Submitted:
29 December 2023
Posted:
03 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Methods
3. Comprehensive Overview
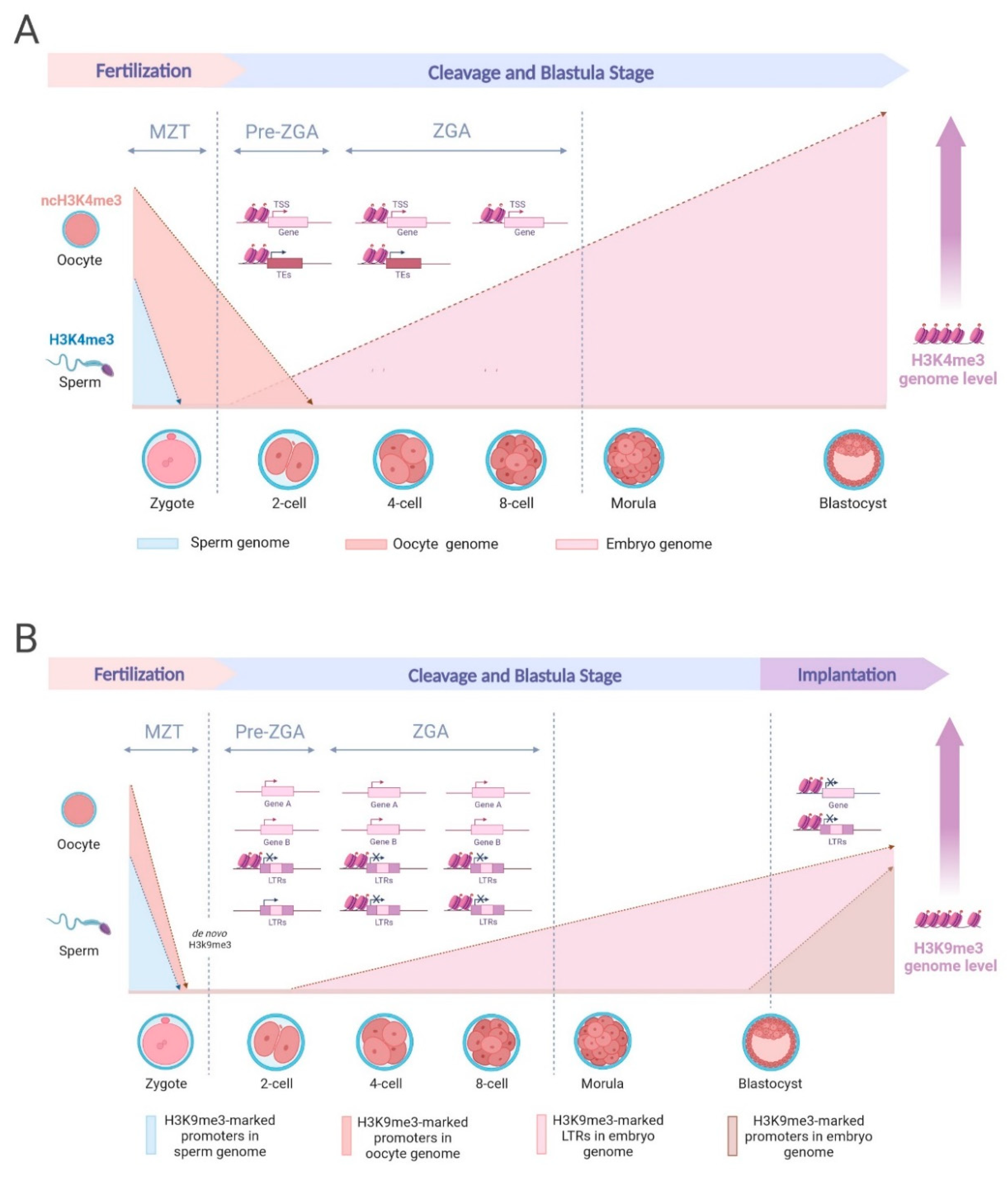
4. H3K4me3 in Zygotic Genome Activation and Gene Expression Regulation
4.1. H3K4me3 at Promoters and Across Species
4.2. Dynamic Reprogramming of H3K4me3
5. Unveiling H3K9me3: Orchestrating Epigenetic Landscapes in Development
5.1. Navigating Reprogramming Challenges and Zygotic Genome Activation
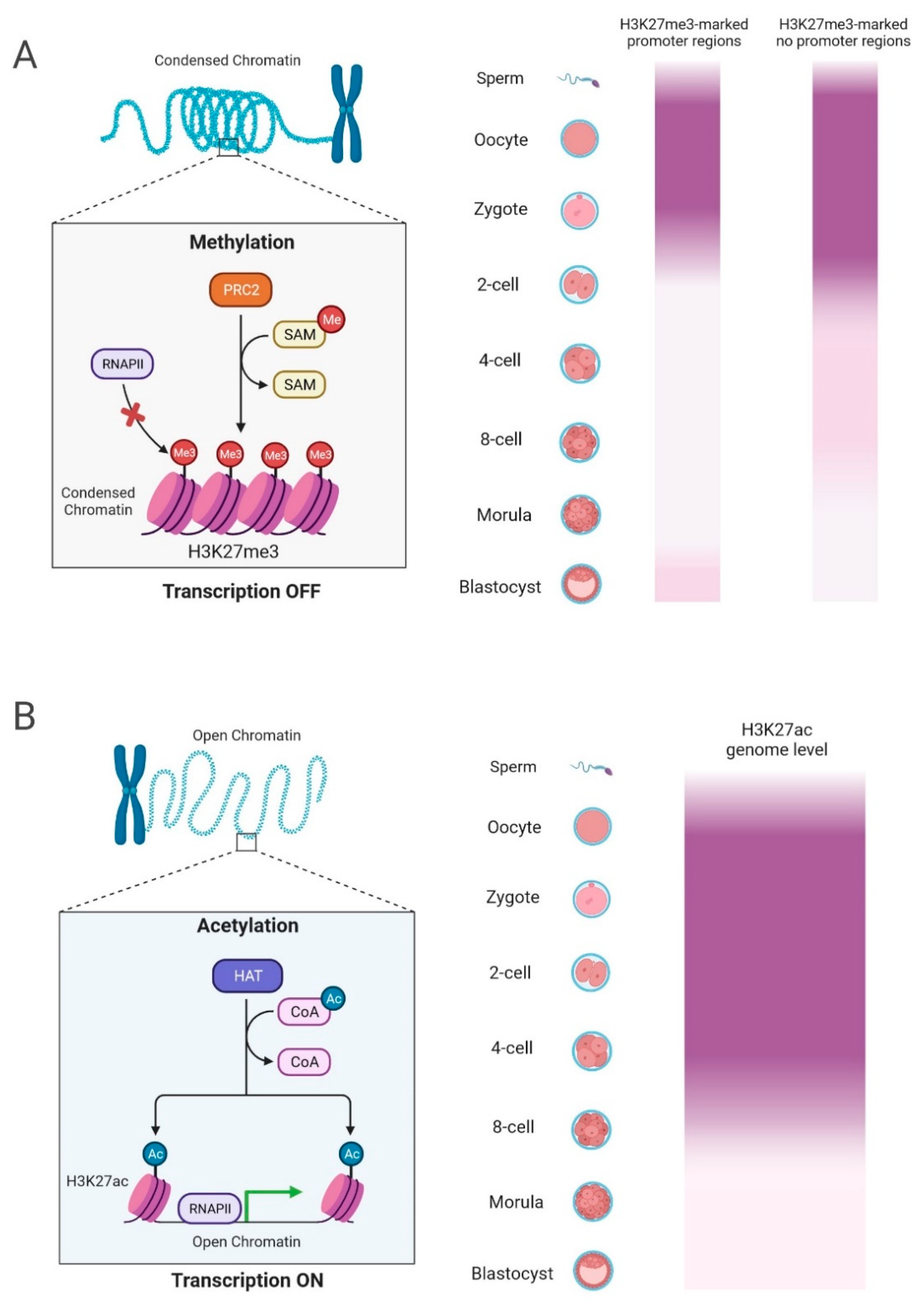
6. H3K27me3 and H3K27ac: Dual Epigenetic Players in Zygotic Genome Activation
6.1. Individual Roles of H3K27me3
6.2. Distinctive Functions of H3K27ac in ZGA
7. Dynamics of Other Histone Modifications in Early Embryonic Development
7.1. H3K36me3 Dynamics Unveiled: Allelic Reprogramming in Early Mouse Embryos
7.2. Histone H3R26me2: Pivotal in Cell Fate Determination in Embryos
8. Functional Diversity of Histone Variants in the Activation of the Zygotic Genome
9. Future Perspectives and Applications of ZGA Histone Modifications in Stem Cell Research
10. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AML | Acute myeloid leukemia |
| BRD2 | Bromodomain-containing protein 2 |
| CARM1 | Coactivator-associated arginine methyltransferase 1 |
| CHAF1A | Chromatin assembly factor 1 subunit A |
| ESCs | Embryonic stem cells |
| ESET | ERG-associated protein with SET domain |
| EZH2 | Enhancer of zeste homolog 2 |
| HATs | Histone acetyltransferases |
| hESCs | Human embryonic stem cells |
| ICM | Inner cell mass |
| ICRs | Imprinted control regions |
| iPSCs | Induced pluripotent stem cells |
| KDM4D | Lysine-specific demethylase 4A |
| KDM6A | Lysine-specific demethylase 6A |
| KDM6B | Lysine-specific demethylase 6B |
| KMT2 | Histone–lysine N-methyltransferase 2 |
| lncRNA | Non-coding RNA |
| LTRs | Long terminal repeats |
| mESCs | Mouse embryonic stem cells |
| MSCs | Mesenchymal stem cells |
| MZT | Maternal-to-zygotic transition |
| NEAT1 | Nuclear Paraspeckle Assembly Transcript 1 |
| NHD | Non-histone domain |
| ntESC | Nuclear-transfer embryonic stem cells |
| NURF | Nucleosome remodeling factor |
| P54NRB | Nuclear RNA-binding protein 54 kDa |
| PMDs | Partially methylated domains |
| PN | Pronuclei |
| PRC2 | Polycomb Repressive Complex 2 |
| PRDM14 | PR domain-containing 14 |
| RRRs | Regions resistant to reprogramming |
| SCNT | Somatic Cell Nuclear Transfer |
| SETD2 | SET domain containing 2 |
| SNPs | Single-nucleotide polymorphisms |
| TE | Trophectoderm |
| TSS | Transcription start site |
| ZGA | Zygotic genome activation |
References
- Siu, K. K.; Serrão, V. H. B.; Ziyyat, A.; Lee, J. E. The Cell Biology of Fertilization: Gamete Attachment and Fusion. Journal of Cell Biology 2021. [CrossRef]
- Bhakta, H. H.; Refai, F. H.; Avella, M. A. The Molecular Mechanisms Mediating Mammalian Fertilization. Development (Cambridge) 2019. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Xie, W. Epigenome in Early Mammalian Development: Inheritance, Reprogramming and Establishment. Trends in Cell Biology 2018. [CrossRef]
- Schulz, K. N.; Harrison, M. M. Mechanisms Regulating Zygotic Genome Activation. Nature Reviews Genetics 2019. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, L.; Guo, F.; Dai, X.; Zhang, X. Epigenetic Reprogramming during the Maternal-to-Zygotic Transition. MedComm 2023. [Google Scholar] [CrossRef]
- Vastenhouw, N. L.; Cao, W. X.; Lipshitz, H. D. The Maternal-to-Zygotic Transition Revisited. Development (Cambridge, England) 2019. [Google Scholar] [CrossRef] [PubMed]
- Lee, M. T.; Bonneau, A. R.; Giraldez, A. J. Zygotic Genome Activation during the Maternal-to-Zygotic Transition. Annual review of cell and developmental biology 2014. [CrossRef]
- Wu, E.; Vastenhouw, N. L. From Mother to Embryo: A Molecular Perspective on Zygotic Genome Activation. In Current Topics in Developmental Biology; 2020; Vol. 140. [CrossRef]
- Vallot, A.; Tachibana, K. The Emergence of Genome Architecture and Zygotic Genome Activation. Current Opinion in Cell Biology 2020. [CrossRef]
- Hackett, J. A.; Azim Surani, M. Regulatory Principles of Pluripotency: From the Ground State Up. Cell Stem Cell 2014. [Google Scholar] [CrossRef]
- Zhou, S.; Li, X.; Liu, Q.; Zhao, Y.; Jiang, W.; Wu, A.; Zhou, D. X. DNA Demethylases Remodel DNA Methylation in Rice Gametes and Zygote and Are Required for Reproduction. Mol Plant 2021, 14. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, J.; Duan, J.; Gao, X.; Zhu, W.; Lu, X.; Yang, L.; Zhang, J.; Li, G.; Ci, W.; et al. Programming and Inheritance of Parental DNA Methylomes in Mammals. Cell 2014, 157. [Google Scholar] [CrossRef]
- Guo, H.; Zhu, P.; Yan, L.; Li, R.; Hu, B.; Lian, Y.; Yan, J.; Ren, X.; Lin, S.; Li, J.; et al. The DNA Methylation Landscape of Human Early Embryos. Nature 2014, 511. [Google Scholar] [CrossRef]
- Robert, V. J. Histone Modifications in Germline Development and Maintenance. In Perinatal and Developmental Epigenetics: Volume 32 in Translational Epigenetics; 2022. [CrossRef]
- Hales, B. F.; Grenier, L.; Lalancette, C.; Robaire, B. Epigenetic Programming: From Gametes to Blastocyst. Birth Defects Research Part A - Clinical and Molecular Teratology, 2011. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, Q.; Tang, F.; Yan, L.; Qiao, J. Epigenetic Regulation and Risk Factors during the Development of Human Gametes and Early Embryos. Annual Review of Genomics and Human Genetics 2019. [Google Scholar] [CrossRef]
- Wu, J.; Xu, J.; Liu, B.; Yao, G.; Wang, P.; Lin, Z.; Huang, B.; Wang, X.; Li, T.; Shi, S.; et al. Chromatin Analysis in Human Early Development Reveals Epigenetic Transition during ZGA. Nature 2018, 557. [Google Scholar] [CrossRef]
- Gorkin, D. U.; Barozzi, I.; Zhao, Y.; Zhang, Y.; Huang, H.; Lee, A. Y.; Li, B.; Chiou, J.; Wildberg, A.; Ding, B.; et al. An Atlas of Dynamic Chromatin Landscapes in Mouse Fetal Development. Nature 2020, 583. [Google Scholar] [CrossRef]
- Gao, L.; Wu, K.; Liu, Z.; Yao, X.; Yuan, S.; Tao, W.; Yi, L.; Yu, G.; Hou, Z.; Fan, D.; et al. Chromatin Accessibility Landscape in Human Early Embryos and Its Association with Evolution. Cell 2018, 173. [Google Scholar] [CrossRef] [PubMed]
- Bonev, B.; Cavalli, G. Organization and Function of the 3D Genome. Nature Reviews Genetics 2016. [Google Scholar] [CrossRef] [PubMed]
- Ke, Y.; Xu, Y.; Chen, X.; Feng, S.; Liu, Z.; Sun, Y.; Yao, X.; Li, F.; Zhu, W.; Gao, L.; et al. 3D Chromatin Structures of Mature Gametes and Structural Reprogramming during Mammalian Embryogenesis. Cell 2017, 170. [Google Scholar] [CrossRef] [PubMed]
- Koyama, M.; Kurumizaka, H. Structural Diversity of the Nucleosome. Journal of Biochemistry 2018. [CrossRef] [PubMed]
- Zhou, K.; Gaullier, G.; Luger, K. Nucleosome Structure and Dynamics Are Coming of Age. Nature Structural and Molecular Biology 2019. [CrossRef] [PubMed]
- Deng, M.; Chen, B.; Liu, Z.; Cai, Y.; Wan, Y.; Zhou, J.; Wang, F. Exchanges of Histone Methylation and Variants during Mouse Zygotic Genome Activation. Zygote 2020, 28. [Google Scholar] [CrossRef] [PubMed]
