Submitted:
04 January 2024
Posted:
04 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Types and features of long read sequencers
3. Application of long-read sequencing to PGT
3.1. Long-read sequencing for PGT-SR
3.2. Long-read sequencing for PGT-M
4.3. Preclinical workup using long-read sequencing for PGT-M conducted at our institution
4.3.1. Materials and Methods
4.3.2. Targeted long-read sequencing using a Nanopore sequencer with adaptive sampling conducted at our institution
4.3.3. Figures of cytogenetical analysis with long-read sequencing and SNP haplotyping
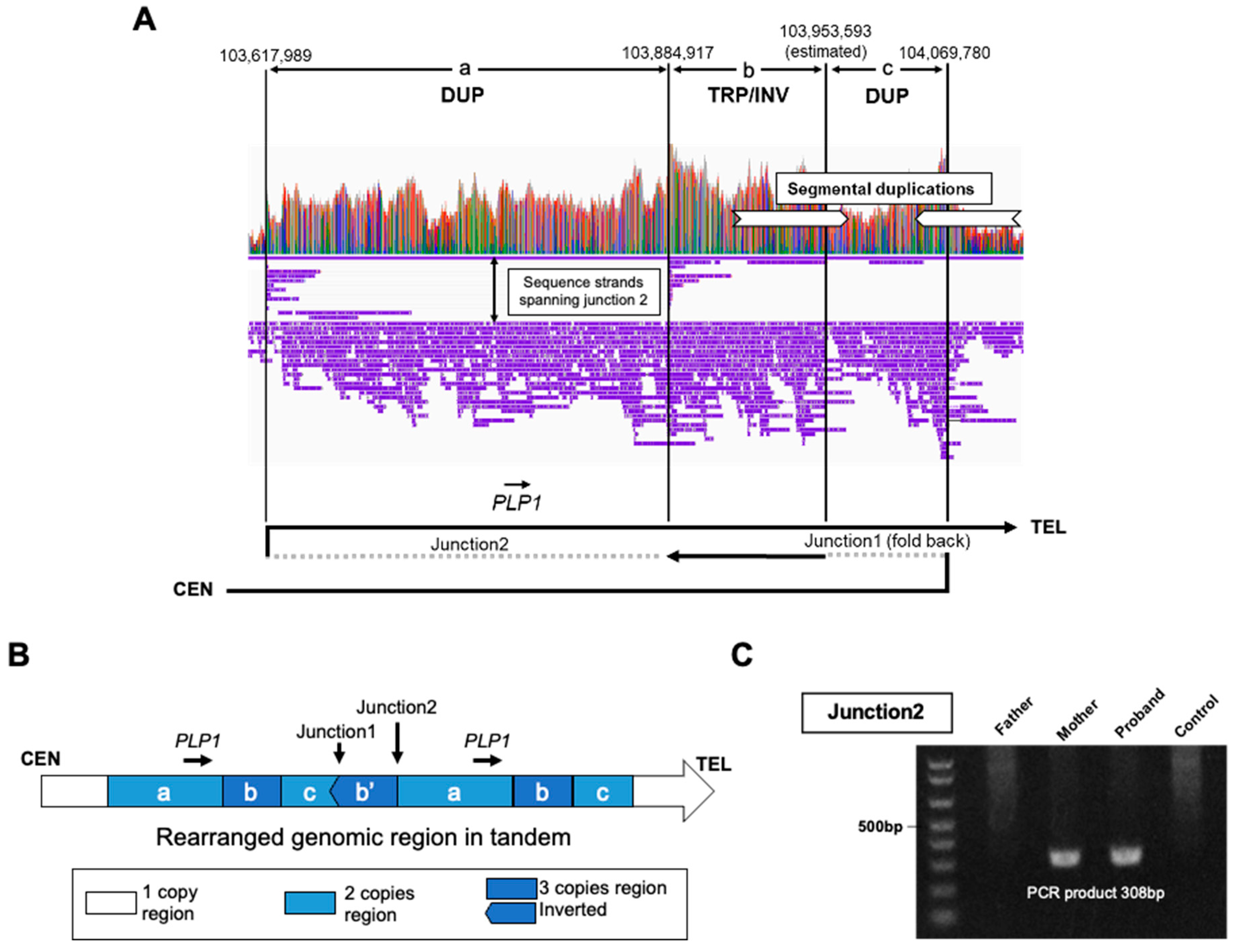
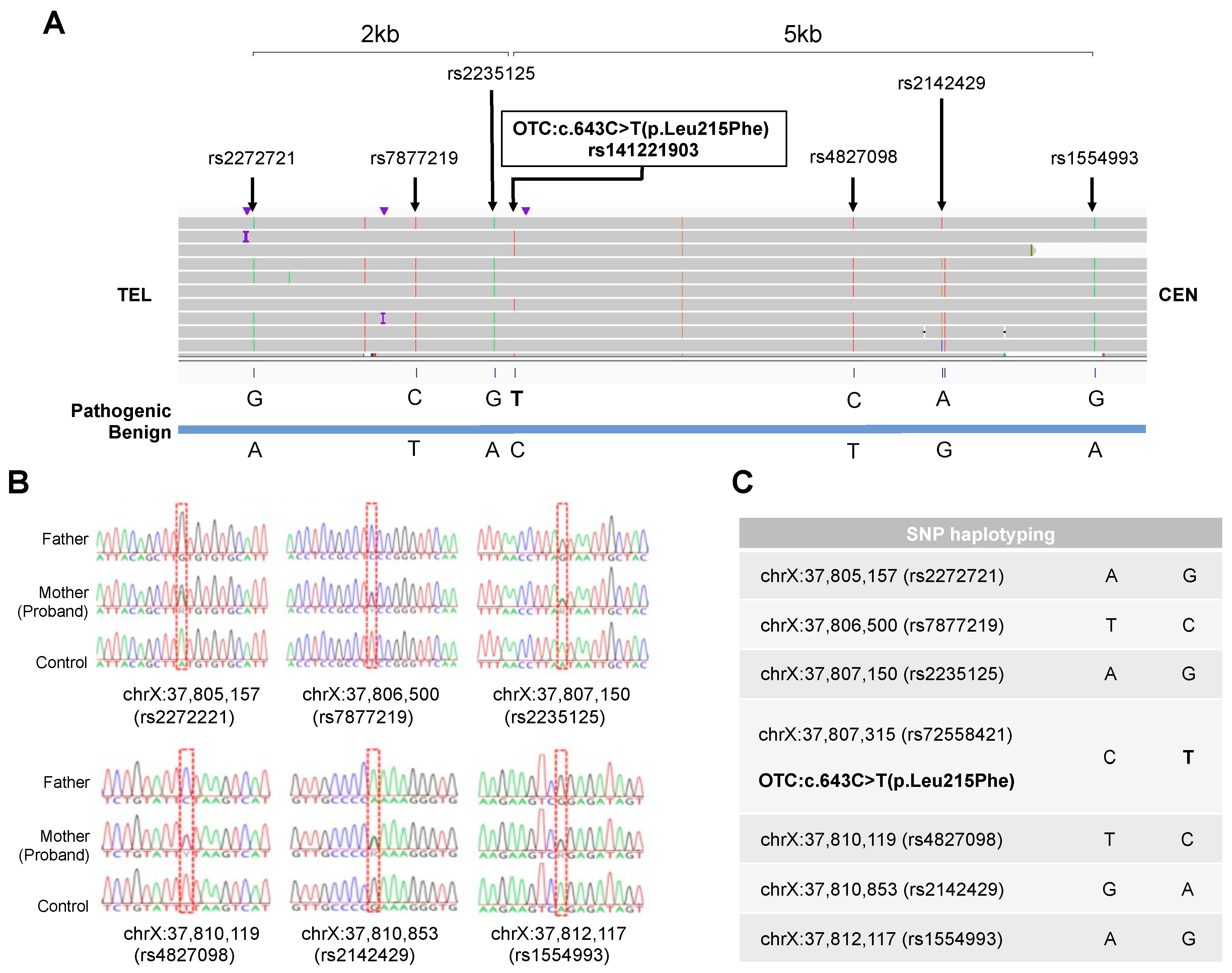
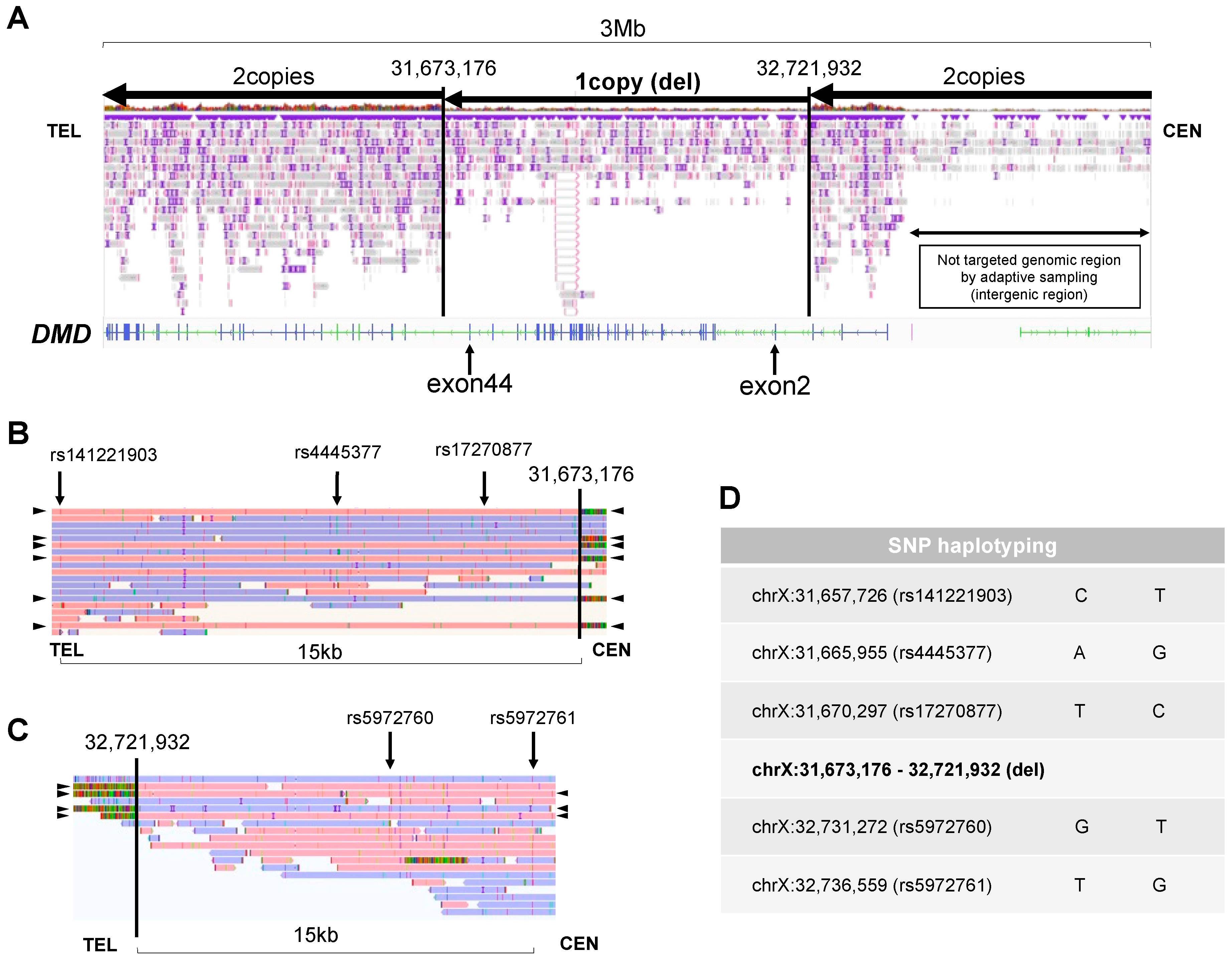
4.4. New nanopore sequencers and PGT-A with the STORK method
5. Conclusion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Logsdon, G. A., M. R. Vollger, and E. E. Eichler. "Long-Read Human Genome Sequencing and Its Applications." Nat Rev Genet 21, no. 10 (2020): 597-614. [CrossRef]
- Zhao, X., R. L. Collins, W. P. Lee, A. M. Weber, Y. Jun, Q. Zhu, B. Weisburd, Y. Huang, P. A. Audano, H. Wang, M. Walker, C. Lowther, J. Fu, Consortium Human Genome Structural Variation, M. B. Gerstein, S. E. Devine, T. Marschall, J. O. Korbel, E. E. Eichler, M. J. P. Chaisson, C. Lee, R. E. Mills, H. Brand, and M. E. Talkowski. "Expectations and Blind Spots for Structural Variation Detection from Long-Read Assemblies and Short-Read Genome Sequencing Technologies." Am J Hum Genet (2021). [CrossRef]
