1. Introduction
Antimicrobial resistance (AMR) has been seriously threatened human public health and global economic development, and the COVID-19 pandemic has further accelerated this problem [
1]. Thereout, new antimicrobial agents desperately need to be developed [
2,
3]. After antimicrobial agents have been used for the treatment of bacterial infection, most of them would bring some side effects to the body, and eventually the resistant mutants to them would be also emerged [
4]. However, some plant secondary metabolites not only have antimicrobial activities, and also show good safety for human body since they exist in all sorts of plant derived foods and beverages [
5,
6]. Among them, plant flavonoids have gradually paid a close attention to [
7,
8,
9,
10,
11].
Flavonoids are an important class of secondary metabolites widely distributed in various plants, and many of them have different degrees of inhibitory activity to many pathogenic bacteria. More importantly, some of them can enhance the antimicrobial activities of some antimicrobial agents, and/or even reverse the AMR [
12,
13]. Various antibacterial mechanisms involving the synthesis inhibitions to DNA, proteins and cell envelope, and the damage of cell membrane were reported for plant flavonoids. Simultaneously, many structure-activity relationships were summarized, while some of them were contradictory [
13,
14,
15]. Recently, Yuan,
et al. established that the antimicrobial quantitative relationship of plant flavonoids to Gram-positive bacteria [
16], using the statistical analysis and validation of large samples. Based on this relationship, the minimum inhibitory concentrations (MICs) of plant flavonoids against Gram-positive bacteria can be calculated. Moreover, it was pointed out that the cell membrane is the major site of plant flavonoids acting on Gram-positive bacteria, and which includes the damage of phospholipid bilayers and likely involves the inhibition of the respiratory chain, or some others [
16,
17]. Next, the interfered experiments with menaquinone-4 or menaquinones extracted from
Staphylococcus aureus further proposed that the quinone pool is a key target [
18].
Generally, it is difficult to discover natural products with remarkable inhibition to Gram-negative bacteria. From above, it was spontaneously wondered whether the antimicrobial activities of plant flavonoids to Gram-negative bacteria are also related to their lipophilicities, and whether the antimicrobial quantitative relationship can be also established, for predicting their MICs and discovering compounds with effectively inhibiting Gram-negative bacteria. Are there are similar antibacterial modes of plant flavonoids against Gram-negative to -positive bacteria? such as their antibacterial effects and mechanisms. Here, the correlation between the physicochemical parameter (ACD/LogP or LogD
7.40) and the MIC, antibacterial quantitative structure-activity relationship, and possible mechanisms of plant flavonoids to Gram-negative bacteria were further explored according to similar procedure reported [
