Submitted:
03 January 2024
Posted:
04 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Applications of AI in Cancer Imaging
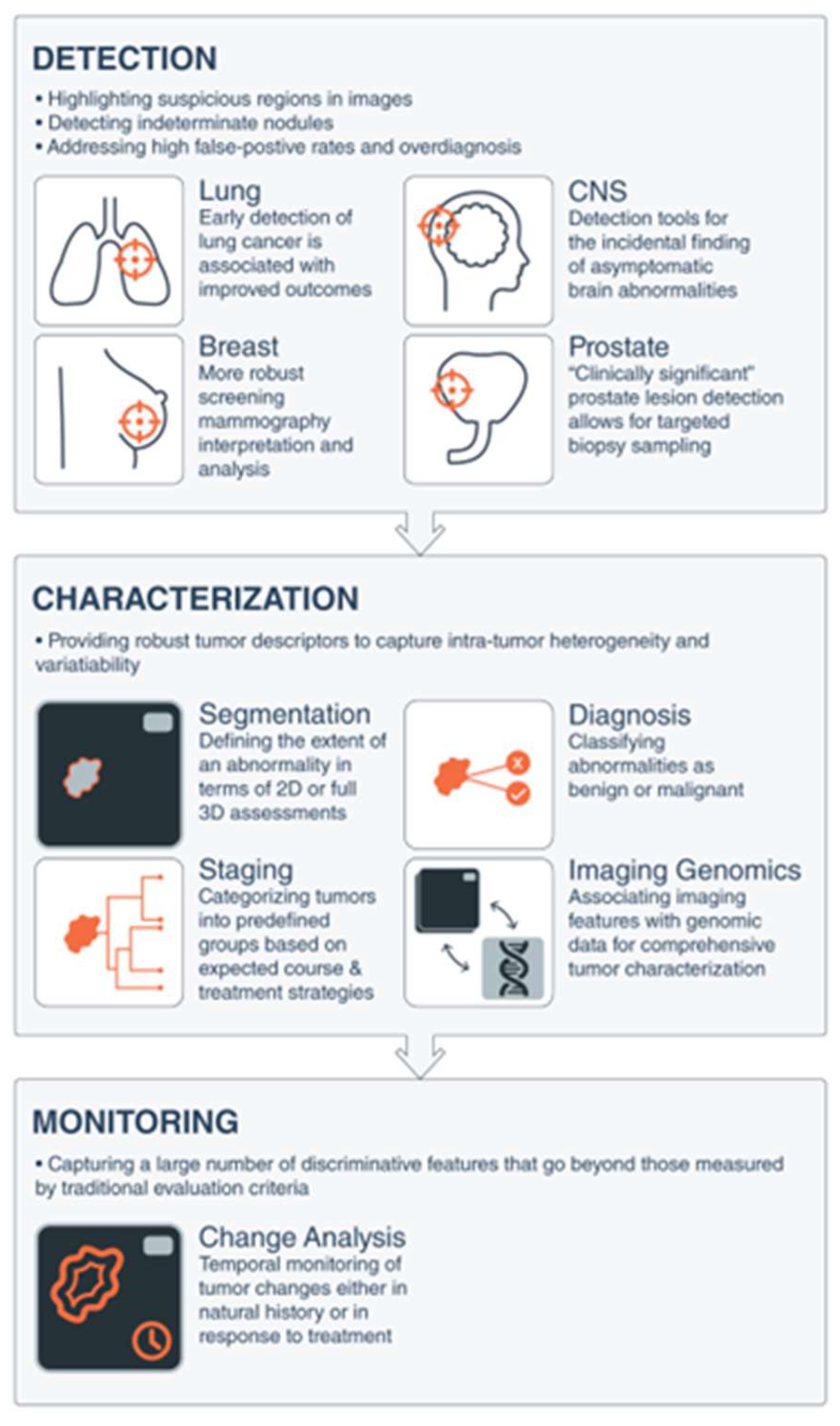
2.1. AI and Radiologic Cancer Screening and Detection
- 1)
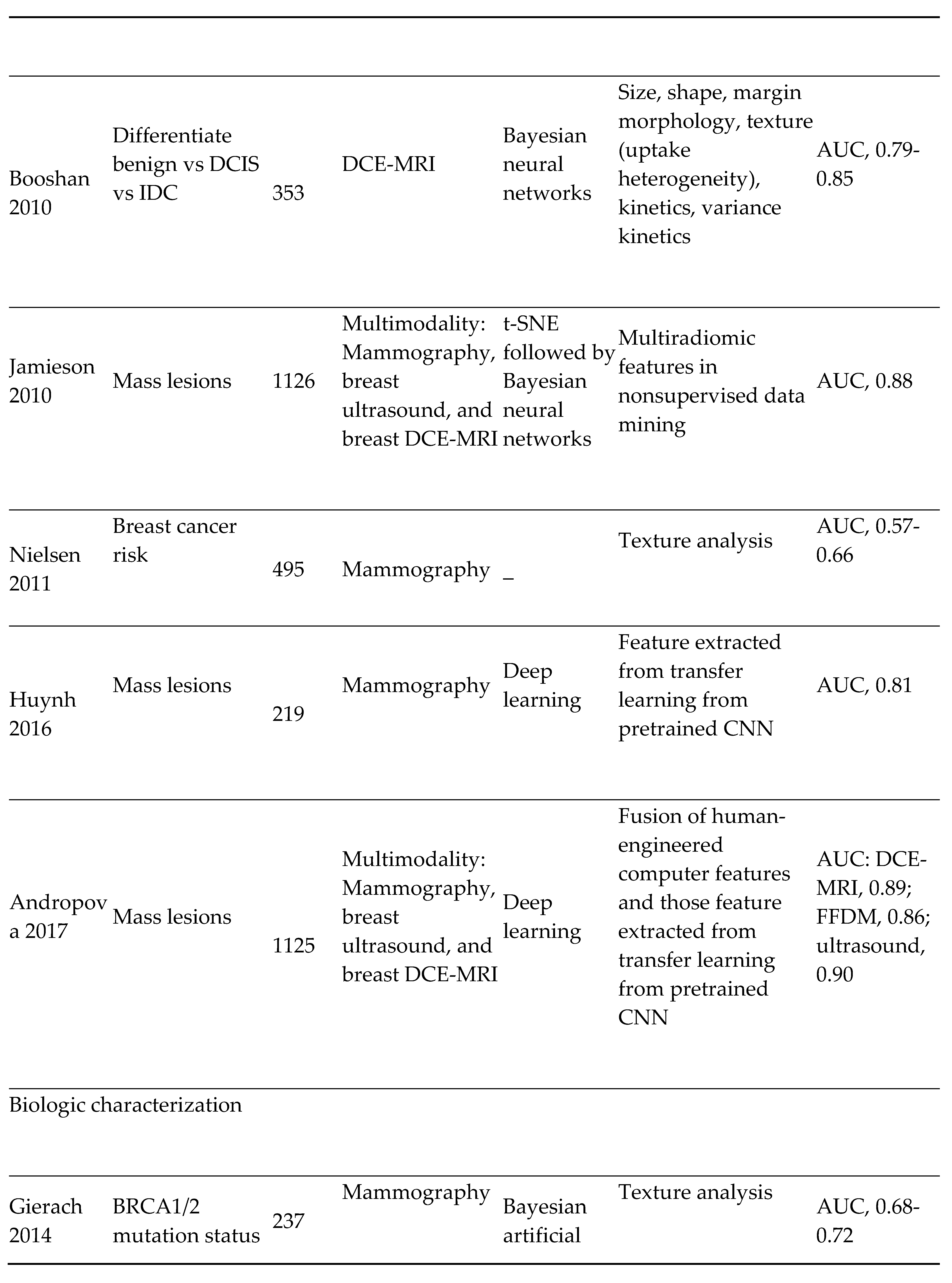
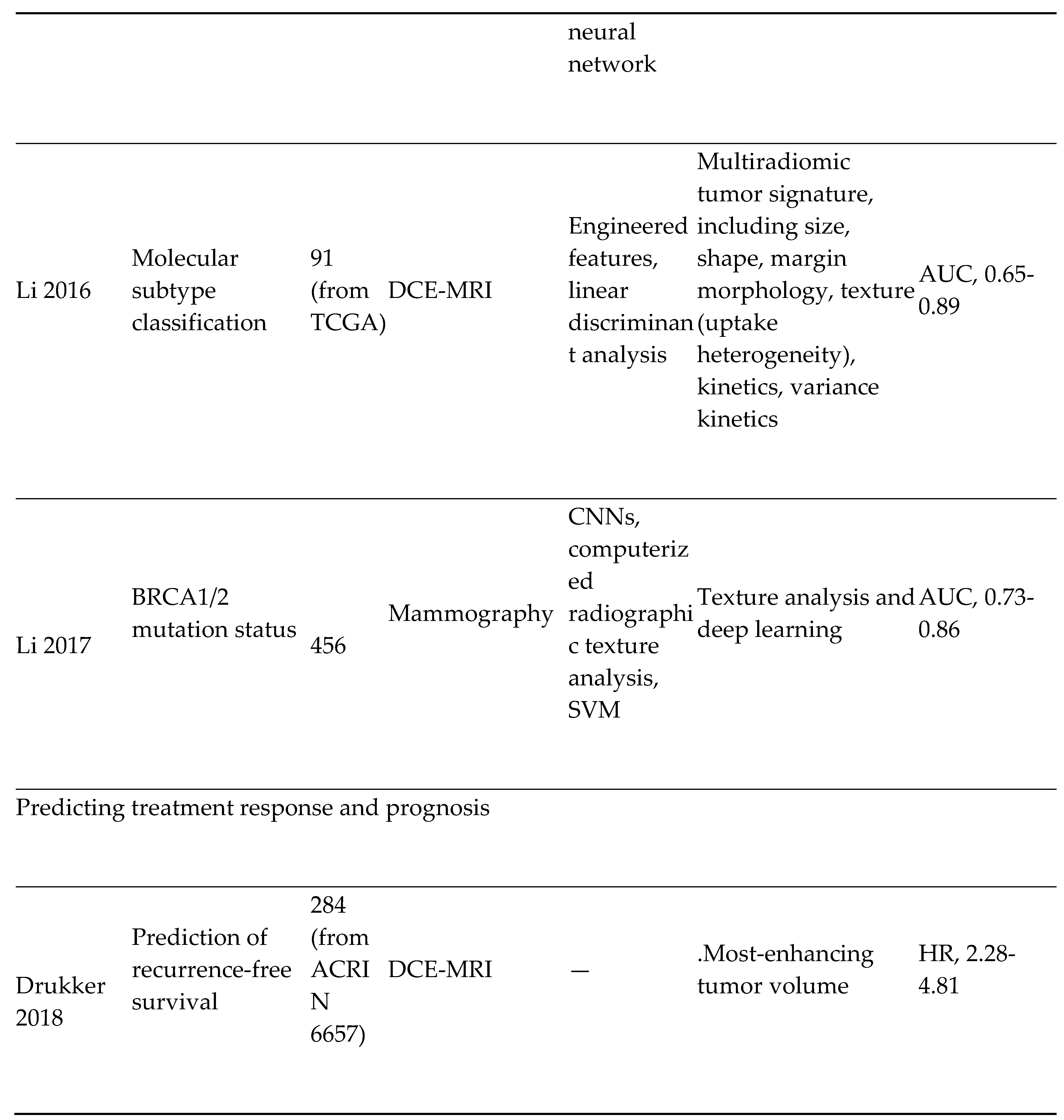
- Breast Cancer Imaging: The most diagnosed cancer among women in the United States is breast cancer. It also accounts for the 2nd highest number of cancer-related deaths. [54] The introduction of mammography for breast cancer screening has significantly improved early cancer detection and decreased morbidity and mortality overall. However, therapeutic response to breast cancer is highly variable and depends on the presence or absence of specific receptors on the tumor. These receptors include; estrogen (ER), progesterone (PR), and Human Epidermal Receptor 2 (HER 2) receptors. Triple receptor-negative breast cancers