1. Introduction
UXS1 (UDP-Glucuronate Decarboxylase 1) is a gene encodes an enzyme located in the perinuclear Golgi that facilitates the production of UDP-xylose, a crucial component in the synthesis of glycosaminoglycans (GAGs) on proteoglycans. These GAG chains become covalently attached to proteoglycans, playing a pivotal role in signaling pathways during the process of development [
1,
2].
UXS plays a key role in the biosynthesis of the polysaccharide xylose (Xyl), an important component of many cellular structures, including glycosaminoglycans [
3]. This enzyme is part of the short-chain dehydrogenase/reductase (SDR) superfamily. The SDR superfamily is a large group of enzymes that share structural similarities and are involved in a diverse range of biological processes, including the metabolism of various compounds [
4].
According studies conducted by Michelle Patricia Cicchini et al. 2023 aimed to validate UXS1 as a synthetic lethal target in UGDH-high cancers. In vitro and in vivo knockout experiments using CRISPR technology were conducted across lung carcinoma cell lines. Their findings suggested that dysregulation of metabolites occurs due to high UGDH expression when UXS1 knockout disrupts the conversion of UDP-glucuronic acid (UDP-GA) to UDP-xylose (UDP-X), leading to a synthetic lethal effect on cell growth [
5]. In addition, the work published by Doshi et al., 2023 reported that UXS1 plays a crucial role in preventing the excessive accumulation of UDP-glucuronic acid (UDPGA), a product of UGDH (UDP-glucuronate dehydrogenase). Beyond simply eliminating UDPGA, UXS1 also exerts control over its production through negative feedback regulation on UGDH. The surplus UDPGA disrupts Golgi morphology and function, impeding the trafficking of surface receptors like EGFR (epidermal growth factor ) to the plasma membrane and diminishing cellular signaling capacity. UGDH is notably upregulated in various tumors, including lung adenocarcinomas, with increased expression observed during chemoresistance. Consequently, tumor cells exhibit a selective dependence on UXS1 for detoxifying UDPGA, revealing a potential vulnerability in tumors characterized by elevated UGDH levels.
The primary objective of this communication was to investigate the potential role of Polydatin, a natural polyphenol known for its low toxicity and anti-tumor effects [
7], through the application of Molecular Docking methods [
8]. Molecular Docking is a computational technique used to predict the binding interactions between small molecules [
9], like Polydatin, and specific target proteins, providing insights into potential therapeutic applications.
2. Material and Methods
UDP-glucuronic acid decarboxylase 1 was performed by Mcule Database [
10] by Autodock Vina [
11].
-UDP-glucuronic acid decarboxylase was taked by Protein Data Bank (PDB Code: 2b69,UXS1_HUMAN); Grid box Coordinates of Binding site center X(50,6115), Y(20,838) Z(23,3764).
3. Results and Discussion
Two recent studies, one led by Michelle Patricia Cicchini et al. in 2023 [
5] and another by Doshi et al. in 2023 [
6], focused on validating UXS1 as a synthetic lethal target in UGDH-high cancers. The studies conducted in vitro and in vivo knockout experiments using CRISPR technology in lung carcinoma cell lines. The findings indicated that dysregulation of metabolites results from high UGDH expression when UXS1 knockout disrupts the conversion of UDP-glucuronic acid (UDP-GA) to UDP-xylose (UDP-X), leading to a synthetic lethal effect on cell growth [
5]. Additionally, the work by Doshi et al. highlighted UXS1's crucial role in preventing the excessive accumulation of UDP-glucuronic acid (UDPGA), a UGDH product. UXS1 not only eliminates UDPGA but also regulates its production through negative feedback on UGDH. The surplus UDPGA disrupts Golgi morphology and function, impacting surface receptor trafficking and reducing cellular signaling in tumors with elevated UGDH levels [
6].
This communication aimed to explore the potential of Polydatin [
7], a natural polyphenol recognized for low toxicity and anti-tumor effects, through Molecular Docking methods. Molecular Docking, a computational technique [
8,
9], helps predict the binding interactions between Polydatin and specific target proteins, offering insights into potential therapeutic applications.