16,
17].
2. Results
2.1. Structure, antibacterial activity, and physicochemical parameter
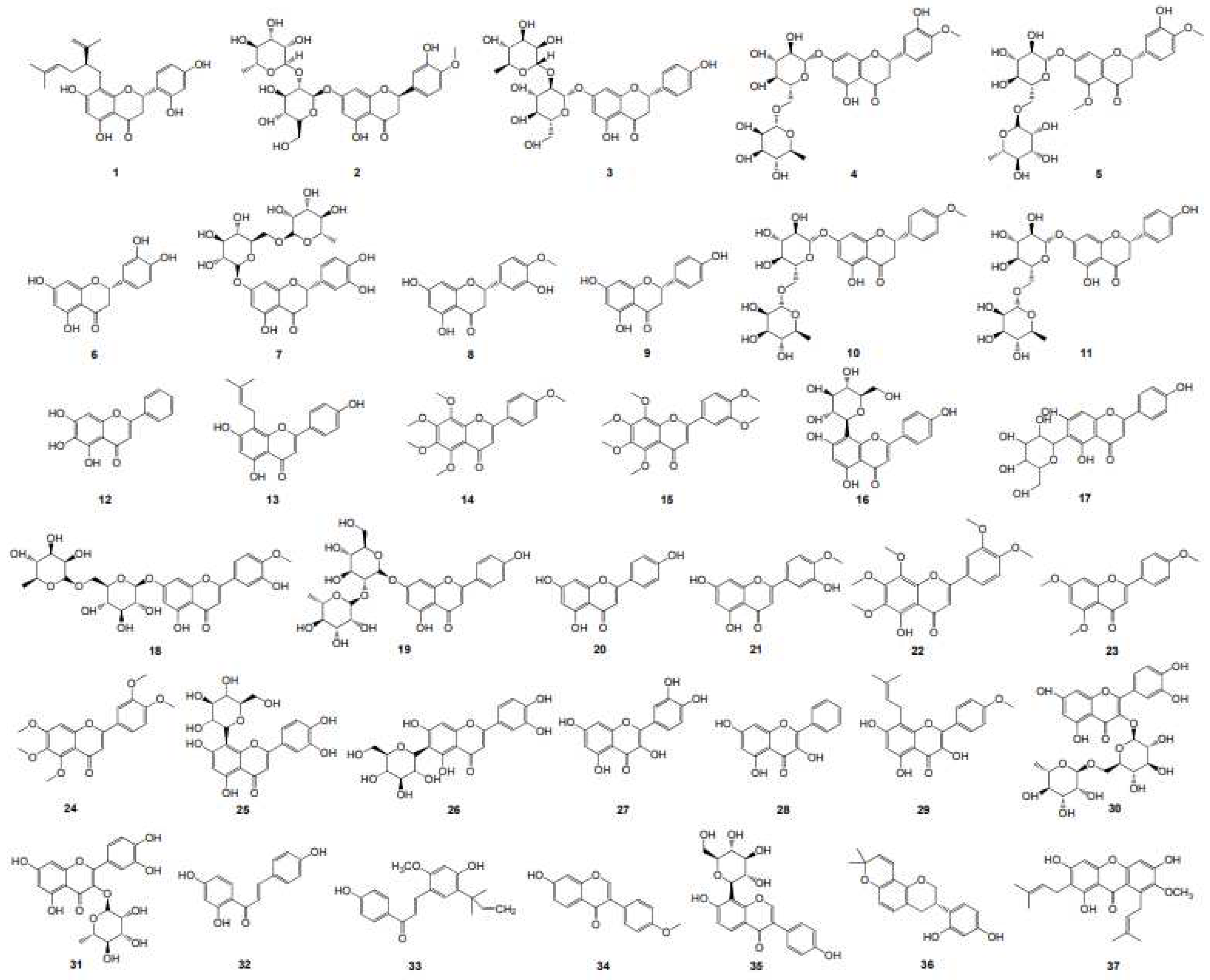
Thirty-seven plant flavonoids (
Table 1 and
Figure 1), with seven structural subtypes including dihydroflavones, flavones, flavonols, chalcones, isoflavones, isoflavanes and xanthones, were selected for the antimicrobial susceptibility assay and the correlation analyses for the antimicrobial activity MIC and the physicochemical parameter LogP or LogD
7.40. Their MIC values (μM) against Escherichia coli and S. aureus were respectively listed in
Table 1, together with their LogP and LogD
7.40 values. The results showed that the antimicrobial activities of all these flavonoids to E. coli are very weak with the MICs ranged from 1206.15 to more than 6820.53 μM. This indicated these plant flavonoids with the LogP and LogD
7.40 values ranged from 1.26 to 6.70 have weak inhibitory activity to Gram-negative bacteria. However, they presented various degrees of antibacterial activities to S. aureus, with the MICs ranged from 9.42 to 13552.14 or more than 7578.45 μM. According to the antimicrobial quantitative relationship reported [
16], the MICs (
Table 1) of these plant flavonoids to Gram-positive bacterial S. aureus were simultaneously calculated by the equation y =-0.1285 x
6 + 0.7944 x
5 + 51.785 x
4-947.64 x
3 + 6638.7 x
2-21,273 x + 26,087 [
16], and the results showed that the calculated MIC values of 81.1% plant flavonoids can be accepted comparing with those tested. This once again verified that the antimicrobial activities of plant flavonoids to Gram-positive bacteria can be effectively calculated or predicted.
Simultaneously, the MIC values of fifty-two plant flavonoids (
Figure S1) against Gram-negative bacterial E. coli were collected from sixteen papers [
19,
20,
21,
22,
23,
24,
25,
26,
27,
28,
29,
30,
31,
32,
33,
34], and showed in
Table 2, for using more data to obtain more reliable conclusions. These flavonoids includes eight structural subtypes of plant flavonoids, such as dihydroflavones, flavones, biflavones, flavonols, chalcones, isoflavones and xanthones. Intuitively observed from
Table 2, these flavonoids, with the LogP values ranged from 1.53 to 8.03, present various degrees of antibacterial activities to E. coli, with the reported MICs ranged from 1.49 to 3308.19 or more than 1722.53 μM.
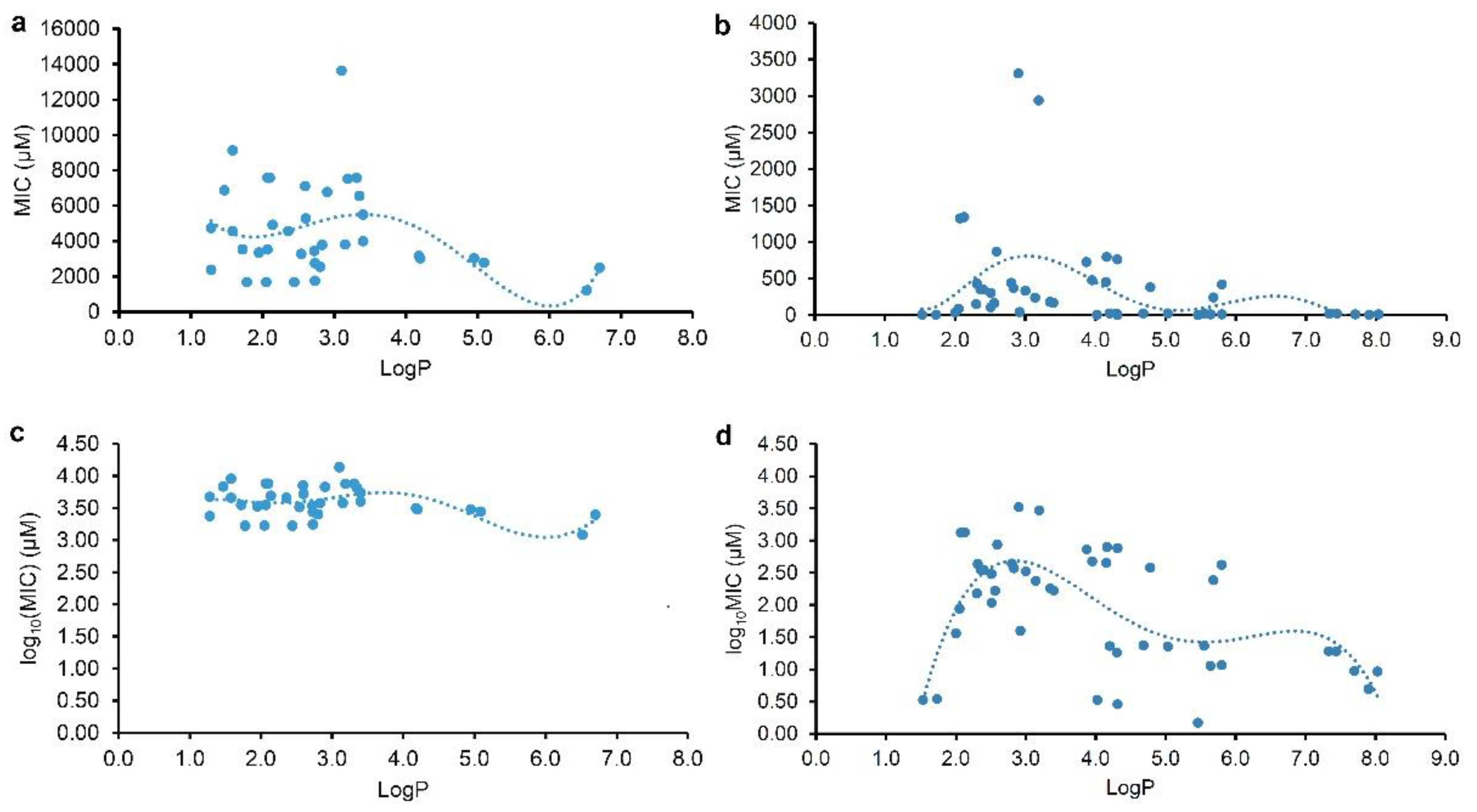
2.2. Correlation and regression analyses for the MICs and the physicochemical parameters
To discover some correlations between the physicochemical parameters (LogP or LogD
7.40) and the MICs of plant flavonoids against Gram-negative, those flavonoids in
Table 1 and
Table 2 were respectively selected for the statistical analyses. As we pointed out [
17], the antimicrobial activities of a compound against different pathogenic bacteria were varied, and even against the same one in different determination conditions. Thereby, the regression analyses were performed for these flavonoids tested and reported, respectively. The regression equations together with their correlation coefficients (r) and coefficients of determination (R