are more difficult to identify on mammography as they lack the typical characteristics of the tumor. [55] Consequently, triple-negative tumors are more likely to be detected later and carry a worse prognosis.
|
- 2)
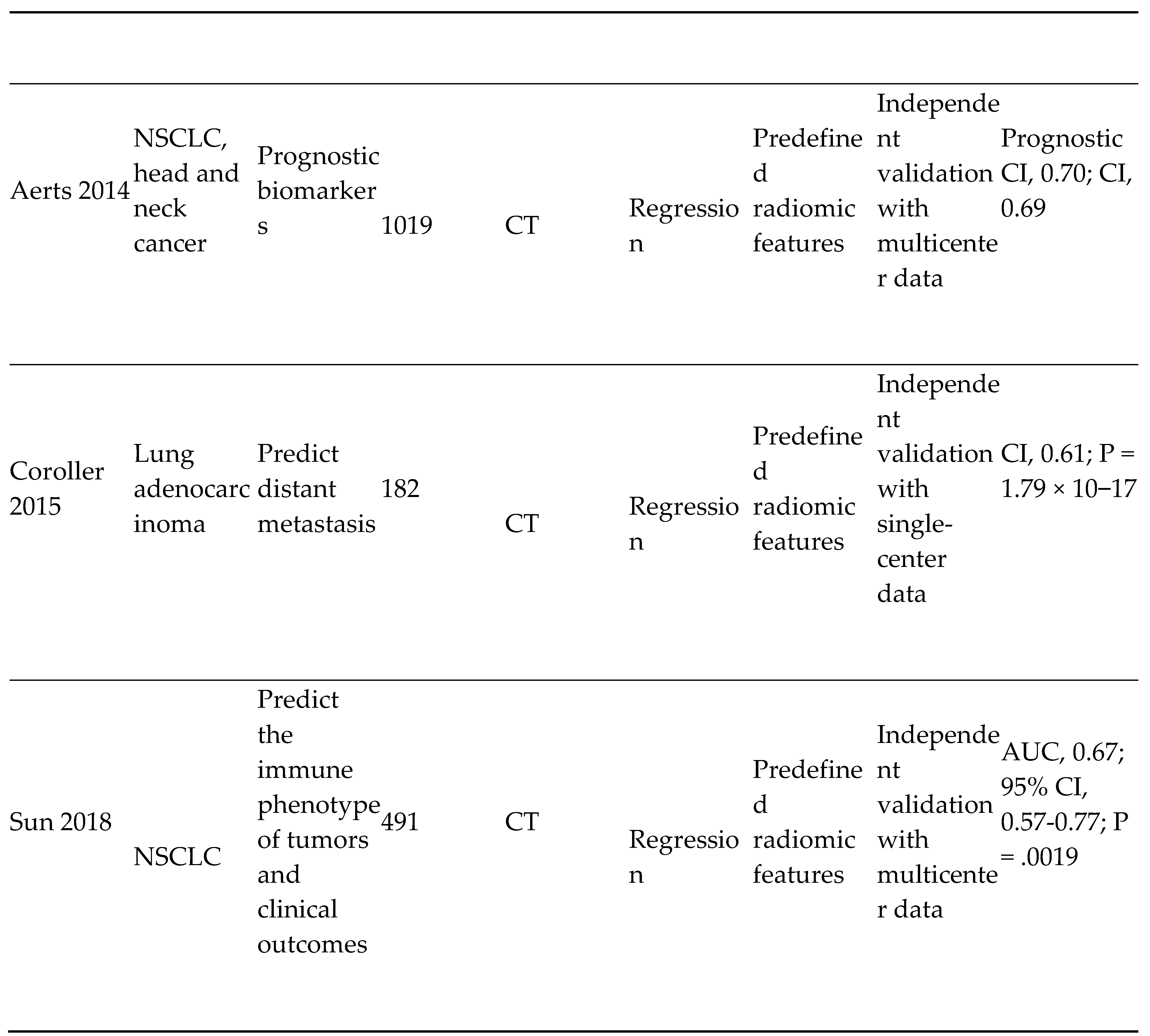
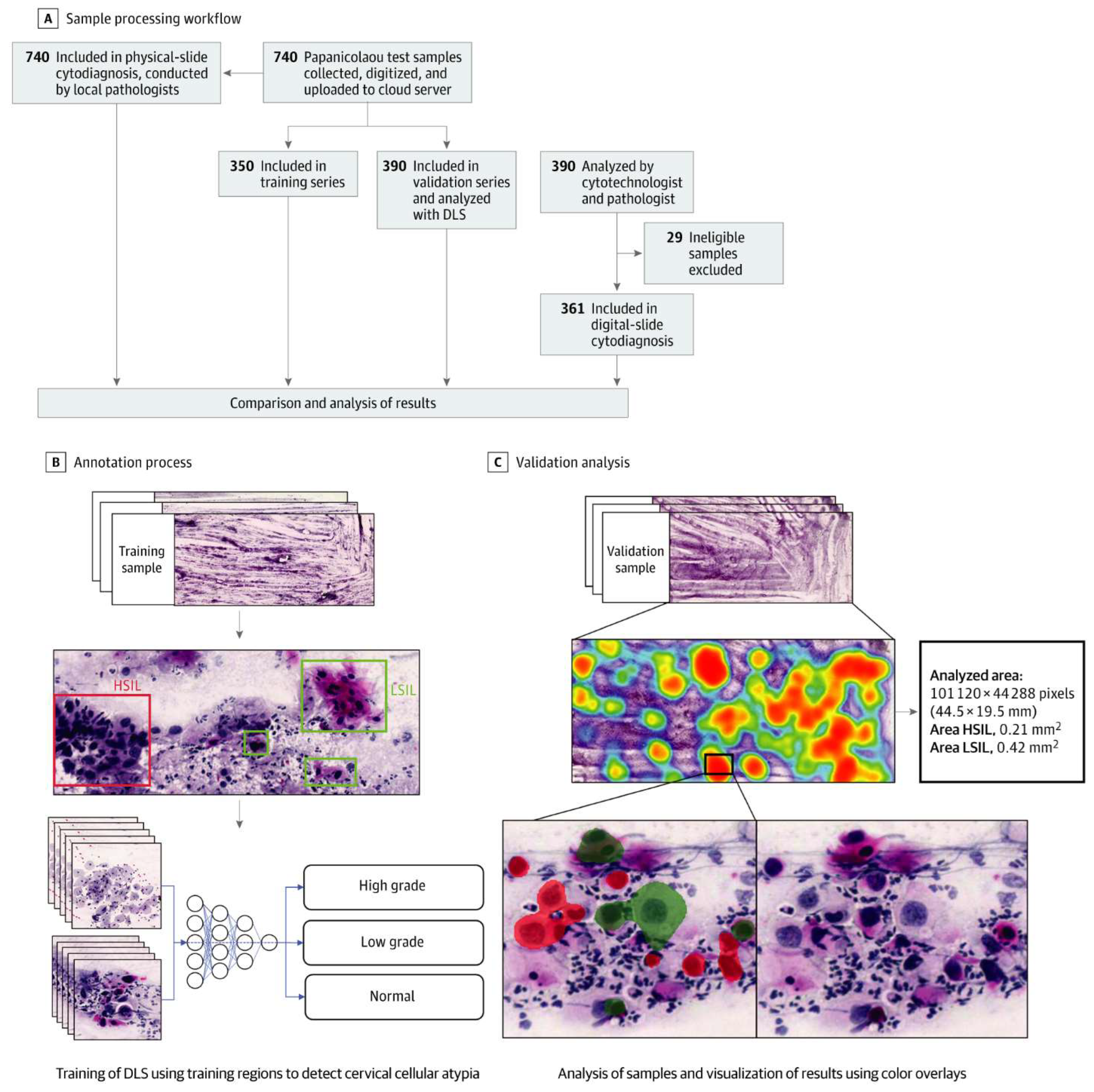
- Cervical cancer screening: Researchers at Karolinska Institute in Sweden detected precursors to cervical cancer in women in resource-limited settings using artificial intelligence and mobile digital microscopy. [10] In this diagnostic study, cervical smears from 740 HIV-positive women aged between 18 and 64 were collected. The smears were then digitized with a portable slide scanner, uploaded to a cloud server using mobile networks, and used to train and validate a deep learning system (DLS) to detect atypical cervical cells. (Figure 3) Sensitivity for detection of atypia was high (96%-100%), with higher specificity for