- Dremsek, P., T. Schwarz, B. Weil, A. Malashka, F. Laccone, and J. Neesen. "Optical Genome Mapping in Routine Human Genetic Diagnostics-Its Advantages and Limitations." Genes (Basel) 12, no. 12 (2021).
- Nurk, Sergey, Sergey Koren, Arang Rhie, Mikko Rautiainen, Andrey V. Bzikadze, Alla Mikheenko, Mitchell R. Vollger, Nicolas Altemose, Lev Uralsky, Ariel Gershman, Sergey Aganezov, Savannah J. Hoyt, Mark Diekhans, Glennis A. Logsdon, Michael Alonge, Stylianos E. Antonarakis, Matthew Borchers, Gerard G. Bouffard, Shelise Y. Brooks, Gina V. Caldas, Haoyu Cheng, Chen-Shan Chin, William Chow, Leonardo G. de Lima, Philip C. Dishuck, Richard Durbin, Tatiana Dvorkina, Ian T. Fiddes, Giulio Formenti, Robert S. Fulton, Arkarachai Fungtammasan, Erik Garrison, Patrick G. S. Grady, Tina A. Graves-Lindsay, Ira M. Hall, Nancy F. Hansen, Gabrielle A. Hartley, Marina Haukness, Kerstin Howe, Michael W. Hunkapiller, Chirag Jain, Miten Jain, Erich D. Jarvis, Peter Kerpedjiev, Melanie Kirsche, Mikhail Kolmogorov, Jonas Korlach, Milinn Kremitzki, Heng Li, Valerie V. Maduro, Tobias Marschall, Ann M. McCartney, Jennifer