2) were listed in
Table 3, and the regression curves from the LogP and MIC or log
10(MIC) were shown on
Figure 2. From
Table 3, it indicated that all the r values for equations (1) to (4) were more than the corresponding critical values of r
0.975 (35) or r
0.975 (44), and the parameter LogP was better than the LogD
7.40 to be selected for the correlation and regression analyses. These indicated that there are correlations between the lipophilic parameter LogP (x) and the MIC or log
10(MIC) (y) of plant flavonoids to gram-negative bacteria especially E. coli, and that the antibacterial activities of plant flavonoids to gram-negative bacteria are related to their lipophilicities, whether resulted from plant flavonoids tested here or reported. However, all the R
2 values for equations (1) to (6) were less than 0.45, far less than 1.0. Generally, the closer the R
2 is to 1, the higher the goodness of fit, and the closer the calculated value is, on the whole, to the actual one [
16]. Thereby, the smaller R
2 values indicated that the goodness of fit of these regression equations were too low to be effectively used for predicting the MIC values of a certain plant flavonoid to Gram-negative bacteria.
From
Figure 2, both curves (Figures 2a and 2c) regressed from the MIC or log
10(MIC) and LogP value of tested plant flavonoids presented biconcave characters, and have two minimum values. However, another two curves (Figures 2b and 2d) regressed from those of reported plant flavonoids have three minimum values. Excluding the difference in the range of LogP value, it was found that all of them showed similar characters when the LogP values of plant flavonoids were less than 7.0. Namely, the left two curves have minimum MIC or log
10(MIC) values when the logP values of tested plant flavonoids were about 2.0 and 6.0, and while the right two curves have minimum MIC or log
10(MIC) values when the logP values of reported plant flavonoids were about 1.5 and 5.5. Differently, the right two curves have the third minimum MIC or log
10(MIC) values. This was mainly attributed to that the LogP values of some reported plant flavonoids were more than 7.0. Although there were a little difference between the curve shapes of
Figure 2a and 2c, another effective fitting equation (6) with a smaller r and R
2 than equation (5) could be also established from the LogP and log
10(MIC) of reported plant flavonoids, and its regression curve (
Figure S2) is similar to that of
Figure 2c. Thereby, these above indicated that that keeping appropriate lipophilicity, probably with the LogP value of about 2.0 (or 1.5), 6.0 (or 5.5), or 7.8, is necessary for plant flavonoids to obtain greater antibacterial activities to gram-negative bacteria, while not like plant flavonoids to gram-positive bacteria that the more the LogP value (from 2.0 to 8.9), the greater the antimicrobial activity presented overall.