high-grade lesions (93%-99%) than for low-grade lesions (82%-86%), and no slides manually classified as the high grade was incorrectly classified as negative.
- 3)
- 4)
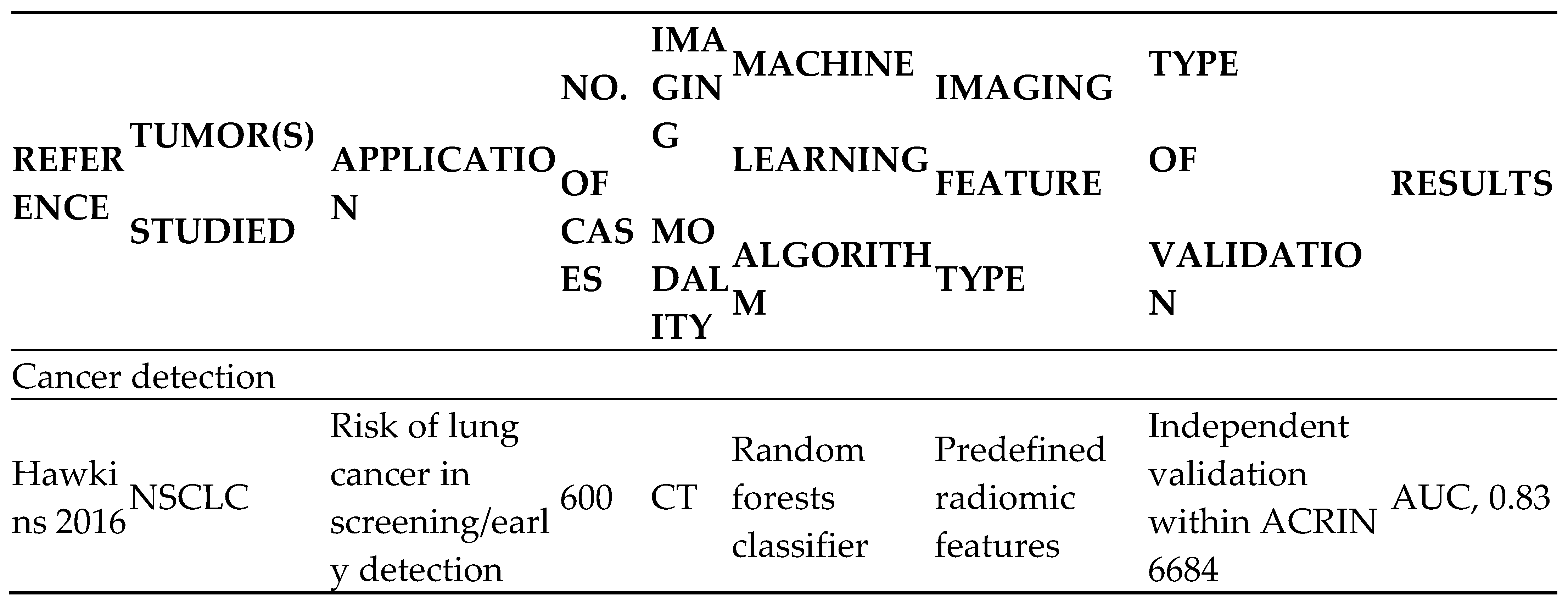
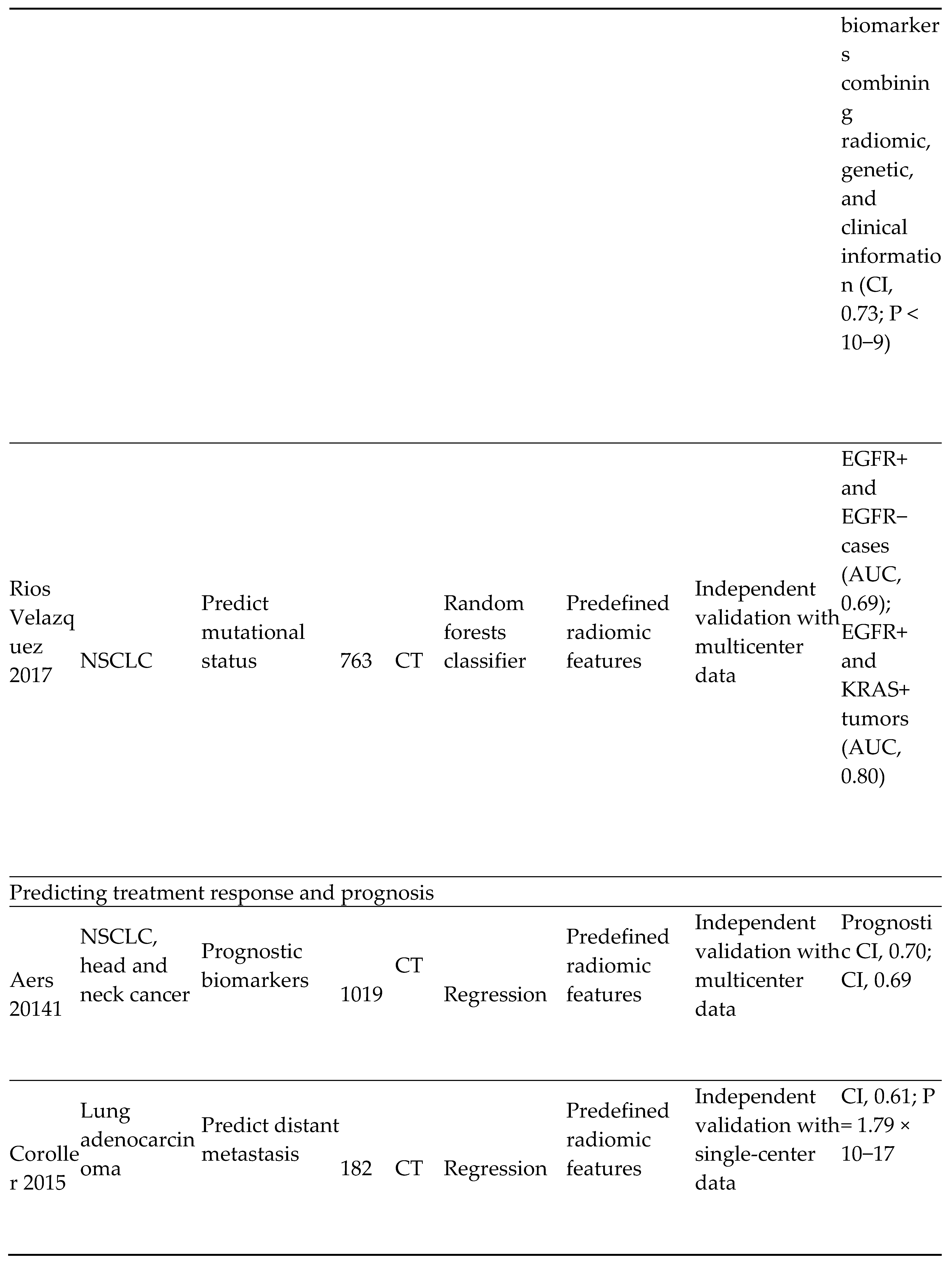
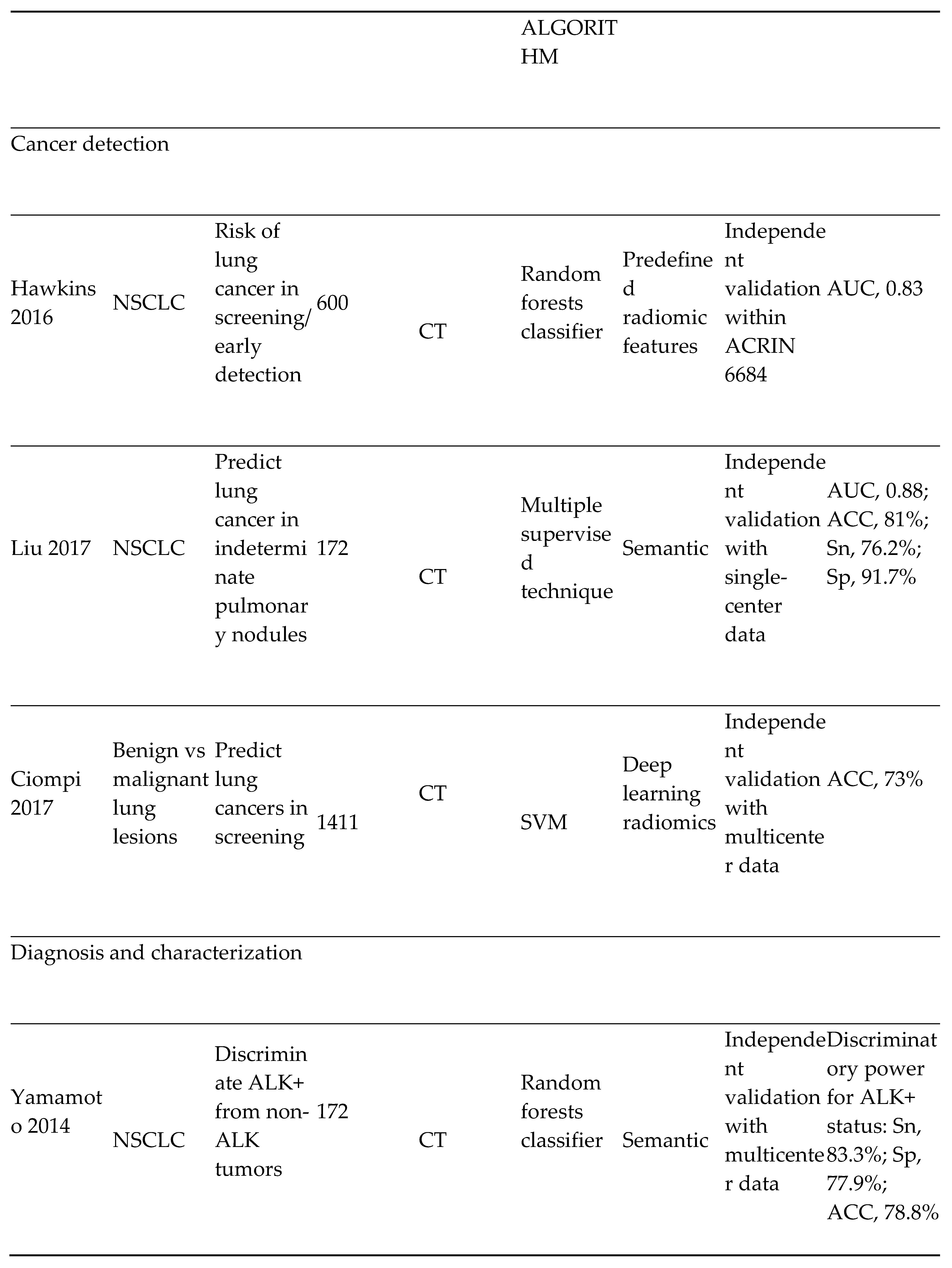
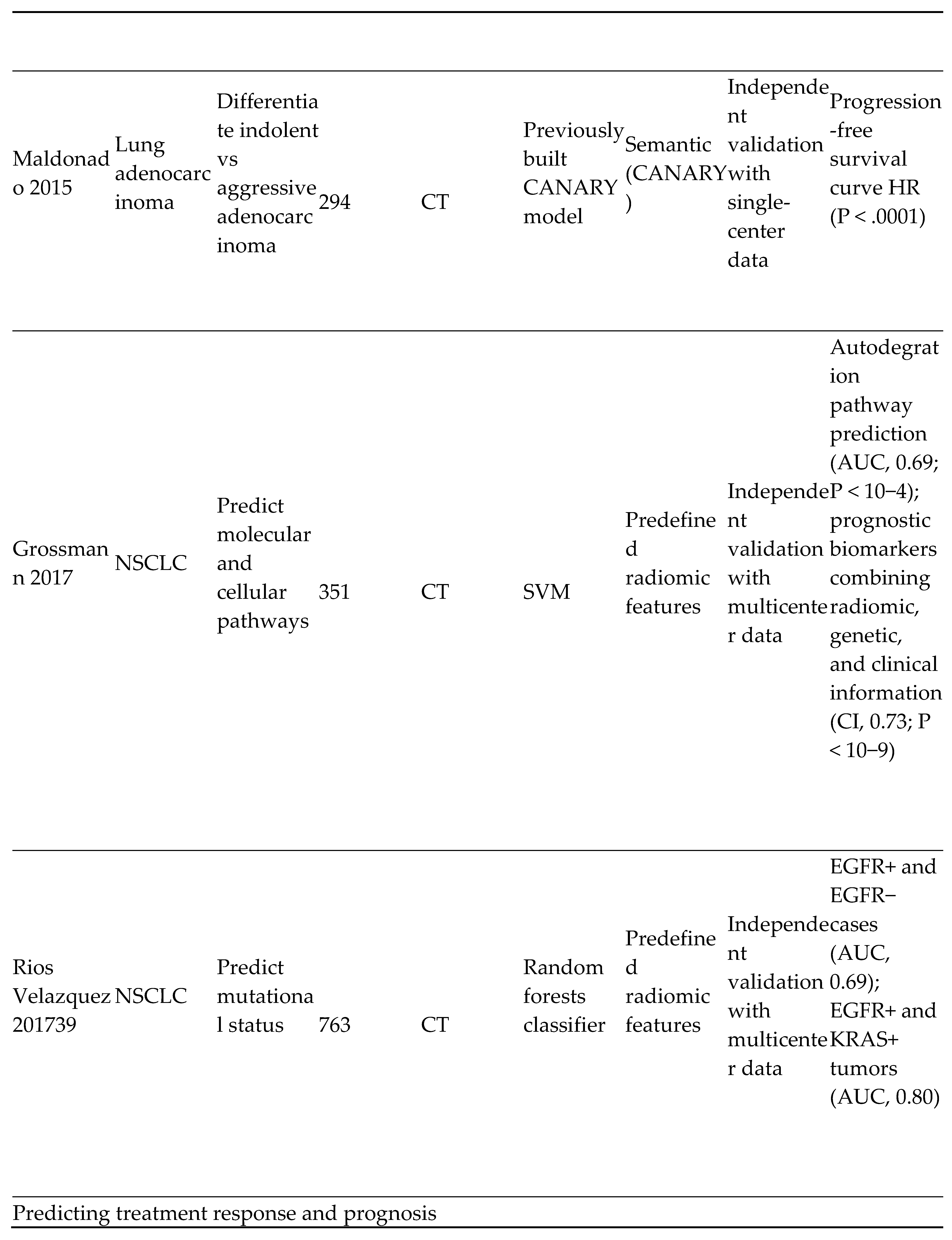
- Lung cancer screening and detection
|
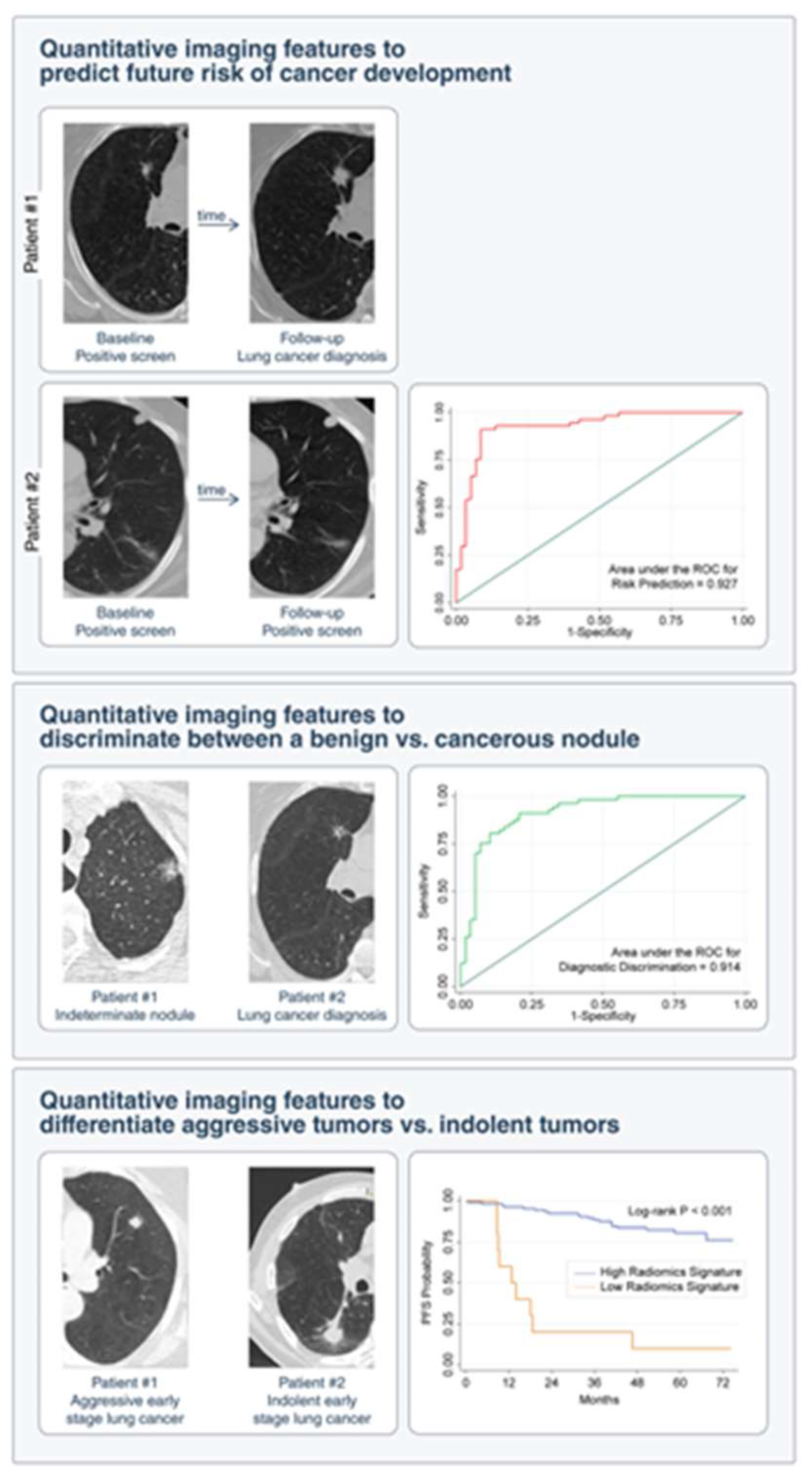
- 5)
- AI and Prostate cancer screening and detection:
- 6)
- Imaging of CNS Tumors:
- A)
- Ensuring that tumor diagnosis is accurate enough to optimize clinical decisions.
- B)
- Ability to distinguish signal characteristics of surrounding neural tissue from those of the primary tumor throughout the clinical surveillance period of the tumor.
- C)
- Ability to map the genotypes of tumors based on their phenotypic manifestations during imaging.
|
- COVER LETTER
Conflicts of Interest
Funding
References