- Bu, G.; Zhu, W.; Liu, X.; Zhang, J.; Yu, L.; Zhou, K.; Wang, S.; Li, Z.; Fan, Z.; Wang, T.; et al. Coordination of Zygotic Genome Activation Entry and Exit by H3K4me3 and H3K27me3 in Porcine Early Embryos. Genome Res 2022, 32. [Google Scholar] [CrossRef] [PubMed]
- Shao, G. B.; Ding, H. M.; Gong, A. H. Role of Histone Methylation in Zygotic Genome Activation in the Preimplantation Mouse Embryo. In Vitro Cell Dev Biol Anim 2008, 44. [Google Scholar] [CrossRef] [PubMed]
- Darbo, E.; Herrmann, C.; Lecuit, T.; Thieffry, D.; van Helden, J. Transcriptional and Epigenetic Signatures of Zygotic Genome Activation during Early Drosophila Embryogenesis. BMC Genomics 2013, 14. [Google Scholar] [CrossRef] [PubMed]
- Pálfy, M.; Joseph, S. R.; Vastenhouw, N. L. The Timing of Zygotic Genome Activation. Current Opinion in Genetics and Development 2017. [Google Scholar] [CrossRef] [PubMed]
- Robertson, S.; Lin, R. The Maternal-to-Zygotic Transition in C. Elegans. In Current Topics in Developmental Biology; 2015; Vol. 113. [CrossRef]
- Blitz, I. L.; Cho, K. W. Y. Control of Zygotic Genome Activation in Xenopus. In Current Topics in Developmental Biology; 2021; Vol. 145. [CrossRef]
- Laue, K.; Rajshekar, S.; Courtney, A. J.; Lewis, Z. A.; Goll, M. G. The Maternal to Zygotic Transition Regulates Genome-Wide Heterochromatin Establishment in the Zebrafish Embryo. Nat Commun 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Jukam, D.; Shariati, S. A. M.; Skotheim, J. M. Zygotic Genome Activation in Vertebrates. Developmental Cell 2017. [Google Scholar] [CrossRef]
- Colonnetta, M. M.; Schedl, P.; Deshpande, G. Germline/Soma Distinction in Drosophila Embryos Requires Regulators of Zygotic Genome Activation. Elife 2023, 12. [Google Scholar] [CrossRef]
- Hamm, D. C.; Harrison, M. M. Regulatory Principles Governing the Maternal-to-Zygotic Transition: Insights from Drosophila Melanogaster. Open Biology 2018. [CrossRef]
- Li, L.; Lu, X.; Dean, J. The Maternal to Zygotic Transition in Mammals. Molecular Aspects of Medicine 2013. [Google Scholar] [CrossRef]
- Aoki, F. Zygotic Gene Activation in Mice: Profile and Regulation. Journal of Reproduction and Development 2022, 68. [Google Scholar] [CrossRef]
- Yuan, S.; Zhan, J.; Zhang, J.; Liu, Z.; Hou, Z.; Zhang, C.; Yi, L.; Gao, L.; Zhao, H.; Chen, Z. J.; et al. Human Zygotic Genome Activation Is Initiated from Paternal Genome. Cell Discov 2023, 9. [Google Scholar] [CrossRef] [PubMed]
- Tesarik, J. Control of Maternal-to-Zygotic Transition in Human Embryos and Other Animal Species (Especially Mouse): Similarities and Differences. International Journal of Molecular Sciences 2022. [CrossRef] [PubMed]
- Yuan, K.; Seller, C. A.; Shermoen, A. W.; O’Farrell, P. H. Timing the Drosophila Mid-Blastula Transition: A Cell Cycle-Centered View. Trends in Genetics 2016. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Fan, D.; Zhao, H.; Liu, Z.; Hou, Z.; Tao, W.; Yu, G.; Yuan, S.; Zhu, X.; Kang, M.; et al. Dynamics of Histone Acetylation during Human Early Embryogenesis. Cell Discov 2023, 9. [Google Scholar] [CrossRef] [PubMed]
- Rossetto, D.; Avvakumov, N.; Côté, J. Histone Phosphorylation. Epigenetics 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Talamillo, A.; Barroso-Gomila, O.; Giordano, I.; Ajuria, L.; Grillo, M.; Mayor, U.; Barrio, R. The Role of SUMOylation during Development. Biochemical Society Transactions 2020. [CrossRef] [PubMed]
- Mattiroli, F.; Penengo, L. Histone Ubiquitination: An Integrative Signaling Platform in Genome Stability. Trends in Genetics 2021. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, Q.; Ding, J.; Yin, T.; Ye, P.; Zhang, Y. The Conceivable Functions of Protein Ubiquitination and Deubiquitination in Reproduction. Frontiers in Physiology 2022. [Google Scholar] [CrossRef]
- Millán-Zambrano, G.; Burton, A.; Bannister, A. J.; Schneider, R. Histone Post-Translational Modifications — Cause and Consequence of Genome Function. Nature Reviews Genetics 2022. [Google Scholar] [CrossRef]
- Liu, R.; Wu, J.; Guo, H.; Yao, W.; Li, S.; Lu, Y.; Jia, Y.; Liang, X.; Tang, J.; Zhang, H. Post-translational Modifications of Histones: Mechanisms, Biological Functions, and Therapeutic Targets. MedComm (Beijing) 2023, 4. [Google Scholar] [CrossRef]
- Smith, B. C.; Denu, J. M. Chemical Mechanisms of Histone Lysine and Arginine Modifications. Biochimica et Biophysica Acta - Gene Regulatory Mechanisms 2009. [Google Scholar] [CrossRef] [PubMed]
- Jambhekar, A.; Dhall, A.; Shi, Y. Roles and Regulation of Histone Methylation in Animal Development. Nature Reviews Molecular Cell Biology 2019. [CrossRef] [PubMed]
- Eberharter, A.; Becker, P. B. Histone Acetylation: A Switch between Repressive and Permissive Chromatin. EMBO Rep 2002, 3. [Google Scholar] [CrossRef]
- Wang, H.; Fan, Z.; Shliaha, P. V.; Miele, M.; Hendrickson, R. C.; Jiang, X.; Helin, K. H3K4me3 Regulates RNA Polymerase II Promoter-Proximal Pause-Release. Nature 2023, 615. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Kim, G. W.; Kwon, S. H.; Lee, J. S. Broad Domains of Histone H3 Lysine 4 Trimethylation in Transcriptional Regulation and Disease. FEBS Journal 2020. [Google Scholar] [CrossRef]
- Wysocka, J.; Swigut, T.; Xiao, H.; Milne, T. A.; Kwon, S. Y.; Landry, J.; Kauer, M.; Tackett, A. J.; Chait, B. T.; Badenhorst, P.; et al. A PHD Finger of NURF Couples Histone H3 Lysine 4 Trimethylation with Chromatin Remodelling. Nature 2006, 442. [Google Scholar] [CrossRef]
- Liu, X.; Wang, C.; Liu, W.; Li, J.; Li, C.; Kou, X.; Chen, J.; Zhao, Y.; Gao, H.; Wang, H.; et al. Distinct Features of H3K4me3 and H3K27me3 Chromatin Domains in Pre-Implantation Embryos. Nature 2016, 537. [Google Scholar] [CrossRef]
- Beacon, T. H.; Delcuve, G. P.; López, C.; Nardocci, G.; Kovalchuk, I.; van Wijnen, A. J.; Davie, J. R. The Dynamic Broad Epigenetic (H3K4me3, H3K27ac) Domain as a Mark of Essential Genes. Clinical Epigenetics 2021. [Google Scholar] [CrossRef]
- Lindeman, L. C.; Andersen, I. S.; Reiner, A. H.; Li, N.; Aanes, H.; Østrup, O.; Winata, C.; Mathavan, S.; Müller, F.; Aleström, P.; et al. Prepatterning of Developmental Gene Expression by Modified Histones before Zygotic Genome Activation. Dev Cell 2011, 21. [Google Scholar] [CrossRef]
- Van Heeringen, S. J.; Akhtar, W.; Jacobi, U. G.; Akkers, R. C.; Suzuki, Y.; Veenstra, G. J. C. Nucleotide Composition-Linked Divergence of Vertebrate Core Promoter Architecture. Genome Res 2011, 21. [Google Scholar] [CrossRef] [PubMed]
- Ardehali, M. B.; Mei, A.; Zobeck, K. L.; Caron, M.; Lis, J. T.; Kusch, T. Drosophila Set1 Is the Major Histone H3 Lysine 4 Trimethyltransferase with Role in Transcription. EMBO Journal 2011, 30. [Google Scholar] [CrossRef] [PubMed]