McDaniel, Danny E. Miller, James C. Mullikin, Eugene W. Myers, Nathan D. Olson, Benedict Paten, Paul Peluso, Pavel A. Pevzner, David Porubsky, Tamara Potapova, Evgeny I. Rogaev, Jeffrey A. Rosenfeld, Steven L. Salzberg, Valerie A. Schneider, Fritz J. Sedlazeck, Kishwar Shafin, Colin J. Shew, Alaina Shumate, Yumi Sims, Arian F. A. Smit, Daniela C. Soto, Ivan Sović, Jessica M. Storer, Aaron Streets, Beth A. Sullivan, Françoise Thibaud-Nissen, James Torrance, Justin Wagner, Brian P. Walenz, Aaron Wenger, Jonathan M. D. Wood, Chunlin Xiao, Stephanie M. Yan, Alice C. Young, Samantha Zarate, Urvashi Surti, Rajiv C. McCoy, Megan Y. Dennis, Ivan A. Alexandrov, Jennifer L. Gerton, Rachel J. O’Neill, Winston Timp, Justin M. Zook, Michael C. Schatz, Evan E. Eichler, Karen H. Miga, and Adam M. Phillippy. "The Complete Sequence of a Human Genome." (2021). [CrossRef]
- Vermeesch, J. R., T. Voet, and K. Devriendt. "Prenatal and Pre-Implantation Genetic Diagnosis." Nat Rev Genet 17, no. 10 (2016): 643-56. [CrossRef]
- Zhou, X., Y. Xu, L. Zhu, Z. Su, X. Han, Z. Zhang, Y. Huang, and Q. Liu. "Comparison of Multiple Displacement Amplification (Mda) and Multiple Annealing and Looping-Based Amplification Cycles (Malbac) in Limited DNA Sequencing Based on Tube and Droplet." Micromachines (Basel) 11, no. 7 (2020).
