1. Introduction
Parkinson’s disease (PD) is the second most common neurodegenerative disorder in the elder population [
1]. The clinical features of Parkinson's disease include not only motor symptoms such as bradykinesia, tremor, and postural rigidity, but also non-motor symptoms such as fatigue, apathy, and cognitive dysfunction [
2]. It was reported that approximately 26.7% of PD patients were diagnosed with mild cognitive impairment (PD-MCI) [
3], and around 20%–60% of PD-MCI may convert to PD with dementia (PDD) within five years after diagnosis [
4]. The incidence rate of developing dementia increased in Patients with PD (PwP) than non-PD control [
5]. The neuronal transmission of fibril alpha-synuclein (α-syn) followed by the formation of Lewy Body in the substantia nigra pars compacta was a well-known biomarker for PwP [
2]. However, the histological and molecular markers for PDD remain inconclusive. Although the widespread of alpha synuclein (α-syn), tau neurofibrillary tangles (NFTs) and amyloid-β (Aβ) plaques were found in most PDD patients, the findings were not consistent [
6]. The motor dysfunctions with demented cognition could increase the economic and psychological burden for caregivers [
7]. The demented PwP will gradually lose their basic living ability and may have shorter lifespan than non-demented PD [
5]. In addition, the efficacy of the approved drug for treating PDD or PD-MCI are still limited [
8]. It could become a serious issue for individual and our society in an aging population [9, 10]. Fortunately, recent study suggested that the impaired cognitive function may be sustained or even rescued after treatments [
1]. Therefore, it is important to identify PD patients with cognitive dysfunction at early stage.
The diagnosis of PD with cognitive impairment and dementia, including executive and visuospatial deficits [
11], is generally based on Level II neuropsychological assessment. Apart from neuropsychological tests, regular examinations includes blood testing and brain imaging such as Magnetic Resonance Imaging (MRI) [12, 13]. However, the entire procedure of cognitive examination is often time-consuming and requires the involvement of multiple medical personnel [12, 13]. Hence, reliable biomarkers that could facilitate the diagnosis and provide quick distinction between PD patients with and without cognitive impairments are the unmet need. Recently, emerging molecular biomarkers regarding inherited genetic mutant or toxic proteomic marker for PDD gradually attracted attention and was recommended for deeper validation in human studies [
1]. Compared with tissue biopsy or biofluid, the non-invasive collection method of plasma makes it become one of the common-studied resource of human biomarkers [
14].
MicroRNA (miRNA) is a single-stranded non-coding RNA with an average length of 22 nucleotides [
15]. MiRNA can mediate post-transcriptional expression via binding with target messenger RNAs (mRNA) [
16] and cease the transcription of the encoded gene. It was reported that miRNA exist not only in cytoplasm but also in extracellular area such as cerebrospinal fluid, blood and other biofluids [16, 17]. The circulating miRNAs may travel across the blood–brain barrier (BBB) due to their short sequence length [
13]. Various studies have shown the association between miRNAs and neurodegenerative diseases, including Alzheimer’s disease (AD) and PD [18-20] and also for patients with cognitive decline [21, 22]. This suggest that miRNAs may be a potential biomarker for PD with cognitive impairment.
It is widely accepted that a miRNA candidate exploration study should include both discovery study phase and followed by validation study phase [
23]. Therefore, we firstly conducted the discovery phase using NGS sequencing and screened for miRNA candidates. Then we conducted the validation phase using ddPCR on a different cohort. We aimed to find out if the miRNA candidates could distinguish between PD with and without cognitive impairment.
2. Results
2.1. Using Small RNA-Seq to Explore the Potential miRNA Cluster that May Differentiate PD with or without Cognitive Impairment
Analyzing miRNA candidates from the NGS profiling in the discovery cohort
We firstly performed a discovery study phase using NGS sequencing (small RNA-seq) to explore the potential miRNA candidates that were associated with cognitive decline. The demographic variables of the discovery cohort were summarized in
Table 1.
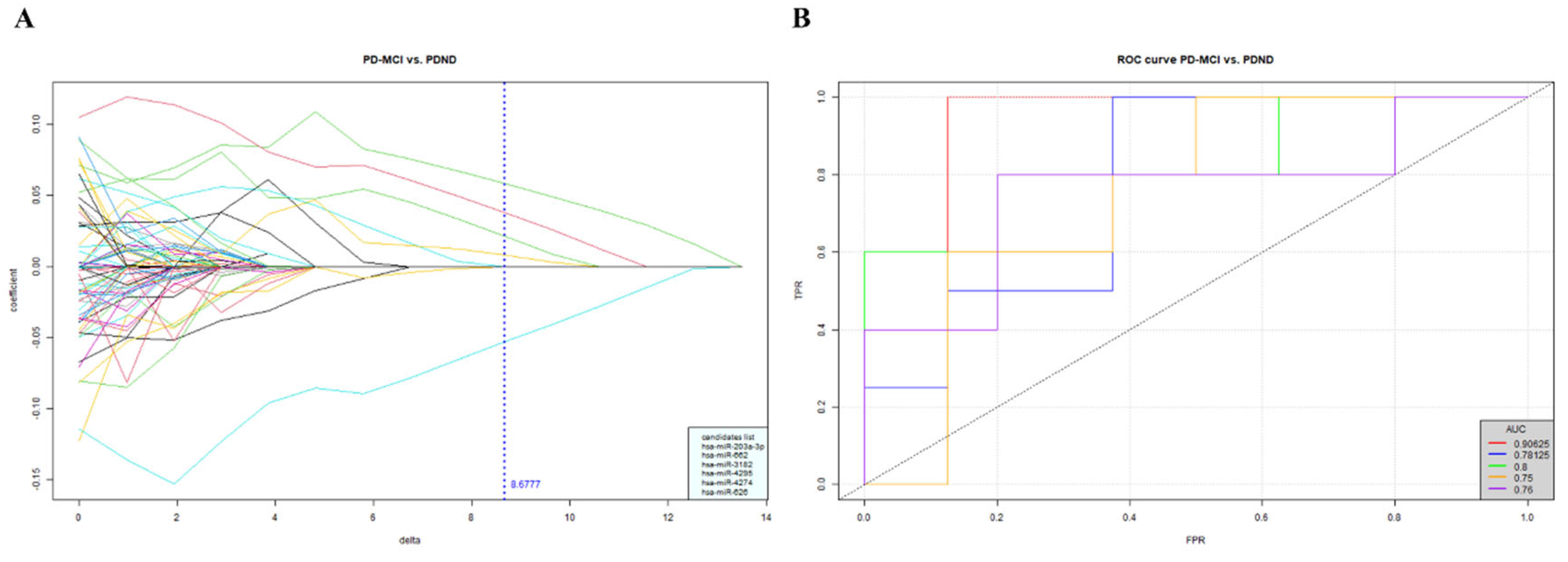
The NGS sequencing results consisting of approximately 2600 miRNAs and were filtered through a series of data processing and statistical analysis via Biomedical Oriented Logistic Dantzig (BOLD) Selector. The BOLD selector was used for miRNA candidate selection in consideration of the supersaturated data that analyzed thousands of miRNA profiling in the relative small sample size. Eventually, 6 miRNA candidates, including miR-203a-3p, miR-626, miR-662, miR-3182, miR-4274 and miR-4295, were clustered as potential biomarkers for identifying PDND from PD-MCI with an average AUC of 0.8 (
Figure 1). The finding encouraged us to carry out a follow-up investigation of these plasma miRNAs in a new PwP cohort with a larger sample size and validated the differential power of the miRNA expression level in PD with different cognitive status.
2.2. Validation of the miRNA Candidates in another PwP Cohort
2.2.1. Measurement of the Selected miRNA Candidates
We then aimed to validate previous findings by recruiting another PwP cohort. We examined the plasma miRNAs using real-time PCR (RT-PCR) to study the expression levels of the 6 miRNAs in the new PwP cohort. The 6 miRNA candidates were first quantified by LNA-based RT-PCR (Qiagen, Germany) and ruled out the possible false positive outcomes via melting curve analysis. However, except for miR-203a-3p, the rest 5 miRNA candidates failed to detect by RT-PCR (Ct>36) or non-overlapped dissociation curves (Data not shown). The outcome may result from the low concentration of total extracted plasma RNA which was lower than the detection limit of Bioanalyzer RNA Nano system (Data not shown).