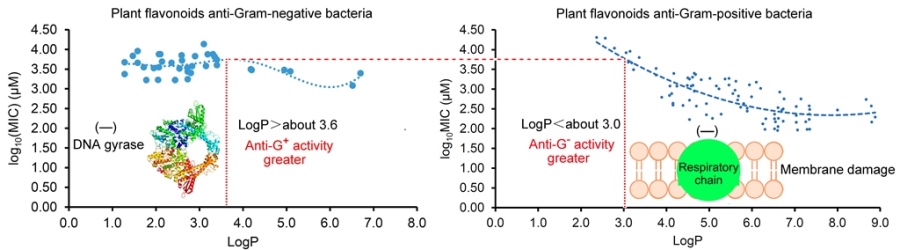
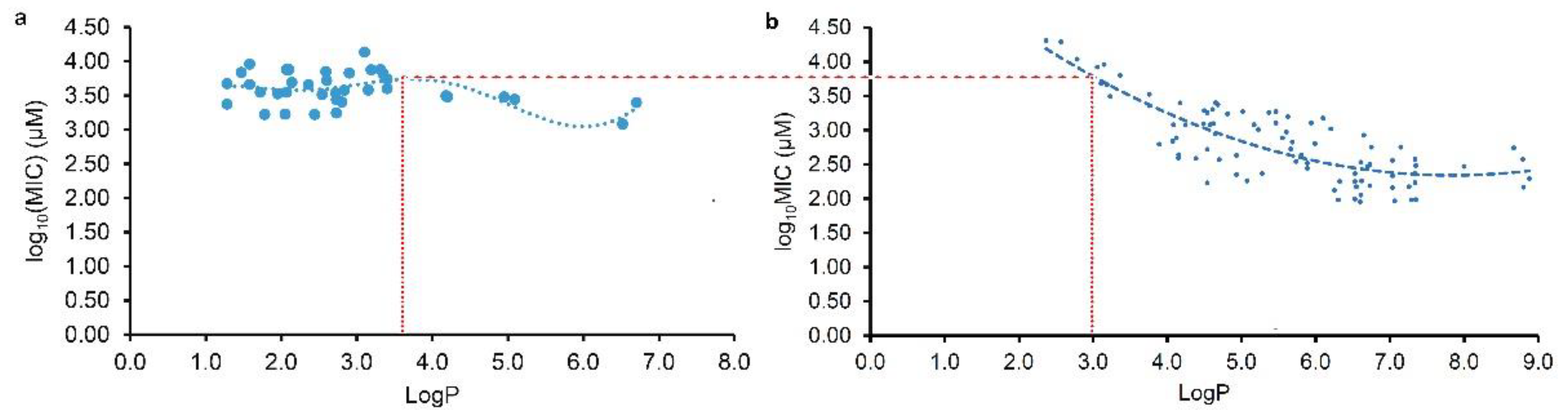
2.3. Different antibacterial modes of plant flavonoids to Gram-negative and Gram-positive bacteria.
As Yuan, et al. mentioned [
16,
17], many factors involving the methods and details of MIC test would have an influence on the experimental results, it is usual fact that the MICs of a compound against the same pathogen are varied large in different labs. Thereby, the MIC data tested in our lab should have better consistency than those collected from the literature, and so the regression curve of
Figure 2c was further selected to compare with the regression curves from the LogP and log
10(MIC) of plant flavonoids against Gram-positive bacteria in our previous work [
16], for discovering their difference. From
Figure 3, the obvious difference of two curves was that most flavonoids likely have greater antimicrobial activities to Gram-negative than Gram-positive bacteria when the LogP value was less than about 3.0. However, most flavonoids likely have greater antimicrobial activities to Gram-positive than Gram-negative bacteria when the LogP value was more than about 3.6. Another, the biconcave (
Figure 3a) or even triple concave (
Figure 2d) character of the regression curve indicated that there are probably multiple action modes of plant flavonoids against Gram-negative bacteria, while a main cell membrane action of plant flavonoids against Gram-positive bacteria only presents one concave character. As both two curves present simialr decreasing character along with the increase of LogP values ranged from about 3.6 to 6.0 (Figure 3), the cell membrane action was probably an important site for some plant flavonoids against Gram-negative bacteria, especially for those with the LogP values ranged from about 3.6 to 6.0 (
Figure 2c or 3a), or from about 2.8 to 5.5 (
Figure 2d).
3. Discussion
To predict the antimicrobial activities of plant flavonoids to Gram-negative bacteria and discover compounds with effectively inhibiting Gram-negative bacteria, the correlation and regression analyses were performed respectively using thirty-seven tested and forty-six reported data pairs composed of the LogP (or LogD7.40) values and MICs of plant flavonoids against Gram-positive bacteria. The results indicated the antibacterial activities of plant flavonoids to Gram-negative bacteria are related to their lipophilicities, and which is like to Gram-positive bacteria. However, the antimicrobial quantitative relationship could not be established for plant flavonoids against Gram-negative bacteria, and so their antimicrobial activities to Gram-negative bacteria could not be predicted from their LogP values. Moreover, different from mainly acting on the cell membrane of Gram-positive bacteria, it was further proposed that there are multiple action modes of plant flavonoids against Gram-negative bacteria, while among them the cell membrane is also an important action site.
As the structural skeletons of plant flavonoids are similar with those of quinolone antimicrobial agents, Ohemeng,
et al. had designed and synthesized fourteen flavonoids [
35]. The evaluation on the DNA-gyrase inhibitory and antibacterial activities showed that the antibacterial activity of these flavonoids did not parallel the potency at the enzyme level, and may be due in part to their inhibition of the DNA-gyrase [
35]. Thereout, the authors concluded that some mechanisms other than DNA gyrase inhibition may play a role in the antibacterial activity of flavonoids. As we interpreted [
16,
17], the antibacterial activities of plant flavonoids to Gram-positive mainly depend on their lipophilicities since the cell membrane is the main site of plant flavonoids acting on Gram-positive bacteria. Inspired by the multiple action modes mentioned above of plant flavonoids acting on Gram-negative bacteria, the ACD/LogP values of those designed flavonoids were calculated for exploring the correlation between their enzyme inhibitory activities and their LogP values from 2.07 to 3.76. Surprisingly, the results (
Tables S2 and S3) showed that there is significant correlation between their enzyme inhibitory activities and LogP values, and the character of obtained regression curve (
Figure S3) was very similar with that of the curve (
Figure 2a or 2c) with the LogP values ranged from 2.07 to 3.76. Furthermore, the antibacterial activities of these flavonoids to Gram-negative bacteria completely paralleled the potency at the enzyme level, especially to
E. coliss. Although no definite MIC values were provided for those flavonoids against some pathogenic Gram-negative bacterial strains, the relative antibacterial activities could be roughly obtained from those when they combined with polymyxin B nonapeptide (PMBN) [
35]. Thereby, it was further suggested that the DNA gyrase is another important target of plant flavonoids against Gram-negative bacteria. This was also supported by other publications [
36,
37,
38].
According to this, the antibacterial activities of plant flavonoids to Gram-positive bacteria should also present a concave feature when the LogP values less than about 3.0 (
Figure 3b). Unfortunately, the LogP value of 3.0 is close to the boundary one, and which possible would cause the curve characteristic to be inconsistent with actual one when the LogP values less than about 3.0 (
Figure 3b). Another, this might be similar with the antibacterial difference of quinolones against Gram-negative from Gram-positive bacteria. Furthermore, their antibacterial activities brought about by the DNA gyrase inhibition were overall weak from
Figure 2c or 3a.