- Li, N., L. Wang, H. Wang, M. Ma, X. Wang, Y. Li, W. Zhang, J. Zhang, D. S. Cram, and Y. Yao. "The Performance of Whole Genome Amplification Methods and Next-Generation Sequencing for Pre-Implantation Genetic Diagnosis of Chromosomal Abnormalities." J Genet Genomics 42, no. 4 (2015): 151-9. [CrossRef]
- Kurahashi, H., T. Kato, J. Miyazaki, H. Nishizawa, E. Nishio, H. Furukawa, H. Miyamura, M. Ito, T. Endo, Y. Ouchi, H. Inagaki, and T. Fujii. "Preimplantation Genetic Diagnosis/Screening by Comprehensive Molecular Testing." Reprod Med Biol 15, no. 1 (2016): 13-19. [CrossRef]
- Eijk-Van Os, P. G., and J. P. Schouten. "Multiplex Ligation-Dependent Probe Amplification (Mlpa®) for the Detection of Copy Number Variation in Genomic Sequences." Methods Mol Biol 688 (2011): 97-126.
- Nakano, Tatsuya, Michiko Ammae, Manabu Satoh, Satoshi Mizuno, Yoshiharu Nakaoka, and Yoshiharu Morimoto. "Analysis of Clinical Outcomes and Meiotic Segregation Modes Following Preimplantation Genetic Testing for Structural Rearrangements Using Acgh/Ngs in Couples with Balanced Chromosome Rearrangement." Reproductive Medicine and Biology 21, no. 1 (2022). [CrossRef]
- Iwasa, T., A. Kuwahara, T. Takeshita, Y. Taniguchi, M. Mikami, and M. Irahara. "Preimplantation Genetic Testing for Aneuploidy and Chromosomal Structural Rearrangement: A Summary of a Nationwide Study by the Japan Society of Obstetrics and Gynecology." Reprod Med Biol 22, no. 1 (2023): e12518. [CrossRef]
- Committee, Eshre Pgt Consortium Steering, F. Carvalho, E. Coonen, V. Goossens, G. Kokkali, C. Rubio, M. Meijer-Hoogeveen, C. Moutou, N. Vermeulen, and M. De Rycke. "Eshre Pgt Consortium Good Practice Recommendations for the Organisation of Pgt." Hum Reprod Open 2020, no. 3 (2020): hoaa021. [CrossRef]
- Zhang, S., F. Liang, C. Lei, J. Wu, J. Fu, Q. Yang, X. Luo, G. Yu, D. Wang, Y. Zhang, D. Lu, X. Sun, Y. Liang, and C. Xu. "Long-Read Sequencing and Haplotype Linkage Analysis Enabled Preimplantation Genetic Testing for Patients Carrying Pathogenic Inversions." J Med Genet 56, no. 11 (2019): 741-49. [CrossRef]
- Chow, J. F. C., H. H. Y. Cheng, E. Y. L. Lau, W. S. B. Yeung, and E. H. Y. Ng. "Distinguishing between Carrier and Noncarrier Embryos with the Use of Long-Read Sequencing in Preimplantation Genetic Testing for Reciprocal Translocations." Genomics 112, no. 1 (2020): 494-500. [CrossRef]
- Liu, S., H. Wang, D. Leigh, D. S. Cram, L. Wang, and Y. Yao. "Third-Generation Sequencing: Any Future Opportunities for Pgt?" J Assist Reprod Genet 38, no. 2 (2021): 357-64.