2.2.2. Motor Function Deterioration Was Associated with Poor Cognitive Status
As shown in
Table 2, demographic variates such as gender, age, education, UPDRS III, MoCA score and LEDD values of each study group were summarized. The percentage of male in HC and PDND were 56.67%, while the percentage of male in PD-MCI and PDD were 53.33% and 46.67%, respectively. The age of PDD were older than HC (p-value<0.0001) and PDND (p-value=0.025), while no significant difference was observed for ages among HC, PDND and PD-MCI. Although the duration of disease and the age of onset showed no significant difference among the PD patients, PDD had significantly higher Hoehn-Yahr stage value compared to PDND (p-value<0.0001) and PD-MCI (p-value=0.0048). Correspondingly, PDD also had higher score of UPDRS III compared to PDND (p-value<0.0001) and PD-MCI (p-value=0.0002). It suggests that the motor and cognitive functions may deteriorate more rapidly in PDD given that the disease duration were similar across PDND, PD-MCI and PDD. No significant difference was observed in the year of formal education among the groups of HC, PDND and PD-MCI. PDD had significantly less years of education than HC (p-value=0.0256) and PDND (p-value=0.0327). Moreover, the value of LEDD was estimated according to the anti-Parkinson drug prescribed to PD patients. The results showed no significant difference in LEDD among PDND, PD-MCI and PDD. Although anticholinergic drugs were excluded from LEDD estimation, it was estimated 5 patients of PDND, 7 patients of PD-MCI and 1 patient of PDD received medication of low dose anticholinergic drug (2-4mg/d) for the management of severe tremor.
2.3. The Expression Level of the Plasma miR-203a-3p/miR-16-5p Validated by ddPCR
Because the total amount of circulating miRNA may differ from person to person and it usually presents a very low concentration, and the quantification with high sensitivity and specificity were required, we used ddPCR for absolutely quantifying the expression level of plasma miR-203a-3p in each fixed volume samples [
24]. Additionally, an endogenous reference miRNA was required for normalization since miR-203a-3p expression may be affected by intrinsic factors [
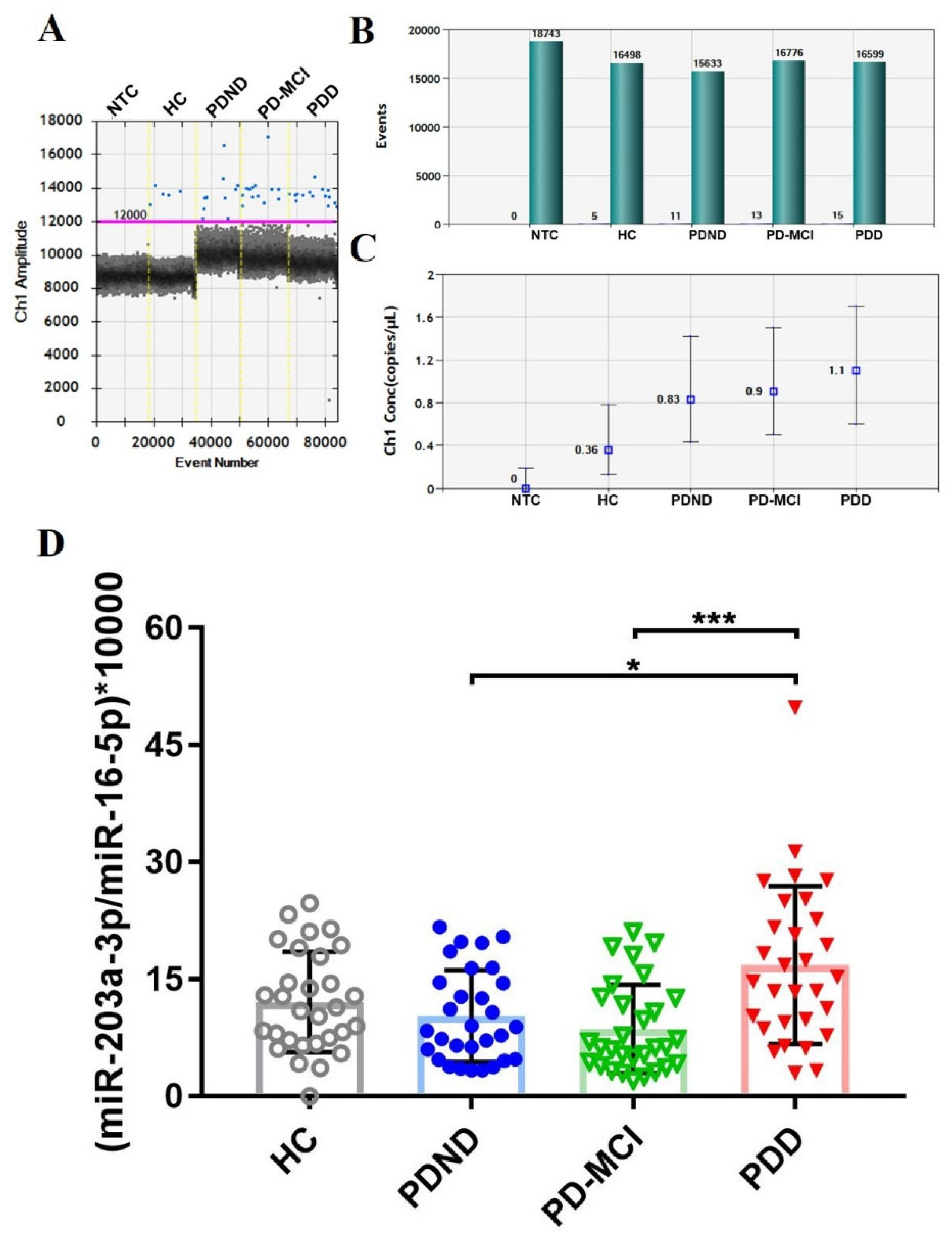
25]. The endogenous miR-16-5p, which is abundant across intracellular or intercellular regions and relatively consistent in biofluid with different age, was used for normalizing miRNA in study of Parkinson’s disease and Multiple System Atrophy [26-28]. Therefore, we combined the detection of our target miRNA and the endogenous reference miRNA, as the ratio of miR-203a-3p/miR-16-5p, to reduce the bias from individual intrinsic factors which may not relate to PD pathologies. An exogenous synthetic oligonucleotide UniSp6 was used as reference of RNA extraction. The ratio of miR-16-5p/ UniSp6 showed no significant difference among the study groups, which suggested the robust extraction efficiency for each sample (
Figure S1.). After performing outlier identification, 4 PD-MCI were excluded from further analysis (data not shown). The examples of ddPCR results and the expression level of miR-203a-3p/miR-16-5p among each study group were visualized in
Figure 2.
To elucidate whether the ratio of miR-203a-3p/miR-16-5p was different among PD patients with cognitive intact and cognitive decline, the nonparametric one-way ANOVA test, Kruskal-Wallis test, was conducted (
Figure 2D.). The result showed that there was significant difference in the ratio of miR-203a-3p/miR-16-5p among the four groups (p-value=0.0009). The mean ratio of miR-203-3p/miR-15-5p was 12.1 (SD, ±6.42), 10.31 (SD, ±5.89), 8.66 (SD, ±5.65) and 16.83 (SD, ±10.12) in each group of HC, PDND, PD-MCI and PDD, respectively. PDD showed statistical significantly increased ratio of miR-203a-3p/miR-16-5p compared to PD-MCI (p-value=0.0006) and PDND (p-value=0.0409). However, the ratio of miRNA had no significant difference in comparison of PDND with PD-MCI. The finding may result from the minimal cognitive decline from PDND to PD-MCI that may be insufficient for detecting the alter expression of plasma miRNA.
2.4. Correlation of the miRNA Expression and the Cognitive Performancer
To understand whether the selected miRNAs are associated with the cognitive impairment, the correlation of the ratio of miR-203a-3p/miR-16-5p and the total score and individual domain score of MoCA were analyzed by Spearman correlation analysis (
Table 3). No significant correlation was observed between the ratio of miR-203a-3p/miR-16-5p and gender, age, education, duration, Hoehn-Yahr stage, UPDRS III, LEDD (p-value>0.05) (data not shown). After Spearman correlation analysis, the ratio of miR-203a-3p/miR-16-5p showed significant negative correlation with total score of MoCA (r=-0.237, p-value=0.024) in PD patients (
Table 3.). To understand the specific cognitive domain that were closely related with the miRNA ratio, different cognitive domains of MoCA, such as visuospatial, naming, attention, language, abstraction, memory and orientation, were analyzed. The results ratio of miR-203a-3p/miR-16-5p had significantly correlation with three MoCA domains, including visuospatial, language and orientation (
Table 3.).