Along with the LogP values increased, the antibacterial activities of plant flavonoids to both Gram-negative and Gram-positive bacteria increased from Figures 3a and 3b. However, their antibacterial activities to Gram-negative bacteria increased more quickly than to Gram-positive ones. This might be attributed to the difference of both Gram-negative and Gram-positive bacteria in cell envelope and respiratory chain (such as the composition of quinone pool) [
18]. As we mentioned [
18], the menaquinones (MKs) are the sole quinones for electron transfer in the respiratory chain of Gram-positive bacteria, and while there are two quinones as MKs and ubiquinones for those of Gram-negative bacteria. This should be an important reason that plant flavonoids show weaker antimicrobial activities to Gram-negative than Gram-negative bacteria when the LogP values were more than approximately 3.6 (
Figure 3a).
Although both regression curves (Figures 2a and 2b) overall presented similar character along with the change of LogP values, it was noted that the MICs of tested plant flavonoids were obviously higher than those of reported plant flavonoids. This was probably attributed to different determination condition, methods, details, and selected pathogenic strains. There were nine same compounds among tested and reported plant flavonoids, and so their antibacterial activities to
E. coli ATCC 25922 were reorganized and listed in
Table S1 for comparing the difference of tested and reported data [
21,
24,
25,
28,
30,
34]. The results indicated that only the MICs of compound
13 (Licoflavone C) presented more than 10 times difference between tested (3026.36 μM) and reported (23.08 μM) values [
25]. Thereby, using broth microdilution method in 96-well plates, the antibacterial activity of this compound to
E. coli ATCC 25922 was determined by six repeats, and all the MICs of compound
13 against
E. coli ATCC 25922 were 3026.36 μM.
As mentioned in section 2.3, the antibacterial activities of plant flavonoids, with the logP value less than about 3.6, to Gram-negative bacteria were statistically more than those to Gram-positive ones, and they would also increase overall along with the increase of LogP values when which more than about 3.6. However, the antibacterial activities of most plant flavonoids to Gram-negative bacteria were weak overall. Probably, it is difficult to discover plant flavonoids with very strong antibacterial activities to Gram-negative bacteria.
4. Materials and Methods
4.1. Materials, Chemicals and Reagents
Thirty-seven plant flavonoids: icaritin (>98%), isoliquiritigenin (98%), formononetin (98%), isoliquiritigenin (98%), galangin (98%), baicalein (98%), diosmin (95%), hesperetin (97%), puerarin (98%), apigenin (≥95%), diosmetin (98%) and naringenin (97%) were purchased from Shanghai Macklin Biochemical Co., Ltd. (Shanghai, China); α-mangostin (>98.0%), licochalcone A (>98.0%), nobiletin (≥98.5%), tangeritin (≥98.5%), quercitrin (98%), sinensetin (98%), narirutin (98%), orientin (99%) and isoorientin (98%) were purchased from Chengdu Push Bio-technology Co., Ltd. (Chengdu, China); naringin (95%), neohesperidin (≥98%) and hesperidin (95%) were purchased from Shanghai Yuanye Bio-Technology Co., Ltd. (Shanghai, China); eriodictyol (≥98%), eriocitrin (≥98%), rhoifolin (≥98%) and licoflavone C (≥98%) were purchased from Wuhan ChemFaces Biochemical Co., Ltd. (Wuhan, China); glabridin (99.8%) and sophoraflavanone G (>98%) were purchased from Shanghai TopScience Co., Ltd. (Shanghai, China); quercetin (97%) was purchased from Meryer (Shanghai) Biochemical Technology Co., Ltd. (Shanghai, China); methyl-hesperidin (95%) was purchased from Shanghai Acmec Biochemical Co.,Ltd. (Shanghai, China); didymin (≥98%), 5-demethylnobiletin (≥98%), 4',5,7-trimethoxyflavone (≥98%), vitexin (≥98%) and isovitexin (≥98%) were purchased from Sichuan Weikeqi Biological Technology Co.,Ltd. (Sichuan, China). All the compounds were stored at −20°C. The stock solutions of above plant flavonoids were prepared by dissolving in a certain volume of dimethyl sulfoxide (DMSO), and diluted with Mueller Hinton broth (MHB) to obtain a concentration of 4096, 8192 or 16384 μg/mL. The stock solution was mixed well, and then diluted to the desired concentrations with MHB immediately before use. In another, the DMSO concentrations in all the test systems were kept to less than 5.0%, and all those in the blank controls were 5.0%.
Casein hydrolysate (Qingdao Hope Bio-Technology Co., Ltd., Qingdao, China), starch soluble, beef extract and agar powder (Sangon Biotech (Shanghai) Co., Ltd., Shanghai, China) were used for preparing the media. Mueller Hinton agar (MHA) consisted of casein hydrolysate 17.5 g/L, starch soluble 1.5 g/L, beef extract 3.0 g/L, and agar powder 17.0 g/L dissolving in purified water, and the pH value of 7.40 ± 0.20. MHB was prepared without agar powder according to the same composition and procedure to MHA. DMSO and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) were purchased from Sangon Biotech (Shanghai) Co., Ltd. (Shanghai, China), and 96-well plates were purchased from Shanghai Excell Biological Technology Co., Ltd. (Shanghai, China). All reagents were analytical or biochemical ones. All TopPette Pipettors (2~20 μL and 20~200 μL) were purchased from DLAB Scientific Co., Ltd., Beijing, China.
4.2. Bacterial Strains and Growth Condition
E. coli ATCC 25922 and S. aureus ATCC 25923 were purchased from American Type Culture Collection, Manassas, VA, USA, and this organism was stored in MicrobankTM microbial storages (PRO-LAB diagnostics, Toronto, Canada) at −20°C. Prior to use, E. coli and S. aureus were cultured onto MHA plate at 37°C, and then pure colonies from the plate were inoculated into MHA at 37°C for 24 h on a rotary shaker (160 rpm). A 1:100 dilution of the overnight culture was made into fresh MHB, and then incubated at 37°C until the exponential phase for the following experiments. MHB was used for the antimicrobial susceptibility tests.