- M, M. Yc, Q. Yu, M. Ma, H. Wang, S. Tian, W. Zhang, M. Jz M, Y. Liu, Q. Yang, X. Pan, H. Liang, L. Wang, D. Leigh, D. S. Cram, and Y. Yao. "Variant Haplophasing by Long-Read Sequencing: A New Approach to Preimplantation Genetic Testing Workups." Fertil Steril 116, no. 3 (2021): 774-83. [CrossRef]
- Xu, J., Z. Zhang, W. Niu, Q. Yang, G. Yao, S. Shi, H. Jin, W. Song, L. Chen, X. Zhang, Y. Guo, Y. Su, L. Hu, J. Zhai, Y. Zhang, F. Dong, Y. Gao, W. Li, S. Bo, M. Hu, J. Ren, L. Huang, S. Lu, X. S. Xie, and Y. Sun. "Mapping Allele with Resolved Carrier Status of Robertsonian and Reciprocal Translocation in Human Preimplantation Embryos." Proc Natl Acad Sci U S A 114, no. 41 (2017): E8695-E702. [CrossRef]
- Xia, Q., S. Li, T. Ding, Z. Liu, J. Liu, Y. Li, H. Zhu, and Z. Yao. "Nanopore Sequencing for Detecting Reciprocal Translocation Carrier Status in Preimplantation Genetic Testing." BMC Genomics 24, no. 1 (2023): 1. [CrossRef]
- Miao, H., J. Zhou, Q. Yang, F. Liang, D. Wang, N. Ma, B. Gao, J. Du, G. Lin, K. Wang, and Q. Zhang. "Long-Read Sequencing Identified a Causal Structural Variant in an Exome-Negative Case and Enabled Preimplantation Genetic Diagnosis." Hereditas 155 (2018): 32. [CrossRef]
- Watson, C. M., D. L. Holliday, L. A. Crinnion, and D. T. Bonthron. "Long-Read Nanopore DNA Sequencing Can Resolve Complex Intragenic Duplication/Deletion Variants, Providing Information to Enable Preimplantation Genetic Diagnosis." Prenat Diagn 42, no. 2 (2022): 226-32.
- Wu, Haitao, Dongjia Chen, Qiang Zhao, Xiaoting Shen, Yongbin Liao, Ping Li, Philip C. N. Chiu, and Canquan Zhou. "Long-Read Sequencing on the Smrt Platform Enables Efficient Haplotype Linkage Analysis in Preimplantation Genetic Testing for Β-Thalassemia." Journal of Assisted Reproduction and Genetics (2022). [CrossRef]
- Tsuiko, O., Y. El Ayeb, T. Jatsenko, J. Allemeersch, C. Melotte, J. Ding, S. Debrock, K. Peeraer, A. Vanhie, A. De Leener, C. Pirard, C. Kluyskens, E. Denayer, E. Legius, J. R. Vermeesch, H. Brems, and E. Dimitriadou. "Preclinical Workup Using Long-Read Amplicon Sequencing Provides Families with De Novo Pathogenic Variants Access to Universal Preimplantation Genetic Testing." Hum Reprod (2023).
- Peng, C., H. Chen, J. Ren, F. Zhou, Y. Li, Y. Keqie, T. Ding, J. Ruan, H. Wang, X. Chen, and S. Liu. "A Long-Read Sequencing and Snp Haplotype-Based Novel Preimplantation Genetic Testing Method for Female Adpkd Patient with De Novo Pkd1 Mutation." BMC Genomics 24, no. 1 (2023): 521. [CrossRef]
- Mariya, T., Y. Shichiri, T. Sugimoto, R. Kawamura, S. Miyai, H. Inagaki, E. Sugihara, K. Ikeda, T. Baba, A. Ishikawa, M. Ammae, Y. Nakaoka, T. Saito, A. Sakurai, and H. Kurahashi. "Clinical Application of Long-Read Nanopore Sequencing in a Preimplantation Genetic Testing Pre-Clinical Workup to Identify the Junction for Complex Xq Chromosome Rearrangement-Related Disease." Prenat Diagn (2023). [CrossRef]
- Neal, S. A., S. J. Morin, J. M. Franasiak, L. R. Goodman, C. R. Juneau, E. J. Forman, M. D. Werner, and R. T. Scott, Jr. "Preimplantation Genetic Testing for Aneuploidy Is Cost-Effective, Shortens Treatment Time, and Reduces the Risk of Failed Embryo Transfer and Clinical Miscarriage." Fertil Steril 110, no. 5 (2018): 896-904. [CrossRef]
- Franasiak, J. M., E. J. Forman, K. H. Hong, M. D. Werner, K. M. Upham, N. R. Treff, and R. T. Scott, Jr. "The Nature of Aneuploidy with Increasing Age of the Female Partner: A Review of 15,169 Consecutive Trophectoderm Biopsies Evaluated with Comprehensive Chromosomal Screening." Fertil Steril 101, no. 3 (2014): 656-63 e1. [CrossRef]
- Reig, A., J. Franasiak, R. T. Scott, Jr., and E. Seli. "The Impact of Age Beyond Ploidy: Outcome Data from 8175 Euploid Single Embryo Transfers." J Assist Reprod Genet 37, no. 3 (2020): 595-602.