- Dahl, J. A.; Jung, I.; Aanes, H.; Greggains, G. D.; Manaf, A.; Lerdrup, M.; Li, G.; Kuan, S.; Li, B.; Lee, A. Y.; et al. Broad Histone H3K4me3 Domains in Mouse Oocytes Modulate Maternal-to-Zygotic Transition. Nature 2016, 537. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Xu, B.; Sun, Y.; Lu, X.; Gu, R.; Wu, L.; Feng, Y.; Xu, C. Dynamic Changes of Histone H3 Trimethylated at Positions K4 and K27 in Human Oocytes and Preimplantation Embryos. Fertil Steril 2012, 98. [Google Scholar] [CrossRef] [PubMed]
- Sendžikaitė, G.; Kelsey, G. The Role and Mechanisms of DNA Methylation in the Oocyte. Essays in Biochemistry 2019. [CrossRef]
- Huang, X.; Gao, X.; Li, W.; Jiang, S.; Li, R.; Hong, H.; Zhao, C.; Zhou, P.; Chen, H.; Bo, X.; et al. Stable H3K4me3 Is Associated with Transcription Initiation during Early Embryo Development. Bioinformatics 2019, 35. [Google Scholar] [CrossRef]
- Brind’Amour, J.; Lorincz, M. C. Profiling Histone Methylation in Low Numbers of Cells. In Methods in Molecular Biology; 2022; Vol 2529. [CrossRef]
- Zhang, B.; Zheng, H.; Huang, B.; Li, W.; Xiang, Y.; Peng, X.; Ming, J.; Wu, X.; Zhang, Y.; Xu, Q.; et al. Allelic Reprogramming of the Histone Modification H3K4me3 in Early Mammalian Development. Nature 2016, 537. [Google Scholar] [CrossRef]
- Ishihara, T.; Griffith, O. W.; Suzuki, S.; Renfree, M. B. Presence of H3K4me3 on Paternally Expressed Genes of the Paternal Genome From Sperm to Implantation. Front Cell Dev Biol 2022, 10. [Google Scholar] [CrossRef]
- Xu, R.; Li, C.; Liu, X.; Gao, S. Insights into Epigenetic Patterns in Mammalian Early Embryos. Protein and Cell 2021. [Google Scholar] [CrossRef]
- Albert, T. K.; Kerl, K. A Histone Tale That EnCOMPASSes Pausing: New Insights into the Functional Repertoire of H3K4me3. Signal Transduction and Targeted Therapy 2023. [Google Scholar] [CrossRef]
- Sha, Q. Q.; Zhang, J.; Fan, H. Y. Function and Regulation of Histone H3 Lysine-4 Methylation During Oocyte Meiosis and Maternal-to-Zygotic Transition. Frontiers in Cell and Developmental Biology 2020. [CrossRef] [PubMed]
- Yoo, J.; Kim, G. W.; Jeon, Y. H.; Kim, J. Y.; Lee, S. W.; Kwon, S. H. Drawing a Line between Histone Demethylase KDM5A and KDM5B: Their Roles in Development and Tumorigenesis. Experimental and Molecular Medicine 2022. [Google Scholar] [CrossRef] [PubMed]
- Xhabija, B.; Kidder, B. L. KDM5B Is a Master Regulator of the H3K4-Methylome in Stem Cells, Development and Cancer. Seminars in Cancer Biology 2019. [CrossRef]
- Wang, Z.; Zhong, C.; Li, H. Histone Demethylase KDM5B Catalyzed H3K4me3 Demethylation to Promote Differentiation of Bone Marrow Mesenchymal Stem Cells into Cardiomyocytes. Mol Biol Rep 2022, 49. [Google Scholar] [CrossRef] [PubMed]
- Kidder, B. L.; Hu, G.; Yu, Z.-X.; Liu, C.; Zhao, K. Extended Self-Renewal and Accelerated Reprogramming in the Absence of Kdm5b. Mol Cell Biol 2013, 33. [Google Scholar] [CrossRef]
- Peaston, A. E.; Evsikov, A. V.; Graber, J. H.; de Vries, W. N.; Holbrook, A. E.; Solter, D.; Knowles, B. B. Retrotransposons Regulate Host Genes in Mouse Oocytes and Preimplantation Embryos. Dev Cell 2004, 7. [Google Scholar] [CrossRef]
- Xia, W.; Xu, J.; Yu, G.; Yao, G.; Xu, K.; Ma, X.; Zhang, N.; Liu, B.; Li, T.; Lin, Z.; et al. Resetting Histone Modifications during Human Parental-to-Zygotic Transition. Science (1979) 2019, 365. [Google Scholar] [CrossRef]
- Reshetnikov, V. V.; Kisaretova, P. E.; Ershov, N. I.; Merkulova, T. I.; Bondar, N. P. Data of Correlation Analysis between the Density of H3K4me3 in Promoters of Genes and Gene Expression: Data from RNA-Seq and ChIP-Seq Analyses of the Murine Prefrontal Cortex. Data Brief 2020, 33. [Google Scholar] [CrossRef]
- Sun, H.; Wang, Y.; Wang, Y.; Ji, F.; Wang, A.; Yang, M.; He, X.; Li, L. Bivalent Regulation and Related Mechanisms of H3K4/27/9me3 in Stem Cells. Stem Cell Reviews and Reports 2022. [Google Scholar] [CrossRef]
- Zhang, J.; Li, X.; Cui, H.; Xiao, S.; Song, E.; Zong, M.; Ling, S.; Rosenwaks, Z.; Gao, S.; Liu, X.; et al. Maternal H3.3-Mediated Paternal Genome Reprogramming Contributes to Minor Zygotic Genome Activation. bioRxiv 2023, 2023.11.07.566007. [Google Scholar] [CrossRef]
- Park, K.; Kim, J. A.; Kim, J. Transcriptional Regulation by the KMT2 Histone H3K4 Methyltransferases. Biochimica et Biophysica Acta - Gene Regulatory Mechanisms 2020. [Google Scholar] [CrossRef] [PubMed]
- Nicetto, D.; Zaret, K. S. Role of H3K9me3 Heterochromatin in Cell Identity Establishment and Maintenance. Current Opinion in Genetics and Development 2019. [Google Scholar] [CrossRef] [PubMed]
- Ninova, M.; Tóth, K. F.; Aravin, A. A. The Control of Gene Expression and Cell Identity by H3K9 Trimethylation. Development (Cambridge) 2019. [Google Scholar] [CrossRef] [PubMed]
- Nicetto, D.; Donahue, G.; Jain, T.; Peng, T.; Sidoli, S.; Sheng, L.; Montavon, T.; Becker, J. S.; Grindheim, J. M.; Blahnik, K.; et al. H3K9me3-Heterochromatin Loss at Protein-Coding Genes Enables Developmental Lineage Specification. Science (1979) 2019, 363. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Zhu, Q.; Zhao, Y.; Chen, M.; Yang, L.; Shen, S.; Yang, G.; Shi, Z.; Zhang, X.; Shi, Q.; et al. Unreprogrammed H3K9me3 Prevents Minor Zygotic Genome Activation and Lineage Commitment in SCNT Embryos. Nat Commun 2023, 14. [Google Scholar] [CrossRef]
- Matoba, S.; Liu, Y.; Lu, F.; Iwabuchi, K. A.; Shen, L.; Inoue, A.; Zhang, Y. Embryonic Development Following Somatic Cell Nuclear Transfer Impeded by Persisting Histone Methylation. Cell 2014, 159. [Google Scholar] [CrossRef]
- Matoba, S.; Zhang, Y. Somatic Cell Nuclear Transfer Reprogramming: Mechanisms and Applications. Cell Stem Cell 2018. [Google Scholar] [CrossRef]
- Srirattana, K.; Kaneda, M.; Parnpai, R. Strategies to Improve the Efficiency of Somatic Cell Nuclear Transfer. International Journal of Molecular Sciences 2022. [CrossRef]
- Moura, M. T. Cloning by SCNT: Integrating Technical and Biology-Driven Advances. In Methods in Molecular Biology; 2023; Vol 2647. [CrossRef]
- Wang, C.; Liu, X.; Gao, Y.; Yang, L.; Li, C.; Liu, W.; Chen, C.; Kou, X.; Zhao, Y.; Chen, J.; et al. Reprogramming of H3K9me3-Dependent Heterochromatin during Mammalian Embryo Development. Nat Cell Biol 2018, 20. [Google Scholar] [CrossRef]
- Sampaio, R. V.; Sangalli, J. R.; De Bem, T. H. C.; Ambrizi, D. R.; del Collado, M.; Bridi, A.; de Ávila, A. C. F. C. M.; Macabelli, C. H.; de Jesus Oliveira, L.; da Silveira, J. C.; et al. Catalytic Inhibition of H3K9me2 Writers Disturbs Epigenetic Marks during Bovine Nuclear Reprogramming. Sci Rep 2020, 10. [Google Scholar] [CrossRef]
- Sankar, A.; Lerdrup, M.; Manaf, A.; Johansen, J. V.; Gonzalez, J. M.; Borup, R.; Blanshard, R.; Klungland, A.; Hansen, K.; Andersen, C. Y.; et al. KDM4A Regulates the Maternal-to-Zygotic Transition by Protecting Broad H3K4me3 Domains from H3K9me3 Invasion in Oocytes. Nat Cell Biol 2020, 22. [Google Scholar] [CrossRef]
- Becker, J. S.; Nicetto, D.; Zaret, K. S. H3K9me3-Dependent Heterochromatin: Barrier to Cell Fate Changes. Trends in Genetics 2016. [Google Scholar] [CrossRef]
- Wu, D. Y.; Li, X.; Sun, Q. R.; Dou, C. L.; Xu, T.; He, H.; Luo, H.; Fu, H.; Bu, G. W.; Luo, B.; et al. Defective Chromatin Architectures in Embryonic Stem Cells Derived from Somatic Cell Nuclear Transfer Impair Their Differentiation Potentials. Cell Death Dis 2021, 12. [Google Scholar] [CrossRef]