4.3. Antimicrobial Susceptibility Assay
According to the standard procedure described by the Clinical and Laboratory Standards Institute (CLSI) [
39], the exponential phase culture was diluted with MHB to achieve a bacterial concentration approximately 1.0×10
6 CFU/mL, and then the susceptibility of plant flavonoids against
E. coli ATCC 25922 or
S. aureus ATCC 25923 was determined using the broth microdilution method on the 96-well plates in triplicate [
4]. Depending on the preliminary MIC values of plant flavonoids, the initial concentration 1024, 2048 or 4096 μg/mL of each compound was respectively set. After the 96-well plate were incubated at 35°C for 24 h, a 20 μL of MTT (4.0 mg/mL) was added into each well, shaking well, and stayed for 30 min at ambient temperature. The MIC, defined as the lowest concentration of compounds that completely inhibited bacterial growth in the micro-wells, was judged from no color change when the bacterial growth in blank wells was sufficient [
18].
4.4. Structures and MICs of plant flavonoids reported
As the antimicrobial activities of a certain compound against different pathogenic strains of Gram-negative bacteria were varied, the data collection focused on plant flavonoids having inhibitory activities to E. coli, a representative Gram-negative bacterium. The structures, the MICs to E. coli, and other related information of plant flavonoids were unsystematically searched from Google academic search engine, and several databases SciFinder, Medline, Elsevier, ACS, ScienceDirect, Wiley Online Library, and Springer-Link, using keywords flavonoid and E. coli, or and antimicrobial, or and antibacterial, and or and Gram-negative bacteria. Furthermore, the relevant references in the obtained literature were also tracked. Next, the structures, the MIC values to E. coli, and other related information of plant flavonoids were collected from the obtained literature. Finally, the structures of collected compounds were drawn using software ChemBioDraw Ultra 14.0.
4.5. Correlation and regression analyses
The physicochemical parameters ACD/LogP and LogD
7.40 of plant flavonoids measured in section 4.3 or previously reported were calculated using software ACD/Labs 6.0. Subsequently, the antimicrobial activities MICs (μM), and the LogP and LogD
7.40 values of flavonoids measured or reported were respectively listed in an excel table. After this, the correlation analyses between the calculated LogP or LogD
7.40 values (x) and the MICs (y) of plant flavonoids in the table were performed using Microsoft Excel software. Simultaneously, the corresponding regression equations were established for further verifications, and the value of correlation coefficient (r) was also calculated. It is noting that those compounds (
Table 2) without accurate MIC values (such as more than the measuring concentration) were not considered for the regression analyses, while their related information can be used for the following discussion.
The correlation between the antimicrobial activities MICs of plant flavonoids to Gram-negative bacteria and their physicochemical parameters LogP and LogD
7.40 was validated by r-test, and the coefficient of determination (R
2) was also calculated for judging the fitting precisions according to equation (5) in previous work [
16].
4.7. Comparision for the characters of regression curves
Referred previous work [
16], the MIC was further transformed to the log
10(MIC), and subsequently the regression analysis between the log
10(MIC) (y) and the LogP or LogD
7.40 (x) was further performed. Subsequently, the characters of their regression curves were compared with those of the cell envelope of Gram-negative and -positive bacteria for exploring possible reasons of the antibacterial difference of plant flavonoids to gram-negative and -positive bacteria.
5. Conclusions
In summary, the antibacterial activities of plant flavonoids to Gram-negative bacteria are related to their lipophilicities, while they cannot be statistically predicted from their ACD/LogP values like to Gram-positive bacteria. Simultaneously, the antibacterial activities of plant flavonoids to Gram-negative bacteria are generally weak, and most are greater than those to Gram-positive ones when the lipophilic parameter LogP values are less than about 3.0, but less than which when their LogP values are greater than about 3.6. Different from mainly acting on the cell membrane of Gram-positive bacteria, there are probably multiple action modes of plant flavonoids against Gram-negative bacteria, and among them the cell membrane is also an important action. Another, the DNA gyrase should be another important target of plant flavonoids against Gram-negative bacteria.
Supplementary Materials
following supporting information can be downloaded at the website of this paper posted on Preprints.org, Figure S1: Chemical structure of fifty-two plant flavonoids reported; Figure S2: Another effective regression curve for equation (6) established from the LogP of reported plant flavonoids and their log
10(MIC) values; Figure S3: The Polynomial regression analyses for the LogP (
x) of plant flavonoids in Table S2 and the log
10(IC
50) (
y) to DNA gyrase; Table S1: Comparison for the tested and reported MIC values of nine compounds;
Table S2: The ACD/LogP values and IC
50 to DNA gyrase of fourteen flavonoids reported, together with their MICs to
E.
coliss [
35]; Table S3: Regression equations established from the LogP (
x) values of flavonoids and the IC
50 or Log
10(IC
50) (μM) values (
y) of these flavonoids to DNA gyrase.