- Yan, Junhao, Yingying Qin, Han Zhao, Yun Sun, Fei Gong, Rong Li, Xiaoxi Sun, Xiufeng Ling, Hong Li, Cuifang Hao, Jichun Tan, Jing Yang, Yimin Zhu, Fenghua Liu, Dawei Chen, Daimin Wei, Juanjuan Lu, Tianxiang Ni, Wei Zhou, Keliang Wu, Yuan Gao, Yuhua Shi, Yao Lu, Ting Zhang, Wei Wu, Xiang Ma, Hailan Ma, Jing Fu, Junqiang Zhang, Qingxia Meng, Heping Zhang, Richard S. Legro, and Zi-Jiang Chen. "Live Birth with or without Preimplantation Genetic Testing for Aneuploidy." New England Journal of Medicine 385, no. 22 (2021): 2047-58. [CrossRef]
- Munné, S., B. Kaplan, J. L. Frattarelli, T. Child, G. Nakhuda, F. N. Shamma, K. Silverberg, T. Kalista, A. H. Handyside, M. Katz-Jaffe, D. Wells, T. Gordon, S. Stock-Myer, and S. Willman. "Preimplantation Genetic Testing for Aneuploidy Versus Morphology as Selection Criteria for Single Frozen-Thawed Embryo Transfer in Good-Prognosis Patients: A Multicenter Randomized Clinical Trial." Fertil Steril 112, no. 6 (2019): 1071-79.e7. [CrossRef]
- Sato, T., M. Sugiura-Ogasawara, F. Ozawa, T. Yamamoto, T. Kato, H. Kurahashi, T. Kuroda, N. Aoyama, K. Kato, R. Kobayashi, A. Fukuda, T. Utsunomiya, A. Kuwahara, H. Saito, T. Takeshita, and M. Irahara. "Preimplantation Genetic Testing for Aneuploidy: A Comparison of Live Birth Rates in Patients with Recurrent Pregnancy Loss Due to Embryonic Aneuploidy or Recurrent Implantation Failure." Hum Reprod 34, no. 12 (2019): 2340-48. [CrossRef]
- Wei, Shan, Alexandre Djandji, Miriam T. Lattin, Odelia Nahum, Nataly Hoffman, Claudia Cujar, Refik Kayali, Cengiz Cinnioglu, Ronald Wapner, Mary D’Alton, Brynn Levy, and Zev Williams. "Rapid Nanopore Sequencing–Based Screen for Aneuploidy in Reproductive Care." New England Journal of Medicine 387, no. 7 (2022): 658-60. [CrossRef]
- Chin, C. S., P. Peluso, F. J. Sedlazeck, M. Nattestad, G. T. Concepcion, A. Clum, C. Dunn, R. O'Malley, R. Figueroa-Balderas, A. Morales-Cruz, G. R. Cramer, M. Delledonne, C. Luo, J. R. Ecker, D. Cantu, D. R. Rank, and M. C. Schatz. "Phased Diploid Genome Assembly with Single-Molecule Real-Time Sequencing." Nat Methods 13, no. 12 (2016): 1050-54. [CrossRef]
- Wang, Y., Y. Zhao, A. Bollas, Y. Wang, and K. F. Au. "Nanopore Sequencing Technology, Bioinformatics and Applications." Nat Biotechnol 39, no. 11 (2021): 1348-65.
- Parker, J., A. J. Helmstetter, D. Devey, T. Wilkinson, and A. S. T. Papadopulos. "Field-Based Species Identification of Closely-Related Plants Using Real-Time Nanopore Sequencing." Sci Rep 7, no. 1 (2017): 8345. [CrossRef]
- Castro-Wallace, S. L., C. Y. Chiu, K. K. John, S. E. Stahl, K. H. Rubins, A. B. R. McIntyre, J. P. Dworkin, M. L. Lupisella, D. J. Smith, D. J. Botkin, T. A. Stephenson, S. Juul, D. J. Turner, F. Izquierdo, S. Federman, D. Stryke, S. Somasekar, N. Alexander, G. Yu, C. E. Mason, and A. S. Burton. "Nanopore DNA Sequencing and Genome Assembly on the International Space Station." Sci Rep 7, no. 1 (2017): 18022. [CrossRef]
- Greninger, A. L., S. N. Naccache, S. Federman, G. Yu, P. Mbala, V. Bres, D. Stryke, J. Bouquet, S. Somasekar, J. M. Linnen, R. Dodd, P. Mulembakani, B. S. Schneider, J. J. Muyembe-Tamfum, S. L. Stramer, and C. Y. Chiu. "Rapid Metagenomic Identification of Viral Pathogens in Clinical Samples by Real-Time Nanopore Sequencing Analysis." Genome Med 7 (2015): 99. [CrossRef]
- Sun, X., L. Song, W. Yang, L. Zhang, M. Liu, X. Li, G. Tian, and W. Wang. "Nanopore Sequencing and Its Clinical Applications." Methods Mol Biol 2204 (2020): 13-32.
- Dhesi, Z., V. I. Enne, J. O'Grady, V. Gant, and D. M. Livermore. "Rapid and Point-of-Care Testing in Respiratory Tract Infections: An Antibiotic Guardian?" ACS Pharmacol Transl Sci 3, no. 3 (2020): 401-17.