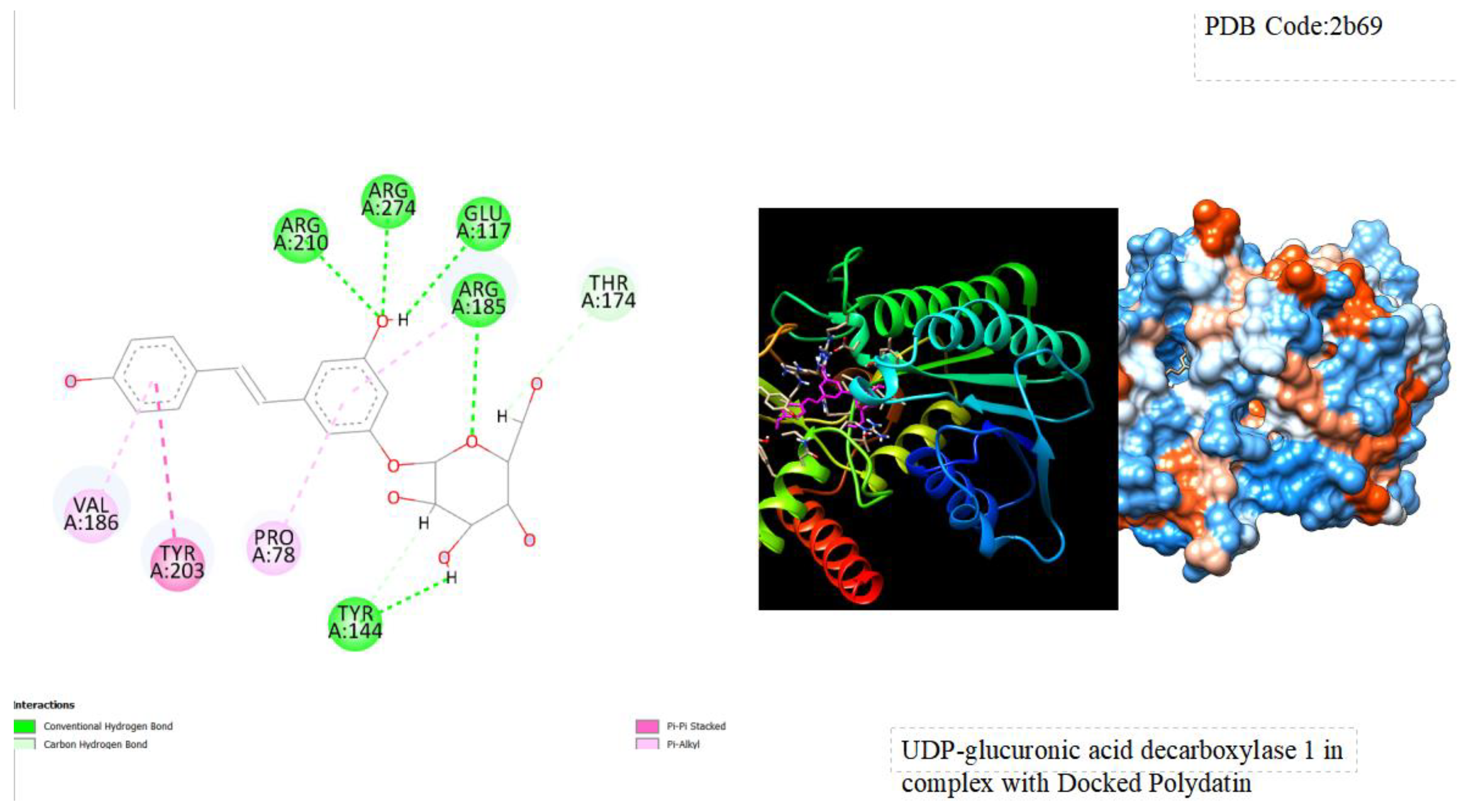
Figure 1.
displays the docking outcomes of Crystal Structure of Human UDP-glucoronic acid decarboxylase in conjunction with Polydatin within the Ligand Binding Site, as analyzed by Autodock Vina through the Mcule Database. On the left side, 2D diagrams illustrate the residue interactions between the protein and Polydatin. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Polydatin.
Figure 1.
displays the docking outcomes of Crystal Structure of Human UDP-glucoronic acid decarboxylase in conjunction with Polydatin within the Ligand Binding Site, as analyzed by Autodock Vina through the Mcule Database. On the left side, 2D diagrams illustrate the residue interactions between the protein and Polydatin. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Polydatin.
