3.1. Target Prediction Model for Reassessment
The first Ames/QSAR International Challenge Project [
4], which was divided into three separate phase challenges, conducted each phase independently. Notably, the Ames test data for compounds used in the external validation sets of a previous phase were disclosed to participants before the start of the subsequent phase. This disclosure allowed participants to adjust their models using the newly available data. As a result, models that shared the same name across different phases might have been adjusted or modified. Furthermore, it was observed that the COVs were often reconfigured for models with the same name across various phases. Therefore, in this reanalysis, all models used in each phase of the first project were treated as independent entities, regardless of whether they shared the same name.
On the other hand,
during the second challenge [
6], a rule was
introduced that
permitted participants to
select which of their
multiple submitted models would be evaluated. This rule
aimed to
reduce bias
toward participants who submitted several models, thereby ensuring a
fairer assessment. However, this approach also
carried the risk of
potentially high-performing models
being excluded from the evaluation process. To
mitigate this, in the current reanalysis, all models submitted were extracted and included in the reassessment.
As a result, the total number of models evaluated
rose to 109 (
Table S1).
3.2. Evaluation Index
Like many toxicity tests, the distribution of positive and negative compounds in the Ames test results is notably skewed. In the first Ames/QSAR International Challenge Project, the external validation set consisted of 12,140 compounds across three phases. Of these, 1,757 (14.5%) were positive and 10,383 (85.5%) were negative. In the second challenge, the external validation set comprised 1,589 compounds, with 236 (14.9%) testing positive and 1,353 (85.1%) testing negative. It’s widely acknowledged that using metrics such as sensitivity, specificity, accuracy, positive predictive value (PPV), and negative predictive value (NPV) for such imbalanced data can often lead to misleading evaluations [
13,
14].
Accuracy (Acc), while a frequently used evaluation metric, is notably vulnerable to skew in datasets that are imbalanced [
15,
16]. Acc is computed using the formula (TP+TN)/(TP+TN+FP+FN), where TP, TN, FP, and FN denote true positives, true negatives, false positives, and false negatives, respectively. This metric is straightforward and easy to comprehend. However, its reliability diminishes in scenarios like the project, where only 14.5% of the cases were positive. If all test compounds were predicted to be negative, the resulting Acc would be 85.5%, which is misleadingly high. Consequently, a model that fails to accurately predict any positive cases may appear to outperform many others across both projects due to this metric’s susceptibility to data imbalance.
In predictive modeling, Sensitivity (also known as Recall) and Specificity often display a trade-off relationship. This is also true for positive predictive value (PPV, also known as Precision) and specificity, as well as Sensitivity and NPV, which are typically inversely related. To assess the robustness of a predictive model’s generalization performance, it’s crucial to achieve a balanced mix of these metrics. As a result, receiver operating characteristic (ROC) curves and precision-recall curves are frequently utilized to examine these relationships [
17,
18]. The area under these curves is an excellent measure for model evaluation. However, these metrics are only applicable to statistical models that can calculate predicted probability values. They were not used in the Ames/QSAR international challenge projects due to their inability to evaluate knowledge-based models. Instead, balanced accuracy (BA), which is the average of sensitivity and specificity, was used as a comprehensive metric to evaluate these two parameters, replacing the ROC curve [
19,
20,
21,
22]. BA is calculated using a specific formula:
Indeed, the formula for BA combines Sensitivity (the true positive rate) and Specificity (the true negative rate), providing a single measure that encapsulates the model’s performance across both positive and negative cases in the dataset. BA is particularly insightful because it reflects the model’s accuracy in identifying classes, regardless of the size of each class in the sample. This makes BA an ideal metric for assessing the overall performance of a model, especially in situations where data classes are imbalanced. Furthermore, the interpretability of this indicator is excellent, offering a clear understanding of model accuracy in a balanced manner.