- What is generative AI? (n.d.). McKinsey & Company. Available online: https://www.mckinsey.com/featured-insights/mckinsey-explainers/what-is-generative-ai.
- Wikipedia contributors. Artificial intelligence. Wikipedia. Available online: https://en.wikipedia.org/wiki/Artificial_intelligence (accessed on 20 February 2023).
- Cancer Imaging Program (CIP). (n.d.). Available online: https://imaging.cancer.gov/imaging_basics/cancer_imaging/uses_of_imaging.htm.
- Can Artificial Intelligence Help See Cancer in New Ways? National Cancer Institute. Available online: https://www.cancer.gov/news-events/cancer-currents-blog/2022/artificial-intelligence-cancer-imaging (accessed on 22 March 2022).
- Cancer Screening Overview (PDQ®)–Patient Version. National Cancer Institute. Available online: https://www.cancer.gov/about-cancer/screening/patient-screening-overview-pdq (accessed on 19 August 2020).
- X, S. AI tool improves breast cancer detection on mammography. Available online: https://medicalxpress.com/news/2020-11-ai-tool-breast-cancer-mammography.html (accessed on 4 November 2020).
- Rawashdeh, M.A.; Lee, W.B.; Bourne, R.M.; Ryan, E.A.; Pietrzyk, M.W.; Reed, W.M.; Heard, R.C.; Black, D.A.; Brennan, P.C. Markers of Good Performance in Mammography Depend on Number of Annual Readings. Radiology 2013, 269, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Pacilè, S.; Lopez, J.; Chone, P.; Bertinotti, T.; Grouin, J.M.; Fillard, P. Improving Breast Cancer Detection Accuracy of Mammography with the Concurrent Use of an Artificial Intelligence Tool. Radiol. Artif. Intell. 2020, 2, e190208. [Google Scholar] [CrossRef] [PubMed]
- Sinichkina, E.; MammoScreen is FDA-cleared and available for sale in the US - MammoScreen®. MammoScreen®. Available online: https://www.mammoscreen.com/mammoscreen-fda-cleared-available-for-sale-us (accessed on 17 November 2020).