- Chung, Y. G.; Matoba, S.; Liu, Y.; Eum, J. H.; Lu, F.; Jiang, W.; Lee, J. E.; Sepilian, V.; Cha, K. Y.; Lee, D. R.; et al. Histone Demethylase Expression Enhances Human Somatic Cell Nuclear Transfer Efficiency and Promotes Derivation of Pluripotent Stem Cells. Cell Stem Cell 2015, 17. [Google Scholar] [CrossRef]
- Geis, F. K.; Goff, S. P. Silencing and Transcriptional Regulation of Endogenous Retroviruses: An Overview. Viruses 2020. [CrossRef]
- Bulut-Karslioglu, A.; DeLaRosa-Velázquez, I. A.; Ramirez, F.; Barenboim, M.; Onishi-Seebacher, M.; Arand, J.; Galán, C.; Winter, G. E.; Engist, B.; Gerle, B.; et al. Suv39h-Dependent H3K9me3 Marks Intact Retrotransposons and Silences LINE Elements in Mouse Embryonic Stem Cells. Mol Cell 2014, 55. [Google Scholar] [CrossRef] [PubMed]
- Yang, B. X.; El Farran, C. A.; Guo, H. C.; Yu, T.; Fang, H. T.; Wang, H. F.; Schlesinger, S.; Seah, Y. F. S.; Goh, G. Y. L.; Neo, S. P.; et al. Systematic Identification of Factors for Provirus Silencing in Embryonic Stem Cells. Cell 2015, 163. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Zhang, Y.; Loh, Y. P.; Tng, J. Q.; Lim, M. C.; Cao, Z.; Raju, A.; Lieberman Aiden, E.; Li, S.; Manikandan, L.; et al. H3K27me3-Rich Genomic Regions Can Function as Silencers to Repress Gene Expression via Chromatin Interactions. Nat Commun 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- van Mierlo, G.; Veenstra, G. J. C.; Vermeulen, M.; Marks, H. The Complexity of PRC2 Subcomplexes. Trends in Cell Biology 2019. [CrossRef] [PubMed]
- Boros, J.; Arnoult, N.; Stroobant, V.; Collet, J.-F.; Decottignies, A. Polycomb Repressive Complex 2 and H3K27me3 Cooperate with H3K9 Methylation To Maintain Heterochromatin Protein 1α at Chromatin. Mol Cell Biol 2014, 34. [Google Scholar] [CrossRef] [PubMed]
- Brumbaugh, J.; Stefano, B. Di; Hochedlinger, K. Reprogramming: Identifying the Mechanisms That Safeguard Cell Identity. Development (Cambridge) 2019. [Google Scholar] [CrossRef] [PubMed]
- Charlet, J.; Duymich, C. E.; Lay, F. D.; Mundbjerg, K.; Dalsgaard Sørensen, K.; Liang, G.; Jones, P. A. Bivalent Regions of Cytosine Methylation and H3K27 Acetylation Suggest an Active Role for DNA Methylation at Enhancers. Mol Cell 2016, 62. [Google Scholar] [CrossRef] [PubMed]
- Andersen, I. S.; Reiner, A. H.; Aanes, H.; Aleström, P.; Collas, P. Developmental Features of DNA Methylation during Activation of the Embryonic Zebrafish Genome. Genome Biol 2012, 13. [Google Scholar] [CrossRef] [PubMed]
- Eckersley-Maslin, M. A.; Alda-Catalinas, C.; Reik, W. Dynamics of the Epigenetic Landscape during the Maternal-to-Zygotic Transition. Nature Reviews Molecular Cell Biology 2018. [CrossRef] [PubMed]
- Liu, C.; Ma, Y.; Shang, Y.; Huo, R.; Li, W. Post-Translational Regulation of the Maternal-to-Zygotic Transition. Cellular and Molecular Life Sciences 2018. [Google Scholar] [CrossRef]
- Zheng, H.; Huang, B.; Zhang, B.; Xiang, Y.; Du, Z.; Xu, Q.; Li, Y.; Wang, Q.; Ma, J.; Peng, X.; et al. Resetting Epigenetic Memory by Reprogramming of Histone Modifications in Mammals. Mol Cell 2016, 63. [Google Scholar] [CrossRef] [PubMed]
- Fraser, R.; Lin, C. J. Epigenetic Reprogramming of the Zygote in Mice and Men: On Your Marks, Get Set, Go! Reproduction 2016. [Google Scholar] [CrossRef]
- Rousseaux, S.; Reynoird, N.; Escoffier, E.; Thevenon, J.; Caron, C.; Khochbin, S. Epigenetic Reprogramming of the Male Genome during Gametogenesis and in the Zygote. Reprod Biomed Online 2008, 16. [Google Scholar] [CrossRef]
- Inoue, A.; Jiang, L.; Lu, F.; Suzuki, T.; Zhang, Y. Maternal H3K27me3 Controls DNA Methylation-Independent Imprinting. Nature 2017, 547. [Google Scholar] [CrossRef]
- Meng, T. G.; Zhou, Q.; Ma, X. S.; Liu, X. Y.; Meng, Q. R.; Huang, X. J.; Liu, H. L.; Lei, W. L.; Zhao, Z. H.; Ouyang, Y. C.; et al. PRC2 and EHMT1 Regulate H3K27me2 and H3K27me3 Establishment across the Zygote Genome. Nat Commun 2020, 11. [Google Scholar] [CrossRef]
- Pailles, M.; Hirlemann, M.; Brochard, V.; Chebrout, M.; Oudin, J. F.; Marks, H.; Jouneau, A.; Bonnet-Garnier, A. H3K27me3 at Pericentromeric Heterochromatin Is a Defining Feature of the Early Mouse Blastocyst. Sci Rep 2022, 12. [Google Scholar] [CrossRef]
- Juan, A. H.; Wang, S.; Ko, K. D.; Zare, H.; Tsai, P. F.; Feng, X.; Vivanco, K. O.; Ascoli, A. M.; Gutierrez-Cruz, G.; Krebs, J.; et al. Roles of H3K27me2 and H3K27me3 Examined during Fate Specification of Embryonic Stem Cells. Cell Rep 2016, 17. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, F.; Kou, Z.; Zhang, Y.; Gao, S. Defective Chromatin Structure in Somatic Cell Cloned Mouse Embryos. Journal of Biological Chemistry 2009, 284. [Google Scholar] [CrossRef]
- Zhou, C.; Wang, Y.; Zhang, J.; Su, J.; An, Q.; Liu, X.; Zhang, M.; Wang, Y.; Liu, J.; Zhang, Y. H3K27me3 Is an Epigenetic Barrier While KDM6A Overexpression Improves Nuclear Reprogramming Efficiency. FASEB Journal 2019, 33. [Google Scholar] [CrossRef]
- Bai, G. Y.; Song, S. H.; Zhang, Y. W.; Huang, X.; Huang, X. W.; Sun, R. Z.; Lei, L. Kdm6a Overexpression Improves the Development of Cloned Mouse Embryos. Zygote 2018, 26. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Song, L.; Liu, X.; Bai, L.; Li, G. KDM 6A and KDM 6B Play Contrasting Roles in Nuclear Transfer Embryos Revealed by MERVL Reporter System. EMBO Rep 2018, 19. [Google Scholar] [CrossRef] [PubMed]
- Matoba, S.; Wang, H.; Jiang, L.; Lu, F.; Iwabuchi, K. A.; Wu, X.; Inoue, K.; Yang, L.; Press, W.; Lee, J. T.; et al. Loss of H3K27me3 Imprinting in Somatic Cell Nuclear Transfer Embryos Disrupts Post-Implantation Development. Cell Stem Cell 2018, 23. [Google Scholar] [CrossRef] [PubMed]
- Raas, M. W. D.; Zijlmans, D. W.; Vermeulen, M.; Marks, H. There Is Another: H3K27me3-Mediated Genomic Imprinting. Trends in Genetics 2022. [Google Scholar] [CrossRef] [PubMed]
- Macrae, T. A.; Fothergill-Robinson, J.; Ramalho-Santos, M. Regulation, Functions and Transmission of Bivalent Chromatin during Mammalian Development. Nature Reviews Molecular Cell Biology 2023. [CrossRef] [PubMed]
- Saha, B.; Home, P.; Ray, S.; Larson, M.; Paul, A.; Rajendran, G.; Behr, B.; Paul, S. EED and KDM6B Coordinate the First Mammalian Cell Lineage Commitment To Ensure Embryo Implantation. Mol Cell Biol 2013, 33. [Google Scholar] [CrossRef] [PubMed]
- Dahl, J. A.; Reiner, A. H.; Klungland, A.; Wakayama, T.; Collas, P. Histone H3 Lysine 27 Methylation Asymmetry on Developmentally-Regulated Promoters Distinguish the First Two Lineages in Mouse Preimplantation Embryos. PLoS One 2010, 5. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y. H.; Yu, J. Epigenetic Disruptions of Histone Signatures for the Trophectoderm and Inner Cell Mass in Mouse Parthenogenetic Embryos. Stem Cells Dev 2015, 24. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Hilbert, L.; Oda, H.; Wan, Y.; Heddleston, J. M.; Chew, T. L.; Zaburdaev, V.; Keller, P.; Lionnet, T.; Vastenhouw, N.; et al. Histone H3K27 Acetylation Precedes Active Transcription during Zebrafish Zygotic Genome Activation as Revealed by Live-Cell Analysis. Development (Cambridge) 2019, 146. [Google Scholar] [CrossRef] [PubMed]