2.5. Using the Ratio of miR-203a-3p/miR-16-5p as Variate for Building Regression Model
The ROC analysis was performed to show the diagnosis power of PD with cognitive decline in PD patients using the ratio of miR-203a-3p/miR-16-5p.
The ROC plots for each comparison group analyzed by the logistic regression with 5-fold cross-validation were summarized in
Table 4. The 95% confidence interval of the sensitivity, specificity and the accuracy of each compared groups were estimated followed by the ROC analysis (
Table 4.). The ROC curve analysis discriminating between PD-MCI and PDD showed an average AUC of 0.716 (95% CI, 0.432-0.951). In addition, the ROC curve analysis discriminating between PDND and PDD showed an average AUC of 0.741 (95% CI, 0.482-0.951). Both ROC analysis supported that the ratio of miR-203a-3p/miR-16-5p could be used for predicting the cognitive status (total score of MoCA≦21).
To understand whether the demographic variables may also serve as confounding factors for detecting PDD (total score of MoCA≤21) from the PwP, the multivariate logistic regression models were performed. The full regression model was developed with the as predictor variates, including age, gender, onset age, year of formal education, score of UPDRS III and the ratio of miR-203a-3p/miR-16-5p that represented as N203 in short. The total score of MoCA was defined as the response variable. The AIC value of each regression model was estimated for their goodness of fit.
The results showed that the reduced model with the three variates, including the ratio of miR-203a-3p/miR-16-5p, age and the score of UPDRS III, represented the best performance for predicting PDD (
Table S1.). The results showed that the ratio of miR-203a-3p/miR-16-5p, age and the score of UPDRS III all have positive associations with PDD. In other words, the elder age and more severe motor symptoms with higher ratio of miR-203a-3p/miR-16-5p could all contribute to the higher risk of being PDD (
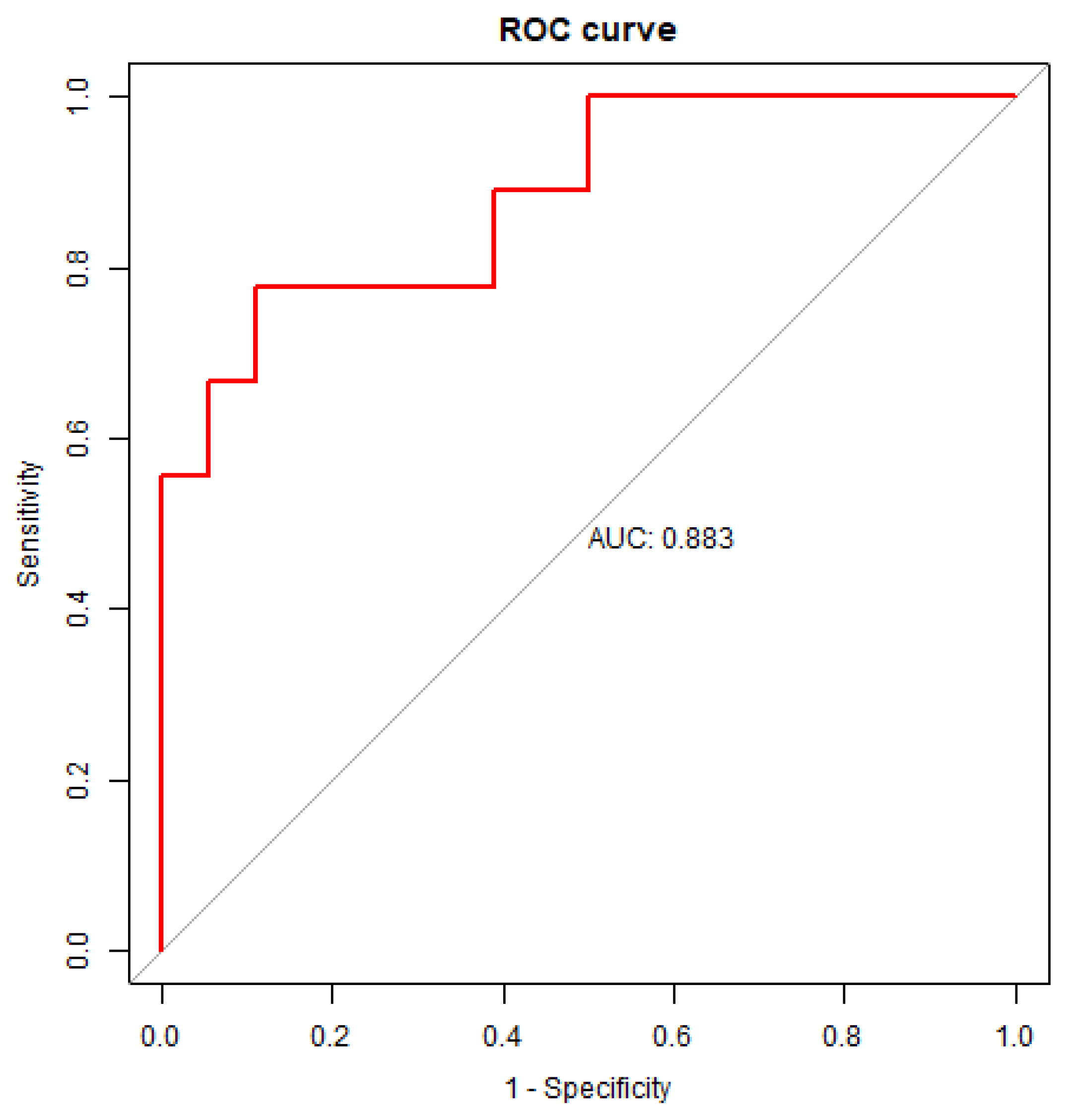
Table S1). After the variable selection, the ROC analysis of test dataset was conducted by a five-fold cross-validation to examine the power of the reduced model and the corresponding ROC curve is shown in
Figure 3. Apart from the abovementioned model, other ROC analysis also performed for the regression models without the variates of N203 to examine whether N203 serve as a predictive parameter for PDD. The ROC analysis and the 95% confidence interval of AUC, specificity, sensitivity and accuracy for the reduced logistic regression model of test dataset were summarized in
Table S2. As a result, the reduced logistic regression model for predicting PDD showed an AUC of 0.8827 (95%CI, 0.7282-0.9938), a sensitivity of 0.7778, specificity of 0.8889, and accuracy of 0.8519 (
Table S2.). The result showed that regression model with the three variates, including N203, age and UPDRS III, had the higher prediction performance (AUC=0.8827) than the regression model with two variates, including age and UPDRS III (AUC=0.8272).
2.6. MiR-203a-3p Associated with Cognitive Related KEGG Pathways
To elucidate the role of miR-203a-3p in the cognitive-related pathological mechanisms of PD, the target genes and the involved molecular pathway were filtered by experimental-based evidence (
Table S3). Five non-cancer related pathways and the predicted target genes of miR-203a-3p were summarized in
Table 5. As a result, KEGG analysis of miR-203a-3p revealed several possible pathways that may associate with the pathology of PD with cognitive dysfunction, including dopaminergic synapse, apoptosis, thyroid hormone signaling pathway, cholinergic synapse and NF-kappa B signaling pathway.