- Lastra, L. S., V. Sharma, N. Farajpour, M. Nguyen, and K. J. Freedman. "Nanodiagnostics: A Review of the Medical Capabilities of Nanopores." Nanomedicine 37 (2021): 102425. [CrossRef]
- Wick, R. R., L. M. Judd, and K. E. Holt. "Performance of Neural Network Basecalling Tools for Oxford Nanopore Sequencing." Genome Biol 20, no. 1 (2019): 129. [CrossRef]
- Lai, H. H., T. H. Chuang, L. K. Wong, M. J. Lee, C. L. Hsieh, H. L. Wang, and S. U. Chen. "Identification of Mosaic and Segmental Aneuploidies by Next-Generation Sequencing in Preimplantation Genetic Screening Can Improve Clinical Outcomes Compared to Array-Comparative Genomic Hybridization." Mol Cytogenet 10 (2017): 14. [CrossRef]
- Fiorentino, F., S. Bono, A. Biricik, A. Nuccitelli, E. Cotroneo, G. Cottone, F. Kokocinski, C. E. Michel, M. G. Minasi, and E. Greco. "Application of Next-Generation Sequencing Technology for Comprehensive Aneuploidy Screening of Blastocysts in Clinical Preimplantation Genetic Screening Cycles." Hum Reprod 29, no. 12 (2014): 2802-13. [CrossRef]
- Shetty, Sachin, Jiny Nair, Jnapti Johnson, Navya Shetty, Ajay Kumar J, Nirmala Thondehalmath, Deepanjali Ganesh, Vidyalakshmi R. Bhat, Sajana M, Anjana R, Rajsekhar Nayak, Devika Gunasheela, Jayarama S. Kadandale, and Swathi Shetty. "Preimplantation Genetic Testing for Couples with Balanced Chromosomal Rearrangements." Journal of Reproduction & Infertility (2022). [CrossRef]
- Miller, D. E., A. Sulovari, T. Wang, H. Loucks, K. Hoekzema, K. M. Munson, A. P. Lewis, E. P. A. Fuerte, C. R. Paschal, T. Walsh, J. Thies, J. T. Bennett, I. Glass, K. M. Dipple, K. Patterson, E. S. Bonkowski, Z. Nelson, A. Squire, M. Sikes, E. Beckman, R. L. Bennett, D. Earl, W. Lee, R. Allikmets, S. J. Perlman, P. Chow, A. V. Hing, T. L. Wenger, M. P. Adam, A. Sun, C. Lam, I. Chang, X. Zou, S. L. Austin, E. Huggins, A. Safi, A. K. Iyengar, T. E. Reddy, W. H. Majoros, A. S. Allen, G. E. Crawford, P. S. Kishnani, Genomics University of Washington Center for Mendelian, M. C. King, T. Cherry, J. X. Chong, M. J. Bamshad, D. A. Nickerson, H. C. Mefford, D. Doherty, and E. E. Eichler. "Targeted Long-Read Sequencing Identifies Missing Disease-Causing Variation." Am J Hum Genet (2021). [CrossRef]
- Mariya, Tasuku, Takema Kato, Takeshi Sugimoto, Syunsuke Miyai, Hidehito Inagaki, Tamae Ohye, Eiji Sugihara, Yukako Muramatsu, Seiji Mizuno, and Hiroki Kurahashi. "Target Enrichment Long-Read Sequencing with Adaptive Sampling Can Determine the Structure of the Small Supernumerary Marker Chromosomes." Journal of Human Genetics (2022). [CrossRef]
- Aganezov, S., S. M. Yan, D. C. Soto, M. Kirsche, S. Zarate, P. Avdeyev, D. J. Taylor, K. Shafin, A. Shumate, C. Xiao, J. Wagner, J. McDaniel, N. D. Olson, M. E. G. Sauria, M. R. Vollger, A. Rhie, M. Meredith, S. Martin, J. Lee, S. Koren, J. A. Rosenfeld, B. Paten, R. Layer, C. S. Chin, F. J. Sedlazeck, N. F. Hansen, D. E. Miller, A. M. Phillippy, K. H. Miga, R. C. McCoy, M. Y. Dennis, J. M. Zook, and M. C. Schatz. "A Complete Reference Genome Improves Analysis of Human Genetic Variation." Science 376, no. 6588 (2022): eabl3533. [CrossRef]
- Li, H. "Minimap2: Pairwise Alignment for Nucleotide Sequences." Bioinformatics 34, no. 18 (2018): 3094-100. [CrossRef]
- Thorvaldsdóttir, H., J. T. Robinson, and J. P. Mesirov. "Integrative Genomics Viewer (Igv): High-Performance Genomics Data Visualization and Exploration." Brief Bioinform 14, no. 2 (2013): 178-92. [CrossRef]
- Loose, M., S. Malla, and M. Stout. "Real-Time Selective Sequencing Using Nanopore Technology." Nat Methods 13, no. 9 (2016): 751-4. [CrossRef]
- Payne, A., N. Holmes, T. Clarke, R. Munro, B. J. Debebe, and M. Loose. "Readfish Enables Targeted Nanopore Sequencing of Gigabase-Sized Genomes." Nat Biotechnol 39, no. 4 (2021): 442-50. [CrossRef]