- Holmström, O.; Linder, N.; Kaingu, H.; Mbuuko, N.; Mbete, J.; Kinyua, F.; Törnquist, S.; Muinde, M.; Krogerus, L.; Lundin, M.; et al. Point-of-Care Digital Cytology With Artificial Intelligence for Cervical Cancer Screening in a Resource-Limited Setting. JAMA Netw. Open 2021, 4, e211740–e211740. [Google Scholar] [CrossRef]
- Shalev-Shwartz, S.; Ben-David, S. Understanding Machine Learning: From Theory to Algorithms; Cambridge University Press: Cambridge, UK, 2014. [Google Scholar]
- Ruffle, J.K.; Farmer, A.D.; Aziz, Q. Artificial Intelligence-Assisted Gastroenterology— Promises and Pitfalls. Am. J. Gastroenterol. 2018, 114, 422–428. [Google Scholar] [CrossRef] [PubMed]
- Hamet, P.; Tremblay, J. Artificial intelligence in medicine. Metabolism 2017, 69, S36–S40. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Maida, M.; Macaluso, F.S.; Ianiro, G.; Mangiola, F.; Sinagra, E.; Hold, G.; Maida, C.; Cammarota, G.; Gasbarrini, A.; Scarpulla, G. Screening of colorectal cancer: present and future. Expert Rev. Anticancer. Ther. 2017, 17, 1131–1146. [Google Scholar] [CrossRef] [PubMed]
- Vining, D.J.; Gelfand, D.W.; Bechtold, R.E.; Scharling, E.S.; Grishaw, E.K.; Shifrin, R.Y. Technical Feasibility of Colon Imaging with Helical CT and Virtual Reality. AJR Am. J. Roentgenol. 1994, 162, 104. [Google Scholar]
- Song, B.; Zhang, G.; Lu, H.; Wang, H.; Zhu, W.; J Pickhardt, P.; Liang, Z. Volumetric texture features from higher-order images for diagnosis of colon lesions via CT colonography. Int. J. Comput. Assist. Radiol. Surg. 2014, 9, 1021–1031. [Google Scholar] [CrossRef]
- Mitsala, A.; Tsalikidis, C.; Pitiakoudis, M.; Simopoulos, C.; Tsaroucha, A.K. Artificial Intelligence in Colorectal Cancer Screening, Diagnosis and Treatment. A New Era. Curr. Oncol. 2021, 28, 1581–1607. [Google Scholar] [CrossRef]
- Taylor, S.A.; Iinuma, G.; Saito, Y.; Zhang, J.; Halligan, S. CT colonography: computer-aided detection of morphologically flat T1 colonic carcinoma. Eur. Radiol. 2008, 18, 1666–1673. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, M. J., Winawer, S. J., Zauber, A. G., Bushey, M. T., Sternberg, S. S., Gottlieb, L. S., Bond, J. H., Waye, J. D., & Schapiro, M. Flat adenomas in the National Polyp Study: Is there an increased risk for high-grade dysplasia initially or during surveillance? Clinical Gastroenterology and Hepatology 2004, 2, 905–911.
- Cancer Imaging Program (CIP). (n.d.). Available online: https://imaging.cancer.gov/imaging_basics/cancer_imaging/digital_mammography.htm.
- Neumann, H.; Kudo, S.; Vieth, M.; Neurath, M.F. Real-time in vivo histologic examination using a probe-based endocytoscopy system for differentiating duodenal polyps. Endoscopy 2013, 45, E53–E54. [Google Scholar] [CrossRef] [PubMed]
- Takeda, K.; Kudo, S.-E.; Mori, Y.; Misawa, M.; Kudo, T.; Wakamura, K.; Katagiri, A.; Baba, T.; Hidaka, E.; Ishida, F.; et al. Accuracy of diagnosing invasive colorectal cancer using computer-aided endocytoscopy. Endoscopy 2017, 49, 798–802. [Google Scholar] [CrossRef] [PubMed]
- Neumann, H.; Kiesslich, R.; Wallace, M.B.; Neurath, M.F. Confocal Laser Endomicroscopy: Technical Advances and Clinical Applications. Gastroenterology 2010, 139, 388–392. [Google Scholar] [CrossRef]
- Ştefănescu, D.; Streba, C.; Cârţână, E.T.; Săftoiu, A.; Gruionu, G.; Gruionu, L.G. Computer Aided Diagnosis for Confocal Laser Endomicroscopy in Advanced Colorectal Adenocarcinoma. PLOS ONE 2016, 11, e0154863–e0154863. [Google Scholar] [CrossRef]
- Svoboda, E. Artificial intelligence is improving the detection of lung cancer. Nature 2020, 587, S20–S22. [Google Scholar] [CrossRef]
- Ardila, D.; Kiraly, A.P.; Bharadwaj, S.; Choi, B.; Reicher, J.J.; Peng, L.; Tse, D.; Etemadi, M.; Ye, W.; Corrado, G.; et al. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat. Med. 2019, 25, 954–961. [Google Scholar] [CrossRef]
- Lung Cancer: Screening. Available online: https://uspreventiveservicestaskforce.org/uspstf/recommendation/lung-cancer-screening (accessed on 9 March 2021).
- Bi, W.L.; Hosny, A.; Schabath, M.B.; Giger, M.L.; Birkbak, N.J.; Mehrtash, A.; Allison, T.; Arnaout, O.; Abbosh, C.; Dunn, I.F.; et al. Artificial intelligence in cancer imaging: Clinical challenges and applications. CA: A Cancer J. Clin. 2019, 69, 127–157. [Google Scholar] [CrossRef]
- Castellino, R.A. Computer aided detection (CAD): an overview. Cancer Imaging 2005, 5, 17–19. [Google Scholar] [CrossRef]
- Liang, M.; Tang, W.; Xu, D.M.; Jirapatnakul, A.C.; Reeves, A.P.; Henschke, C.I.; Yankelevitz, D. Low-Dose CT Screening for Lung Cancer: Computer-aided Detection of Missed Lung Cancers. Radiology 2016, 281, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Ambrosini R, Wang P, Kolar B, O’Dell W. Computer-aided detection of metastatic brain tumors using automated 3-D template matching [abstract]. Proc Intl Soc Mag Reson Med. 2008, 16, 3414.
- Cheng, H.; Cai, X.; Chen, X.; Hu, L.; Lou, X. Computer-aided detection and classification of microcalcifications in mammograms: a survey. Pattern Recognit. 2003, 36, 2967–2991. [Google Scholar] [CrossRef]
- Nishikawa, RM. Computer-aided detection and diagnosis. In: Bick U, Diekmann F, eds. Digital Mammography. Berlin, Germany: Springer, 2010, 85-106. Available online: https://link.springer.com/chapter/10.1007/978-3-540-78450-0_6.
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA: A Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- National Lung Screening Trial Research Team; Aberle, D. R.; Adams, A.M.; Berg, C.D.; Black, W.C.; Clapp, J.D.; Fagerstrom, R.M.; Gareen, I.F.; Gatsonis, C.; Marcus, P.M.; et al. Reduced Lung-Cancer Mortality with Low-Dose Computed Tomographic Screening. N. Engl. J. Med. 2011, 365, 395–409. [Google Scholar] [CrossRef]
- McKee, B.J.; Regis, S.; Borondy-Kitts, A.K.; Hashim, J.A.; French, R.J.; Wald, C.; McKee, A.B. NCCN Guidelines as a Model of Extended Criteria for Lung Cancer Screening. J. Natl. Compr. Cancer Netw. 2018, 16, 444–449. [Google Scholar] [CrossRef]
- American College of Radiology. Lung-RADS. Reston, VA: American College of Radiology; 2018. acr.org/Clinical-Resources/Reporting-and-Data-Systems/Lung-Rads. Accessed May 15, 2018.
- MacMahon, H.; Naidich, D.P.; Goo, J.M.; Lee, K.S.; Leung, A.N.C.; Mayo, J.R.; Mehta, A.C.; Ohno, Y.; Powell, C.A.; Prokop, M.; et al. Guidelines for Management of Incidental Pulmonary Nodules Detected on CT Images: From the Fleischner Society 2017. Radiology 2017, 284, 228–243. [Google Scholar] [CrossRef] [PubMed]
- Kaggle Inc. Data Science Bowl 2017. San Francisco, CA: Kaggle Inc; 2017. kaggle.com/c/data-science-bowl-2017. Accessed May 14, 2018.