Positive predictive value (PPV, also known as Precision) represents the proportion of items predicted as positive that are actually positive. On the other hand, Sensitivity (or Recall) signifies the proportion of actual positive items that are correctly identified as such. The F1 Score, which is the harmonic mean of PPV and sensitivity, is commonly used for a comprehensive evaluation of these two metrics [
23,
24]. It is important to note that Sensitivity and PPV are also referred to as recall and precision, respectively. The F1 Score is calculated using the following formula:
In the second Ames/QSAR International Challenge Project [
6], the F1 Score was introduced as an additional evaluation metric, supplementing those used in the first project. The F1 Score is particularly beneficial in achieving a balance between PPV and Sensitivity. A higher F1 Score indicates a superior model, as it represents a strong balance between predictive precision and recall. This is particularly important in scenarios such as Ames test predictions, where accurately detecting positives and minimizing false positives are equally important.
The MCC is another metric that is related to the chi-square statistic in a 2x2 contingency table. It incorporates a significant amount of information by considering the balance ratio of the four categories in the confusion matrix: true positives (TP), true negatives (TN), false positives (FP), and false negatives (FN) [
25,
26]. The MCC is expressed as follows:
Indeed, BA, the MCC, and the F1 Score each have their own unique statistical properties. However, they all serve as valuable integrative indicators for evaluating models, particularly when dealing with imbalanced data. In this study, a novel approach was adopted to streamline the performance evaluation of predictive models. This approach involved combining these diverse indices using principal component analysis. This method offers a more consolidated and definitive evaluation metric, thereby simplifying the otherwise complex task of model assessment.
3.3. Principal Component Analysis
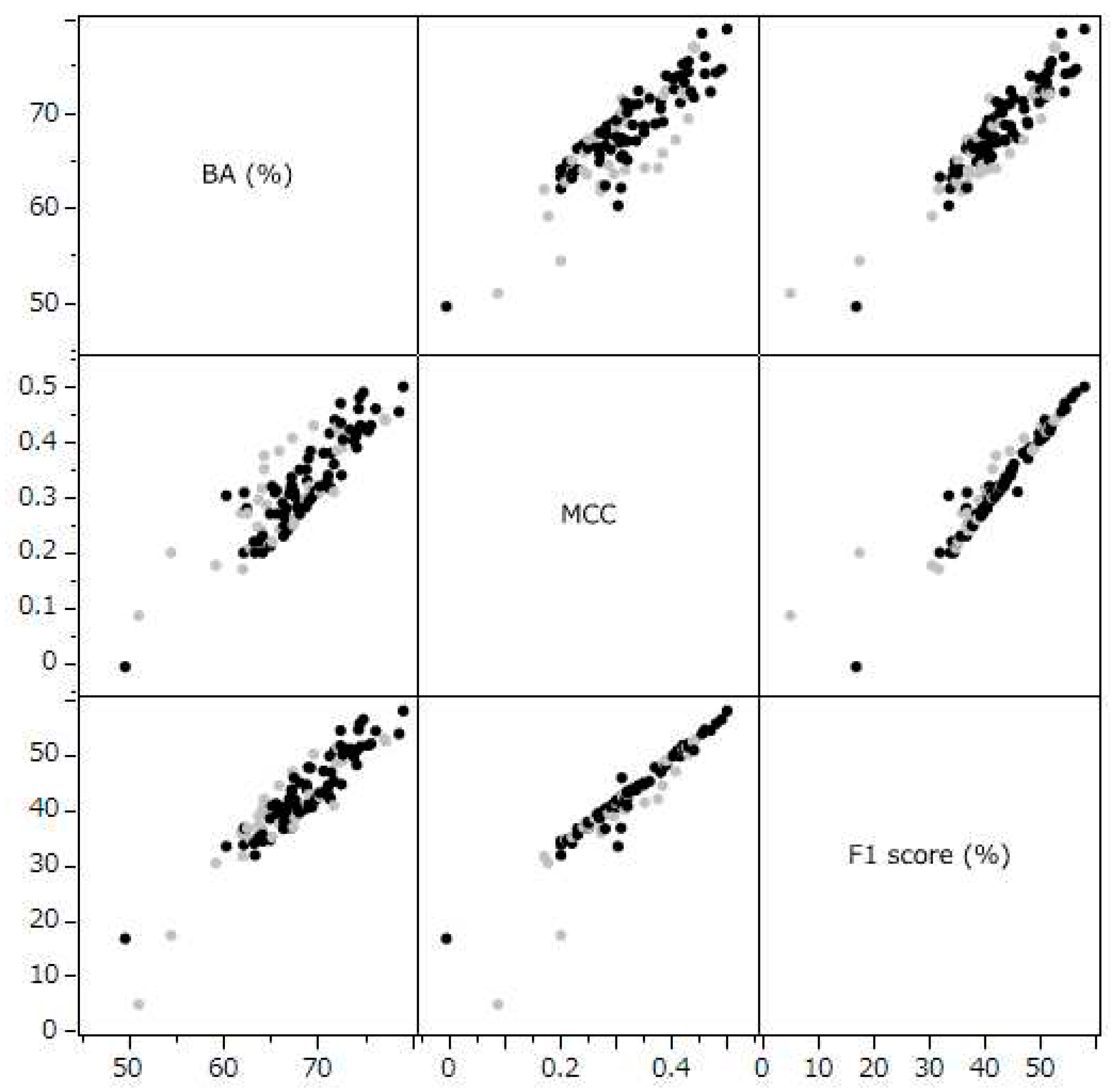
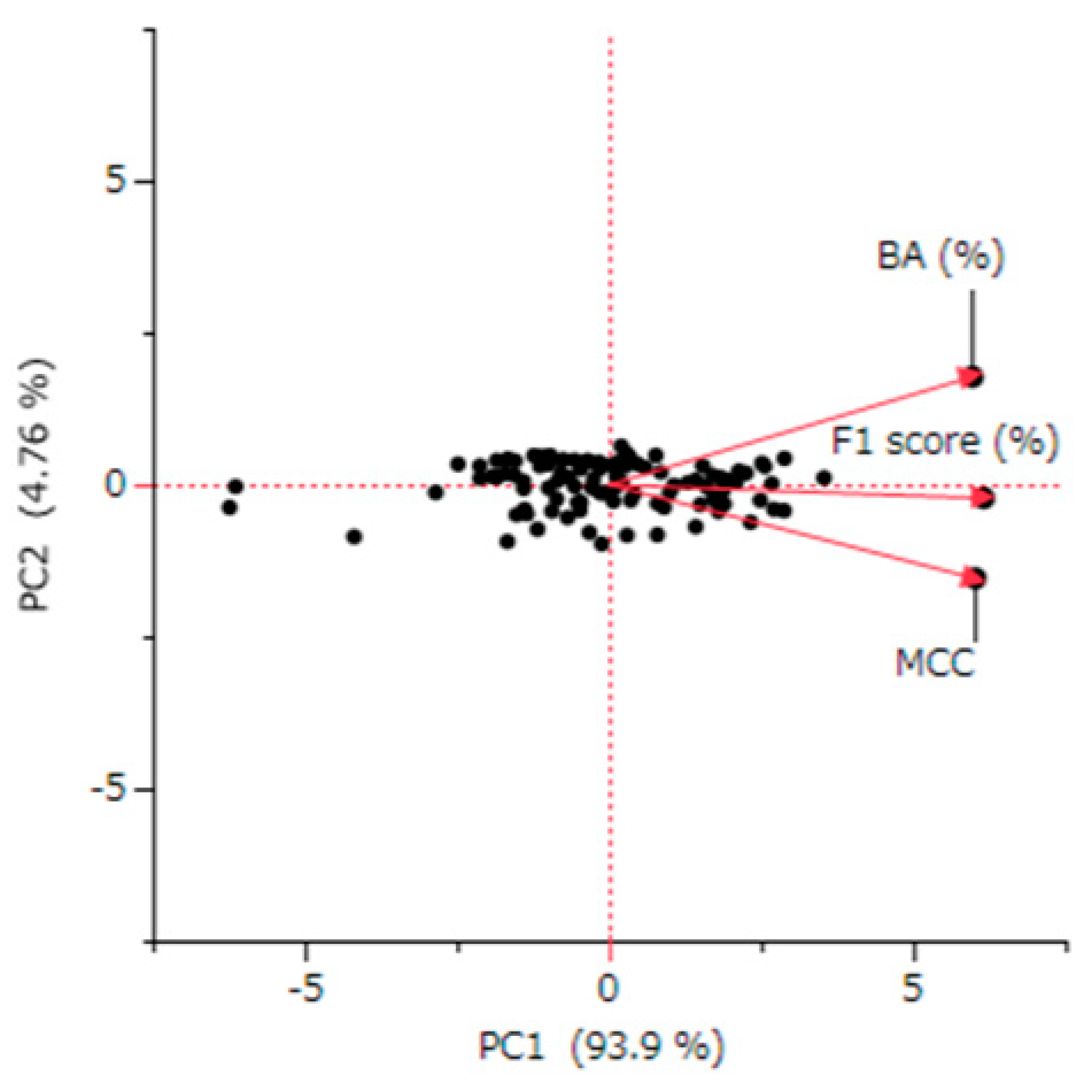
The Principal component analysis (PCA) conducted in this study revealed a strong consolidation among BA, the MCC, and the F1 Score. These metrics accounted for 94% of the variance in the same principal component direction (
Figure 3). This pattern suggests a significant collinearity among these metrics. Indeed, the correlation coefficients between these evaluation indicators showed strong correlations, ranging between 0.859 and 0.943 (
Figure 2). This finding highlights the utility of the PC1, which integrates these metrics, as a common and definitive indicator of integrated predictive performance.
