smolka, M.B., Albuquerque, C.P., Chen, S., & Zhou, H. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proceedings of the National Academy of Sciences 2007, 104, 10364–10369. [CrossRef]
- Wang, W., Chen, J., Hu, D., Pan, P., Liang, L., Wu, W., Tang, Y., Huang, X.R., Yu, X., Wu, J., & Lan, H.Y. SARS-CoV-2 N Protein Induces Acute Kidney Injury via Smad3-Dependent G1 Cell Cycle Arrest Mechanism. Advanced Science 2022, 9, 2103248. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).