- Jamal-Hanjani, M.; Wilson, G.A.; McGranahan, N.; Birkbak, N.J.; Watkins, T.B.K.; Veeriah, S.; Shafi, S.; Johnson, D.H.; Mitter, R.; Rosenthal, R.; et al. Tracking the Evolution of Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2017, 376, 2109–2121. [Google Scholar] [CrossRef]
- Aerts HJ, Velazquez ER, Leijenaar RT, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach [serial online]. Nat Commun. 2014, 5, 4006. [Google Scholar] [CrossRef]
- Coroller, T.P.; Grossmann, P.; Hou, Y.; Rios Velazquez, E.; Leijenaar, R.T.H.; Hermann, G.; Lambin, P.; Haibe-Kains, B.; Mak, R.H.; Aerts, H.J.W.L. CT-based radiomic signature predicts distant metastasis in lung adenocarcinoma. Radiother. Oncol. 2015, 114, 345–350. [Google Scholar] [CrossRef]
- Tunali, I.; Stringfield, O.; Guvenis, A.; Wang, H.; Liu, Y.; Balagurunathan, Y.; Lambin, P.; Gillies, R.J.; Schabath, M.B. Radial gradient and radial deviation radiomic features from pre-surgical CT scans are associated with survival among lung adenocarcinoma patients. Oncotarget 2017, 8, 96013–96026. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Kim, J.; Balagurunathan, Y.; Qi, J.; Liu, Y.; Latifi, K.; Moros, E.G.; Schabath, M.B.; Ye, Z.; Gillies, R.J.; et al. CT imaging features associated with recurrence in non-small cell lung cancer patients after stereotactic body radiotherapy. Radiat. Oncol. 2017, 12, 158–158. [Google Scholar] [CrossRef] [PubMed]
- Wu W, Parmar C, Grossmann P, et al. Exploratory study to identify radiomics classifiers for lung cancer histology [serial online]. Front Oncol. 2016, 6, 71.
- Parmar C, Grossmann P, Bussink J. Lambin P Aerts HJ. Machine learning methods for quantitative radiomic biomarkers [serial online]. Sci Rep. 2015, 5, 13087.
- Coroller, T.P.; Grossmann, P.; Hou, Y.; Rios Velazquez, E.; Leijenaar, R.T.H.; Hermann, G.; Lambin, P.; Haibe-Kains, B.; Mak, R.H.; Aerts, H.J.W.L. CT-based radiomic signature predicts distant metastasis in lung adenocarcinoma. Radiother. Oncol. 2015, 114, 345–350. [Google Scholar] [CrossRef] [PubMed]
- Rios Velazquez E, Parmar C, Liu Y, et al. Somatic mutations drive distinct imaging phenotypes in lung cancer. Cancer Res. 2017, 77, 3922–3930. [Google Scholar] [CrossRef]
- Grossmann P, Stringfield O, El-Hachem N, et al. Defining the biological basis of radiomic phenotypes in lung cancer [serial online]. Elife. 2017, 6, 323421. [Google Scholar]
- Gatenby, R.A.; Grove, O.; Gillies, R.J. Quantitative Imaging in Cancer Evolution and Ecology. Radiology 2013, 269, 8–14. [Google Scholar] [CrossRef]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef]
- National Lung Screening Trial Research Team, Aberle DR, Adams AM, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med. 2011, 365, 395–409. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Zhang, H.; Zhang, S.-D.; Cheng, X.-Y.; Zheng, S.-M.; Sun, Y.-H.; Zhang, D.-W.; Jiang, Y.; Tian, J.-W. Mammographic and clinicopathological features of triple-negative breast cancer. Br. J. Radiol. 2014, 87, 20130496. [Google Scholar] [CrossRef] [PubMed]
- Hamdy, F.C.; Donovan, J.L.; Lane, J.A.; Mason, M.; Metcalfe, C.; Holding, P.; Davis, M.; Peters, T.J.; Turner, E.L.; Martin, R.M.; et al. 10-Year Outcomes after Monitoring, Surgery, or Radiotherapy for Localized Prostate Cancer. N. Engl. J. Med. 2016, 375, 1415–1424. [Google Scholar] [CrossRef] [PubMed]
- Loeb, S.; Bjurlin, M.A.; Nicholson, J.; Tammela, T.L.; Penson, D.F.; Carter, H.B.; Carroll, P.; Etzioni, R. Overdiagnosis and Overtreatment of Prostate Cancer. Eur. Urol. 2014, 65, 1046–1055. [Google Scholar] [CrossRef]
- Ahmed, H.U.; El-Shater Bosaily, A.; Brown, L.C.; Gabe, R.; Kaplan, R.; Parmar, M.K.; Collaco-Moraes, Y.; Ward, K.; Hindley, R.G.; Freeman, A.; et al. Diagnostic accuracy of multi-parametric MRI and TRUS biopsy in prostate cancer (PROMIS): A paired validating confirmatory study. Lancet 2017, 389, 815–822. [Google Scholar] [CrossRef] [PubMed]
- Kasivisvanathan, V.; Rannikko, A.S.; Borghi, M.; Panebianco, V.; Mynderse, L.A.; Vaarala, M.H.; Briganti, A.; Budäus, L.; Hellawell, G.; Hindley, R.G.; et al. ECISION Study Group Collaborators. MRI-Targeted or Standard Biopsy for Prostate-Cancer Diagnosis. New Engl. J. Med. 2018, 378, 1767–1777. [Google Scholar] [CrossRef] [PubMed]
- Liu L, Tian Z, Zhang Z, Fei B. Computer-aided detection of prostate cancer with MRI: technology and applications. Acad Radiol. 2016, 23, 1024–1046. [Google Scholar] [CrossRef] [PubMed]
- Giannini, V.; Mazzetti, S.; Armando, E.; Carabalona, S.; Russo, F.; Giacobbe, A.; Muto, G.; Regge, D. Multiparametric magnetic resonance imaging of the prostate with computer-aided detection: experienced observer performance study. Eur. Radiol. 2017, 27, 4200–4208. [Google Scholar] [CrossRef]
- Hambrock, T.; Vos, P.C.; de Kaa, C.A.H.; Barentsz, J.O.; Huisman, H.J. Prostate Cancer: Computer-aided Diagnosis with Multiparametric 3-T MR Imaging—Effect on Observer Performance. Radiology 2013, 266, 521–530. [Google Scholar] [CrossRef]
- E Seltzer, S.; Getty, D.J.; Tempany, C.M.; Pickett, R.M.; Schnall, M.D.; McNeil, B.J.; A Swets, J. Staging prostate cancer with MR imaging: a combined radiologist-computer system. Radiology 1997, 202, 219–226. [Google Scholar] [CrossRef]
- Seah, J., Tang, J. H., & Kitchen, A. (2017). Detection of prostate cancer on multiparametric MRI. Proceedings of SPIE. Available online: https://www.spiedigitallibrary.org/conference-proceedings-of-spie/10134/1/Detection-of-prostate-cancer-on-multiparametric-MRI/10.1117/12.2277122.short.
- Liu, S., Zheng, H., Feng, Y., & Li, W. (2017). Prostate cancer diagnosis using deep learning with 3D multiparametric MRI. Proceedings of SPIE.
- Chen, Q., Xu, X., Hu, S., Li, X., Zou, Q., & Li, Y. (2017). A transfer learning approach for classification of clinical significant prostate cancers from mpMRI scans. Proceedings of SPIE. Available online: https://www.spiedigitallibrary.org/conference-proceedings-of-spie/10134/1/A-transfer-learning-approach-for-classification-of-clinical-significant-prostate/10.1117/12.2279021.short.
- Kiraly AP, Nader CA, Tyusuzoglu A, et al.Deep convolutional encoder-decoders for prostate cancer detection and classification. In: Descoteaux M, Maier-Hein L, Franz AM, Jannin P, Collins L, Duchesne S, eds. Medical Image Computing and Computer-Assisted Intervention-MICCAI 2017. 20th International Conference, Quebec City, QC, Canada, September 11–13, 2017. Proceedings, pt III. Cham, Switzerland: Springer International Publishing AG, 2017, 489-497. Available online: https://link.springer.com/chapter/10.1007/978-3-319-66179-7_56?utm_source=getftr&utm_medium=getftr&utm_campaign=getftr_pilot.