- Lavarone, E.; Barbieri, C. M.; Pasini, D. Dissecting the Role of H3K27 Acetylation and Methylation in PRC2 Mediated Control of Cellular Identity. Nat Commun 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Vavouri, T.; Lehner, B. Human Genes with CpG Island Promoters Have a Distinct Transcription-Associated Chromatin Organization. Genome Biol 2012, 13. [Google Scholar] [CrossRef]
- Zhang, T.; Zhang, Z.; Dong, Q.; Xiong, J.; Zhu, B. Histone H3K27 Acetylation Is Dispensable for Enhancer Activity in Mouse Embryonic Stem Cells. Genome Biol 2020, 21. [Google Scholar] [CrossRef] [PubMed]
- DiFiore, J. V.; Ptacek, T. S.; Wang, Y.; Li, B.; Simon, J. M.; Strahl, B. D. Unique and Shared Roles for Histone H3K36 Methylation States in Transcription Regulation Functions. Cell Rep 2020, 31. [Google Scholar] [CrossRef]
- Sen, P.; Dang, W.; Donahue, G.; Dai, J.; Dorsey, J.; Cao, X.; Liu, W.; Cao, K.; Perry, R.; Lee, J. Y.; et al. H3K36 Methylation Promotes Longevity by Enhancing Transcriptional Fidelity. Genes Dev 2015, 29. [Google Scholar] [CrossRef]
- Leung, C. S.; Douglass, S. M.; Morselli, M.; Obusan, M. B.; Pavlyukov, M. S.; Pellegrini, M.; Johnson, T. L. H3K36 Methylation and the Chromodomain Protein Eaf3 Are Required for Proper Cotranscriptional Spliceosome Assembly. Cell Rep 2019, 27. [Google Scholar] [CrossRef]
- Kim, S.; Kim, H.; Fong, N.; Erickson, B.; Bentley, D. L. Pre-MRNA Splicing Is a Determinant of Histone H3K36 Methylation. Proc Natl Acad Sci U S A 2011, 108. [Google Scholar] [CrossRef]
- Li, F.; Mao, G.; Tong, D.; Huang, J.; Gu, L.; Yang, W.; Li, G. M. The Histone Mark H3K36me3 Regulates Human DNA Mismatch Repair through Its Interaction with MutSα. Cell 2013, 153. [Google Scholar] [CrossRef]
- Sun, Z.; Zhang, Y.; Jia, J.; Fang, Y.; Tang, Y.; Wu, H.; Fang, D. H3K36me3, Message from Chromatin to DNA Damage Repair. Cell and Bioscience 2020. [CrossRef]
- Wang, L.; Niu, N.; Li, L.; Shao, R.; Ouyang, H.; Zou, W. H3K36 Trimethylation Mediated by SETD2 Regulates the Fate of Bone Marrow Mesenchymal Stem Cells. PLoS Biol 2018, 16. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Workman, J. L. Regulation of SETD2 Stability Is Important for the Fidelity of H3K36me3 Deposition. Epigenetics Chromatin 2020, 13. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Zhu, B. Roles of H3K36-Specific Histone Methyltransferases in Transcription: Antagonizing Silencing and Safeguarding Transcription Fidelity. Biophys Rep 2018, 4. [Google Scholar] [CrossRef]
- Jha, D. K.; Strahl, B. D. An RNA Polymerase II-Coupled Function for Histone H3K36 Methylation in Checkpoint Activation and DSB Repair. Nat Commun 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, D. N.; Papillon-Cavanagh, S.; Chen, H.; Yue, Y.; Chen, X.; Rajagopalan, K. N.; Horth, C.; McGuire, J. T.; Xu, X.; Nikbakht, H.; et al. The Histone Mark H3K36me2 Recruits DNMT3A and Shapes the Intergenic DNA Methylation Landscape. Nature 2019, 573. [Google Scholar] [CrossRef]
- Yano, S.; Ishiuchi, T.; Abe, S.; Namekawa, S. H.; Huang, G.; Ogawa, Y.; Sasaki, H. Histone H3K36me2 and H3K36me3 Form a Chromatin Platform Essential for DNMT3A-Dependent DNA Methylation in Mouse Oocytes. Nat Commun 2022, 13. [Google Scholar] [CrossRef]
- Xu, Q.; Xiang, Y.; Wang, Q.; Wang, L.; Brind’Amour, J.; Bogutz, A. B.; Zhang, Y.; Zhang, B.; Yu, G.; Xia, W.; et al. SETD2 Regulates the Maternal Epigenome, Genomic Imprinting and Embryonic Development. Nat Genet 2019, 51. [Google Scholar] [CrossRef]
- Hu, M.; Sun, X. J.; Zhang, Y. L.; Kuang, Y.; Hu, C. Q.; Wu, W. L.; Shen, S. H.; Du, T. T.; Li, H.; He, F.; et al. Histone H3 Lysine 36 Methyltransferase Hypb/Setd2 Is Required for Embryonic Vascular Remodeling. Proc Natl Acad Sci U S A 2010, 107. [Google Scholar] [CrossRef]
- Aoshima, K.; Inoue, E.; Sawa, H.; Okada, Y. Paternal H3K4 Methylation Is Required for Minor Zygotic Gene Activation and Early Mouse Embryonic Development. EMBO Rep 2015, 16. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, Y. Maternal H3K27me3-Dependent Autosomal and X Chromosome Imprinting. Nature Reviews Genetics 2020. [Google Scholar] [CrossRef]
- Igolkina, A. A.; Zinkevich, A.; Karandasheva, K. O.; Popov, A. A.; Selifanova, M. V.; Nikolaeva, D.; Tkachev, V.; Penzar, D.; Nikitin, D. M.; Buzdin, A. H3K4me3, H3K9ac, H3K27ac, H3K27me3 and H3K9me3 Histone Tags Suggest Distinct Regulatory Evolution of Open and Condensed Chromatin Landmarks. Cells 2019, 8. [Google Scholar] [CrossRef]
- Lismer, A.; Lambrot, R.; Lafleur, C.; Dumeaux, V.; Kimmins, S. ChIP-Seq Protocol for Sperm Cells and Embryos to Assess Environmental Impacts and Epigenetic Inheritance. STAR Protoc 2021, 2. [Google Scholar] [CrossRef]
- Lismer, A.; Kimmins, S. Emerging Evidence That the Mammalian Sperm Epigenome Serves as a Template for Embryo Development. Nat Commun 2023, 14. [Google Scholar] [CrossRef]
- Cockrum, C. S.; Strome, S. Maternal H3K36 and H3K27 HMTs Protect Germline Development via Regulation of the Transcription Factor LIN-15B. Elife 2022, 11. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Zhang, C.; Zhang, Y. Epigenetic Regulation of Mouse Preimplantation Embryo Development. Current Opinion in Genetics and Development 2020. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Hu, B.; Wang, Z.; Wu, X.; Luo, L.; Li, S.; Wang, S.; Zhang, K.; Wang, H. Functional Role of GATA3 and CDX2 in Lineage Specification during Bovine Early Embryonic Development. Reproduction 2023, 165. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Guo, G.; Yuan, P.; Ralston, A.; Sun, L.; Huss, M.; Mistri, T.; Pinello, L.; Ng, H. H.; Yuan, G.; et al. The Role of Cdx2 as a Lineage Specific Transcriptional Repressor for Pluripotent Network during the First Developmental Cell Lineage Segregation. Sci Rep 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Home, P.; Ray, S.; Dutta, D.; Bronshteyn, I.; Larson, M.; Paul, S. GATA3 Is Selectively Expressed in the Trophectoderm of Peri-Implantation Embryo and Directly Regulates Cdx2 Gene Expression. Journal of Biological Chemistry 2009, 284. [Google Scholar] [CrossRef] [PubMed]
- Rebuzzini, P.; Zuccotti, M.; Garagna, S. Building Pluripotency Identity in the Early Embryo and Derived Stem Cells. Cells 2021. [CrossRef]
- Niwa, H. How Is Pluripotency Determined and Maintained? Development 2007. [Google Scholar] [CrossRef] [PubMed]
- Allègre, N.; Chauveau, S.; Dennis, C.; Renaud, Y.; Meistermann, D.; Estrella, L. V.; Pouchin, P.; Cohen-Tannoudji, M.; David, L.; Chazaud, C. NANOG Initiates Epiblast Fate through the Coordination of Pluripotency Genes Expression. Nat Commun 2022, 13. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Jho, E. H. The History and Regulatory Mechanism of the Hippo Pathway. BMB Reports 2018. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Guan, K. L. Hippo Signaling in Embryogenesis and Development. Trends in Biochemical Sciences 2021. [CrossRef] [PubMed]