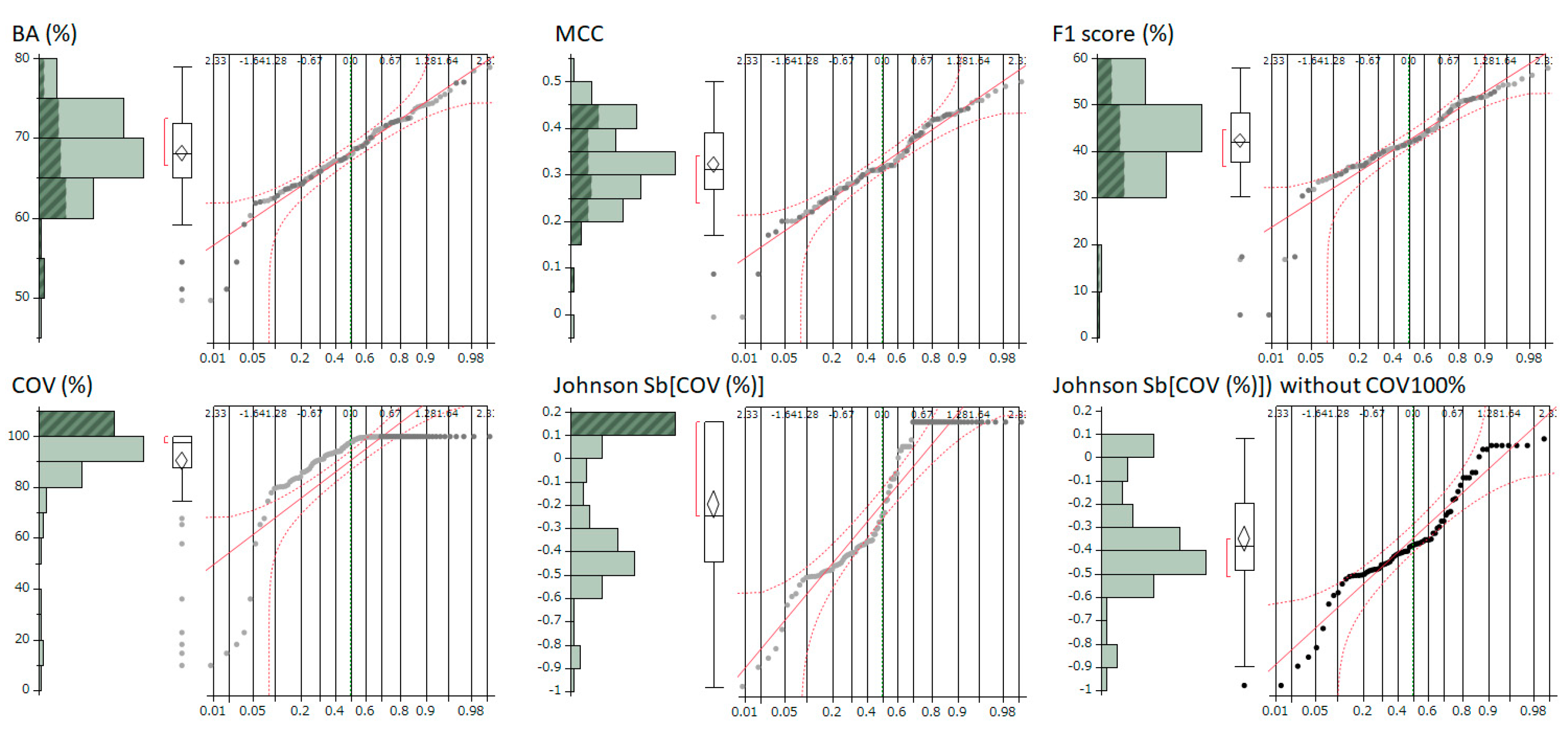
A crucial aspect of this study was the quantitative correlation analysis conducted to assess the impact of the COV on the evaluation indicators BA, MCC, F1, and PC1. Correlation analysis using Pearson’s correlation coefficient is generally most reliable when the variables under consideration follow a normal distribution [
27]. However, when the variables deviate from normality, there is an increased risk of the analysis being influenced by outliers or an overestimation of the degree of correlation. Moreover, if the assumption of normal