3. Discussion
Common biomarker for differentiating PDD involved in genetic and proteomic biomarkers. It was suggested that genetic mutations such as GBA, MAPT, LRRK2, ApoE may have contribute to increased risk of developing PD or rapid cognitive decline from PDND to PDD [
29] . However, the prediction power of genetic mutation alone remained elusive because the onset age of PwP with known genetic mutation was uncertain and the genetic marker itself may not serve as the prognosis indicator. Hence, the proteomic markers such as circulating pathological proteins, including α-syn, β-Amyloid, tau, NfL, were considered to be the progressing indicators for motor and cognitive performance of PwP by recent studies [30-32]. A one year follow-up study also showed an elevated expression of plasma EV-derived alpha-synuclein, tau and β-amyloid were correlated with the motor and cognition decline in PD [
30]. The general techniques for examining CSF or plasma proteomic targets were based on high-affinity protein purifying column or immunostaining kits such as ELISA assay [
33]. However, it is expensive and requires calibrate management for examining proteomic markers compared to genetic markers. Additionally, the controversy of the protein expression level in different motor and cognition severity of PwP remains to be an unsolved problem. On the other hand, plasma miRNA may provide benefits as state-specific biomarker for the dynamic motor and cognitive status. The alteration of plasma miRNA may also consider as the prognosis indicator for evaluating the predicted pharmacological changes of treatment [
34].
miR-203a-3p was suggested to binding to the 3’UTR of human DJ-1, which is a Parkinson’s disease-related gene and may prevent neurons from cytotic oxidative damage [34-36]. The overexpressed miR-203a-3p was suggested to cause deficiency of DJ-1 and further result in the oxidative stress-induced cell death [34, 35], the microglia-regulated neuronal injury [
37] and promoted neurodegenerative phenotype in vivo [
38]. MiR-203a-3p was also assumed to binding to the 3’UTR of SNCA, encoded for α-syn, which was well-known as elevating risk of developing PD [39, 40]. The aforementioned findings may support our hypothesis for selecting miR-203a-3p as biomarker for PDD. However, reported miRNAs for demented PD are limited due to the poor prognosis and the loss of follow-up patient numbers. The lack of experimental standardized protocol for examining plasma miRNA in human biopsy was an unsolved problem as well.
In the current study, our results indicated that PDD may correlate with severer motor and cognitive dysfunctions under the similar age of onset and duration of disease after diagnosis (
Table 2). It was consistent with previous study that the later age of onset was associated with the rapid progression from non-demented PD to PDD [
41]. The ddPCR detection showed that the ratio of miR-203a-3p/miR-16-5p was significantly increased in PDD compared to PD-MCI and PDND. In addition, the ratio of miR-203a-3p/miR-16-5p had significantly correlation with the total score and the three MoCA domains, including visuospatial, language and orientation. According to previous studies, the three cognitive domains were associated with frontal lobe functions, which corresponded to the pathological brain region of PD with cognitive dysfunction [
41]. Apart from poor executive function, the diminished visuospatial and language functions in PD-MCI and PDD were also highlighted as the features of motor and cognitive decline symptoms [11, 42]. Overall, the findings support our hypothesis that miR-203a-3p may serve as a dynamic biomarker for recognizing global and domain-specific cognitive decline in PwP.
MiR-203a-3p belongs to the miRNA family of miR-203. The underlying genes regulation via miR-203a-3p have been proposed to be related with the cognitive decline in PwP. The KEGG analysis showed multiple target genes of miR-203a-3p consisted in pathways, including apoptosis and NF-kappa B signaling pathway. It is noteworthy that PDD was characterized not only by the aggregation of fibril α-syn but also tau and amyloid plaque pathology [
6]. The correlation of the upregulated miR-203 and the activated apoptotic pathway was first reported by Swarup et al. [
38]. Evidences suggested that miR-203 dysregulation was correlated with tauopathy such as frontal temporal dementia (FTD), AD and progressive supranuclear palsy (PSP). The authors suggested that the downregulation of neurodegeneration-associated synaptic (NAS) module and the upregulation of apoptotic pathway detected by caspase-8 protein expression were resulted from the overexpressed miR-203 in both of the primary cortical mouse neuronal cultures and the Tg4510 tau transgenic mice. Li et al. also reported that the overexpressed miR-203 in both BV2 cells and mouse hippocampus resulted in the reduced protein expression of 14-3-3θ. As the inhibitor of NF-κB signaling and the target of miR-203, 14-3-3θ may inhibit TLR2-induced NF-κB signaling [
43]. The overexpressed miR-203 may also resulted in neuroinflammation and neuronal cell death in the hippocampus of mice which led to spatial learning and memory dysfunction in the Barnes maze test. Taken together, the upregulated miR-203 may cause the activation of inflammation and apoptotic pathway. While the decreased expression of miR-203a-3p may provide the neuron protecting effect in neurodegenerative disorder.
In addition to the genes involved in the apoptotic and inflammation regulation, KEGG analysis also revealed dopaminergic synapse, thyroid hormone signaling pathway and cholinergic synapse were associated with miR-203a-3p. The loss of dopaminergic synapse in the substantia nigra was assumed to be the hallmark for progressed motor symptoms in PwP [
2]. The evidence may support our findings that the upregulated miR-203a-3p/miR-16-5p found in PDD compared to PDND, which indicated the relationship of increased miR-203a-3p and the worse dopaminergic neuron loss. Dysregulated thyroid hormone signaling was also considered as one of the potential cause of cognitive dysfunction in PD. The thyroid disturbance combined with the age above 70 was assumed to be the potential risk factor of developing PD according to the interconnection of the hypothalamic-pituitary-thyroid axis [
44]. Additionally, patients with subclinical hypothyroidism generally had difficulty in gait, which represented the similar clinical motor symptoms in PwP [45, 46]. Degeneration of cholinergic system was assumed to play an important role in multiple neurodegenerative disorders. The dysfunction of cholinergic synapses was generally recognized in Alzheimer’s disease and the loss of basal forebrain cholinergic system was reported in the human post-mortem evidence of PDD as well [
47]. The alteration of cholinergic system may not only change the motor function but also non-motor symptoms [
48]. The severe loss of cholinergic synapse or cholinergic receptor was reported in PD with cognitive decline compared to that with intact cognition [49, 50]. Based on the prior finding, cholinergic drugs were developed to treat cognitive decline in PD. Notably, cholinergic inhibitors such as cholinesterase inhibitor rivastigmine was approved to treat dementia and other related cognitive dysfunctions in PD and AD [51, 52]. While the subtype-specific muscarinic acetylcholine receptors (mAChR) antagonists was proposed as an alternative treatment for cognitive impairment [
49].
There were some limitations in this study. The sample size was limited in this study. It has been observed that plasma examination may be interfered by the mood, medical and surgical therapies of participants [
53], hence a larger sample size and a follow-up study would be needed. Furthermore, since PDD was associated with higher age in our study (
Table 2 and
Table 4), the number of elder HC and PDND should be increase to rule out the aging effect. Although level II neuropsychological assessment was suggested to gain better sensitivity and accuracy when diagnosing PD-MCI, the global cognitive tests MoCA may perform benefit of prediction for conversion of PD-MCI to PDD [
54]. Moreover, this study only measured plasma miRNA, so the exosome-derived miRNA may not be detected via our extraction method. The exosome-derived miRNA extraction would normally requires more preparation than plasma miRNA due to the low yields and extra separation and purification steps [
55]. Hence, cell-free plasma miRNA was preferred when the amount of sample is limited.
4. Materials and Methods
4.1. Plasma miRNA Profiling in the Discovery Cohort
4.1.1. Recruitment of Participants
All patients with PD achieved the inclusion criteria proposed by UK Parkinson’s Disease Society Brain Bank Criteria. Total 174 participants including 40 HC, 51 MSA, 37 PDND, 23 PD-MCI and 23 PDD were recruited. The current study focused on the findings of patients with PD, the analysis of patients with MSA would be discussed in another publication.
4.1.2. Plasma Collection
10 ml of blood was collected in the BD Vacutainer® K2E (EDTA) Plus Blood Collection Tubes (Becton Dickinson, USA) and centrifuged at 2200xg for 15 minutes at room temperature (swinging bucket, KUBOTA 4000, Japan) within 3 hours after blood collection. The plasma layer was transferred, mixed via pipetting and stored at -80℃ until the follow-up experiments.
4.1.3. Plasma miRNA Sequencing
The small RNAs (<200 nucleotides) containing samples from 200-400 μl human plasma were prepared using the Qiagen miRNeasy Mini kit (Qiagen, #217004). The small RNA-Seq library was constructed using the QIAseq miRNA Library Kit (Qiagen, #331502). All single-ended small RNA-Seq was performed on Illumina NextSeq. The resulting small RNA-Seq datasets were generated. The sequencing reads of each microRNAs from two batches of subjects (75 and 99 respectively) were normalized by Trimmed Mean of the M-values (TMM) for each batch first.