- Yang, J.; Jiang, W. Dynamic Changes in Epigenetic Modifications During Mammalian Early Embryo Development. In Handbook of Epigenetics: The New Molecular and Medical Genetics, Third Edition; 2022. [CrossRef]
- Torres-Padilla, M. E.; Parfitt, D. E.; Kouzarides, T.; Zernicka-Goetz, M. Histone Arginine Methylation Regulates Pluripotency in the Early Mouse Embryo. Nature 2007, 445. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Bruce, A. W.; Jedrusik, A.; Ellis, P. D.; Andrews, R. M.; Langford, C. F.; Glover, D. M.; Zernicka-Goetz, M. CARM1 Is Required in Embryonic Stem Cells to Maintain Pluripotency and Resist Differentiation. Stem Cells 2009, 27. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Lyu, X.; Qin, N.; Liu, H.; Zhang, M.; Lai, Y.; Dong, B.; Lu, P. Coactivator-Associated Arginine Methyltransferase 1: A Versatile Player in Cell Differentiation and Development. Genes and Diseases 2023. [Google Scholar] [CrossRef]
- Hupalowska, A.; Jedrusik, A.; Zhu, M.; Bedford, M. T.; Glover, D. M.; Zernicka-Goetz, M. CARM1 and Paraspeckles Regulate Pre-Implantation Mouse Embryo Development. Cell 2018, 175. [Google Scholar] [CrossRef]
- Franek, M.; Legartová, S.; Suchánková, J.; Milite, C.; Castellano, S.; Sbardella, G.; Kozubek, S.; Bártová, E. CARM1 Modulators Affect Epigenome of Stem Cells and Change Morphology of Nucleoli. Physiol Res 2015, 64. [Google Scholar] [CrossRef]
- Wang, J.; Wang, L.; Feng, G.; Wang, Y.; Li, Y.; Li, X.; Liu, C.; Jiao, G.; Huang, C.; Shi, J.; et al. Asymmetric Expression of LincGET Biases Cell Fate in Two-Cell Mouse Embryos. Cell 2018, 175. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Jiang, J.; Xu, C.; Wang, Y.; Sun, L.; Guo, X.; Liu, H. MicroRNA-181 Regulates CARM1 and Histone Aginine Methylation to Promote Differentiation of Human Embryonic Stem Cells. PLoS One 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Goolam, M.; Scialdone, A.; Graham, S. J. L.; MacAulay, I. C.; Jedrusik, A.; Hupalowska, A.; Voet, T.; Marioni, J. C.; Zernicka-Goetz, M. Heterogeneity in Oct4 and Sox2 Targets Biases Cell Fate in 4-Cell Mouse Embryos. Cell 2016, 165. [Google Scholar] [CrossRef]
- Zhao, H. yong; Zhang, Y. jun; Dai, H.; Zhang, Y.; Shen, Y. fei. CARM1 Mediates Modulation of Sox2. PLoS One 2011, 6. [Google Scholar] [CrossRef]
- Cao, Z.; Tong, X.; Yin, H.; Zhou, N.; Zhang, X.; Zhang, M.; Wang, X.; Liu, Q.; Yan, Y.; Ma, Y.; et al. Histone Arginine Methyltransferase CARM1-Mediated H3R26me2 Is Essential for Morula-to-Blastocyst Transition in Pigs. Front Cell Dev Biol 2021, 9. [Google Scholar] [CrossRef]
- Burton, A.; Muller, J.; Tu, S.; Padilla-Longoria, P.; Guccione, E.; Torres-Padilla, M. E. Single-Cell Profiling of Epigenetic Modifiers Identifies PRDM14 as an Inducer of Cell Fate in the Mammalian Embryo. Cell Rep 2013, 5. [Google Scholar] [CrossRef]
- Wu, J.; Belmonte, J. C. I. The Molecular Harbingers of Early Mammalian Embryo Patterning. Cell 2016. [Google Scholar] [CrossRef] [PubMed]
- Nakaki, F.; Saitou, M. PRDM14: A Unique Regulator for Pluripotency and Epigenetic Reprogramming. Trends in Biochemical Sciences 2014. [CrossRef]
- Yao, C.; Zhang, W.; Shuai, L. The First Cell Fate Decision in Pre-Implantation Mouse Embryos. Cell Regeneration 2019. [Google Scholar] [CrossRef]
- Zhang, Y.; Duan, E. LncRNAs and Paraspeckles Predict Cell Fate in Early Mouse Embryo. Biol Reprod 2019, 100. [Google Scholar] [CrossRef]
- Grosch, M.; Ittermann, S.; Shaposhnikov, D.; Drukker, M. Chromatin-Associated Membraneless Organelles in Regulation of Cellular Differentiation. Stem Cell Reports 2020. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Wang, T.; Zhao, Z.; Wei, W.; Xin, W.; Yang, X.; Wang, X. Novel Insights into the Emerging Role of Neat1 and Its Effects Downstream in the Regulation of Inflammation. Journal of Inflammation Research 2022. [Google Scholar] [CrossRef] [PubMed]
- Henikoff, S.; Smith, M. M. Histone Variants and Epigenetics. Cold Spring Harb Perspect Biol 2015, 7. [Google Scholar] [CrossRef] [PubMed]
- Talbert, P. B.; Henikoff, S. Histone Variants at a Glance. J Cell Sci 2021, 134. [Google Scholar] [CrossRef] [PubMed]
- Martire, S.; Banaszynski, L. A. The Roles of Histone Variants in Fine-Tuning Chromatin Organization and Function. Nature Reviews Molecular Cell Biology 2020. [CrossRef]
- Buschbeck, M.; Hake, S. B. Variants of Core Histones and Their Roles in Cell Fate Decisions, Development and Cancer. Nature Reviews Molecular Cell Biology 2017. [CrossRef]
- Herchenröther, A.; Wunderlich, T. M.; Lan, J.; Hake, S. B. Spotlight on Histone H2A Variants: From B to X to Z. Seminars in Cell and Developmental Biology 2023. [CrossRef]
- Oberdoerffer, P.; Miller, K. M. Histone H2A Variants: Diversifying Chromatin to Ensure Genome Integrity. Seminars in Cell and Developmental Biology 2023. [CrossRef]
- Chakravarthy, S.; Gundimella, S. K. Y.; Caron, C.; Perche, P.-Y.; Pehrson, J. R.; Khochbin, S.; Luger, K. Structural Characterization of the Histone Variant MacroH2A. Mol Cell Biol 2005, 25. [Google Scholar] [CrossRef]
- Buschbeck, M.; Uribesalgo, I.; Wibowo, I.; Rué, P.; Martin, D.; Gutierrez, A.; Morey, L.; Guigó, R.; López-Schier, H.; Di Croce, L. The Histone Variant MacroH2A Is an Epigenetic Regulator of Key Developmental Genes. Nat Struct Mol Biol 2009, 16. [Google Scholar] [CrossRef]
- Duthie, S. M. Mechanisms of X-inactivation. In eLS; 2001. [CrossRef]
- Rasmussen, T. P.; Mastrangelo, M. A.; Eden, A.; Pehrson, J. R.; Jaenisch, R. Dynamic Relocalization of Histone MacroH2A1 from Centrosomes to Inactive X Chromosomes during X Inactivation. Journal of Cell Biology 2000, 150. [Google Scholar] [CrossRef]
- Chang, C. C.; Ma, Y.; Jacobs, S.; Tian, X. C.; Yang, X.; Rasmussen, T. P. A Maternal Store of MacroH2A Is Removed from Pronuclei Prior to Onset of Somatic MacroH2A Expression in Preimplantation Embryos. Dev Biol 2005, 278. [Google Scholar] [CrossRef]
- Angelov, D.; Molla, A.; Perche, P. Y.; Hans, F.; Côté, J.; Khochbin, S.; Bouvet, P.; Dimitrov, S. The Histone Variant MacroH2A Interferes with Transcription Factor Binding and SWI/SNF Nucleosome Remodeling. Mol Cell 2003, 11. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C. J.; Meers, O.; Buschbeck, M.; Heidel, F. H. The Role of Macroh2a Histone Variants in Cancer. Cancers 2021. [CrossRef] [PubMed]
- Mermoud, J. E.; Tassin, A. M.; Pehrson, J. R.; Brockdorff, N. Centrosomal Association of Histone MacroH2A1.2 in Embryonic Stem Cells and Somatic Cells. Exp Cell Res 2001, 268. [Google Scholar] [CrossRef] [PubMed]
- Paull, T. T.; Rogakou, E. P.; Yamazaki, V.; Kirchgessner, C. U.; Gellert, M.; Bonner, W. M. A Critical Role for Histone H2AX in Recruitment of Repair Factors to Nuclear Foci after DNA Damage. Current Biology 2000, 10. [Google Scholar] [CrossRef]