Author Contributions
Conceptualization, G.Y.; methodology, G.Y.; software, G.Y.; validation, G.Y., Y.Y. and X.X.; formal analysis, G.Y., Y.Y. and X.X.; investigation, Y.Y., X.X., A.F., L.Z., G.Y., F.L. and Y.W.; resources, G.Y.; data curation, Y.Y., X.X., L.Z., G.Y., F.L. and Y.W.; writing—original draft preparation, G.Y., Y.Y., A.F. and X.X.; writing—review and editing, G.Y., Y.Y., A.F., L.Z. and X.X.; visualization, G.Y. and Y.W.; supervision, G.Y.; project administration, G.Y.; funding acquisition, G.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Natural Science Foundation of China, Grant numbers 82360691, 82073745 and 81960636.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Acknowledgments
Authors are thankful to the financial supports from National Natural Science Foundation of China, Grant numbers 82360691, 82073745 and 81960636.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Wool, P.R.E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Laxminarayan, R.; Sridhar, D.; Blaser, M.; Wang, M.; Woolhouse, M. Achieving global targets for antimicrobial resistance. Science 2016, 353, 874–875. [Google Scholar] [CrossRef]
- Kurosu, M.; Siricilla, S.; Mitachi, K. Advances in MRSA drug discovery: where are we and where do we need to be? Exp. Opin. Drug Discov. 2013, 8, 1095–1116. [Google Scholar] [CrossRef]
- Xu, X.; Xu, L.; Yuan, G.; Wang, Y.; Qu, Y.; Zhou, M. Synergistic combination of two antimicrobial agents closing each other's mutant selection windows to prevent antimicrobial resistance. Sci. Rep. 2018, 8, 7237. [Google Scholar] [CrossRef] [PubMed]
- Liang, M.; Ge, X.; Xua, H.; Ma, K.; Zhang, W.; Zan, Y.; Efferth, T.; Xue, Z.; Hua, X. Phytochemicals with activity against methicillin-resistant Staphylococcus aureus. Phytomedicine 2022, 100, 154073. [Google Scholar] [CrossRef] [PubMed]
- Panda, L; Duarte-Sierra, A. Recent advancements in enhancing antimicrobial activity of plant-derived polyphenols by biochemical means. Horticulturae 2022, 8, 401. [Google Scholar] [CrossRef]
- Xie, Y.; Yang, W.; Tang, F.; Chen, X.; Ren, L. Antibacterial activities of flavonoids: structure-activity relationship and mechanism. Curr. Med. Chem. 2015, 22, 132–149. [Google Scholar] [CrossRef]
- Tan, Z.; Deng, J.; Ye, Q.; Zhang, Z. The antibacterial activity of natural-derived flavonoids. Curr. Top. Med. Chem. 2022, 22, 1009–1019. [Google Scholar] [CrossRef]
- Song, L.; Hu, X.; Ren, X.; Liu, J.; Liu, X. Antibacterial modes of herbal flavonoids combat resistant bacteria. Front. Pharmacol. 2022, 13, 873374. [Google Scholar] [CrossRef]
- Wu, S.C.; Yang, Z.Q.; Liu, F.; Peng, W.J.; Qu, S.Q.; Li, Q.; Song, X.B.; Zhu, K.; Shen, J.Z. Antibacterial effect and mode of action of flavonoids from licorice against methicillin-resistant Staphylococcus aureus. Front. Microbiol. 2019, 10, 2489. [Google Scholar] [CrossRef]
- Zhou, K.; Yang, S.; Li, S.M. Naturally occurring prenylated chalcones from plants: structural diversity, distribution, activities and biosynthesis. Nat. Prod. Rep. 2021, 38, 2236–2260. [Google Scholar] [CrossRef] [PubMed]
- Górniak, I.; Bartoszewski, R.; Króliczewski, J. Comprehensive review of antimicrobial activities of plant flavonoids. Phytochem. Rev. 2019, 18, 241–272. [Google Scholar] [CrossRef]
- Farhadi, F.; Khameneh, B.; Iranshahi, M.; Iranshahy, M. Antibacterial activity of flavonoids and their structure-activity relationship: An update review. Phytother. Res. 2019, 33, 13–40. [Google Scholar] [CrossRef]
- Panda, L.; Duarte-Sierra, A. Recent advancements in enhancing antimicrobial activity of plant-derived polyphenols by biochemical means. Horticulturae 2022, 8, 401. [Google Scholar] [CrossRef]
- Shamsudin, N.F.; Ahmed, Q.U.; Mahmood, S.; Ali Shah, S.A.; Khatib, A.; Mukhtar, S.; Alsharif, M.A.; Parveen, H.; Zakaria, Z.A. Antibacterial effects of flavonoids and their structure-activity relationship study: A comparative interpretation. Molecules 2022, 27, 1149. [Google Scholar] [CrossRef] [PubMed]
- Yuan, G.; Xia, X.; Guan, Y.; Yi, H.; Lai, S.; Sun, Y.; Cao, S. Antimicrobial quantitative relationship and mechanism of plant flavonoids to gram-positive bacteria. Pharmaceuticals 2022, 15, 1190. [Google Scholar] [CrossRef] [PubMed]
- Yuan, G.; Guan, Y.; Yi, H.; Lai, S.; Sun, Y.; Cao, S. Antibacterial activity and mechanism of plant flavonoids to gram-positive bacteria predicted from their lipophilicities. Sci. Rep. 2021, 11, 10471. [Google Scholar] [CrossRef]
- Zhang, L.; Yan, Y.; Zhu, J.; Xia, X.; Yuan, G.; Li, S.; Deng, B.; Luo, X. Quinone pool, a key target of plant flavonoids inhibiting gram-positive bacteria. Molecules 2023, 28, 4972. [Google Scholar] [CrossRef]
- Mbaveng, A.T.; Sandjo, L.P.; Tankeo, S.B.; Ndifor, A.R.; Pantaleon, A.; Nagdjui, B.T.; Kuete, V. Antibacterial activity of nineteen selected natural products against multi-drug resistant Gram-negative phenotypes. SpringerPlus 2015, 4, 823. [Google Scholar] [CrossRef]