4.1.4. BOLD Selector Included Data Analytic Scheme
In miRNA data analysis, we undertake a three-stage process. The initial stage involves data preprocessing. The missing value is approximated as 0. To merge the two datasets (of 75 and 99, surrogate variable analysis in R (SVA; V.3.48) was used to normalize and remove batch effects, the union of the lowest expressed 10% microRNAs from both batches were trimmed out before statistical analysis. The BOLD selector included data analytic scheme [
56], narrowing down the ranking of miRNAs with an adjusted parameter δ, was suitable for clustering biomarker selection in supersaturated data. The second stage focuses on cross-validation for the dataset using the BOLD Selector. Prior to commencing this stage, we standardize the expression matrix X and center the response variable Y, originally coded as 0 and 1. We choose the best δ from 15 uniformly spaced cut points within the interval from 0 to the maximum absolute value of max|X^T Y|. Data are split into 5 parts for cross-validation, with 80% used for training and the remaining part for testing in each fold. We applied the BOLD selector to the training data and constructing a logistic regression formula based on the selected microRNA candidates for predicting the testing data. The model's fitness across 5 testing folds is assessed using the average area under the receiver operating characteristic curve (AUC), enabling us to select the optimal tuning parameter with the highest average AUC. The final stage entails the full data analysis on the tuning parameters falling between the best δ and max|X^T Y|. During this phase, we rank and identify the most important factors, which are subsequently employed to construct a final logistic regression formula.
4.2. Validating Plasma miRNA Candidates in New PD Cohort
4.2.1. Sample Size Estimation
Two miRNAs were measured in the 4 study groups, including Parkinson’s disease with no dementia (PDND), Parkinson’s disease with mild cognitive impairment (PD-MCI), Parkinson’s disease with dementia (PDD) and healthy control (HC, as a control group). Prior sample size estimation was performed by G-power 3.1.9.4 [
57] with the statistic F test of ANOVA (Fixed effects, special, main effects and interactions), effect size=0.5, power=0.8 and α=0.05. Total sample size was suggested to be or above 48. Considering the greater power of statistic and avoid the heterogeneity within each study group, total 120 participants were recruited and examined.
4.2.2. Recruitment of Participants
All PD patients achieved the same inclusion and exclusion criteria as the recruitment of the discovery phase. Participants who received treatments or had history of the following criteria were excluded from this study: (1) cancer, (2) server cardiovascular disease, renal disease, brain injury, (3) autoimmune disease, (4) psychiatric disorder such as schizophrenia, (5) deep brain stimuli surgery, (6) carrying known genetic mutation associated with Parkinson’s disease, (7) having difficulty in blood clotting and (8) other possible cognitive-affected neurological, musculoskeletal disorder or secondary and atypical Parkinsonism, including Multiple System Atrophy, Progressive Supranuclear Palsy, Corticobasal Degeneration, Dementia with Lewy Bodies. In addition, Unified Parkinson’s disease Rating Scale (UPDRS) part III was used to evaluate the motor function of PDND, PD-MCI and PDD. PD-MCI, PDD should achieved the criteria of Montreal Cognitive Assessment (MoCA), the level I cognitive assessment proposed by MDS Task Force. Levodopa equivalent daily dose (LEDD) was calculated via LEDD conversion factor for PD patients [
58]. Moreover, demographic variables included gender, age, duration of disease, score of UPDRS III, total score of MoCA, year of formal education and LEDD were collected for each participant. Anticholinergic drugs, including Akinfree and Biperiden, were estimated by daily dose for each PD patients if treated.
Eventually, 30 PDND, 30 PD-MCI, 30 PDD were recruited from Parkinson disease center in National Taiwan University Hospital. Moreover, 30 HC were recruited from National Taiwan University Hospital and the Shixiang Community in Taiwan. All subjects gave and signed their informed consent for inclusion before they participated in the study.
4.2.3. Cognitive Assessments
All participants received Montreal Cognitive Assessment (MoCA) [
4] to quickly scanning their cognitive performance by Y.F. Hsu. The cognitive domains, including visuospatial, naming, attention, language, abstraction, memory and orientation, were evaluated via MoCA and the total score was used for grouping. HC and PDND should meet the total score of MoCA equal or above 26. PD-MCI should meet the total score of MoCA ranged from 22 to 25. PDD should meet the total score of MoCA equal or below 21.
4.2.4. Plasma Collection
Plasma collection followed the protocol mentioned in the discovery cohort.
4.2.5. RNA Extraction
Small RNAs were extracted from 200 μl plasma using miRNeasy Serum/Plasma Advanced kit (Qiagen, Germany). The extraction process generally followed the guideline of manufacturer’s instructions with several modifications listed below. To test whether the extraction efficiency was robust, the exogenous synthetic spike-in UniSp6 (Qiagen, Germany) was added to the lysis buffer (
Figure S1). However, UniSp6 is recommended for measuring extraction efficiency but not suitable for normalization of miRNA expression level according to the manufacturer’s instructions. Thawed plasma samples underwent a series of centrifugation procedures: centrifuged at 12000xg at 4℃ for 3 minutes (fixed-angled, KUBOTA 6200, Japan) at first, and further centrifuged at 12000xg (fixed-angled, KUBOTA 3300T, Japan) at room temperature for 30 seconds, 30 seconds, 30 seconds, 2 minutes and 5 minutes, respectively. Twenty-two microliters of 55℃ pre-warmed RNase-free water (Invitrogen, Thermo Fisher) was added for RNA elution. Additionally, the eluted RNA was transferred and incubated on UCP MiniElute column (Qiagen, Germany) again for 10 minutes at room temperature. After centrifuging at 12000xg for 1 minute (fixed-angled, KUBOTA 3300T, Japan), the final RNA was placed immediately on ice for reverse transcription (RT). The cDNA synthesis was manipulated by the instructions of miRCURY LNA miRNA SYBR Green kit (Qiagen, Germany). A mixture prepared for cDNA synthesis contained 10 μl extracted RNA, 2μl 10x miRCURY RT Enzyme Mix, 4 μl 5x miRCURY SYBR Green RT Reaction Buffer and 4 μl RNase-free water (Invitrogen, Thermo Fisher). After the thermal reaction cycle complete, the cDNA sample was stored at -20℃ until ddPCR examination.
4.2.6. Droplet Digital PCR
For examination of miR-203a-3p and miR-16-5p, the cDNA samples were diluted by 1:10 and 1:320, respectively. One Non-RT control (no transcriptase added during RT) and one non-template control (no cDNA template added during ddPCR amplification; NTC) were quantified to make sure no genomic DNA remained and no false positive result detected. The miRNA was quantified using the ddPCR system (Bio-Rad, USA). First, total volume of 20 μl mixture was prepared with 9 μl the diluted cDNA, 10 μl digital PCR™ Eva-Green supermix (Bio-Rad, USA) and 1μl LNA miRCURY miRNA PCR Assay (for miR-203a-3p and miR-16-5p) (Qiagen, Germany). Second, 70 μl QX100 Droplet Generation oil (Bio-Rad, USA) and the 20 μl mixture were loaded into a cartridge. After processing in QX200 Droplet Generator (Bio-Rad, USA), the droplet-contained liquid was gently transferred into a 96-well plate and started the cDNA amplification in the T100 thermal cycler (Bio-Rad, USA). The thermal cycling was modified from the manufacturer’s instructions (Bio-Rad, USA). The annealing temperature (Tm) was adjusted individually for miR-203a-3p (Tm=55℃) and miR-16-5p (Tm=56℃). The PCR products were quantified by the QX200 Droplet Reader (Bio-Rad, USA) and QuantaSoft software (Bio-Rad, USA). The quality control of ddPCR result was suggested to meet the criteria of total droplet numbers above 10000 copies/μl and total positive droplets above 3 copies/μl. The copies number per microliter of miR-203a-3p divided by that of miR-16-5p was then multiplied by 10000 to obtain the ratio of miRNA.
4.2.7. Pathway Prediction
As a well-known database which integrates up-to-date genomic information, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis provides graphic maps for genes of interest to interpret and predict the molecule cascades and potential function [59-61]. Hence, the target genes of miR-203a-3p were predicted using KEGG pathway database using miRPathDB 2.0 and the predicted pathways were filtered by the experimental(any) data [
62].