- Moradi, M.; Abolmaesumi, P.; Siemens, D.R.; Sauerbrei, E.E.; Boag, A.H.; Mousavi, P. Augmenting Detection of Prostate Cancer in Transrectal Ultrasound Images Using SVM and RF Time Series. IEEE Trans. Biomed. Eng. 2008, 56, 2214–2224. [Google Scholar] [CrossRef] [PubMed]
- Imani, F.; Abolmaesumi, P.; Gibson, E.; Khojaste, A.; Gaed, M.; Moussa, M.; Gomez, J.A.; Romagnoli, C.; Leveridge, M.; Chang, S.; et al. Computer-Aided Prostate Cancer Detection Using Ultrasound RF Time Series: In Vivo Feasibility Study. IEEE Trans. Med Imaging 2015, 34, 2248–2257. [Google Scholar] [CrossRef]
- Azizi, S.; Bayat, S.; Yan, P.; Tahmasebi, A.; Nir, G.; Kwak, J.T.; Xu, S.; Wilson, S.; Iczkowski, K.A.; Lucia, M.S.; et al. Detection and grading of prostate cancer using temporal enhanced ultrasound: combining deep neural networks and tissue mimicking simulations. Int. J. Comput. Assist. Radiol. Surg. 2017, 12, 1293–1305. [Google Scholar] [CrossRef]
- Parwani, A.V. Next generation diagnostic pathology: use of digital pathology and artificial intelligence tools to augment a pathological diagnosis. Diagn. Pathol. 2019, 14, 1–3. [Google Scholar] [CrossRef]
- Mandong, B.M. Diagnostic oncology: role of the pathologist in surgical oncology--a review article. . 2009, 81–88. [Google Scholar]
- Amin, W.; Srintrapun, S.J.; Parwani, A.V.; Md; D, P. Automated whole slide imaging. Expert Opin. Med Diagn. 2008, 2, 1173–1181. [Google Scholar] [CrossRef] [PubMed]
- Abels, E.; Pantanowitz, L.; Aeffner, F.; Zarella, M.D.; van der Laak, J.; Bui, M.M.; Vemuri, V.N.; Parwani, A.V.; Gibbs, J.; Agosto-Arroyo, E.; et al. Computational pathology definitions, best practices, and recommendations for regulatory guidance: a white paper from the Digital Pathology Association. J. Pathol. 2019, 249, 286–294. [Google Scholar] [CrossRef]
- Abels, E.; Pantanowitz, L.; Aeffner, F.; Zarella, M.D.; van der Laak, J.; Bui, M.M.; Vemuri, V.N.; Parwani, A.V.; Gibbs, J.; Agosto-Arroyo, E.; et al. Computational pathology definitions, best practices, and recommendations for regulatory guidance: a white paper from the Digital Pathology Association. J. Pathol. 2019, 249, 286–294. [Google Scholar] [CrossRef] [PubMed]
- Zarella, M.D.; Bowman, D.; Aeffner, F.; Farahani, N.; Xthona, A.; Absar, S.F.; Parwani, A.; Bui, M.; Hartman, D.J. A Practical Guide to Whole Slide Imaging: A White Paper From the Digital Pathology Association. Arch. Pathol. Lab. Med. 2018, 143, 222–234. [Google Scholar] [CrossRef]
- Zhao, C.; Wu, T.; Ding, X.; Parwani, A.V.; Chen, H.; McHugh, J.; Piccoli, A.; Xie, Q.; Lauro, G.R.; Feng, X.; et al. International telepathology consultation: Three years of experience between the University of Pittsburgh Medical Center and KingMed Diagnostics in China. J. Pathol. Informatics 2015, 6, 63. [Google Scholar] [CrossRef]
- Amin, S.; Mori, T.; Itoh, T. A validation study of whole slide imaging for primary diagnosis of lymphoma. Pathol. Int. 2019, 69, 341–349. [Google Scholar] [CrossRef] [PubMed]
- Azizi, S.; Bayat, S.; Yan, P.; Tahmasebi, A.; Nir, G.; Kwak, J.T.; Xu, S.; Wilson, S.; Iczkowski, K.A.; Lucia, M.S.; et al. Detection and grading of prostate cancer using temporal enhanced ultrasound: combining deep neural networks and tissue mimicking simulations. Int. J. Comput. Assist. Radiol. Surg. 2017, 12, 1293–1305. [Google Scholar] [CrossRef] [PubMed]
- Bauer TW, et al. Validation of whole slide imaging for primary diagnosis in surgical pathology. Arch Pathol Lab Med. 2013, 137, 518–524.
- Buck, T.P.; Dilorio, R.; Havrilla, L.; O’neill, D.G. Validation of a whole slide imaging system for primary diagnosis in surgical pathology: A community hospital experience. J. Pathol. Informatics 2014, 5, 43. [Google Scholar] [CrossRef]
- Beck, A.H.; Sangoi, A.R.; Leung, S.; Marinelli, R.J.; Nielsen, T.O.; van de Vijver, M.J.; West, R.B.; van de Rijn, M.; Koller, D. Systematic Analysis of Breast Cancer Morphology Uncovers Stromal Features Associated with Survival. Sci. Transl. Med. 2011, 3, 108ra113–108ra113. [Google Scholar] [CrossRef] [PubMed]
- Nagpal K, et al. Development and validation of a deep learning algorithm for improving Gleason scoring of prostate cancer. NPJ Digit Med. 2019, 2, 48.
- Hegde, N.; Hipp, J.D.; Liu, Y.; Emmert-Buck, M.; Reif, E.; Smilkov, D.; Terry, M.; Cai, C.J.; Amin, M.B.; Mermel, C.H.; et al. Similar image search for histopathology: SMILY. npj Digit. Med. 2019, 2, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Fraggetta, F.; Yagi, Y.; Garcia-Rojo, M.; Evans, A.J.; Tuthill, J.M.; Baidoshvili, A.; Hartman, D.J.; Fukuoka, J.; Pantanowitz, L. The Importance of eSlide Macro Images for Primary Diagnosis with Whole Slide Imaging. J. Pathol. Informatics 2018, 9, 46. [Google Scholar] [CrossRef]
- Vodovnik A, Aghdam MRF. Complete routine remote digital pathology services. J Pathol Inform. 2018, 9, 36. [Google Scholar] [CrossRef]
- Aganj I, Harisinghani MG, Weissleder R, Fischl B. Unsupervised medical image segmentation based on the local center of mass [serial online]. Sci Rep. 2018, 8, 13012. [Google Scholar] [CrossRef]
- Jamaludin A, Kadir T,Zisserman A. Self-supervised learning for spinal MRIs. In: Cardoso J, Argbel T, Carniero G, et al. eds. Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support. Third International Workshop, DLMIA 2017, and 7th International Workshop, ML-CDS 2017; Held in Conjunction with MICCAI 2017, Quebec City, QC, Canada, September 14, Proceedings. Cham, Switzerland: Springer International Publishing AG, 2017, 294-302.
- Shin, H.-C.; Tenenholtz, N.A.; Rogers, J.K.; Schwarz, C.G.; Senjem, M.L.; Gunter, J.L.; Andriole, K.P.; Michalski, M. Medical Image Synthesis for Data Augmentation and Anonymization Using Generative Adversarial Networks. In International Workshop on Simulation and Synthesis in Medical Imaging; Springer: Cham, Switzerland, 2018; pp. 1–11. [Google Scholar]
- Purushotham, S.; Meng, C.; Che, Z.; Liu, Y. Benchmarking deep learning models on large healthcare datasets. J. Biomed. Informatics 2018, 83, 112–134. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.; Balachandar, N.; Lam, C.; Yi, D.; Brown, J.; Beers, A.; Rosen, B.; Rubin, D.L.; Kalpathy-Cramer, J. Distributed deep learning networks among institutions for medical imaging. J. Am. Med Informatics Assoc. 2018, 25, 945–954. [Google Scholar] [CrossRef] [PubMed]
- Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Sumers RM. ChestX-ray8: hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In: Institute of Electrical and Electronics Engineers (IEEE) and IEEE Computer Society, eds. 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). Los Alamitos, CA: IEEE Computer Society; 2017. Available online: https://ieeexplore.ieee.org/document/8099852/keywords.
- Barrett, S.R.H.; Speth, R.L.; Eastham, S.D.; Dedoussi, I.C.; Ashok, A.; Malina, R.; Keith, D.W. Impact of the Volkswagen emissions control defeat device on US public health. Environ. Res. Lett. 2015, 10, 114005. [Google Scholar] [CrossRef]
- Char, D.S.; Shah, N.H.; Magnus, D. Implementing Machine Learning in Health Care — Addressing Ethical Challenges. New Engl. J. Med. 2018, 378, 981–983. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