Table 1.
shows the comparison of predicted toxicity parameters with Polydatin through pKCSM Server.
Table 1.
shows the comparison of predicted toxicity parameters with Polydatin through pKCSM Server.
| Compounds |
AMES
toxicity |
Max.
tolerated dose(human) (logmg/kg/day) |
hERG I inhibitor |
hERG II inhibitor |
Oral Rat Acute Toxicity (LD50) (mol/kg) |
Oral Rat Chronic Toxicity (LOAEL)
(log mg/kg_bw/day) |
Hepatotoxicity |
| Polydatin |
no |
0.569 |
no |
no |
2.516
|
4.473 |
no |
4. Conclusion
Polydatin, a polyphenolic compound derived from diverse natural sources, has attracted interest due to its promising anti-tumor properties coupled with minimal toxicity. In this study utilizing Molecular Docking methods, we sought to unravel the molecular interactions between Polydatin and the Crystal Structure of Human UDP-glucuronic acid decarboxylase (UXS1), shedding light on the mechanisms contributing to its anti-tumor effects. The study revealed a notable binding energy value of approximately -10 kcal/mol between Polydatin and the target UXS1, indicating a strong and favorable binding interaction. This finding suggests that Polydatin holds significant potential in the realm of cancer research, offering insights into its ability to interact with and potentially modulate the activity of UXS1. The study's emphasis on molecular docking provides a computational perspective on the favorable binding between Polydatin and UXS1, supporting its consideration as a candidate for further exploration in cancer therapeutic strategies.
References
- Kuang, B.; Zhao, X.; Zhou, C.; Zeng, W.; Ren, J.; Ebert, B.; Beahan, C.T.; Deng, X.; Zeng, Q.; Zhou, G.; Doblin, M.S. Role of UDP-glucuronic acid decarboxylase in xylan biosynthesis in Arabidopsis. Molecular plant 2016, 9, 1119–1131. [Google Scholar] [CrossRef] [PubMed]
- Coyne, M.J.; Fletcher, C.M.; Reinap, B.; Comstock, L.E. UDP-glucuronic acid decarboxylases of Bacteroides fragilis and their prevalence in bacteria. Journal of bacteriology 2011, 193, 5252–5259. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Watanabe, K.; Masumura, T.; Kitamura, S. Characterization of soluble and putative membrane-bound UDP-glucuronic acid decarboxylase (OsUXS) isoforms in rice. Archives of biochemistry and biophysics 2004, 431, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Kallberg, Y.; Oppermann, U.; Persson, B. Classification of the short-chain dehydrogenase/reductase superfamily using hidden Markov models. The FEBS journal 2010, 277, 2375–2386. [Google Scholar] [CrossRef] [PubMed]
- Cicchini, M.P.; Wu, S.; Peng, Y.; Rao, Y. UXS1 is a synthetic lethal target in cancer. Cancer Research 2023, 83 (Suppl. 7), 3707. [Google Scholar] [CrossRef]
- Doshi, M.B.; Lee, N.; Tseyang, T.; Ponomarova, O.; Goel, H.L.; Spears, M.; Li, R.; Zhu, L.J.; Ashwood, C.; Simin, K.; Jang, C. Disruption of sugar nucleotide clearance is a therapeutic vulnerability of cancer cells. Nature 2023, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Wu, Y.; Du, D. Polydatin inhibits cell proliferation, invasion and migration, and induces cell apoptosis in hepatocellular carcinoma. Brazilian Journal of Medical and Biological Research 2018, 51. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.Y.; Zhang, H.X.; Mezei, M.; Cui, M. Molecular docking: a powerful approach for structure-based drug discovery. Current computer-aided drug design 2011, 7, 146–157. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, S.; Mehrotra, R.J.J.C. An overview of molecular docking. JSM chem 2016, 4, 1024–1028. [Google Scholar]
- Odhar, H.A.; Rayshan, A.M.; Ahjel, S.W.; Hashim, A.A.; Albeer, A.A.M.A. Molecular docking enabled updated screening of the matrix protein VP40 from Ebola virus with millions of compounds in the MCULE database for potential inhibitors. Bioinformation 2019, 15, 627. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of computational chemistry 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Pires, D.E.; Blundell, T.L.; Ascher, D.B. pkCSM: predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. Journal of medicinal chemistry 2015, 58, 4066–4072. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).