4.2.8. Statistical Analysis
The demographic variables and the ratio of miR-203a-3p/miR-16-5p were analyzed by Kruskal-Wallis test via GraphPad Prism. Demographic variables analyzed by non-parametric one-way ANOVA Krustall-walli tests included gender, age, total score of MoCA, duration of disease and the score of UPDRS III, year of formal education and LEDD. After performing Kruskal-Wallis tests, Dunn's multiple comparisons tests were conducted for post-hoc analysis to reveal whether significant differences between which study groups. Additionally, outliers were identified and excluded from analysis if it was larger than Q3 +1.5IQR or smaller than Q1 -1.5IQR. Spearman correlation tests, a nonparametric correlation analysis, were applied for the ratio of miR-203a-3p/miR-16-5p, different cognitive domains (visuospatial, naming, attention, language, abstraction, memory and orientation) and the demographic variables. To understand the diagnosis power of PD with or without cognition decline using the ratio of miR-203a-3p/miR-16-5p, the 4 study groups were compared and analyzed by the receiver operating characteristic (ROC) curve analysis using R software (version 4.3.2), package pROC (version 1.18.5) [
63] and package caret (version 6.0-94) [
64] . In addition, the multivariate logistic regression model was developed consisting of the demographic variates and the ratio of miRNA factors. The demographic and miRNA data of 90 PD patients were divided by 70% for model training and 30% for model testing. The reduced logistic regression model was determined via the smallest Akaike Information Criteria (AIC). The predicted values of average AUC obtained by 5-fold cross-validation were used in the test dataset evaluation. All coordinates of ROC curves, including the area under curve (AUC), sensitivity, specificity and accuracy, were estimated by the maximal sum (sensitivity + specificity). Additionally, the 95% confidence interval (CI) were also calculated for AUC, sensitivity, specificity and accuracy using bootstrapping (boot runs = 2000).
5. Conclusions
In summary, plasma miR-203a-3p was significantly increased in PDD compared to PD-MCI and PDND. Moreover, the ratio of miR-203a-3p/miR-16-5p was significantly correlated to MoCA with its total score and multiple domain such as visuospatial, language and orientation. Combining with age, ratio of miRNA and UPDRS III, the logistic regression model (AUC = 0.883) may facilitate the diagnosis of PDD from PwP. Therefore, the ratio of plasma miR-203a-3p/miR-16-5p may be a novel biofluid marker for patients of PD with cognitive dysfunction.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org.
Author Contributions
Conceptualization, Y.-F.H., S.-P.L. and R.-M.W.; Methodology, Y.-F.H. and Y.-T.Y.; Formal Analysis, Y.-F.H., Y.-T.C. and F.K.H.P.; Investigation, Y.-F.H.; Resources, R.-M.W.; Writing – Original Draft Preparation, Y.-F.H. and R.-M.W.; Visualization, Y.-F.H.; Supervision, R.-M.W.; Funding Acquisition, S.-P.L. and R.-M.W. All authors have read and agreed to the published version of the manuscript before submission.
Funding
This research was funded by National Taiwan University and National Taiwan University Hospital, grant number 111-UN0007.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the National Taiwan University Hospital Research Ethics Committee, Taipei, Taiwan. (NTUH-REC No. 201711054RINA, date: 7 February 2018; NTUH-REC No. 201912129RINB, date: 17 January 2020; NTUH-REC No. 201905113RINC, date: 18 September 2019; NTUH-REC No. 202011020RINC, date: 21 December 2020).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to the privacy of participants.
Acknowledgments
We thank Prof. Takahiro Ochiya for technical support during the discovery phase. We thank Miss Jing-Wen Huang for developing BOLD selector data analytic scheme and Mr. Yan-Ru Ju for statistically analyzing the NGS sequencing dataset. We thank the support from the Centre for Parkinson and Movement Disorders during the participant recruitment process. We also thank the staff of the Second and the Fifth Core Lab, Department of Medical Research, National Taiwan University Hospital for technical support during the study. We thank Dr. Sam Chi-Hao, Liu for English editing of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Aarsland, D.; Batzu, L.; Halliday, G. M.; Geurtsen, G. J.; Ballard, C.; Ray Chaudhuri, K.; Weintraub, D., Parkinson disease-associated cognitive impairment. Nat Rev Dis Primers 2021, 7, (1), 47. [CrossRef] [PubMed]
- Armstrong, M. J.; Okun, M. S., Diagnosis and Treatment of Parkinson Disease: A Review. JAMA 2020, 323, (6), 548-560. [CrossRef] [PubMed]
- Yu, R. L.; Wu, R. M.; Tai, C. H.; Lin, C. H.; Cheng, T. W.; Hua, M. S., Neuropsychological profile in patients with early stage of Parkinson's disease in Taiwan. Parkinsonism Relat Disord 2012, 18, (10), 1067-72. [CrossRef] [PubMed]
- Litvan, I.; Goldman, J. G.; Troster, A. I.; Schmand, B. A.; Weintraub, D.; Petersen, R. C.; Mollenhauer, B.; Adler, C. H.; Marder, K.; Williams-Gray, C. H.; Aarsland, D.; Kulisevsky, J.; Rodriguez-Oroz, M. C.; Burn, D. J.; Barker, R. A.; Emre, M., Diagnostic criteria for mild cognitive impairment in Parkinson's disease: Movement Disorder Society Task Force guidelines. Mov Disord 2012, 27, (3), 349-56. [CrossRef] [PubMed]
- Åström, D. O.; Simonsen, J.; Raket, L. L.; Sgarbi, S.; Hellsten, J.; Hagell, P.; Norlin, J. M.; Kellerborg, K.; Martinez-Martin, P.; Odin, P., High risk of developing dementia in Parkinson's disease: a Swedish registry-based study. Sci Rep 2022, 12, (1), 16759. [CrossRef] [PubMed]
- Irwin, D. J.; Lee, V. M.; Trojanowski, J. Q., Parkinson's disease dementia: convergence of alpha-synuclein, tau and amyloid-beta pathologies. Nat Rev Neurosci 2013, 14, (9), 626-36. [CrossRef] [PubMed]
- Dorsey, E. R.; Bloem, B. R., The Parkinson Pandemic-A Call to Action. JAMA Neurol 2018, 75, (1), 9-10. [CrossRef] [PubMed]
- Elkouzi, A.; Vedam-Mai, V.; Eisinger, R. S.; Okun, M. S., Emerging therapies in Parkinson disease - repurposed drugs and new approaches. Nat Rev Neurol 2019, 15, (4), 204-223. [CrossRef] [PubMed]
- Kao, Y. H.; Hsu, C. C.; Yang, Y. H., A Nationwide Survey of Dementia Prevalence in Long-Term Care Facilities in Taiwan. J Clin Med 2022, 11, (6). [CrossRef]
- Lin, Y.-Y.; Huang, C.-S., Aging in Taiwan: Building a Society for Active Aging and Aging in Place. The Gerontologist 2015, 56, (2), 176-183. [CrossRef]
- Huertas, I.; Jesús, S.; García-Gómez, F. J.; Lojo, J. A.; Bernal-Bernal, I.; Bonilla-Toribio, M.; Martín-Rodriguez, J. F.; García-Solís, D.; Gómez-Garre, P.; Mir, P., Genetic factors influencing frontostriatal dysfunction and the development of dementia in Parkinson's disease. PLoS One 2017, 12, (4), e0175560. [CrossRef]