- Mastrorosa, F. K., D. E. Miller, and E. E. Eichler. "Applications of Long-Read Sequencing to Mendelian Genetics." Genome Med 15, no. 1 (2023): 42. [CrossRef]
- Miller, D. E., L. Lee, M. Galey, R. Kandhaya-Pillai, M. Tischkowitz, D. Amalnath, A. Vithlani, K. Yokote, H. Kato, Y. Maezawa, A. Takada-Watanabe, M. Takemoto, G. M. Martin, E. E. Eichler, F. M. Hisama, and J. Oshima. "Targeted Long-Read Sequencing Identifies Missing Pathogenic Variants in Unsolved Werner Syndrome Cases." J Med Genet (2022). [CrossRef]
- Yamaguchi, K., R. Kasajima, K. Takane, S. Hatakeyama, E. Shimizu, R. Yamaguchi, K. Katayama, M. Arai, C. Ishioka, T. Iwama, S. Kaneko, N. Matsubara, Y. Moriya, T. Nomizu, K. Sugano, K. Tamura, N. Tomita, T. Yoshida, K. Sugihara, Y. Nakamura, S. Miyano, S. Imoto, Y. Furukawa, and T. Ikenoue. "Application of Targeted Nanopore Sequencing for the Screening and Determination of Structural Variants in Patients with Lynch Syndrome." J Hum Genet (2021). [CrossRef]
- Filser, M., M. Schwartz, K. Merchadou, A. Hamza, M. C. Villy, A. Decees, E. Frouin, E. Girard, S. M. Caputo, V. Renault, V. Becette, L. Golmard, N. Servant, D. Stoppa-Lyonnet, O. Delattre, C. Colas, and J. Masliah-Planchon. "Adaptive Nanopore Sequencing to Determine Pathogenicity of Brca1 Exonic Duplication." J Med Genet 60, no. 12 (2023): 1206-09. [CrossRef]
- Martin, S., D. Heavens, Y. Lan, S. Horsfield, M. D. Clark, and R. M. Leggett. "Nanopore Adaptive Sampling: A Tool for Enrichment of Low Abundance Species in Metagenomic Samples." Genome Biol 23, no. 1 (2022): 11. [CrossRef]
- Beck, C. R., C. M. Carvalho, L. Banser, T. Gambin, D. Stubbolo, B. Yuan, K. Sperle, S. M. McCahan, M. Henneke, P. Seeman, J. Y. Garbern, G. M. Hobson, and J. R. Lupski. "Complex Genomic Rearrangements at the Plp1 Locus Include Triplication and Quadruplication." PLoS Genet 11, no. 3 (2015): e1005050. [CrossRef]
- Bahrambeigi, V., X. Song, K. Sperle, C. R. Beck, H. Hijazi, C. M. Grochowski, S. Gu, P. Seeman, K. J. Woodward, C. M. B. Carvalho, G. M. Hobson, and J. R. Lupski. "Distinct Patterns of Complex Rearrangements and a Mutational Signature of Microhomeology Are Frequently Observed in Plp1 Copy Number Gain Structural Variants." Genome Med 11, no. 1 (2019): 80. [CrossRef]
- Choi, J. H., B. H. Lee, J. H. Kim, G. H. Kim, Y. M. Kim, J. Cho, C. K. Cheon, J. M. Ko, J. H. Lee, and H. W. Yoo. "Clinical Outcomes and the Mutation Spectrum of the Otc Gene in Patients with Ornithine Transcarbamylase Deficiency." J Hum Genet 60, no. 9 (2015): 501-7. [CrossRef]
- Kido, J., K. Sugawara, T. Sawada, S. Matsumoto, and K. Nakamura. "Pathogenic Variants of Ornithine Transcarbamylase Deficiency: Nation-Wide Study in Japan and Literature Review." Front Genet 13 (2022): 952467. [CrossRef]
- "Oxford Nanopore Tech Update: New Duplex Method for Q30 Nanopore Single Molecule Reads, Promethion 2, and More." @nanopore, https://nanoporetech.com/about-us/news/oxford-nanopore-tech-update-new-duplex-method-q30-nanopore-single-molecule-reads-0 (.
- Marx, V. "Method of the Year: Long-Read Sequencing." Nat Methods 20, no. 1 (2023): 6-11. [CrossRef]
- Wei, S., Z. R. Weiss, P. Gaur, E. Forman, and Z. Williams. "Rapid Preimplantation Genetic Screening Using a Handheld, Nanopore-Based DNA Sequencer." Fertil Steril 110, no. 5 (2018): 910-16.e2.
- Baslan, T., S. Kovaka, F. J. Sedlazeck, Y. Zhang, R. Wappel, S. Tian, S. W. Lowe, S. Goodwin, and M. C. Schatz. "High Resolution Copy Number Inference in Cancer Using Short-Molecule Nanopore Sequencing." Nucleic Acids Res (2021). [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).