distribution is not met, the accuracy in determining significance levels may be compromised. Therefore, it is crucial to verify the normality of each dataset before performing correlation analysis. Upon checking the normal distribution of these parameters using normal quantile plots, a distribution heavily skewed toward COV was observed. As a result, the Johnson normal distribution method [
11,
12] was employed to correct the COV distribution to a normal form, achieving effective normalization.
3.4. Impact of COV on Metrics
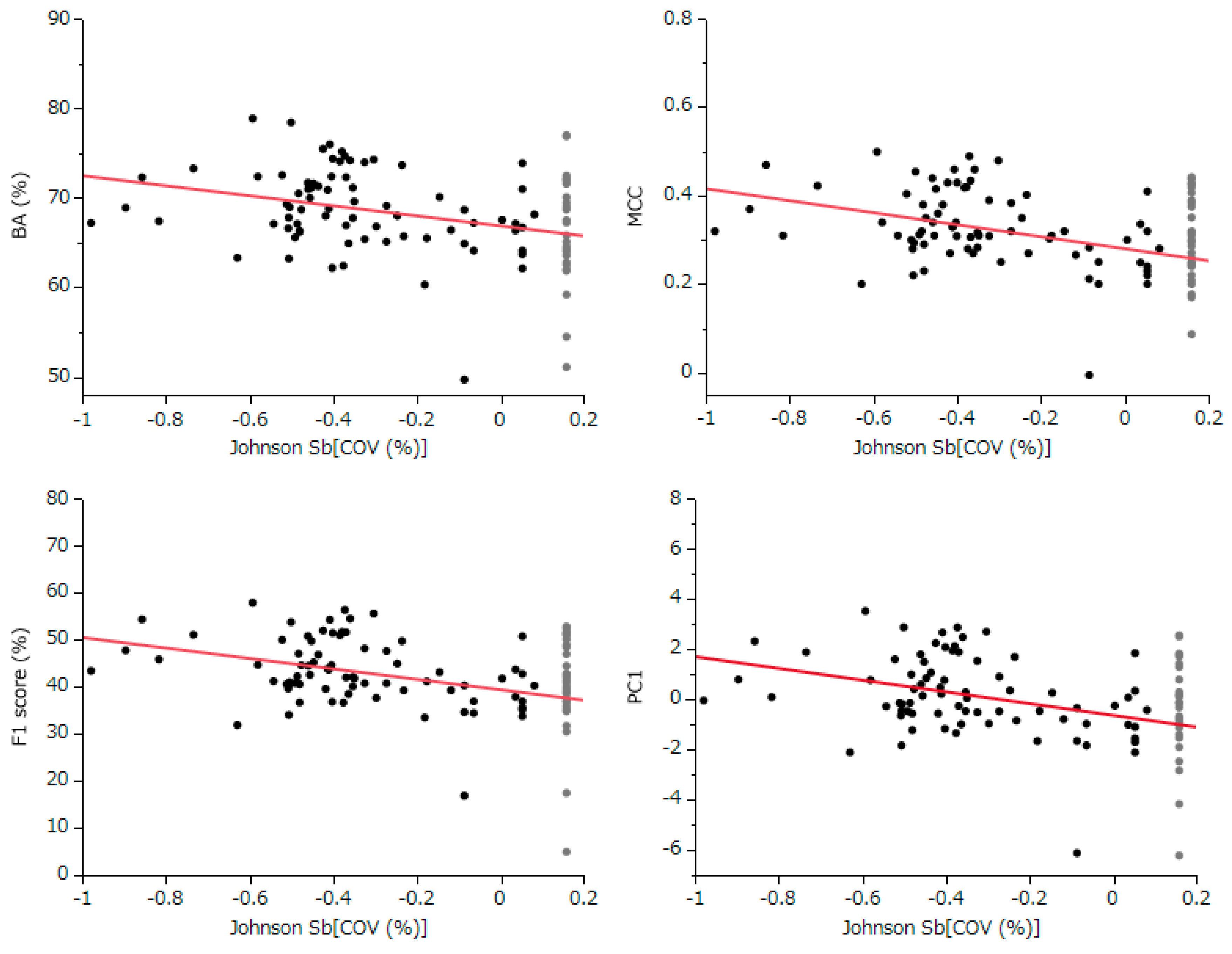