- Lanskey, J. H.; McColgan, P.; Schrag, A. E.; Acosta-Cabronero, J.; Rees, G.; Morris, H. R.; Weil, R. S., Can neuroimaging predict dementia in Parkinson's disease? Brain 2018, 141, (9), 2545-2560. [CrossRef] [PubMed]
- Dubois, B.; Burn, D.; Goetz, C.; Aarsland, D.; Brown, R. G.; Broe, G. A.; Dickson, D.; Duyckaerts, C.; Cummings, J.; Gauthier, S.; Korczyn, A.; Lees, A.; Levy, R.; Litvan, I.; Mizuno, Y.; McKeith, I. G.; Olanow, C. W.; Poewe, W.; Sampaio, C.; Tolosa, E.; Emre, M., Diagnostic procedures for Parkinson's disease dementia: recommendations from the movement disorder society task force. Mov Disord 2007, 22, (16), 2314-24. [CrossRef] [PubMed]
- Htike, T. T.; Mishra, S.; Kumar, S.; Padmanabhan, P.; Gulyas, B., Peripheral Biomarkers for Early Detection of Alzheimer's and Parkinson's Diseases. Mol Neurobiol 2019, 56, (3), 2256-2277. [CrossRef] [PubMed]
- O'Brien, J.; Hayder, H.; Zayed, Y.; Peng, C., Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front Endocrinol (Lausanne) 2018, 9, 402. [CrossRef] [PubMed]
- Kuo, M. C.; Liu, S. C.; Hsu, Y. F.; Wu, R. M., The role of noncoding RNAs in Parkinson's disease: biomarkers and associations with pathogenic pathways. J Biomed Sci 2021, 28, (1), 78. [CrossRef] [PubMed]
- Weber, J. A.; Baxter, D. H.; Zhang, S.; Huang, D. Y.; Huang, K. H.; Lee, M. J.; Galas, D. J.; Wang, K., The microRNA spectrum in 12 body fluids. Clin Chem 2010, 56, (11), 1733-41. [CrossRef] [PubMed]
- Chen, L.; Yang, J.; Lu, J.; Cao, S.; Zhao, Q.; Yu, Z., Identification of aberrant circulating miRNAs in Parkinson's disease plasma samples. Brain Behav 2018, 8, (4), e00941. [CrossRef] [PubMed]
- Arshad, A. R.; Sulaiman, S. A.; Saperi, A. A.; Jamal, R.; Mohamed Ibrahim, N.; Abdul Murad, N. A., MicroRNAs and Target Genes As Biomarkers for the Diagnosis of Early Onset of Parkinson Disease. Front Mol Neurosci 2017, 10, 352. [CrossRef] [PubMed]
- Batistela, M. S.; Josviak, N. D.; Sulzbach, C. D.; de Souza, R. L., An overview of circulating cell-free microRNAs as putative biomarkers in Alzheimer's and Parkinson's Diseases. Int J Neurosci 2017, 127, (6), 547-558. [CrossRef]
- Chiu, P. Y.; Yang, F. C.; Chiu, M. J.; Lin, W. C.; Lu, C. H.; Yang, S. Y., Relevance of plasma biomarkers to pathologies in Alzheimer's disease, Parkinson's disease and frontotemporal dementia. Sci Rep 2022, 12, (1), 17919. [CrossRef]
- Fan, T. S.; Liu, S. C.; Wu, R. M., Alpha-Synuclein and Cognitive Decline in Parkinson Disease. Life (Basel, Switzerland) 2021, 11, (11). [CrossRef]
- Pritchard, C. C.; Cheng, H. H.; Tewari, M., MicroRNA profiling: approaches and considerations. Nature Reviews Genetics 2012, 13, (5), 358-369. [CrossRef] [PubMed]
- Hindson, C. M.; Chevillet, J. R.; Briggs, H. A.; Gallichotte, E. N.; Ruf, I. K.; Hindson, B. J.; Vessella, R. L.; Tewari, M., Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat Methods 2013, 10, (10), 1003-5. [CrossRef] [PubMed]
- Faraldi, M.; Gomarasca, M.; Banfi, G.; Lombardi, G., Free Circulating miRNAs Measurement in Clinical Settings: The Still Unsolved Issue of the Normalization. Advances in clinical chemistry 2018, 87, 113-139. [CrossRef] [PubMed]
- Fehlmann, T.; Lehallier, B.; Schaum, N.; Hahn, O.; Kahraman, M.; Li, Y.; Grammes, N.; Geffers, L.; Backes, C.; Balling, R.; Kern, F.; Kruger, R.; Lammert, F.; Ludwig, N.; Meder, B.; Fromm, B.; Maetzler, W.; Berg, D.; Brockmann, K.; Deuschle, C.; von Thaler, A. K.; Eschweiler, G. W.; Milman, S.; Barziliai, N.; Reichert, M.; Wyss-Coray, T.; Meese, E.; Keller, A., Common diseases alter the physiological age-related blood microRNA profile. Nat Commun 2020, 11, (1), 5958. [CrossRef] [PubMed]
- Ravanidis, S.; Bougea, A.; Papagiannakis, N.; Maniati, M.; Koros, C.; Simitsi, A. M.; Bozi, M.; Pachi, I.; Stamelou, M.; Paraskevas, G. P.; Kapaki, E.; Moraitou, M.; Michelakakis, H.; Stefanis, L.; Doxakis, E., Circulating Brain-enriched MicroRNAs for detection and discrimination of idiopathic and genetic Parkinson's disease. Mov Disord 2020, 35, (3), 457-467. [CrossRef] [PubMed]
- Marques, T. M.; Kuiperij, H. B.; Bruinsma, I. B.; van Rumund, A.; Aerts, M. B.; Esselink, R. A. J.; Bloem, B. R.; Verbeek, M. M., MicroRNAs in Cerebrospinal Fluid as Potential Biomarkers for Parkinson's Disease and Multiple System Atrophy. Mol Neurobiol 2017, 54, (10), 7736-7745. [CrossRef] [PubMed]
- Ding, J.; Zhang, J.; Wang, X.; Zhang, L.; Jiang, S.; Yuan, Y.; Li, J.; Zhu, L.; Zhang, K., Relationship between the plasma levels of neurodegenerative proteins and motor subtypes of Parkinson's disease. J Neural Transm (Vienna) 2017, 124, (3), 353-360. [CrossRef]
- Chan, L.; Chung, C. C.; Hsieh, Y. C.; Wu, R. M.; Hong, C. T., Plasma extracellular vesicle tau, beta-amyloid, and alpha-synuclein and the progression of Parkinson's disease: a follow-up study. Ther Adv Neurol Disord 2023, 16, 17562864221150329. [CrossRef]
- Huang, Y.; Huang, C.; Zhang, Q.; Shen, T.; Sun, J., Serum NFL discriminates Parkinson disease from essential tremor and reflect motor and cognition severity. BMC neurology 2022, 22, (1), 39. [CrossRef] [PubMed]
- Lin, W. T.; Shaw, J. S.; Cheng, F. Y.; Chen, P. H., Plasma total tau predicts executive dysfunction in Parkinson's disease. Acta neurologica Scandinavica 2022, 145, (1), 30-37. [CrossRef]
- Coughlin, D. G.; Irwin, D. J., Fluid and Biopsy Based Biomarkers in Parkinson's Disease. Neurotherapeutics : the journal of the American Society for Experimental NeuroTherapeutics 2023, 20, (4), 932-954. [CrossRef]
- Tsoporis, J. N.; Ektesabi, A. M.; Gupta, S.; Izhar, S.; Salpeas, V.; Rizos, I. K.; Kympouropoulos, S. P.; Dos Santos, C. C.; Parker, T. G.; Rizos, E., A longitudinal study of alterations of circulating DJ-1 and miR203a-3p in association to olanzapine medication in a sample of first episode patients with schizophrenia. J Psychiatr Res 2022, 146, 109-117. [CrossRef]
- Antipova, D.; Bandopadhyay, R., Expression of DJ-1 in Neurodegenerative Disorders. Advances in experimental medicine and biology 2017, 1037, 25-43. [CrossRef] [PubMed]
- Biosa, A.; Sandrelli, F.; Beltramini, M.; Greggio, E.; Bubacco, L.; Bisaglia, M., Recent findings on the physiological function of DJ-1: Beyond Parkinson's disease. Neurobiology of disease 2017, 108, 65-72. [CrossRef]
- He, C.; Li, Z.; Yang, M.; Yu, W.; Luo, R.; Zhou, J.; He, J.; Chen, Q.; Song, Z.; Cheng, S., Non-Coding RNA in Microglia Activation and Neuroinflammation in Alzheimer's Disease. J Inflamm Res 2023, 16, 4165-4211. [CrossRef]
- Swarup, V.; Hinz, F. I.; Rexach, J. E.; Noguchi, K. I.; Toyoshiba, H.; Oda, A.; Hirai, K.; Sarkar, A.; Seyfried, N. T.; Cheng, C.; Haggarty, S. J.; Grossman, M.; Van Deerlin, V. M.; Trojanowski, J. Q.; Lah, J. J.; Levey, A. I.; Kondou, S.; Geschwind, D. H., Identification of evolutionarily conserved gene networks mediating neurodegenerative dementia. Nat Med 2019, 25, (1), 152-164. [CrossRef]