- Chukwujekwu, J.C.; Van Heerden, F.R.; Van Staden, J. Antibacterial activity of flavonoids from the stem bark of Erythrina caffra Thunb. Phytother. Res. 2011, 25, 46–48. [Google Scholar] [CrossRef]
- Mandalari, G.; Bennett, R.N.; Bisignano, G.; Trombetta, D.; Saija, A.; Faulds, C.B.; Gasson, M.J.; Narbad, A. Antimicrobial activity of flavonoids extracted from bergamot (Citrus bergamia Risso) peel, a byproduct of the essential oil industry. J. Appl. Microbiol. 2007, 103, 2056–2064. [Google Scholar] [CrossRef] [PubMed]
- Martini, N.D.; Katerere, D.R.; Eloff, J.N. Biological activity of five antibacterial flavonoids from Combretum erythrophyllum (Combretaceae). J. Ethnopharmacol. 2004, 93, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Bagla, V.P.; McGaw, L.J.; Elgorashi, E.E.; Eloff, J.N. Antimicrobial activity, toxicity and selectivity index of two biflavonoids and a flavone isolated from Podocarpus henkelii (Podocarpaceae) leaves. BMC Complement. Altern. Med. 2014, 14, 383. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Zang, X.; He, M.; Pan, S.; Xu, X. Structure-activity relationship of flavonoids on their anti-Escherichia coli activity and inhibition of DNA gyrase. J. Agric. Food. Chem. 2013, 61, 8185–8190. [Google Scholar] [CrossRef] [PubMed]
- Edziri, H.; Mastouri, M.; Mahjoub, M.A.; Mighri, Z.; Mahjoub, A.; Verschaeve, L. Antibacterial, antifungal and cytotoxic activities of two flavonoids from Retama raetam flowers. Molecules 2012, 17, 7284–7293. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, M.S.; Abdelkader, K.; Gomaa, H.A.M.; Batubara, A.S.; Gamal, M.; Sayed, A.M. Mechanistic study of the antibacterial potential of the prenylated flavonoid auriculasin against Escherichia coli. Arch. Pharm. (Weinheim) 2022, 355, e2200360. [Google Scholar] [CrossRef] [PubMed]
- Eumkeb, G.; Siriwong, S.; Thumanu, K. Synergistic activity of luteolin and amoxicillin combination against amoxicillin-resistant Escherichia coli and mode of action. J. Photochem. Photobiol. B. 2012, 117, 247–253. [Google Scholar] [CrossRef] [PubMed]
- Jaisinghani, R.N. Antibacterial properties of quercetin. Microbiol. Res. 2017, 8, 6877. [Google Scholar] [CrossRef]
- Dzoyem, J.P.; Melong, R.; Tsamo, A.T.; Tchinda, A.T.; Kapche, D.G.; Ngadjui, B.T.; McGaw, L.J.; Eloff, J.N. Cytotoxicity, antimicrobial and antioxidant activity of eight compounds isolated from Entada abyssinica (Fabaceae). BMC Res. Notes 2017, 10, 118. [Google Scholar] [CrossRef]
- Eumkeb, G.; Siriwong, S.; Phitaktim, S.; Rojtinnakorn, N.; Sakdarat, S. Synergistic activity and mode of action of flavonoids isolated from smaller galangal and amoxicillin combinations against amoxicillin-resistant Escherichia coli. J. Appl. Microbiol. 2012, 112, 55–64. [Google Scholar] [CrossRef]
- Ozçelik, B.; Orhan, I.; Toker, G. Antiviral and antimicrobial assessment of some selected flavonoids. Z. Naturforsch. C J. Biosci. 2006, 61, 632–638. [Google Scholar] [CrossRef] [PubMed]
- Kuete, V.; Simo, I.K.; Ngameni, B.; Bigoga, J.D.; Watchueng, J.; Kapguep, R.N.; Etoa, F.X.; Tchaleu, B.N.; Beng, V.P. Antimicrobial activity of the methanolic extract, fractions and four flavonoids from the twigs of Dorstenia angusticornis Engl. (Moraceae). J. Ethnopharmacol. 2007, 112, 271–277. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, M. de los A.; Zarelli, V.E.; Pappano, N.B.; Debattista, N.B. Bacteriostatic action of synthetic polyhydroxylated chalcones against Escherichia coli. Biocell. 2004, 28, 31–34. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Liu, Y.; Li, T.; Liu, X.; Hao, Z.; Ding, S.; Panichayupakaranant, P.; Zhu, K.; Shen, J. Plant natural flavonoids against multidrug resistant pathogens. Adv. Sci. 2021, 8, e2100749. [Google Scholar] [CrossRef] [PubMed]
- Ohemeng, K.A.; Schwender, C.F.; Fu, K.P.; Barrett, J.F. DNA gyrase inhibitory and antibacterial activity of some flavones (l). Bioorg. Med. Chem. Lett. 1993, 3, 225–230. [Google Scholar] [CrossRef]
- Fang, Y.; Lu, Y.; Zang, X.; Wu, T.; Qi, X.; Pan, S.; Xu, X. 3D-QSAR and docking studies of flavonoids as potent Escherichia coli inhibitors. Sci. Rep. 2016, 6, 23634. [Google Scholar] [CrossRef] [PubMed]
- Verghese, J.; Nguyen, T.; Oppegard, L.M.; Seivert, L.M.; Hiasa, H.; Ellis, K.C. Flavone-based analogues inspired by the natural product simocyclinone D8 as DNA gyrase inhibitors. Bioorg. Med. Chem. Lett. 2013, 23, 5874–5877. [Google Scholar] [CrossRef]
- Wu, T.; Zang, X.; He, M.; Pan, S.; Xu, X. Structure-activity relationship of flavonoids on their anti-Escherichia coli activity and inhibition of DNA gyrase. J. Agric. Food. Chem. 2013, 61, 8185–8190. [Google Scholar] [CrossRef]
- Clinical and Laboratory and Standards Institute (CLSI). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, 10th ed.; Approved Standards, CLSI document M07-A10; Clinical and Laboratory and Standards Institute: Wayne, PA, USA, 2015. [Google Scholar]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).