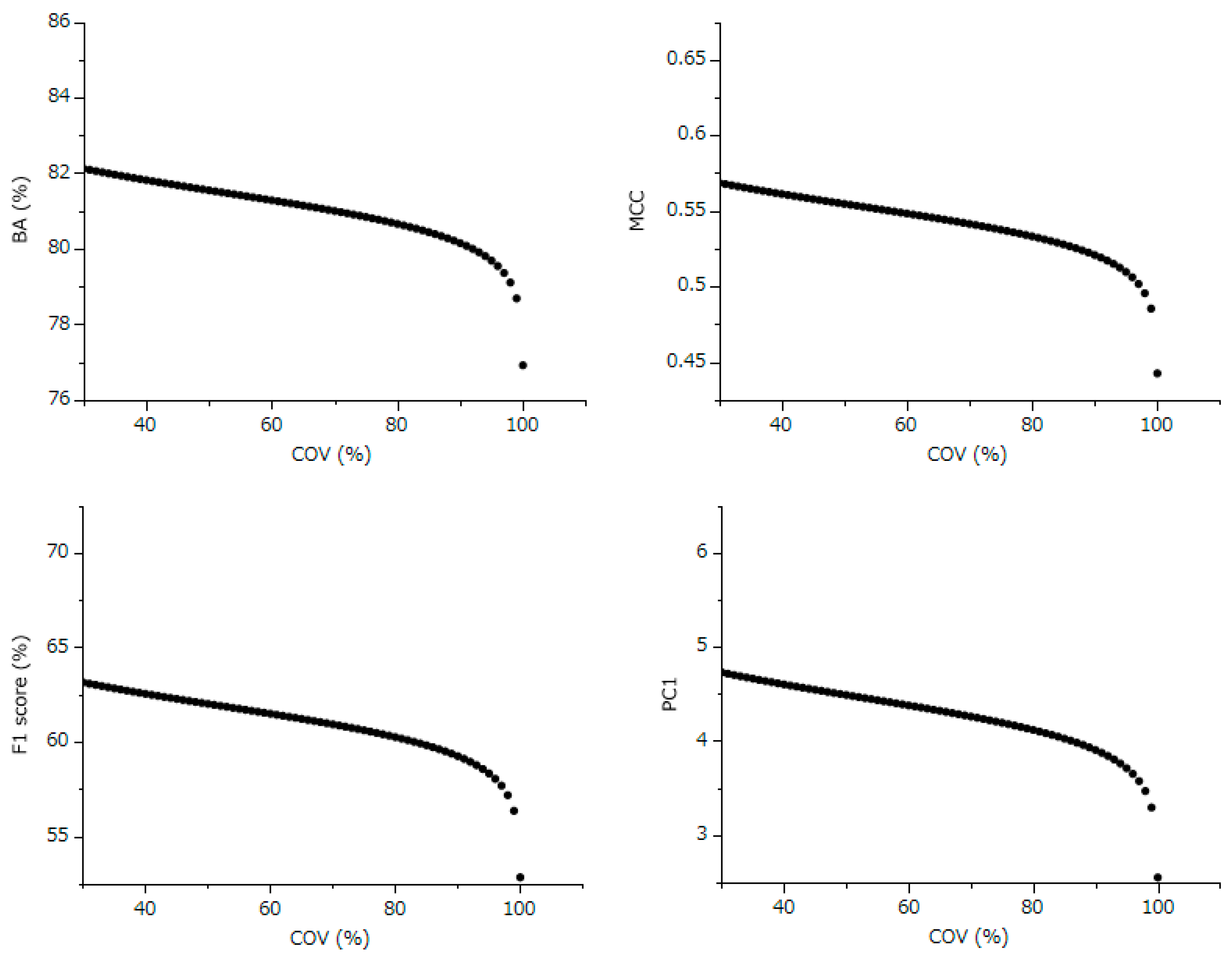
The conducted correlation analysis, which was between the normally COV and comprehensive evaluation indicators BA, MCC, F1 Score, and PC1 unveiled statistically significant negative correlations across all metrics. This finding implies that models with better predictive performance are likely to have lower COV settings. Despite the fact that each participant uniquely determined the COV settings using various techniques, it is still possible to estimate the standard influence of COV across all models from the slope of the least squares line.