- Collins, P. L.; Purman, C.; Porter, S. I.; Nganga, V.; Saini, A.; Hayer, K. E.; Gurewitz, G. L.; Sleckman, B. P.; Bednarski, J. J.; Bassing, C. H.; et al. DNA Double-Strand Breaks Induce H2Ax Phosphorylation Domains in a Contact-Dependent Manner. Nat Commun 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Sun, K. Y.; Guo, S. M.; Cheng, G. P.; Yin, Y.; He, X.; Zhou, L. Q. Cleavage-Embryo Genes and Transposable Elements Are Regulated by Histone Variant H2a.x. Journal of Reproduction and Development 2021, 67. [Google Scholar] [CrossRef] [PubMed]
- Schmücker, A.; Lei, B.; Lorković, Z. J.; Capella, M.; Braun, S.; Bourguet, P.; Mathieu, O.; Mechtler, K.; Berger, F. Crosstalk between H2A Variant-Specific Modifications Impacts Vital Cell Functions. PLoS Genet 2021, 17. [Google Scholar] [CrossRef] [PubMed]
- Kinner, A.; Wu, W.; Staudt, C.; Iliakis, G. Gamma-H2AX in Recognition and Signaling of DNA Double-Strand Breaks in the Context of Chromatin. Nucleic acids research 2008. [Google Scholar] [CrossRef]
- Nishiyama, A.; Xin, L.; Sharov, A. A.; Thomas, M.; Mowrer, G.; Meyers, E.; Piao, Y.; Mehta, S.; Yee, S.; Nakatake, Y.; et al. Uncovering Early Response of Gene Regulatory Networks in ESCs by Systematic Induction of Transcription Factors. Cell Stem Cell 2009, 5. [Google Scholar] [CrossRef]
- Wu, T.; Liu, Y.; Wen, D.; Tseng, Z.; Tahmasian, M.; Zhong, M.; Rafii, S.; Stadtfeld, M.; Hochedlinger, K.; Xiao, A. Histone Variant H2A.X Deposition Pattern Serves as a Functional Epigenetic Mark for Distinguishing the Developmental Potentials of IPSCs. Cell Stem Cell 2014, 15. [Google Scholar] [CrossRef]
- Wu, B. J.; Dong, F. L.; Ma, X. S.; Wang, X. G.; Lin, F.; Liu, H. L. Localization and Expression of Histone H2A Variants during Mouse Oogenesis and Preimplantation Embryo Development. Genetics and Molecular Research 2014, 13. [Google Scholar] [CrossRef]
- Sugie, K.; Funaya, S.; Kawamura, M.; Nakamura, T.; Suzuki, M. G.; Aoki, F. Expression of Dux Family Genes in Early Preimplantation Embryos. Sci Rep 2020, 10. [Google Scholar] [CrossRef]
- Ren, W.; Gao, L.; Mou, Y.; Deng, W.; Hua, J.; Yang, F. DUX: One Transcription Factor Controls 2-Cell-like Fate. International Journal of Molecular Sciences 2022. [CrossRef]
- Ibarra-Morales, D.; Rauer, M.; Quarato, P.; Rabbani, L.; Zenk, F.; Schulte-Sasse, M.; Cardamone, F.; Gomez-Auli, A.; Cecere, G.; Iovino, N. Histone Variant H2A.Z Regulates Zygotic Genome Activation. Nat Commun 2021, 12. [Google Scholar] [CrossRef]
- Giaimo, B. D.; Ferrante, F.; Herchenröther, A.; Hake, S. B.; Borggrefe, T. The Histone Variant H2A.Z in Gene Regulation. Epigenetics and Chromatin 2019. [Google Scholar] [CrossRef]
- Semer, M.; Bidon, B.; Larnicol, A.; Caliskan, G.; Catez, P.; Egly, J. M.; Coin, F.; Le May, N. DNA Repair Complex Licenses Acetylation of H2A.Z.1 by KAT2A during Transcription. Nat Chem Biol 2019, 15. [Google Scholar] [CrossRef]
- Tsukii, K.; Takahata, S.; Murakami, Y. Histone Variant H2A.Z Plays Multiple Roles in the Maintenance of Heterochromatin Integrity. Genes to Cells 2022, 27. [Google Scholar] [CrossRef]
- Cole, L.; Kurscheid, S.; Nekrasov, M.; Domaschenz, R.; Vera, D. L.; Dennis, J. H.; Tremethick, D. J. Multiple Roles of H2A.Z in Regulating Promoter Chromatin Architecture in Human Cells. Nat Commun 2021, 12. [Google Scholar] [CrossRef]
- Rudnizky, S.; Bavly, A.; Malik, O.; Pnueli, L.; Melamed, P.; Kaplan, A. H2A.Z Controls the Stability and Mobility of Nucleosomes to Regulate Expression of the LH Genes. Nat Commun 2016, 7. [Google Scholar] [CrossRef]
- Sales-Gil, R.; Kommer, D. C.; de Castro, I. J.; Amin, H. A.; Vinciotti, V.; Sisu, C.; Vagnarelli, P. Non-redundant Functions of H2A.Z.1 and H2A.Z.2 in Chromosome Segregation and Cell Cycle Progression. EMBO Rep 2021, 22. [Google Scholar] [CrossRef]
- Iouzalen, N.; Moreau, J.; Méchali, M. H2A.ZI, a New Variant Histone Expressed during Xenopus Early Development Exhibits Several Distinct Features from the Core Histone H2A. Nucleic Acids Res 1996, 24. [Google Scholar] [CrossRef]
- Clarkson, M. J.; Wells, J. R. E.; Gibson, F.; Saint, R.; Tremethick, D. J. Regions of Variant Histone His2AvD Required for Drosophila Development. Nature 1999, 399. [Google Scholar] [CrossRef]
- Shen, T.; Ji, F.; Wang, Y.; Lei, X.; Zhang, D.; Jiao, J. Brain-Specific Deletion of Histone Variant H2A.z Results in Cortical Neurogenesis Defects and Neurodevelopmental Disorder. Nucleic Acids Res 2018, 46. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, J.; Zhou, J.; Bu, G.; Zhu, W.; He, H.; Sun, Q.; Yu, Z.; Xiong, W.; Wang, L.; et al. Hierarchical Accumulation of Histone Variant H2A.Z Regulates Transcriptional States and Histone Modifications in Early Mammalian Embryos. Advanced Science 2022, 9. [Google Scholar] [CrossRef]
- Dryhurst, D.; Ishibashi, T.; Rose, K. L.; Eirín-López, J. M.; McDonald, D.; Silva-Moreno, B.; Veldhoen, N.; Helbing, C. C.; Hendzel, M. J.; Shabanowitz, J.; et al. Characterization of the Histone H2A.Z-1 and H2A.Z-2 Isoforms in Vertebrates. BMC Biol 2009, 7. [Google Scholar] [CrossRef]
- Mylonas, C.; Lee, C.; Auld, A. L.; Cisse, I. I.; Boyer, L. A. A Dual Role for H2A.Z.1 in Modulating the Dynamics of RNA Polymerase II Initiation and Elongation. Nat Struct Mol Biol 2021, 28. [Google Scholar] [CrossRef]
- Scacchetti, A.; Schauer, T.; Reim, A.; Apostolou, Z.; Sparr, A. C.; Krause, S.; Heun, P.; Wierer, M.; Becker, P. B. Drosophila SWR1 and NuA4 Complexes Are Defined by DOMINO Isoforms. Elife 2020, 9. [Google Scholar] [CrossRef]
- Scacchetti, A.; Becker, P. B. Variation on a Theme: Evolutionary Strategies for H2A.Z Exchange by SWR1-Type Remodelers. Current Opinion in Cell Biology 2021. [Google Scholar] [CrossRef]
- Fujii, T.; Ueda, T.; Nagata, S.; Fukunaga, R. Essential Role of P400/MDomino Chromatin-Remodeling ATPase in Bone Marrow Hematopoiesis and Cell-Cycle Progression. Journal of Biological Chemistry 2010, 285. [Google Scholar] [CrossRef]
- Dickinson, M. E.; Flenniken, A. M.; Ji, X.; Teboul, L.; Wong, M. D.; White, J. K.; Meehan, T. F.; Weninger, W. J.; Westerberg, H.; Adissu, H.; et al. High-Throughput Discovery of Novel Developmental Phenotypes. Nature 2016, 537. [Google Scholar] [CrossRef]
- Faast, R.; Thonglairoam, V.; Schulz, T. C.; Beall, J.; Wells, J. R. E.; Taylor, H.; Matthaei, K.; Rathjen, P. D.; Tremethick, D. J.; Lyons, I. Histone Variant H2A.Z Is Required for Early Mammalian Development. Current Biology 2001, 11. [Google Scholar] [CrossRef]
- McHaourab, Z. F.; Perreault, A. A.; Venters, B. J. ChIP-Seq and ChIP-Exo Profiling of Pol II, H2A.Z, and H3K4me3 in Human K562 Cells. Sci Data 2018, 5. [Google Scholar] [CrossRef]
- Loppin, B.; Berger, F. Histone Variants: The Nexus of Developmental Decisions and Epigenetic Memory. Annual Review of Genetics 2020. [Google Scholar] [CrossRef]
- Kumar, V. C.; Pai, R. Genes of the Month: H3.3 Histone Genes: H3F3A and H3F3B. Journal of clinical pathology 2021. [Google Scholar] [CrossRef]
- Ishiuchi, T.; Abe, S.; Inoue, K.; Yeung, W. K. A.; Miki, Y.; Ogura, A.; Sasaki, H. Reprogramming of the Histone H3.3 Landscape in the Early Mouse Embryo. Nat Struct Mol Biol 2021, 28. [Google Scholar] [CrossRef]