- Marchese, D.; Botta-Orfila, T.; Cirillo, D.; Rodriguez, J. A.; Livi, C. M.; Fernández-Santiago, R.; Ezquerra, M.; Martí, M. J.; Bechara, E.; Tartaglia, G. G., Discovering the 3' UTR-mediated regulation of alpha-synuclein. Nucleic Acids Res 2017, 45, (22), 12888-12903. [CrossRef] [PubMed]
- Nies, Y. H.; Mohamad Najib, N. H.; Lim, W. L.; Kamaruzzaman, M. A.; Yahaya, M. F.; Teoh, S. L., MicroRNA Dysregulation in Parkinson's Disease: A Narrative Review. Front Neurosci 2021, 15, 660379. [CrossRef]
- Phongpreecha, T.; Cholerton, B.; Mata, I. F.; Zabetian, C. P.; Poston, K. L.; Aghaeepour, N.; Tian, L.; Quinn, J. F.; Chung, K. A.; Hiller, A. L.; Hu, S. C.; Edwards, K. L.; Montine, T. J., Multivariate prediction of dementia in Parkinson's disease. NPJ Parkinsons Dis 2020, 6, 20. [CrossRef]
- Auclair-Ouellet, N.; Lieberman, P.; Monchi, O., Contribution of language studies to the understanding of cognitive impairment and its progression over time in Parkinson's disease. Neurosci Biobehav Rev 2017, 80, 657-672. [CrossRef] [PubMed]
- Li, S.; Li, L.; Li, J.; Liang, X.; Song, C.; Zou, Y., miR-203, fine-tunning neuroinflammation by juggling different components of NF-kappaB signaling. J Neuroinflammation 2022, 19, (1), 84. [CrossRef]
- Kim, J. H.; Lee, H. S.; Ahn, J. H.; Oh, J. K.; Chang, I. B.; Song, J. H.; Wee, J. H.; Min, C. Y.; Yoo, D. M.; Choi, H. G., Association Between Thyroid Diseases and Parkinson's Disease: A Nested Case-Control Study Using a National Health Screening Cohort. J Parkinsons Dis 2021, 11, (1), 211-220. [CrossRef]
- Iadecola, C., The Neurovascular Unit Coming of Age: A Journey through Neurovascular Coupling in Health and Disease. Neuron 2017, 96, (1), 17-42. [CrossRef]
- Bano, A.; Chaker, L.; Darweesh, S. K.; Korevaar, T. I.; Mattace-Raso, F. U.; Dehghan, A.; Franco, O. H.; van der Geest, J. N.; Ikram, M. A.; Peeters, R. P., Gait patterns associated with thyroid function: The Rotterdam Study. Sci Rep 2016, 6, 38912. [CrossRef]
- Bohnen, N. I.; Albin, R. L., The cholinergic system and Parkinson disease. Behavioural brain research 2011, 221, (2), 564-73. [CrossRef]
- Sanjari Moghaddam, H.; Zare-Shahabadi, A.; Rahmani, F.; Rezaei, N., Neurotransmission systems in Parkinson's disease. Reviews in the neurosciences 2017, 28, (5), 509-536. [CrossRef]
- Bohnen, N. I.; Yarnall, A. J.; Weil, R. S.; Moro, E.; Moehle, M. S.; Borghammer, P.; Bedard, M. A.; Albin, R. L., Cholinergic system changes in Parkinson's disease: emerging therapeutic approaches. Lancet Neurol 2022, 21, (4), 381-392. [CrossRef]
- Colloby, S. J.; Nathan, P. J.; Bakker, G.; Lawson, R. A.; Yarnall, A. J.; Burn, D. J.; O'Brien, J. T.; Taylor, J. P., Spatial Covariance of Cholinergic Muscarinic M(1) /M(4) Receptors in Parkinson's Disease. Mov Disord 2021, 36, (8), 1879-1888. [CrossRef] [PubMed]
- Hampel, H.; Mesulam, M. M.; Cuello, A. C.; Farlow, M. R.; Giacobini, E.; Grossberg, G. T.; Khachaturian, A. S.; Vergallo, A.; Cavedo, E.; Snyder, P. J.; Khachaturian, Z. S., The cholinergic system in the pathophysiology and treatment of Alzheimer's disease. Brain 2018, 141, (7), 1917-1933. [CrossRef] [PubMed]
- Kandiah, N.; Pai, M. C.; Senanarong, V.; Looi, I.; Ampil, E.; Park, K. W.; Karanam, A. K.; Christopher, S., Rivastigmine: the advantages of dual inhibition of acetylcholinesterase and butyrylcholinesterase and its role in subcortical vascular dementia and Parkinson's disease dementia. Clinical interventions in aging 2017, 12, 697-707. [CrossRef]
- Litvan, I.; Kieburtz, K.; Troster, A. I.; Aarsland, D., Strengths and challenges in conducting clinical trials in Parkinson's disease mild cognitive impairment. Mov Disord 2018, 33, (4), 520-527. [CrossRef]
- Boel, J. A.; de Bie, R. M. A.; Schmand, B. A.; Dalrymple-Alford, J. C.; Marras, C.; Adler, C. H.; Goldman, J. G.; Troster, A. I.; Burn, D. J.; Litvan, I.; Geurtsen, G. J.; Disease, M. D. S. S. G. M. C. I. i. P. s., Level I PD-MCI Using Global Cognitive Tests and the Risk for Parkinson's Disease Dementia. Mov Disord Clin Pract 2022, 9, (4), 479-483. [CrossRef] [PubMed]
- Li, X.; Corbett, A. L.; Taatizadeh, E.; Tasnim, N.; Little, J. P.; Garnis, C.; Daugaard, M.; Guns, E.; Hoorfar, M.; Li, I. T. S., Challenges and opportunities in exosome research-Perspectives from biology, engineering, and cancer therapy. APL bioengineering 2019, 3, (1), 011503. [CrossRef] [PubMed]
- Huang, J. W.; Lin, Y. H.; Phoa, F. K. H.; Lin, S. P.; Tsai, Y. T.; Kuo, M. C.; Ueda, K.; Wu, R. M., Differentiating Patient Group across Parkinsonism Spectrum via the Biomedical Oriented Logistic Dantzig Selector (BOLD Selector). (manuscript in submission) 2023.
- Faul, F.; Erdfelder, E.; Buchner, A.; Lang, A. G., Statistical power analyses using G*Power 3.1: tests for correlation and regression analyses. Behavior research methods 2009, 41, (4), 1149-60. [CrossRef]
- Jost, S. T.; Kaldenbach, M. A.; Antonini, A.; Martinez-Martin, P.; Timmermann, L.; Odin, P.; Katzenschlager, R.; Borgohain, R.; Fasano, A.; Stocchi, F.; Hattori, N.; Kukkle, P. L.; Rodriguez-Violante, M.; Falup-Pecurariu, C.; Schade, S.; Petry-Schmelzer, J. N.; Metta, V.; Weintraub, D.; Deuschl, G.; Espay, A. J.; Tan, E. K.; Bhidayasiri, R.; Fung, V. S. C.; Cardoso, F.; Trenkwalder, C.; Jenner, P.; Ray Chaudhuri, K.; Dafsari, H. S.; International, P.; Movement Disorders Society Non-Motor Parkinson Disease Study, G., Levodopa Dose Equivalency in Parkinson's Disease: Updated Systematic Review and Proposals. Mov Disord 2023, 38, (7), 1236-1252. [CrossRef]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Kawashima, M.; Ishiguro-Watanabe, M., KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res 2022. [CrossRef] [PubMed]
- Kanehisa, M., Toward understanding the origin and evolution of cellular organisms. Protein science : a publication of the Protein Society 2019, 28, (11), 1947-1951. [CrossRef]
- Kanehisa, M.; Goto, S., KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000, 28, (1), 27-30. [CrossRef] [PubMed]
- Kehl, T.; Kern, F.; Backes, C.; Fehlmann, T.; Stöckel, D.; Meese, E.; Lenhof, H. P.; Keller, A., miRPathDB 2.0: a novel release of the miRNA Pathway Dictionary Database. Nucleic Acids Res 2020, 48, (D1), D142-d147. [CrossRef]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J. C.; Müller, M., pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC bioinformatics 2011, 12, 77. [CrossRef]
- Kuhn, M., Building Predictive Models in R Using the caret Package. Journal of Statistical Software 2008, 28, (5), 1 - 26. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).