Following this, we used the slope of this linear relationship and the residuals from each evaluation index to estimate the values of these indices, assuming a COV of 100% for each model. This method effectively shifts the evaluation values of each model along the slope of the straight line to a point where the COV equals 100% (
Figure 4). As a result, this calculation allows for a correction of all models to the evaluation indices at 100% COV, thereby enabling a fair comparison and evaluation across all models (
Figure 4, and
Table 1).
3.5. Best Models
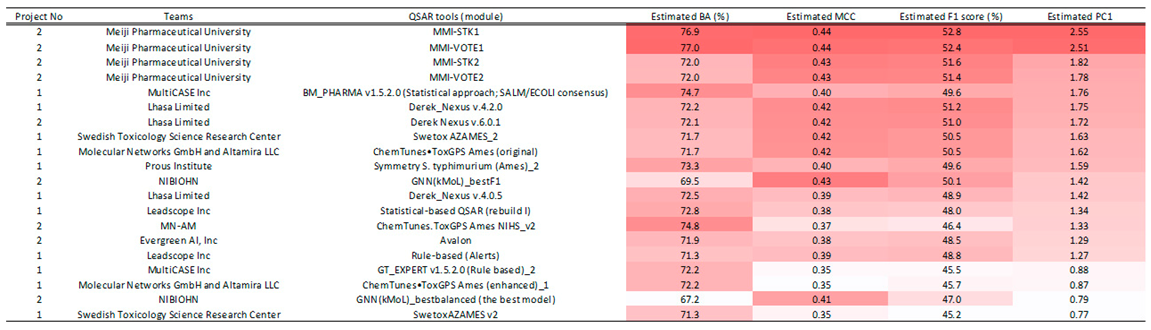
The analysis, adjusted to a 100% COV for all models (
Table 1,
Table S1), unveiled that the most efficacious predictive model, utilizing the integrated comprehensive index PC1, was MMI-STK1. This particular model, submitted by Meiji Pharmaceutical University during the second project, demonstrated superior performance. It’s noteworthy that MMI-STK1’s training data exclusively encompassed the phase 1, 2, and 3 datasets from the NIHS. For an in-depth understanding of the methodology contributing to its excellence, the algorithm employed for MMI-STK1 is elucidated in the supplementary file of the report [
6]. “Ninety-nine stacking models were constructed using multiple descriptors (Dragon, MOE, Mordred) and various machine learning algorithms (light GBM, XG-Boost, deep learning, graph convolutional network). The ultimate prediction was determined through a majority vote from all prediction results. The descriptors utilized were computed through Dragon, MOE, and Mordred. “
My analysis identified MMI-VOTE1 as the second most effective model Like MMI-STK1, MMI-VOTE1 was submitted by Meiji Pharmaceutical University during the second project and exclusively trained on NIHS-provided phase 1, 2, and 3 data. The algorithmic approach for MMI-VOTE1 is extensively detailed in the supplementary file of the report [
6]. This supplementary information provides a comprehensive insight into the methodologies and principles that underscore the success of MMI-VOTE1. “Nine stacking models were developed utilizing a combination of descriptors (Dragon, MOE, Mordred) and diverse machine learning algorithms (light GBM, XG-boost, deep learning, graph convolutional network). The final prediction was determined through a majority vote based on all prediction results. Descriptors were computed using Dragon, MOE, and Mordred.”