- Kong, Q.; Banaszynski, L. A.; Geng, F.; Zhang, X.; Zhang, J.; Zhang, H.; O’Neill, C. L.; Yan, P.; Liu, Z.; Shido, K.; et al. Histone Variant H3.3–Mediated Chromatin Remodeling Is Essential for Paternal Genome Activation in Mouse Preimplantation Embryos. Journal of Biological Chemistry 2018, 293. [Google Scholar] [CrossRef]
- Wen, D.; Rosenwaks, Z. Activation of the Maternal Genome Through Asymmetric Distribution of Oocyte-Genome-Associated Histone H3.3. bioRxiv 2023, 2023.11.01.565208. [Google Scholar] [CrossRef]
- Bao, J.; Bedford, M. T. Epigenetic Regulation of the Histone-to-Protamine Transition during Spermiogenesis. Reproduction 2016. [Google Scholar] [CrossRef]
- Wen, D.; Banaszynski, L. A.; Liu, Y.; Geng, F.; Noh, K. M.; Xiang, J.; Elemento, O.; Rosenwaks, Z.; David Allis, C.; Rafii, S. Histone Variant H3.3 Is an Essential Maternal Factor for Oocyte Reprogramming. Proc Natl Acad Sci U S A 2014, 111. [Google Scholar] [CrossRef]
- Lin, Y.; Qiu, T.; Wei, G.; Que, Y.; Wang, W.; Kong, Y.; Xie, T.; Chen, X. Role of Histone Post-Translational Modifications in Inflammatory Diseases. Frontiers in Immunology 2022. [Google Scholar] [CrossRef]
- Park, J.; Lee, K.; Kim, K.; Yi, S. J. The Role of Histone Modifications: From Neurodevelopment to Neurodiseases. Signal Transduction and Targeted Therapy 2022. [Google Scholar] [CrossRef]
- Fitzsimons, C. P.; Van Bodegraven, E.; Schouten, M.; Lardenoije, R.; Kompotis, K.; Kenis, G.; Van Den Hurk, M.; Boks, M. P.; Biojone, C.; Joca, S.; et al. Epigenetic Regulation of Adult Neural Stem Cells: Implications for Alzheimer’s Disease. Molecular Neurodegeneration 2014. [Google Scholar] [CrossRef]
- Audesse, A. J.; Webb, A. E. Mechanisms of Enhanced Quiescence in Neural Stem Cell Aging. Mech Ageing Dev 2020, 191. [Google Scholar] [CrossRef]
- Basavarajappa, B. S.; Subbanna, S. Histone Methylation Regulation in Neurodegenerative Disorders. International Journal of Molecular Sciences 2021. [CrossRef]
- Ritchie, F. D.; Lizarraga, S. B. The Role of Histone Methyltransferases in Neurocognitive Disorders Associated with Brain Size Abnormalities. Frontiers in Neuroscience 2023. [Google Scholar] [CrossRef]
- Vitorakis, N.; Piperi, C. Insights into the Role of Histone Methylation in Brain Aging and Potential Therapeutic Interventions. Int J Mol Sci 2023, 24. [Google Scholar] [CrossRef]
- Ding, Y.; Liu, C.; Zhang, Y. Aging-Related Histone Modification Changes in Brain Function. Ibrain 2023. [Google Scholar] [CrossRef]
- Berson, A.; Nativio, R.; Berger, S. L.; Bonini, N. M. Epigenetic Regulation in Neurodegenerative Diseases. Trends in Neurosciences 2018. [Google Scholar] [CrossRef]
- Omole, A. E.; Fakoya, A. O. J. Ten Years of Progress and Promise of Induced Pluripotent Stem Cells: Historical Origins, Characteristics, Mechanisms, Limitations, and Potential Applications. PeerJ, 2018, 2018 (MAY). [CrossRef]
- Papp, B.; Plath, K. Epigenetics of Reprogramming to Induced Pluripotency. Cell 2013. [Google Scholar] [CrossRef]
- Huang, M.; Xiao, X.; Ji, G.; Wu, Q. Histone Modifications in Neurodifferentiation of Embryonic Stem Cells. Heliyon 2022. [Google Scholar] [CrossRef]
- Castillo Bautista, C. M.; Sterneckert, J. Progress and Challenges in Directing the Differentiation of Human IPSCs into Spinal Motor Neurons. Frontiers in Cell and Developmental Biology 2023. [CrossRef]
- Ganesan, A.; Arimondo, P. B.; Rots, M. G.; Jeronimo, C.; Berdasco, M. The Timeline of Epigenetic Drug Discovery: From Reality to Dreams. Clinical Epigenetics 2019. [Google Scholar] [CrossRef]
- Holdgate, G. A.; Bardelle, C.; Lanne, A.; Read, J.; O’Donovan, D. H.; Smith, J. M.; Selmi, N.; Sheppard, R. Drug Discovery for Epigenetics Targets. Drug Discovery Today 2022. [Google Scholar] [CrossRef]
- Montecino, M.; Carrasco, M. E.; Nardocci, G. Epigenetic Control of Osteogenic Lineage Commitment. Frontiers in Cell and Developmental Biology. Frontiers Media S.A. January 8, 2021. [CrossRef]
- Sepulveda, H.; Aguilar, R.; Prieto, C. P.; Bustos, F.; Aedo, S.; Lattus, J.; van Zundert, B.; Palma, V.; Montecino, M. Epigenetic Signatures at the RUNX2-P1 and Sp7 Gene Promoters Control Osteogenic Lineage Commitment of Umbilical Cord-Derived Mesenchymal Stem Cells. J Cell Physiol 2017, 232. [Google Scholar] [CrossRef]
- Jin, M. L.; Jeong, K. W. Histone Modifications in Drug-Resistant Cancers: From a Cancer Stem Cell and Immune Evasion Perspective. Experimental and Molecular Medicine 2023. [Google Scholar] [CrossRef]
- Bajbouj, K.; Al-ali, A.; Ramakrishnan, R. K.; Saber-ayad, M.; Hamid, Q. Histone Modification in Nsclc: Molecular Mechanisms and Therapeutic Targets. International Journal of Molecular Sciences 2021. [CrossRef]
- Markouli, M.; Strepkos, D.; Piperi, C. Impact of Histone Modifications and Their Therapeutic Targeting in Hematological Malignancies. International Journal of Molecular Sciences 2022. [CrossRef]
- Monaghan, L.; Massett, M. E.; Bunschoten, R. P.; Hoose, A.; Pirvan, P. A.; Liskamp, R. M. J.; Jørgensen, H. G.; Huang, X. The Emerging Role of H3K9me3 as a Potential Therapeutic Target in Acute Myeloid Leukemia. Frontiers in Oncology 2019. [Google Scholar] [CrossRef]
- He, L. R.; Liu, M. Z.; Li, B. K.; Rao, H. L.; Liao, Y. J.; Guan, X. Y.; Zeng, Y. X.; Xie, D. Prognostic Impact of H3K27me3 Expression on Locoregional Progression after Chemoradiotherapy in Esophageal Squamous Cell Carcinoma. BMC Cancer 2009, 9. [Google Scholar] [CrossRef]
- Ngollo, M.; Lebert, A.; Daures, M.; Judes, G.; Rifai, K.; Dubois, L.; Kemeny, J. L.; Penault-Llorca, F.; Bignon, Y. J.; Guy, L.; et al. Global Analysis of H3K27me3 as an Epigenetic Marker in Prostate Cancer Progression. BMC Cancer 2017, 17. [Google Scholar] [CrossRef]
- Belhocine, M.; Simonin, M.; Abad Flores, J. D.; Cieslak, A.; Manosalva, I.; Pradel, L.; Smith, C.; Mathieu, E. L.; Charbonnier, G.; Martens, J. H. A.; et al. Dynamics of Broad H3K4me3 Domains Uncover an Epigenetic Switch between Cell Identity and Cancer-Related Genes. Genome Res 2022, 32. [Google Scholar] [CrossRef]
- Dutta, H.; Jain, N. Post-Translational Modifications and Their Implications in Cancer. Frontiers in Oncology 2023. [Google Scholar] [CrossRef]
- Guan, X.; Deng, H.; Choi, U. L.; Li, Z.; Yang, Y.; Zeng, J.; Liu, Y.; Zhang, X.; Li, G. EZH2 Overexpression Dampens Tumor-Suppressive Signals via an EGR1 Silencer to Drive Breast Tumorigenesis. Oncogene 2020, 39. [Google Scholar] [CrossRef]
- Hirukawa, A.; Smith, H. W.; Zuo, D.; Dufour, C. R.; Savage, P.; Bertos, N.; Johnson, R. M.; Bui, T.; Bourque, G.; Basik, M.; et al. Targeting EZH2 Reactivates a Breast Cancer Subtype-Specific Anti-Metastatic Transcriptional Program. Nat Commun 2018, 9. [Google Scholar] [CrossRef]
- Sun, Y.; Jiang, X.; Xu, Y.; Ayrapetov, M. K.; Moreau, L. A.; Whetstine, J. R.; Price, B. D. Histone H3 Methylation Links DNA Damage Detection to Activation of the Tumour Suppressor Tip60. Nat Cell Biol 2009, 11. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