The third most effective predictive model identified in the study is MMI-STK2, submitted by Meiji Pharmaceutical University during the second project. In contrast to previous models, the training data for MMI-STK2 included not only the phase 1, 2, and 3 data provided by the NIHS but also Hansen’s data. The detailed algorithmic approach for MMI-STK2 is expounded upon in the supplementary file of the report [
6]. “MMI-STK2 is a stacking model constructed using Light GBM, deep learning, and graph convolutional network algorithms, with descriptors calculated using Dragon and MOE.”
The fourth best-performing model identified in the analysis was MMI-VOTE2, another model submitted by Meiji Pharmaceutical University during the second project. Similar to MMI-STK2, the training data for MMI-VOTE2 not only included the phase 1, 2, and 3 data from the NIHS, but also incorporated Hansen’s data. More detailed information about the algorithm used for MMI-VOTE2, including its approach to integrating these diverse datasets, is available in the supplementary file [
6]. “MMI-VOTE2 is a majority voting model constructed using Light GBM, Deep Learning, Random Forest, and graph convolutional network algorithms. The descriptors used were calculated using Dragon, MOE, and DNA docking simulations.”
In this project, Meiji Pharmaceutical University registered the four types of models previously mentioned. Impressively, all these models ranked as the top performers among the 109 models evaluated in this study. These models did not undergo adjustments to their coverage rate by altering their applicability domain. Given that setting a model’s applicability domain is a highly technical process and varies significantly from model to model, it is reasonable to infer that the estimated values of the metrics used for evaluating prediction models at COV levels other than 100% might contain considerable error. Despite this, the results mentioned above demonstrate the current technical capabilities of Ames prediction, underlining both the advances and potential limitations in this field.
3.6. Effect of Coverage on the Best Model
For MMI-STK1, which was identified as the top-performing model in terms of MCC, F1, and the integrated index PC1, we estimated how BA, MCC, F1, and PC1 would change with alterations in the COV (refer to
Figure 5). For example, while the F1 Score was at 52.8% with a COV of 100%, it was projected to increase to 59.2%, 61.5%, and 63.1% when the COV was adjusted to 90%, 60%, and 30%, respectively. This trend indicates that the predictive performance of the model can significantly improve even with a 90% COV, which involves excluding only 10% of the compounds from prediction. This observation implies that the external validation set included a small proportion of compounds that this model found particularly challenging to predict accurately.
These results lead to an important conclusion: when applying the Ames prediction model in real-world scenarios, it is crucial to consider the applicability domain while setting the COV [
7,
8,
9,
10]. Although the estimated values presented here might contain substantial errors, the performance of the model could be further improved if the COV settings are based on appropriately defined applicability domains.
3.7. Evaluation of Adaptive Domain Setting Technology and Future Prospects
This study represents the first instance in Ames/QSAR challenge projects where it has been explicitly shown that the performance of predictive models can be improved by adjusting COV settings. Notably, it also pinpointed the model with the highest predictive performance by taking into account COV. This significant discovery, achieved in a highly competitive environment, highlights the current limitations of QSAR technology in predicting Ames test outcomes. However, the enhanced prediction performance attributed to COV settings depends on the accurate definition of the models applicability domain. The performance evaluation at 100% COV presented in this study is essentially a projection based on standard COV settings. With careful consideration of the applicability domain settings, there is potential to exceed these standard performance levels.
While the technology for setting applicability domains-evaluated based on compound similarities and predicted probabilities derived from models-is advanced, research on optimal methodologies for setting applicability domains is still in its early stages [
7,
8,
9,
10]. This reanalysis had limited capacity to assess this specific aspect of model performance. However, as technologies for systematically determining suitable applicability domains for each model advance, a combination of diverse models, like those presented in this project, could lead to improved prediction accuracy. Although this project was primarily a competition evaluating the standalone performance of various models, future enhancements in prediction rates are expected, especially with the application of ensemble and consensus methods based on advanced techniques for estimating applicability domains [
28].